CircLZIC regulates ox-LDL-induced HUVEC cell proliferation and apoptosis via Micro-330-5p/NOTCH2 axis in atherosclerosis
Abstract
Atherosclerosis (AS) is a major chronic non-communicable disease and a primary cause of cardiovascular disease. Recent studies have shown that circRNAs are potential epigenetic factors that regulate vascular endothelial inflammatory responses and AS progression. Therefore, identification of the circRNAs that regulate ox-LDL levels is a critical step to understanding the pathology of AS. Our study is aim to investigate how circLZIC regulates atherosclerosis (AS) via the Micro-330-5p/NOTCH2 regulatory axis. The results showed that CircLZIC and NOTCH2 are highly expressed in human AS clinical samples, while Micro-330-5p is expressed locally. The CCK-8 experiment results showed that circLZIC promotes the proliferation of HUVECS cells. Flow cytometry analysis showed that circLZIC act as an inhibitor of HUVEC cell apoptosis. The expression level of Micro-330-5p can be up-regulated by transfection of small interfering RNA against circLZIC. Further, Starbase predicted that Micro-330-5p could target and regulate NOTCH2. Next, we confirmed that overexpression of Micro-330-5p could significantly reduce the expression of fluorescein using the double Luciferase reporter assay. RIP-qRT-PCR experiment showed that Micro-330-5p and NOTCH2 mRNAs are effectively enriched by ago2 protein. Further, we found that knocking down circLZIC increases the expression of Micro-330-5p and promotes cell apoptosis, while inhibiting the expression of NOTCH2 and cell activity. On the other hand, co-transfection of Micro-330-5p inhibitor decreases Micro-330-5p expression and inhibit cell apoptosis, while increasing NOTCH2 expression and cell activity. In conclusion, CircLZIC regulates HUVEC cell activity by the Micro-330-5p/NOTCH2 signaling pathway, suggesting that circLZIC plays a key role in atherosclerosis development.
1Introduction
Atherosclerosis (AS) is a major chronic non-communicable disease and a primary cause of cardiovascular disease [1]. In addition, inflammation has been shown to be crucial throughout atherosclerosis from endothelial dysfunction to plaque rupture and thrombosis [2]. AS is characterized by the thickening of artery intima, especially in large and medium-sized arteries [3]. Patients with AS are more prone to develop lipid metabolism disorders as AS patients have significantly lower lipid metabolism possibly due to underlying inflammation [4, 5]. However, the pathogenesis of atherosclerosis is not clear. Low-density lipoprotein (LDL) has been reported to be a risk factor for AS, and oxidatively modified LDL (ox-LDL) further exacerbates AS [6]. Ox-LDL can also inhibit vascular endothelial cell proliferation and induce apoptosis, causing inflammation and oxidative stress. Further studies have shown that activation of the cell death effector CPP32 by oxLDL may provide a mechanistic clue to the “response-to-injury” hypothesis of AS [7]. In addition, excessive apoptosis of vascular endothelium leads to dysfunction of endothelial cells [8–10]. These results suggest that ox-LDL is correlated with AS development [11]. Recent studies have shown that circRNAs are potential epigenetic factors that regulate vascular endothelial inflammatory responses and AS progression [12]. These circRNAs include circRNA-PTPRA [13], circSCRG1 [14], and circ_0026218 [15] and so on. For example, circ_0093887 can regulate ox-LDL induced human aortic endothelial cells viability, apoptosis, and inflammation through modulating miR-758-3p/BAMBI axis in AS [16]. Circ_CHMP5 aggravates ox-LDL induced damage to human umbilical vein endothelial cells through miR-516b-5p/TGFßR2 axis [17]. In addition, circIRAK1 can aggravate ox-LDL-induced endothelial cell injury in AS via TRIM14 upregulation by binding to miR-330-5p [4]. Therefore, identification of the circRNAs that regulate ox-LDL levels is a critical step to understanding the pathology of AS. Here, we found that circLZIC expression was downregulated in AS by a high-throughput sequencing set of the GEO database. Bioinformatics analysis revealed that circLZIC targets Micro-330-5p and Micro-330-5p bind to the 3’-UTR of NOTCH2. Collectively, our findings elucidated a novel regulatory axis involving circLZIC/Micro-330-5p/NOTCH2 in AS development, providing new insights into the pathology of AS.
Here, we found that circLZIC expression was downregulated in AS by a high-throughput sequencing set of the GEO database. Bioinformatics analysis revealed that circLZIC targets Micro-330-5p and Micro-330-5p bind to the 3’-UTR of NOTCH2. Collectively, our findings elucidated a novel regulatory axis involving circLZIC/Micro-330-5p/NOTCH2 in AS development, providing new insights into the pathology of AS.
2Materials and methods
2.1Bioinformatics and clinical sample collection
The gene expression dataset GSE161913 [18], including circRNA expression profiles in 5 atherosclerotic patients and 4 normal volunteers, was downloaded from the GEO dataset. The clinical samples were collected from 20 AS patients and 25 volunteers in the outpatient and inpatient departments of our hospital from 2021 to 2023. The participants were informed of the aim of this study and consented to provide samples. This study was approved by our Hospital’s ethics committee.
2.2Cell culture
Cells were cultured in DMEM containing 10% FBS and 1% penicillomycin, at 37°C in an incubator containing 5% carbon dioxide. After conventional resuscitation, HUVECs were inoculated into DMEM medium for culturing. When the cell growth reached to the logarithmic phase, passage or other treatments were performed. Passage 4–8 were used for experiment [19]. HUVECs were cultured with various concentrations of ox-LDL (0, 25, 50, and 100μg/ml) for 24 h at 37°C [20].
2.3Transfection
HUVEC were seeded in 6-well plates (Merck, USA) (5×105 cells/well), and transfection was performed when the cell confluence was more than 80%. The cells were transfected with the constructs (ox-LDL+sh-NC, ox-LDL+sh-circLZIC, ox-LDL+sh-circLZIC+miR-NC, or ox-LDL+sh-circLZIC+Micro-330-5p inhibitor using Lipofectamine 3000 (Invitrogen). si-LZIC was transfected at a concentration of 50 nmol/L and Micro-330-5p inhibitor at a concentration of 100 nmol/L. After 6 h of transfection, the cell culture medium was replaced by a fresh medium, and the transfection efficiency was measured by RT-qPCR.
2.4CCK-8 assay
The HUVECs were digested and suspended the day before the experiment. The cells were seeded in a 96-well cell culture plate (Merck, USA) at 5000 cells per cell. After 24 hours, the cells in each well were preheated. After 1-4 hours, the proliferation level of HUVEC was detected by an enzyme plate analyzer.
2.5Flow cytometry determination of apoptosis
The AnnexinV-FITC/PI kit was used to prepare the cell samples for the flow cytometry analysis. In brief, HUVEC (4 x 105) were collected and suspended in the binding buffer, and 5 uL AnnexinV-FITC and 5μL PI reagents were added. The reaction was performed for 5-15 min without light, and the apoptosis rate was analyzed by flow cytometer and FlowJ software.
2.6Dual luciferase reporter gene assay
The binding sites of Micro-330-5p in circLZIC and NOTCH2 were predicted by analysis using the bioinformatics StarBasev2.0 database. The sequences of WT and circLZIC-MUT were cloned into pGL3 vector to construct circLZIC-WT and circLZIC-MUT plasmids. Similarly, NOTCH2-WT and NOTCH2-MUT plasmids were constructed. HUVECS were inoculated in 24-well plates (Merck, USA) and cultured until the cell confluence reached to 80%. After that, the cells were transfected with recombinant luciferase reporter constructs plus Micro-330-5p mimic or miR-NC using Lipofectamine 3000 transfection reagent. After 48 h transfection, the cells were lysed with PLB cell lysis buffer. Then, the activity of firefly luciferase and renilla luciferase in HUVECs lysates was detected by a dual luciferase reporter gene system.
2.7qRT-PCR
Trizol was used to extract total RNA from tissues and HUVECs. The reverse transcription was performed with the TaKaRa PrimeScript RT Reagent Kit. ABI One-Step PLUS real-time PCR system was used for detection. 2-δT method was used to analyze the mRNA expression level of genes. The primers are shown in Table 1.
Table 1
Primer sequences for RT-PCR
| Forward primer (5’-3’) | Reverse primer (5’-3’) | |
| LZIC | GGCACTGTGCAGAGATCAGA | GCACCTGCATTTGCTGACAA |
| NOTCH2 | GTGTGTTGATGGCCTGGGTA | AGACAATGCCCTGGATGGAAA |
| CircLZIC | GATGAGGCCTTCTTGTCAGC | CTGCTAACCTTGTCCGAAGC |
| GAPDH | AATGGGCAGCCGTTAGGAAA | GCGCCCAATACGACCAAATC |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| miR-330-5p | TCTCTGGGCCTGTGTCTTAGGC | CTAAGACACAGCCCAGAGATT |
2.8Western blot
Proteins were extracted from HUVECs using RIPA lysate buffer containing protease inhibition and were quantified using a BCA kit. Then, the proteins were separated on a 10% PAGE gel followed by the transfer to a PVDF membrane. The membrane was blocked in 5% skim milk followed by treatment with primary antibodies against PCNA, KI-67, Bcl-2, Bax, Cleaved caspase3, NOTCH or GAPDH for 12 hours. Next, the membrane was washed and treated with an HRP secondary antibody. The blot was developed by an ECL protein luminescence detection kit, and the band density was quantified using Image J software.
2.9Statistical analysis
All statistical analyses were performed using SPSS statistical software and GraphPad Prism 8. The student’s t-test was used to compare the results between two groups.
3Results
3.1CircLZIC is upregulated in AS serum and vascular endothelial cells treated with ox-LDL
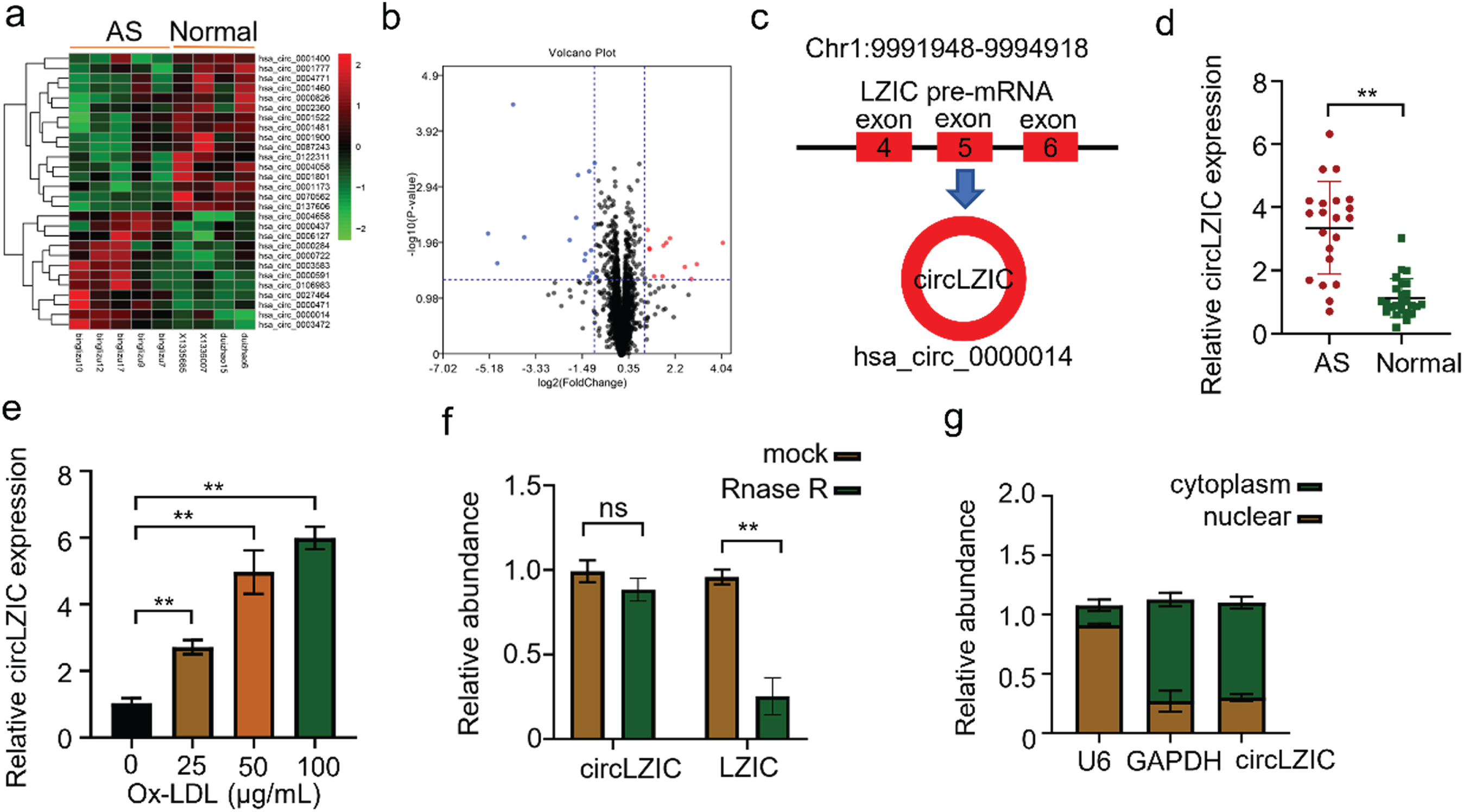
First, we analyzed the differentially expressed circRNAs in 5 atherosclerotic patients and 4 normal volunteers in the GSE161913 dataset and found 12 up-regulated and 16 down-regulated circRNAs, among which circLZIC was significantly up-regulated one (Fig. 1A-B). CircLZIC was located at Chr1:9991948-9994918, which was formed by splicing the first and last exon of 4, 5 and 6 of the parent gene LZIC (Fig. 1C). To calidate the expression levels of circLZIC in clinical samples, we collected blood samples from 20 AS patients and 25 healthy volunteers. Using qRT-CPR, we found that circLZIC was highly expressed in the samples of atherosclerotic patients (Fig. 1D). Subsequently, to explore the roles of circLZIC in ox-LDL induced HUVECs, we examined the mRNA expression of circLZIC in HUVECs after treatment different concentration of ox-LDL (0, 25, 50, and 100μg/ml) for 24 h using qRT-PCR. The expression of circLZIC in HUVECs was increased by ox-LDL in a dose dependent manner (Fig. 1E), and reached maximum at 100μg/ml. Therefore, 100μg/ml was used as the ox-LDL induction concentration for subsequent experiments. In addition, we also examined the stability of circLZIC by RNase digestion. Our results showed that RNase R could effectively digested LZIC linear mRNA but not circLZIC (Fig. 1F). We isolated cytoplasmic and nuclear fractions of HUVECs to detected the subcellular localization of circLZIC. We found that circLZIC was mainly localized in the cytoplasm (Fig. 1G).
Fig. 1
CircLZIC is upregulated in atherosclarsis serum and ox-LD-treated vascular endothelial cells. (A) Heat map showing circRNA expression levels in GEO database. (B) Volcano plot showing circRNA expression pattern. (C) The diagram of CircLZIC formation. (D) qRT-PCR was performed to detect circLZIC expression in serum samples of AS patients and normal volunteers. (E) QRT-PCR was implemented to detect circLZIC expression in each group of cells. (F) qRT-PCR was used to quantify linear LZIC and circLZIC. (G) qRT-PCR was used to quantify circLZIC in the cytoplasm and nucleus of HUVECs.
3.2CircLZIC regulate HUVECS cells proliferation and apoptosis induced by ox-LDL
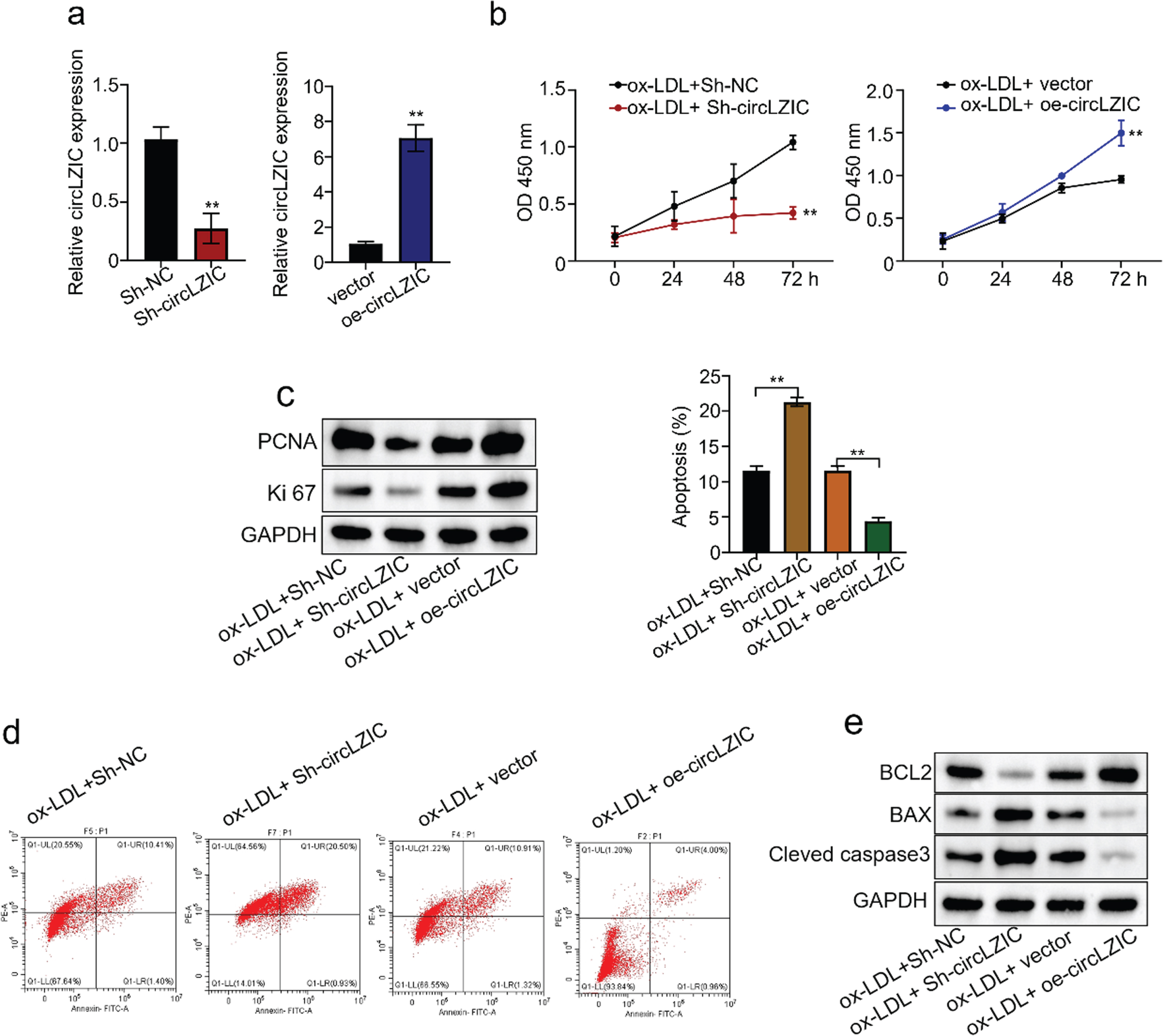
To investigate whether circLZIC can regulate ox-LDL-induced proliferation and apoptosis in HUVECS cells, we constructed shRNAs to knock down and overexpression vectors to overexpress circLZIC in HUVECs. QRT-PCR confirmed the knockdown/overexpression efficiency (Fig. 2A). Next, the CCK-8 results showed that knockdown of circLZIC significantly decreased cell activity, whereas overexpression of circLZIC significantly increased cell activity (Fig. 2B). Meanwhile, we performed Western blot to measure the expression level of key proteins related to proliferation. The results showed that the expression of PCNA and ki67 was reduced after circLZIC knockdown and increased after circLZIC overexpression (Fig. 2C). Further, we examined the apoptosis of ox-LDL-treated HUVECS cells after knockdown/overexpression of circLZIC by flow cytometric assay. We found that the apoptosis rate was increased after circLZIC knockdown and was decreased after circLZIC overexpression (Fig. 2D). In addition, we performed Western blot to detect the expression of proteins related to cell proliferation. The results showed that knockdown of circLZIC decreased BCL2 expression and increased the expression of BAX and cleaved caspase3, whereas overexpression of circLZIC increased BCL2 expression and decreased the expression of Bax and cleaved caspase3 (Fig. 2E). Together, our data suggest that circLZIC acts as an efficient regulator of OX-LDL-induced proliferation and apoptotic processes in HUVECs.
Fig. 2
CircLZIC regulates ox-LDL-induced vascular endothelial cell proliferation and apoptosis. (A) qRT-PCR was used to detect the knockdown/overexpression efficiency of circLZIC in HUVECs. (B) The CCK-8 experiment detected the changes in cell activity of HUVECs treated with ox-LDL after circLZIC knockdown or overexpression. (C) PCNA and ki67 expression in HUVECs treated with ox-LDL after circLZIC knockdown or overexpression was detected by Western blot. (D) Cell apoptosis in HUVECs treated with ox-LDL after knockdown/overexpression of circLZIC was assessed by flow cytometry analysis. (E) The expression of apoptosis-related proteins in HUVECs treated with ox-LDL after knocking down/overexpressing circLZIC was determined by Western blot.
3.3CircLZIC directly targets Micro-330-5p in HUVECs
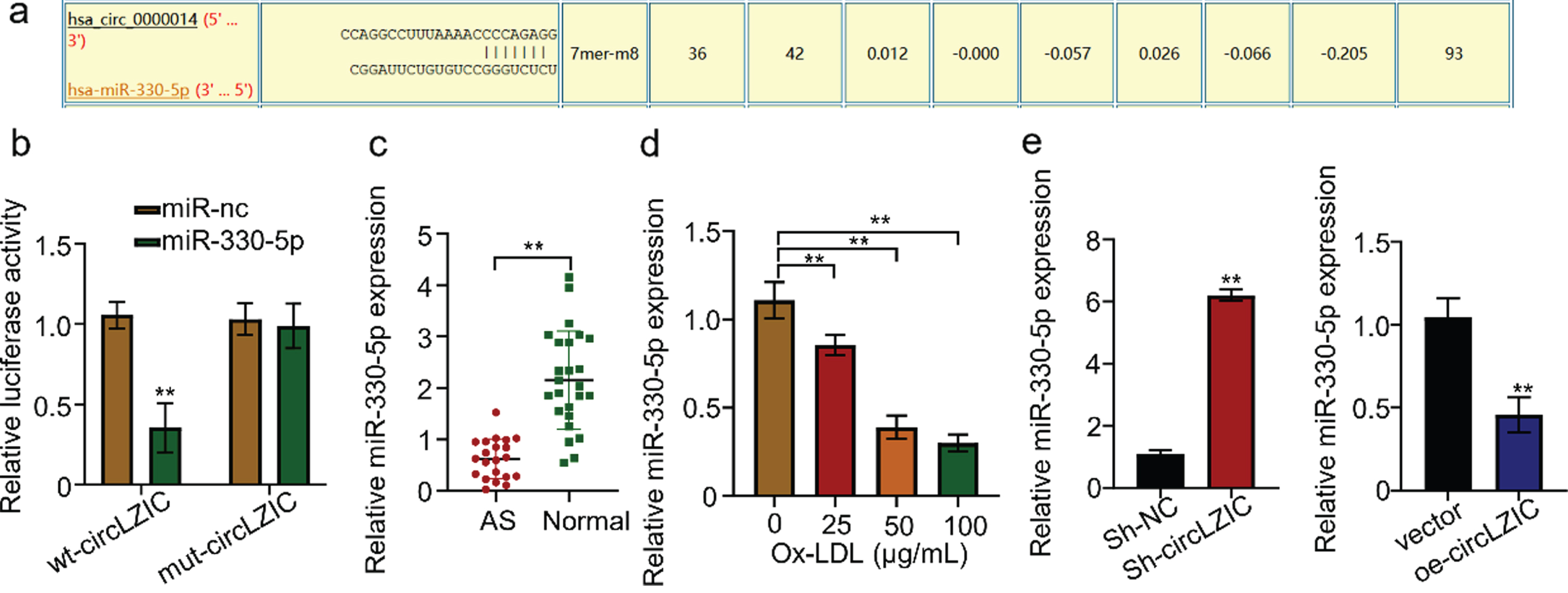
The CircInteractome was used to predict potential target miRNAs that are targeted by circLZIC, and Micro-330-5p was found to be a putative target (Fig. 3A). Luciferase reporter gene experiments revealed that transfection of HUVECs containing circLZIC with the construct carrying Micro-330-5p binding site or mutated binding site showed that overexpression of Micro-330-5p significantly reduced luciferin expression in the wild-type reporter gene, while not affecting the expression in the mutant reporter gene (Fig. 3B). This suggests that Micro-330-5p is a target of circLZIC. We further analyzed whether Micro-330-5p showed the same effect in clinical samples. Micro-330-5p expression was significantly reduced in AS samples (Fig. 3C). Consistently, we detected that Micro-330-5p expression was decreased by ox-LDL treatment in a concentration-dependent manner in HUVECs (Fig. 3D). To demonstrate the relationship between Micro-330-5p and circLZIC, we used qRT-PCR to detect the change of Micro-330-5p expression in HUVECs after circLZIC knockdown or overexpression. The results showed that Micro-330-5p expression was elevated when circLZIC expression was knocked down, while decreased by circLZIC overexpression (Fig. 3E). The above results confirmed that circLZIC directly targets Micro-330-5p in HUVECs.
Fig. 3
CircLZIC directly targets Micro-330-5p in HUVECs. (A) Circular interaction prediction revealed Micro-330-5p as a putative target ofcircLZIC.(B) The interaction between circLZIC and Micro-330-5p was investigated by the luciferase fluorescent reporter assay. (C) qRT-PCR was used to detect the expression of Micro-330-5p in the serum of 20 AS patients and 25 normal volunteers. (D) qRT-PCR was used to detect the expression of Micro-330-5p in each group of cells. (E) qRT-PCR was used to detect the expression of Micro-330-5p in HUVECs after knocking down or overexpressing circLZIC.
3.4NOTCH2 is a target gene of Micro-330-5p
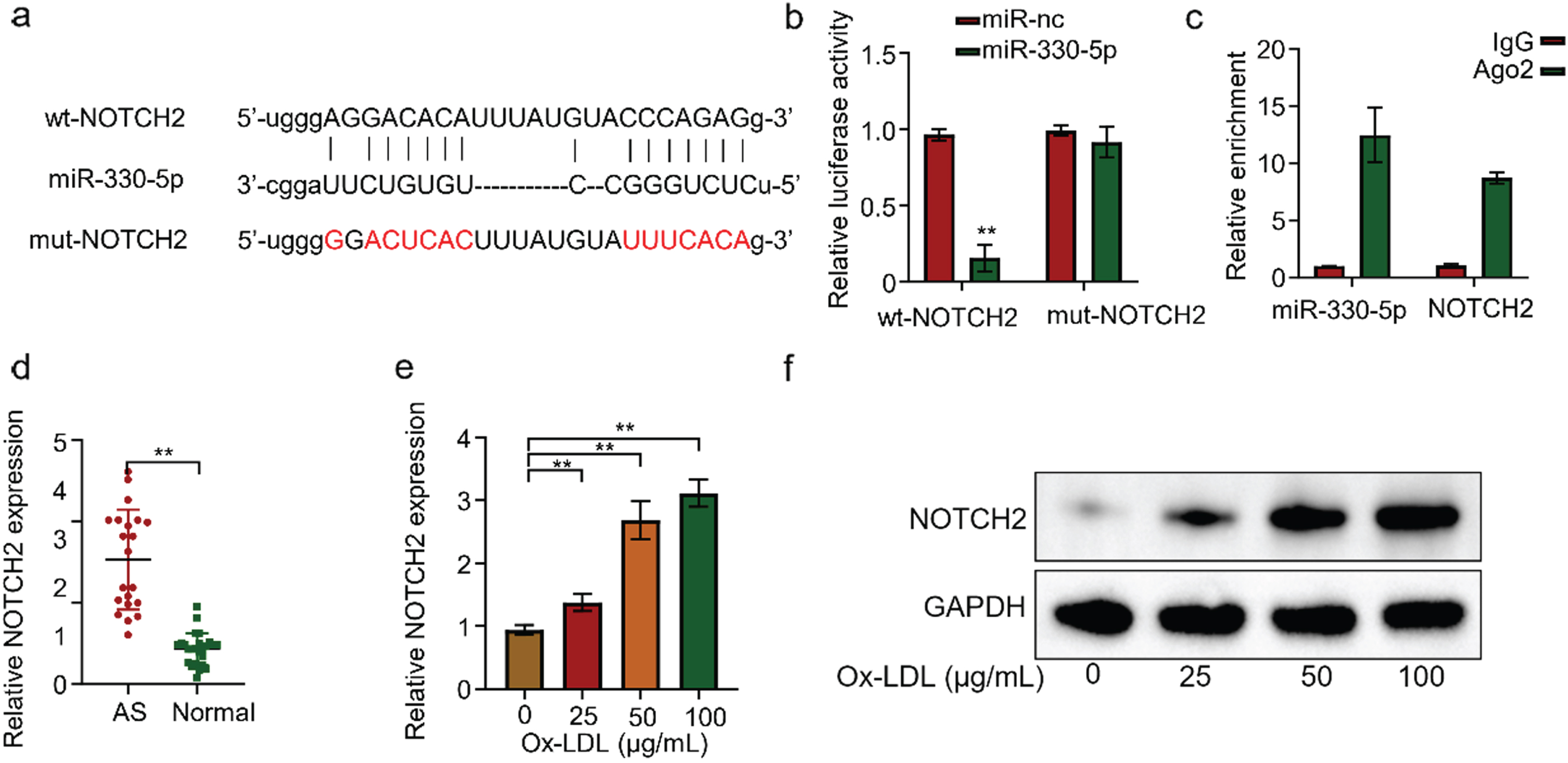
The StarBase prediction revealed a complementary nucleotide sequence between Micro-330-5p and NOTCH2 (Fig. 4A). Then we used the luciferase reporter gene experiment to validate the interaction between Micro-330-5p and NOTCH2. We transfected NOTCH2-containing HUVECs with WT or mutated 3’-UTR-region of NOTCH2. Micro-330-5p overexpression significantly reduced luciferin expression in the wild-type reporter gene, while luciferin expression in the mutant reporter gene was not affected (Fig. 4B). This suggests that TNOTCH2 is a target of Micro-330-5p action. RIP-qRT-PCR experiments in HUVECs showed that both Micro-330-5p and NOTCH2 mRNA could be efficiently enriched by the ago2 protein, but not by the negative control, IgG antibody (Fig. 4C). Similarly, we examined NOTCH2 expression in the serum of 20 AS and 25 normal volunteers by qRT-PCR. The results showed that NOTCH2 was generally highly expressed in AS with statistically significant differences (Fig. 4D). To further validate the results, we treated HUVECS cells with different concentrations of ox-LDL for 24 h. qRT-PCR detected that NOTCH2 expression in HUVECs gradually increased with the increase of ox-LDL concentration (Fig. 4E). Similarly, at the protein level, WB also showed the same pattern (Fig. 4F). Collectively, the above results confirmed that NOTCH2 is a target gene of Micro-330-5p.
Fig. 4
NOTCH2 is a target gene of Micro-330-5p. (A) Starbase predicted NOTCH2 as a candidate gene targeted by Micro-330-5p. (B) Luciferase fluorescent reporter assay was performed to validate the binding of Micro-330-5p tothe 3’-UTR region of NOTCH2. (C) RIP-qRT-PCR experiments were conducted using Ago2 antibodies in HUVECS cells. (D) qRT-PCR was used to detect NOTCH2 expression in the serum of AS and healthy controls. (E) qRT-PCR was used to detect NOTCH2 expression in cells of each group. (F) Western blot was performed to assess NOTCH2 expression in each group of cells.
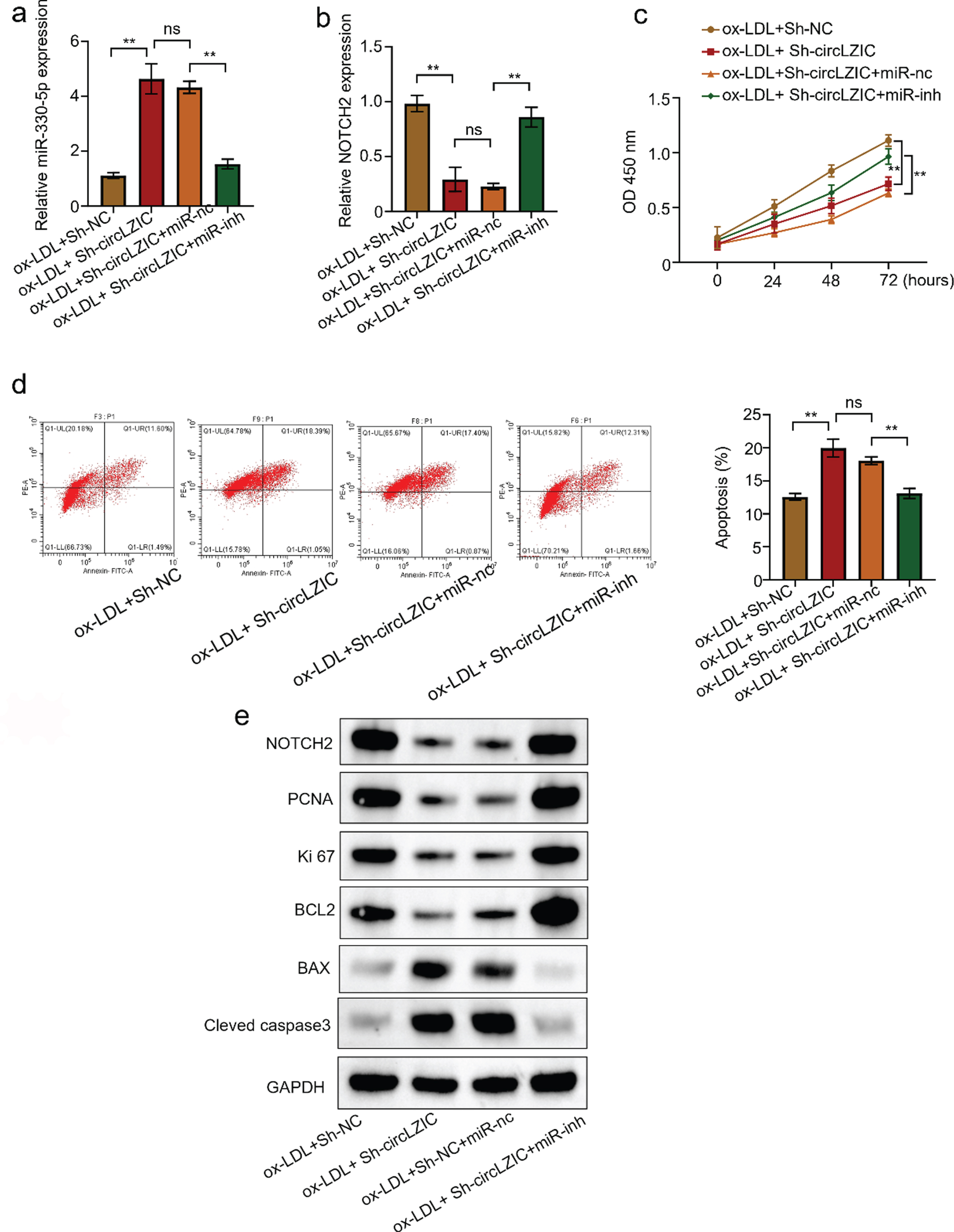
3.5CircLZIC regulates HUVECs proliferation and apoptosis induced by ox-LDL via Micro-330-5p/NOTCH2 axis
To further investigate whether circLZIC directly targets the Micro-330-5p/NOTCH2 axis, we examined the regulation of expression of Micro-330-5p and NOTCH2 by circLZIC. The results showed that knockdown of circLZIC elevated the expression of Micro-330-5p, while co-transfection of Micro-330-5p inhibitor decreased the expression (Fig. 5A). CircLZIC knockdown decreased the expression of NOTCH2, and co-transfection with Micro-330-5p inhibitor, restored the expression of NOTCH2 (Fig. 5B). Further, CCK-8 analysis showed that knockdown of circLZIC reduced cell activity, which was rescued by co-transfection with Micro-330-5p inhibitor (Fig. 5 C). Moreover, we assessed the effect of circLZIC knockdown on the apoptosis percentage of HUVECs cells by flow-through analysis. We found that the cell apoptosis rate of HUVECs was increased, and co-transfection with Micro-330-5p inhibitor abolished this effect (Fig. 5D). Furthermore, WB results revealed the expression of key proteins in cell proliferation and apoptosis was in line with the results above (Fig. 5E). The above results confirm that circLZIC regulates HUVECs cell proliferation and apoptosis via the Micro-330-5p/NOTCH2 axis.
Fig. 5
CircLZIC regulates ox-LDL-induced vascular endothelial cell proliferation and apoptosis via Micro-330-5p/NOTCH2 axis. (A) qRT PCR was used to detect the expression of Micro-330-5p in different groups. (B) qRT PCR was used to detect the expression of NOTCH2 in different groups. (C) CCK-8 detected the changes in the activity of HUVECS cells in different groups. (D) Flow cytometry apoptosis assay was used to detect the changes in the proportion of HUVECs undergoing apoptosis in different groups. (E) Western blot was used to detect the expression of proliferation- and apoptosis-related proteins in different groups of HUVECs.
4Discussion
AS is a chronic inflammatory lesion, serving as the main pathological basis of adverse cardiovascular events [21]. With the aging of the population, the total number of people with cardiovascular disease is now more than 587 million worldwide [22]. Currently, clinical treatment of AS mainly includes pharmacological therapy and revascularization therapy. Ox-LDL induces the proliferation and migration of smooth muscle cells and is considered to be a key factor in AS development [23]. Moreover, Ox-LDL is an important pathogenic factor that induces excessive oxidative stress and apoptosis in vascular endothelial cells, which ultimately promoted AS development [5]. In the present study, the apoptosis rate of HUVECs was increased, and the cell proliferation capacity was reduced after 24 h of treatment with 100 µg/mL ox-LDL, suggesting that ox-LDL induction enhanced the apoptosis but inhibited the proliferation of HUVEC. There is growing evidence that dysregulation of circRNA expression is associated with AS lesion formation, and circRNAs are thought to be potential drug targets. For example, circUSP36 competitively binds to miR-637, regulates the expression level of member 4 of the wingless-type mammary tumor virus integration site family in mice, and exacerbates the damage to HUVECs caused by ox-LDL [24]. CircSCRG1 is associated with angiogenesis in HUVECs by regulating the expression of miR-1268b/NR4A1 axis [14]. Circ_0033596 depletion ameliorates ox-LDL-induced human umbilical vein endothelial cell damage by miR-637/GRB2 pathway, providing a therapeutic target for AS [25]. In addition, circ_0029589 promotes ox-LDL-induced endothelial cell injury through regulating RAB22A by serving as a sponge of miR-1197 [26]. However, our understanding of the mechanism by which circRNAs regulate OX-LDL is still flimsy. In this study, the GEO database analysis revealed that circLZIC was significantly upregulated in AS, and circLZIC expression was upregulated in AS clinical serum samples. Overexpression of circLZIC enhanced the proliferative activity and inhibited the apoptotic capacity of HUVECs, whereas inhibition of circLZIC expression inhibited the proliferative activity of HUVECs and enhanced the apoptotic capacity of HUVECs. These results suggest that circLZIC expression may be associated with AS plaque formation, and its downregulation could promote AS development.
There is growing evidence that ceRNA crosstalk plays a crucial role in the pathology of AS [27]. The present study implicated circLZIC in AS development. Bioinformatic analysis predicted a mutual binding site between Micro-330-5p and circLZIC. Nucleoplasmic separation experiments showed that circLZIC is existed with Micro-330-5p in the cytoplasm of HUVECs. Dual luciferase reporter gene assay confirmed that circLZIC was able to interact with Micro-330-5p. MiR -330-5p expression was down-regulated in serum samples from AS patients. After ox-LDL treatment, either inhibition of circLZIC or overexpression of Micro-330-5p increased HUVEC proliferation and decreased apoptosis, while overexpression of circLZIC and inhibition of Micro-330-5p decreased HUVEC proliferation and increased apoptosis. The above results suggests that Micro-330-5p play a regulatory role in AS development. In addition, the proportion of macrophage apoptosis in oronary heart disease treated with HDL was significantly upregulated, which skewed the coordinated expression of cleaved caspase 3, BCL-2 and BAX proteins related to apoptosis [28].
Wnt is an important signal that regulates cell growth, chemotaxis, apoptosis, migration, and differentiation [29, 30]. There is a strong correlation between the Notch pathway and Wnt in regulating blood vessels. The study found that ox-LDL can facilitate Wnt signal activation, up-regulate WNT5A expression, promote the transformation of VSMC into proliferative foam cells, and trigger strong inflammation, all of which lead to excessive proliferation, migration and invasion of VSMC, ultimately promoting the occurrence and progress of AS. Inhibiting the Wnt pathway can alleviate inflammation, inhibit VSMC overgrowth, induce apoptosis, and reduce VSMC invasion and migration ability [31]. This also suggests that the Notch pathway has a certain regulatory role in regulating smooth muscle cell apoptosis and proliferation. Notch2, located on chromosome 19p12, has been confirmed to be abnormally expressed in diseases [32]. The Notch signaling pathway is involved in cell proliferation, differentiation and apoptosis. Notch and its related genes, including Notch receptors including Notch1-4 and Notch ligands including Jagged1, Jagged2 and DLL1, 3, 4 in mammals. are widely expressed on the surface of various immune cells [33, 34]. As an inflammatory immune disease, AS is initiated by the activation of the natural immune response, and macrophages play an important role in this process [35]. Activation of the Notch signaling pathway in macrophages has been hypothesized to be related to AS. Reportedly, overexpression of DLL4 induced by VEGFA downstream of Notch 2 decreased the migration capacity of endothelial cells [36]. Davis-Knowlton et al. have reported that Jagged1 inhibits cell proliferation through Notch2 in the cells with high growth potential, but not in the cells with low growth potential [37]. The current study confirmed that Jagged1 did not promote contractile smooth muscle actin and had no significant effect on the mature differentiation phenotype. However, another study reported that deletion of Notch2 gene in SMC in APOE-/-deficient mice did not promote AS progression [38]. In the study, circLZIC-overexpressing HUVECs treated with ox-LDL showed a significant decrease in Micro-330-5p expression and a significant increase in NOTCH2 expression, suggesting that circLZIC may reduce the proliferation of HUVECs induced by ox-LDL through downregulating Micro-330-5p expression and overexpressing NOTCH2 expression. The present study found that inhibition of miR-124 expression reversed the mitigating effect of circLZIC on inflammatory injury in HUVECs induced by ox-LDL, further confirming that circLZIC suppresses Micro-330-5p expression and attenuates ox-LDL-induced injury in HUVECs.
Because AS is usually asymptomatic, a feature that significantly complicates the diagnosis of this disease and undermines the efficacy of treatment. Therefore, although significant progress has been made in understanding AS pathogenesis and developing clinical treatment, it is still urgent to identify new targets for early diagnosis and therapeutic intervention of AS.
Acknowledgments
Not applicable.
Author contributions
Xingping Men, Aizhen Hu and Tingting Xu conceived this study; Xingping Men and Aizhen Hu carried out experiments, analyzed the data, Xingping Men prepared the first draft of this manuscript; Tingting Xu revised this manuscript critically. All authors have read and approved the final manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the institutional review board of Yantai Yuhuangding Hospital, and written informed consent was obtained from all participants.
Consent for publication
All patients in this study provided their consent for publication.
Conflict of interest
The authors declare that they have no competing interests.
Funding
No funding.
References
[1] | Gao X , Lv T , Li G , Tse G , Liu T . Association Between Atherosclerosis-Related Cardiovascular Disease and Uveitis: A Systematic Review and Meta-Analysis. Diagnostics (Basel). 12: (12). |
[2] | Corrado E , Rizzo M , Coppola G , Fattouch K , Novo G , Marturana I , Ferrara F , Novo S . An update on the role of markers of inflammation in atherosclerosis. J Atheroscler Thromb. 17: (1):1–11. |
[3] | Zhao Y , Malik S , Criqui MH , Allison MA , Budoff MJ , Sandfort V , Wong ND . Coronary calcium density in relation to coronary heart disease and cardiovascular disease in adults with diabetes or metabolic syndrome: the Multi-ethnic Study of Atherosclerosis (MESA) BMC Cardiovasc Disord. 22: (1):536. |
[4] | Liu F , Gao B , Wang Y CircIRAK1 aggravates ox-LDL-induced endothelial cell injury in atherosclerosis via TRIM14 upregulation by binding to miR-330-5p1. Clin Hemorheol Microcirc. 2022; doi:10.3233/ch-221551. |
[5] | Wang Q , Wang T , Liang S , Zhou L . Ox-LDL-Induced Vascular Smooth Muscle Cell Dysfunction Partly Depends on the Circ_0044073/miR-377-3p/AURKA Axis in Atherosclerosis. Int Heart J. 64: (2):252–262. |
[6] | Guo X , Guo Y , Wang Z , Cao B , Zheng C , Zeng Z , Wei Y Reducing the Damage of Ox-LDL/LOX-1 Pathway to Vascular Endothelial Barrier Can Inhibit Atherosclerosis. Oxid Med Cell Longev. 2022;7541411. |
[7] | Dimmeler S , Haendeler J , Galle J , Zeiher AM . Oxidized low-density lipoprotein induces apoptosis of human endothelial cells by activation of CPP32-like proteases. A mechanistic clue to the ‘response to injury’ hypothesis. Circulation. 95: (7):1760–1763. |
[8] | Sola-Riera C , Garcia M , Ljunggren HG , Klingstrom J . Hantavirus inhibits apoptosis by preventing mitochondrial membrane potential loss through up-regulation of the pro-survival factor BCL-2. PLoS Pathog. 16: (2):e1008297. |
[9] | Ou M , Li X , Zhao S , Cui S , Tu J . Long non-coding RNA CDKN2B-AS1 contributes to atherosclerotic plaque formation by forming RNA-DNA triplex in the CDKN2B promoter. EBioMedicine. 55: :102694. |
[10] | Krüger-Genge A , Blocki A , Franke RP , Jung F . Vascular Endothelial Cell Biology: An Update. Int J Mol Sci (2019) ;20: (18). |
[11] | Tomar A , Sahoo S , Aathi M , Kuila S , Khan MA , Ravi GRR , Jeyaraman J , Mehta JL , Varughese KI , Arockiasamy A . Exploring the druggability of oxidized low-density lipoprotein (ox-LDL) receptor, LOX-1, a proatherogenic drug target involved in atherosclerosis. Biochem Biophys Res Commun. 623: :59–65. |
[12] | Yang J , Rong S . The Emerging Role of CircRNAs in Atherosclerosis. Curr Vasc Pharmacol. 21: (1):26–41. |
[13] | Luo X , Zhou X . CircRNA-PTPRA Knockdown Inhibits Atherosclerosis Progression by Repressing ox-LDL-Induced Endothelial Cell Injury via Sponging of miR-671-5 Biochem Genet. 61: (1):187–201. |
[14] | Yuan R , Xin Q , Ma X , Yu M , Miao Y , Chen K , Cong W . Identification of a Novel Angiogenesis Signalling circSCRG1/miR-1268b/NR4A1 Pathway in Atherosclerosis and the Regulatory Effects of TMP-PF In Vitro. Molecules. 28: (3). |
[15] | Yang L , Chen W , Li B , Hu Y , Lu H , Zhang P , Yang H , Zhang M , Pan D . Circular RNA circ_0026218 Suppressed Atherosclerosis Progression via miR-338-3p/SIRT6 Axis. Biomed Res Int. 2023;5647758. |
[16] | Wang Y , Chen X , Lu Z , Lai C . Circ_0093887 regulated ox-LDL induced human aortic endothelial cells viability, apoptosis, and inflammation through modulating miR-758-3p/BAMBI axis in atherosclerosis. Clin Hemorheol Microcirc (2022) :81: (4):343–358. |
[17] | Wang Y , Liu P , Chen X , Yang W . Circ CHMP5 aggravates oxidized low-density lipoprotein-induced damage to human umbilical vein endothelial cells through miR-516b-5p/TGFβR2 axis. Clin Hemorheol Microcirc. 2023. doi: 10.3233/ch-231722. |
[18] | Dong Z , Deng L , Peng Q , Pan J , Wang Y . CircRNA expression profiles and function prediction in peripheral blood mononuclear cells of patients with acute ischemic stroke. J Cell Physiol. 235: (3):2609–2618. |
[19] | Yu F , Zhang Y , Wang Z , Gong W , Zhang C . Hsa_circ_0030042 regulates abnormal autophagy and protects atherosclerotic plaque stability by targeting eIF4A3. Theranostics. 11: (11):5404–5417. |
[20] | Cai Y , Xu L , Xu C , Wang Y , Fan C . Hsa_circ_0001445 inhibits ox-LDL-induced HUVECs inflammation, oxidative stress and apoptosis by regulating miRNA-640. Perfusion. 37: (1);86–94. |
[21] | Yu W , Liu Z , La Y , Feng C , Yu B , Wang Q , Liu M , Li Z , Feng Y , Ciren L , Zeng Q , Zhou J , Zhao X , Jia P , Yang S . Associations between residential greenness and the predicted 10-year risk for atherosclerosis cardiovascular disease among Chinese adults. Sci Total Environ. 868: :161643. |
[22] | Panopoulos S , Drosos GC , Konstantonis G , Sfikakis PP , Tektonidou MG . Generic and disease-adapted cardiovascular risk scores as predictors of atherosclerosis progression in SLE. Lupus Sci Med. 10: (1). |
[23] | Chen F , Ning Y , Liu J , Lian M , Wang J , Dan H . miRNA miR-147a targets ZEB2 to regulate ox-LDL-induced monocyte adherence to HUVECs, atherosclerotic plaque formation and stability in atherosclerosis. J Biol Chem. doi:10.1016/j.jbc.2023.104657104657. |
[24] | Huang JG , Tang X , Wang JJ , Liu J , Chen P , Sun Y . A circular RNA, circUSP36, accelerates endothelial cell dysfunction in atherosclerosis by adsorbing miR-637 to enhance WNT4 expression. Bioengineered. 12: (1):6759–6770. |
[25] | Teng Y , Ren F , Wang Y , Xu H , Song H . Circ_0033596 depletion ameliorates oxidized low-density lipoprotein-induced human umbilical vein endothelial cell damage. Clin Hemorheol Microcirc (2023) ;84: (1):53–70. |
[26] | He D , Li Z , Chen Y , Huang M . Circular RNA circ_0029589 promotes ox-LDL-induced endothelial cell injury through regulating RAB22A by serving as a sponge of miR-1197. Clin Hemorheol Microcirc. (2023) ;83: (4):359–376. |
[27] | Liu Y , Liu N , Liu Q . Constructing a ceRNA-immunoregulatory network associated with the development and prognosis of human atherosclerosis through weighted gene co-expression network analysis. Aging (Albany NY). 13: (2):3080–3100. |
[28] | Holdt LM , Stahringer A , Sass K , Pichler G , Kulak NA , Wilfert W , Kohlmaier A , Herbst A , Northoff BH , Nicolaou A , Gabel G , Beutner F , Scholz M , Thiery J , Musunuru K , Krohn K , Mann M , Teupser D . Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 7: :12429. |
[29] | Dietrich B , Haider S , Meinhardt G , Pollheimer J , Knofler M . WNT and NOTCH signaling in human trophoblast development and differentiation. Cell Mol Life Sci. 79: (6):292. |
[30] | Weng M , Zhao R , Niu Q , Zeng Y , Wang X , Gao X , Han Z , Bing J , Xi C , Liu J , Xu J , Yang S , Zeng S . Adenovirus-mediated effects of Wnt and Notch signalling pathways on hair cell regeneration in mice. Biochem Biophys Res Commun. 658: :44–54. |
[31] | Zhang L , Cheng H , Yue Y , Li S , Zhang D , He R . H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/beta-catenin in ox-LDL -stimulated vascular smooth muscle cells. J Biomed Sci. 25: (1):11. |
[32] | Peymanfar Y , Su YW , Hassanshahi M , Xian CJ . Therapeutic Targeting Notch2 Protects Bone Micro-Vasculatures from Methotrexate Chemotherapy-Induced Adverse Effects in Rats. Cells. 11: (15) |
[33] | Gaudette BT , Roman CJ , Ochoa TA , Gomez Atria D , Jones DD , Siebel CW , Maillard I , Allman D . Resting innate-like B cells leverage sustained Notch2/mTORC1 signaling to achieve rapid and mitosis-independent plasma cell differentiation. J Clin Invest.. 131: (20) |
[34] | Jiao WE , Xu S , Qiao YL , Kong YG , Sun L , Deng YQ , Yang R , Tao ZZ , Hua QQ , Chen SM . Notch2-dependent GATA3+Treg cells alleviate allergic rhinitis by suppressing the Th2 cell response, Int Immunopharmacol. 112 109261. |
[35] | Lin X , Wang S , Sun M , Zhang C , Wei C , Yang C , Dou R , Liu Q , Xiong B . miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol. 12: (1):20. |
[36] | Duarte A , Hirashima M , Benedito R , Trindade A , Diniz P , Bekman E , Costa L , Henrique D , Rossant J . Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 18: (20):2474–2478. |
[37] | Davis-Knowlton J , Turner JE , Turner A , Damian-Loring S , Hagler N , Henderson T , Emery IF , Bond K , Duarte CW , Vary CPH , Eldrup-Jorgensen J , Liaw L . Characterization of smooth muscle cells from human atherosclerotic lesions and their responses to Notch signaling. Lab Invest. 99: (3):290–304. |
[38] | Davis-Knowlton J , Turner JE , Harrington A , Liaw L . Smooth Muscle Cell Notch2 Is Not Required for Atherosclerotic Plaque Formation in ApoE Null Mice. J Vasc Res. 59: (5):261–274. |




