Hsa_ circ_0006867 regulates ox-LDL-induced endothelial injury via the miR-499a-3p/ADAM10 axis
Abstract
Circular RNAs (circRNAs) have been reported to participate in the development of various diseases. In this study, we investigated the potential mechanism underlying the role of circRNAs in atherosclerosis. Human umbilical vein endothelial cells (HUVECs) were treated with 100 μg/mL oxidized low-density lipoprotein (ox-LDL) to simulate atherosclerosis. We observed that hsa_circ_0006867 (circ_0006867), a circRNA markedly increased in ox-LDL-treated endothelial cells, acted as a molecular sponge of miR-499a-3p and regulated its expression. This interaction led to changes in the downstream target gene ADAM10, thus affecting cell apoptosis and migration. Thus, our study suggests that circ_0006867 regulates ox-LDL-induced endothelial injury via the circ_0006867/miR-499a-3p/ADAM10 axis, indicating its potential as an exploitable therapeutic target for atherosclerosis.
1Introduction
Atherosclerosis (AS) is a complex process involving lipoprotein metabolism, inflammation, oxidative stress and endothelial injury, which is generally considered to be the common pathological mechanism of many cardiovascular diseases [1]. Under physiological conditions, endothelial cells are key regulators of vascular homeostasis [2]. Oxidized low-density lipoprotein (ox-LDL)-evoked endothelial cell injury is considered to be an important factor in the pathogenesis of AS [3]. However, the pathogenesis of AS is still unclear.
MicroRNAs (miRNAs) are short (approximately 22 nucleic acids) single-stranded noncoding RNAs found in almost all living organisms and have been reported to participate in the progression of many diseases [4–7]. Several studies have highlighted the potential role of abnormally expressed miRNAs as new biomarkers and intervention targets in atherosclerosis [8, 9]. For example, downregulation of miR-211-5p in AS patients indicates a poor prognosis [10]. MiR-455-5p inhibits cell proliferation and migration by targeting SOCS3 and is considered to be a reliable predictor of atherosclerosis risk. MiR-34a was found to influence the progression of atherosclerosis by regulating Bcl-2 [11].
MiR-499a-3p has been detected at high levels in AS patient serum and could promote endothelial cell proliferation and migration by directly regulating myocyte enhancement factor 2C (MEF2C) [12]. In contrast, our present study showed that miR-499a-3p expression was significantly decreased in ox-LDL-induced endothelial cells, which prompted us to further explore the regulatory mechanism.
Circular RNAs (circRNAs), which are covalent closed-loop single-stranded RNA without 5’ and 3’ poly A tails, are widely distributed throughout the human transcriptome and function as competitive endogenous RNAs (ceRNAs) by sponging miRNAs, thereby regulating gene expression and influencing disease progression [13–15]. For example, silencing circIRAK1 promotes viability and angiogenesis of ox-LDL-stimulated endothelial cells by regulating the miR-330-5p/TRIM14 axis [16]. It has been reported that down-regulation of circ_0091822 protects endothelial cells against ox-LDL-induced injury by sponging miR-661 [17]. Wei et al. demonstrated that knockdown of circ_HECW2 protected endothelial cells against ox-LDL-evoked cytotoxicity via sponging miR-942-5p to regulate the level of TLR4 [18]. In the present study, we used circBank online software to predict that circ_0006867 binds to miR-499a-3p and is highly expressed in ox-LDL-treated endothelial injury. Circ_0006867 was reported to be aberrantly expressed in inflammatory factor-stimulated chondrocyte injury [19]. Our study revealed for the first time the functions of circ_0006867 in endothelial cells and its influence on the expression level of a disintegrin and metalloprotease 10 (ADAM10) by competing for miR-499-3p, and thus protecting endothelial cells from ox-LDL-induced injury. Our findings may provide a new therapeutic candidate for AS.
2Materials and methods
2.1Cell culture
Human umbilical vein endothelial cells (HUVECs) were obtained from the Cell Bank of the Shanghai Research Institute, Chinese Academy of Medical Sciences (Shanghai, China). The cells were cultured at 37°C in endothelial cell culture medium containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin, 100 μg/ml streptomycin and 1% endothelial cell growth supplement (ECGS) in a humidified atmosphere of 5% CO2. At approximately 70% confluence, the HUVECs were treated with ox-LDL (Solarbio, Beijing, China) at a concentration of 100 mg/L for 24 h for further experiments.
2.2Cell transfection
Circ_0006867 siRNA (si-circ), miR-499a-3p mimic (miR-499a-3p), miR-499a-3p inhibitor (miR-499a-3p inhi) and their respective negative control were purchased from Han Heng (Shanghai, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen, China) according to the manufacturer’s protocols. After 48 h of culture, the cells were used in the experiments.
2.3Cell counting kit-8 (CCK-8) assay
Cell viability was assayed using a CCK-8 kit (Dojindo, Shanghai, China) according to the manufacturer’s protocols. Cells were seeded into 96-well plates at a density of 5×103 cells/well before 10 μl of CCK-8 reagent was added to each well. After 2 h of incubation at 37°C, the optical density (OD) at 450 nm was then measured with a microplate reader (Thermo Fisher, MA, USA).
2.4Flow cytometry
Cell apoptosis was determined by flow cytometry with an annexin V-fluorescein isothiocyanate (annexin V-FITC)/PI double-staining kit (BestBio, Shanghai, China). Cells (1×105) were suspended in the binding buffer containing 5 μL each of annexin V-FITC and PI. and incubated away from light at room temperature. After 30 min, the apoptotic cells were determined using a flow cytometer (BD Biosciences, San Jose, CA, USA).
2.5Migration assay
Cell migration ability was measured by Transwell assay (8 μm; BD Biosciences). Briefly, cells (5×104 cells) suspended in 100 μl of serum-free medium were incubated in the upper chamber, while the lower chamber was filled with 20% FBS to induce migration. After incubation for 24 h, the cells on the lower side of the chamber were fixed in 4% paraformaldehyde for 20 min, stained with 0.1% crystal violet staining solution (Beyotime, Nantong, China) for 10 min, and then counted and photographed in five representative fields.
2.6Wound healing assay
Cell motility was evaluated in wound healing assays. Briefly, cells were seeded into 6-well plates. At 80%–90% confluence, a sterile 200-μL pipette tip was used to introduce a straight line scratch (wound) into the cell monolayer, and the cell debris was washed away with PBS. The damaged monolayer was allowed to heal under standard conditions for 24 h. The cells were observed under a microscope at 0 and 24 h and the scratch width was measured using ImageJ software (NIH, Bethesda, MD, USA).
2.7Dual-luciferase reporter assay
The relationship between circ_0006867, miR-499a-3p and ADAM10 was verified by a dual luciferase reporting assay. The wild-type luciferase vectors encoding circ_0006867 and ADAM10 (circ_0006867-wt and ADAM10-wt) and their mutant luciferase vectors (circ_0006867-mu and ADAM10-mu) were synthesized by Han Heng Co., Ltd. (Shanghai, China). The luciferase vector and miR-499a-3p mimic or mimic NC were cotransfected into 293T cells for 48 h by Lipofectamine 3000. Subsequently, the luciferase activity was examined using a dual luciferase reporter detection system (Promega Corp., Madison, WI, USA) according to the manufacturer’s instructions.
2.8RNA immunoprecipitation (RIP)
RIP assays were performed with a Magna RIP Kit (Millipore, MA, US) according to the manufacturer’s guidelines. RT-qPCR was used to detect the enrichment of circ_0006867 and miR-499a-3p. IgG and Ago2 antibodies for RIP were purchased from Abcam.
2.9Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from cultured HUVECs with TRIzol reagent (Sigma, USA), and 1 μg of total RNA was reverse transcribed into cDNA using PrimeScriptTM RT Master Mix (TaKaRa). RT-qPCR was then performed using SYBR Premix Ex TaqTM II (TaKaRa) to detect the expression levels of circ_0006867 and ADAM10 using the Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems, CA, USA). GAPDH was used as an internal reference. The relative gene expression was analyzed by 2-ΔΔCT method. MiR-499a-3p expression was examined by TaqMan miRNA assay, and U6 snRNA was used as an internal reference. The primers were as follows:
| Circ_0006867 | F: 5’-ATCTTACCTTGCCCACCA-3’ |
| R: 5’-GATTGATCGTATCTCTGTGAACA-3’ | |
| LRBA | F: 5’-TATCCAGGATGTGACGCTGGAA-3’ |
| R: 5’-GTCACCATCAGTAGCGGGTTTG-3’ | |
| ADAM10 | F: 5’-CGATTGGTTATATACTGGGCACTGA-3’ |
| R: 5’-AACCAAGTATGACACCTGCCAAAG-3’ | |
| GAPDH | F: 5’- GGGAAACTGTGGCGTGAT-3’ |
| R: 5’-GAGTGGGTGTCGCTGTTGA -3’ | |
| MiR-499a-3p | F: 5’-AACATCACAGCAAGTCTGTGCT-3’ |
| R: 5’-AGTGCAGGGTCCGAGGTATT-3’ | |
| U6 | F: 5′-GCTTCGGCA GCACATATACTAAAAT-3’ |
| R: 5’-CGCTTCACGAATTTGCGTGTCAT-3’ |
2.10RNase R treatment
RNA samples (2.5 μg) were treated with or without 10 units RNase R (Geneseed, Guang Zhou, China; Cat. no. R0301) for 30 min at room temperature, and the expression levels of circ_0006867 and LPS responsive beige-like anchor (LRBA) mRNA were detected by RT-qPCR.
2.11RNA fluorescence in situ hybridization (FISH)
FISH assays were performed using a Fluorescent In Situ Hybridization Kit (RiboBio, Guangzhou, China) according to the manufacturer’s protocols. The Cy3-labeled probes were observed by a confocal microscopy (Leica, Germany).
2.12Western blot analysis
Total proteins were extracted from the transfected HUVECs by radioimmunoprecipitation assay buffer (RIPA, Beyotime, China). The proteins were then separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. After being blocked by immersion in 10% skimmed milk at room temperature for 2 h, the membranes were incubated overnight at 4°C with the following primary antibodies: anti-Bax (1:1,000), anti-Bcl-2 (1:1,000), anti-c-caspase 3 (1:1,000) and anti-ADAM10 (1:500). All the antibodies were purchased from Abcam. The membranes were then washed three times with TBST and incubated with horseradish peroxidase-conjugated secondary antibodies at room temperature for 2 h. Finally, the protein bands were visualized with an ECL kit (Pierce, Thermo Fisher Scientific, IL, USA).
2.13Statistical analysis
Data were presented as the mean±standard deviation (SD). Comparisons between groups were analyzed by Student’s t-test and one-way analysis of variance (ANOVA) with P < 0.05 set as the threshold for statistical significance. All statistical analyses and graph construction were performed using SPSS 22.0 and GraphPad Prism 7.
3Results
3.1Ox-LDL induced the inhibition of endothelial cell viability, migration and the promotion of apoptosis
Since ox-LDL can act as an inducer of AS, ox-LDL induced endothelial cell injury was dose - and time-dependent, with a concentration of 100 mg/L treated for 24 hours as the optimal treatment condition [20, 21]. Therefore, endothelial cells exposed to a concentration of 100 mg/L treatment for 24 h was chose as experimental condition. We first analyzed the effect of ox-LDL on endothelial cells. CCK-8 assay showed that after treatment with ox- LDL, cell viability was significantly inhibited (Fig. 1A). Then the apoptosis rate in the ox-LDL treatment group was significantly higher than that in the control group (Fig. 1B). Furthermore, the protein levels of Bcl-2 in HUVECs were significantly inhibited, while Bax and cleaved caspase 3 were increased due to the stimulation of ox-LDL (Fig. 1C). In addition, cell migration detected by Wound Healing (Fig. 1D) and Transwell (Fig. 1E) assays was inhibited by ox-LDL. These data indicated that ox-LDL inhibitd cell viability and migration while promoting apoptosis of HUVECs.
Fig. 1
Ox-LDL induced the inhibition of endothelial cell viability, migration and the promotion of apoptosis. Cell viability (A), apoptosis (B), and expression of the apoptosis-related proteins Bcl-2, Bax, and cleaved-caspase 3 (C) detected by CCK-8 assay, flow cytometry, and Western blot analysis, respectively, in HUVECs treated with Control or 100 mg/L ox-LDL. Wound Healing (E) and Transwell (F) assays of cell migration ability. *P < 0.05.
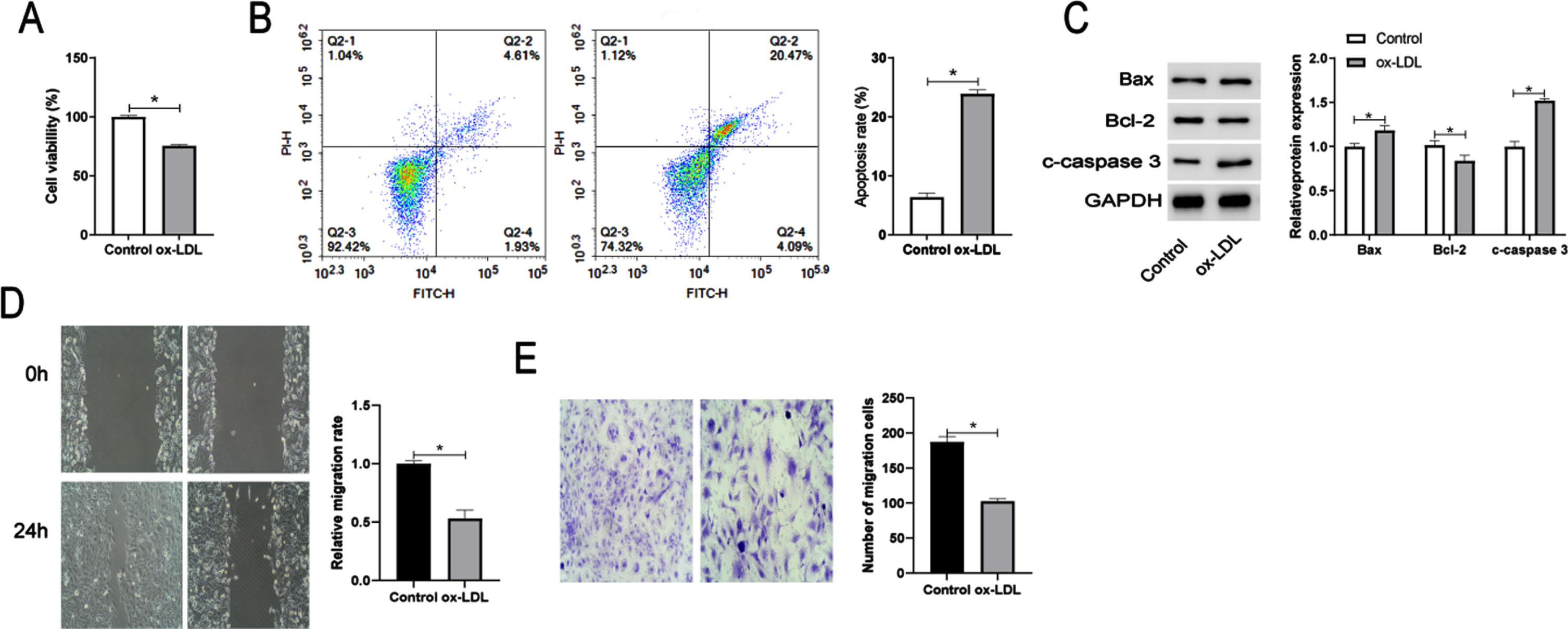
3.2MiR-499a-3p inhibits the proliferation and migration but promotes apoptosis of ox-LDL-induced HUVECs
After HUVECs were exposed to 100 mg/L ox-LDL, RT-qPCR analysis showed that miR-499a-3p expression was down-regulated (Fig. 2A). Next, we explored the functions of miR-499a-3p in regulating ox-LDL-induced endothelial cells by transfecting HUVECs with miR-499a-3p mimic and inhibitor. The miR-499a-3p mimic significantly elevated the expression of miR-499a-3p in ox-LDL-induced HUVECs, while the miR-499a-3p inhibitor markedly reduced its expression (Fig. 2B). CCK-8 assays demonstrated that ectopic expression of miR-499a-3p inhibited cell viability and this effect was prevented by inhibition of miR-499a-3p (Fig. 2C). Flow cytometric analysis indicated that miR-499a-3p enhanced cell apoptosis induced by ox-LDL (Fig. 2D). In addition, Western blot analysis of the levels of the apoptosis-related proteins Bax, Bcl-2 and c-caspase 3 further confirmed that miR-499a-3p_induced apoptosis in ox-LDL-treated HUVECs (Fig. 2E). Wound healing and Transwell assays demonstrated that the migration ability of ox-LDL-induced HUVECs was suppressed by miR-499a-3p upregulation, promoted by miR-499a-3p downregulation (Fig. 2F, G). From these results, we concluded that miR-499a-3p inhibited the proliferation and migration of ox-LDL-induced HUVECs.
Fig. 2
Expression and functions of miR-499a-3p in ox-LDL-induced HUVECs. (A) Relative miR-499q-3p expression level in ox-LDL-induced HUVECs. (B-G) HUVECs treated with ox-LDL were transfected with miR-NC, miR-499a-3p mimic, inhi-NC and miR-499a-3p inhi. (B) RT-qPCR verification of the efficiency of transfection of HUVECs. Detection of cell viability (C), apoptosis (D), and expression of apoptosis-related proteins Bcl-2, Bax, and cleaved-caspase 3 (E) by CCK-8 assay, flow cytometry, and WB analysis. Cell migration ability was detected by wound healing (F) and Transwell (G) assays. *P < 0.05.
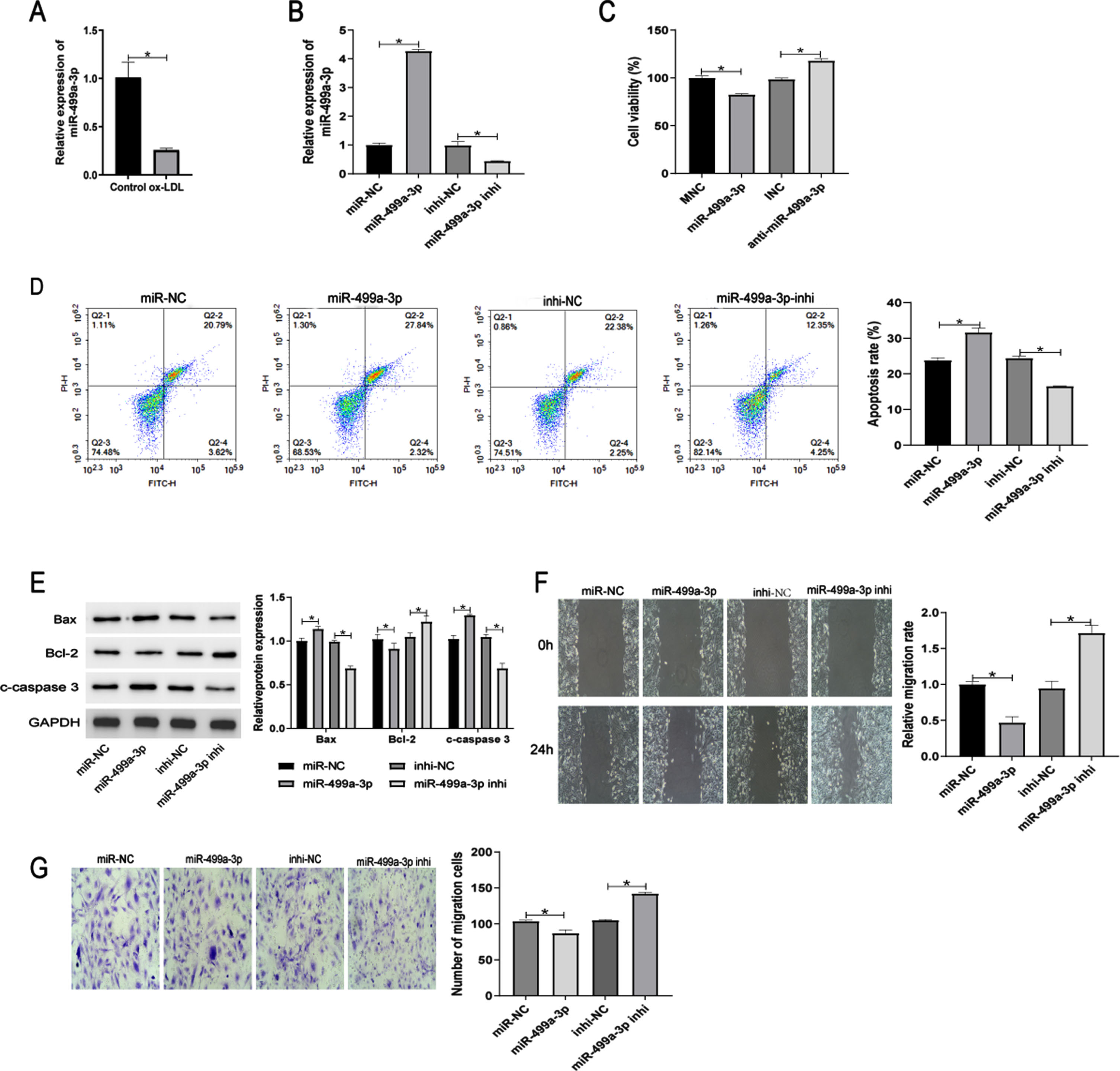
3.3ADAM10 is a functional target gene of miR-499a-3p
We next investigated the functional target gene of miR-499a-3. TargetScan and RNAhybrid online software predicted that the gene, ADAM10 binds directly to the miR-499a-3p seed sequence (Fig. 3A). This relationship was confirmed by dual-luciferase reporter assays (Fig. 3B), which showed that the miR-499a-3p mimic significantly inhibited the luciferase activity of ADAM10-Wt plasmid, but had no effect on the luciferase activity of ADAM10-Mut plasmid. Furthermore, RT-qPCR analysis revealed that ADAM10 mRNA expression was suppressed by the miR-499a-3p mimic, but elevated by the miR-499a-3p inhibitor (Fig. 3C). Western blot analysis confirmed this pattern of changed in the expression of ADAM10 at the protein level (Fig. 3D). These results showed that ADAM10 is a target gene of miR-499a-3p.
Fig. 3
ADAM10 is a target gene of miR-499a-3p. MiR-499a-3p and ADAM10 binding sites predicted by TargetScan and RNAhybrid. (B) Confirmation of the predicted binding by dual-luciferase reporter assay. (C-D) ADAM10 mRNA and protein expression in ox-LDL HUVECs after transfection with miR-499a-3p mimic or inhibitor. *P < 0.05.
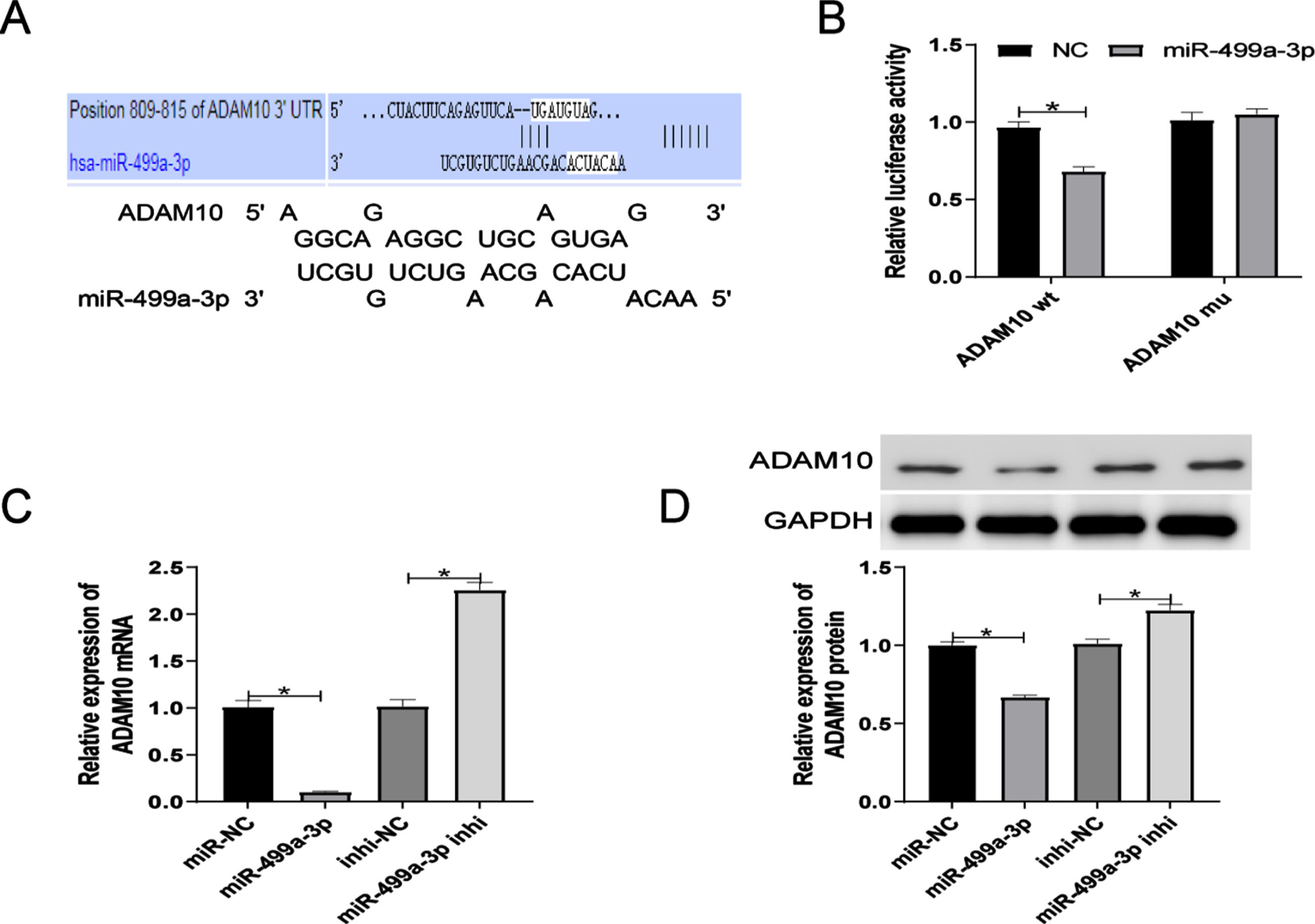
3.4Identification of circ_0006867 as a novel upregulated circRNA in ox-LDL-induced HUVECs and validation as a molecular sponge of miR-499a-3p
Circ_0006867 was significantly upregulated in HUVECs treated with ox-LDL (Fig. 4A). We also found that circ_0006867 was derived from LRBA on chromosome 4. LRBA deficiency has been shown to be associated with vesicular transport, autophagy and apoptosis [22]. Specific primers were designed and the amplified products were verified by Sanger sequencing (Fig. 4B). FISH assay revealed that circ_0006867 was mainly located in the cytoplasm of HUVECs (Fig. 4C). Furthermore, RNase R digestion assays showed the structural stability of circ_0006867 (Fig. 4D). Overall, circ_0006867 was found to be upregulated in ox-LDL-induced HUVECs, suggesting that it plays an important role in endothelial cell injury induced by ox-LDL.
Fig. 4
Identification of circ_0006867 as a novel upregulated circRNA in ox-LDL-induced HUVECs and validation as a molecular sponge of miR-499a-3p. Relative expression of circ_0006867 in ox-LDL-induced HUVECs. (B) Validation of circ_0006867 by Sanger sequencing. (C) FISH localization of circ_0006867. (D) Relative expression of circ_0006867 and LRBA mRNA after RNase R treatment. (E) The predicted binding sites between circ_0006867 and miR-499a-3p. (F-G). Verification of the binding between circ_0006867 and miR-499a-3p by dual-luciferase reporter assay and RNA immunoprecipitation assays. (H). RT-qPCR analysis of the effect of circ_0006867 knockdown on miR-499q-3p expression. *P < 0.05.
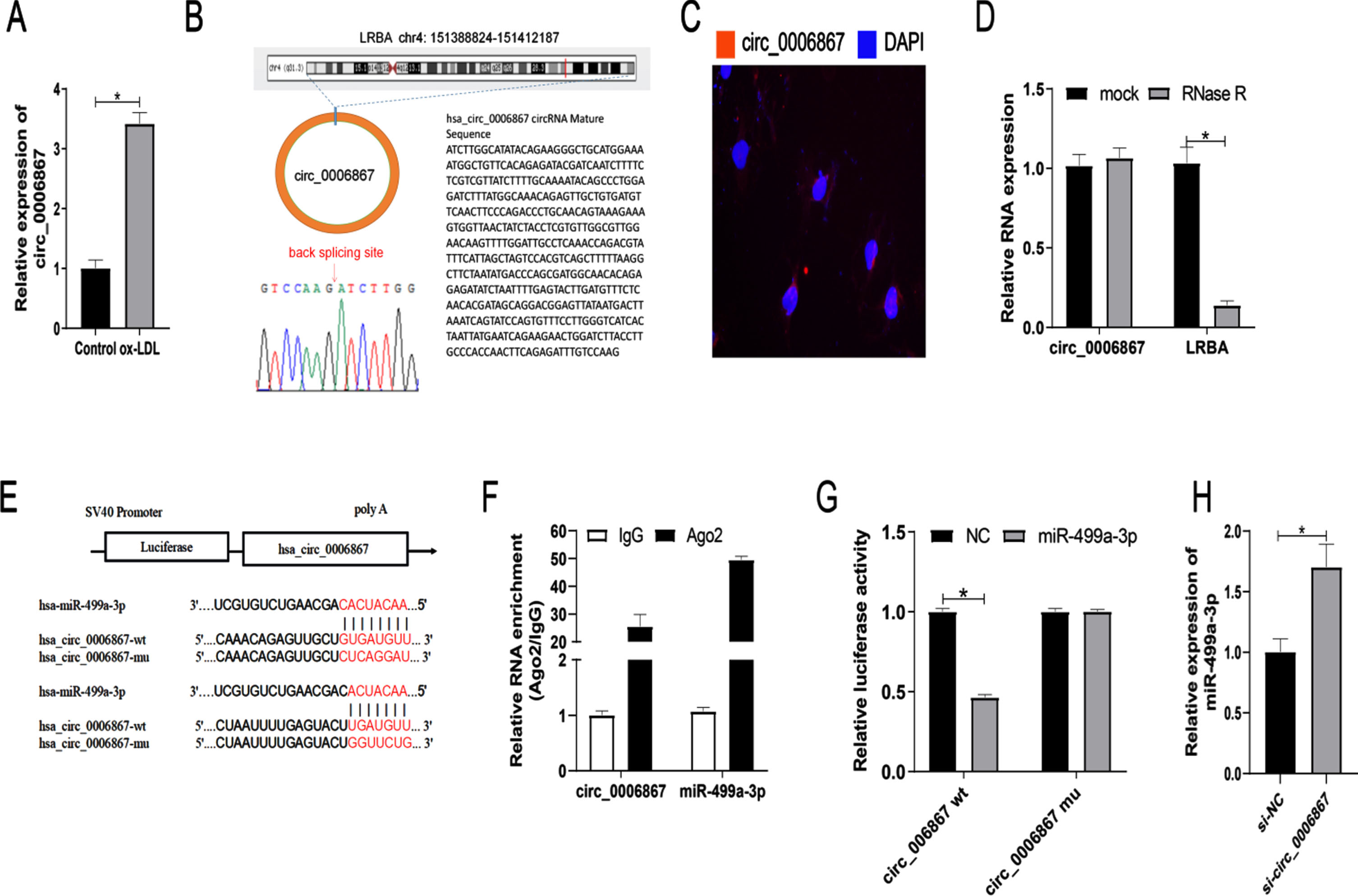
Next, we investigated the relationship between circ_0006867 and miR-499a-3p. Bioinformatic analysis by circbank demonstrated that circ_0006867 contains complementary binding sites for miR-499a-3p (Fig. 4E). This binding was further confirmed by RNA immunoprecipitation and dual luciferase reporter assay (Fig. 4F, G). The luciferase activity mediated by wild-type circ_0006867 vector was significantly decreased by cotransfection with miR-499a-3p mimic. Additionally, circ_0006867 silencing increased the expression of miR-499a-3p in ox-LDL-induced HUVECs (Fig. 4H). These results indicated that circ_0006867 acts as a molecular sponge of miR-499a-3p to negatively regulate its expression.
3.5Circ_0006867 regulates the apoptosis and migration of ox-LDL-induced HUVECs via the miR-499a-3p/ADAM10 axis
Having shown that circ_0006867 targets miR-499a-3p and identified ADAM10 as a target gene of miR-499a-3p, we decided to further investigate whether circ_0006867 functions by regulating the miR-499a-3p/ADAM10 axis. We designed two small interfering RNAs (si-RNAs) against backsplice sequences and then detected the interfering efficiency by RT-qPCR before finally selecting si-circ_0006867-1 for follow-up experiments (Supplementary Figure 1A). Circ_0006867 silencing suppressed ADAM10 expression at both the transcriptional and translational levels, while this effect was reversed by the miR-499a-3p inhibitor (Fig. 5A, B). CCK-8 assays showed that the miR-499a-3p inhibitor largely alleviated the suppression of ox-LDL-treated HUVEC viability mediated by circ_0006867 downregulation (Fig. 5C). Additionally, flow cytometric analysis of apoptosis and Western blot analysis of apoptosis-related proteins demonstrated that si-circ_0006867 enhanced the apoptosis of HUVECs induced by ox-LDL and this effect was alleviated by the miR-499a-3p inhibitor (Fig. 5D, E). The effects of circ_0006867 on HUVEC migration were attenuated by the miR-499a-3p inhibitor (Fig. 5F, G). These results suggest that circ_0006867 functions as a cytoprotective factor in atherosclerosis by acting as a decoy for miR-499a-3p to regulate the expression of ADAM10 (Fig. 6).
Fig. 5
Circ_0006867 functions via the miR-499a-3p/ADAm10 axis. (A–G) HUVECs treated with ox-LDL were transfected with si-NC, si-circ, si-circ+inhi-NC and si-circ+miR-499a-3p inhi. RT-qPCR and Western blot analyses of ADAM10 mRNA (A) and protein levels (B), respectively. Cell viability (C), apoptosis (D), and expression of the apoptosis-related proteins Bcl-2, Bax, and cleaved-caspase 3 (E) detected by CCK-8 assay, flow cytometry, and Western blot analysis, respectively. Wound Healing (F) and Transwell (G) assays of cell migration ability. *P < 0.05.
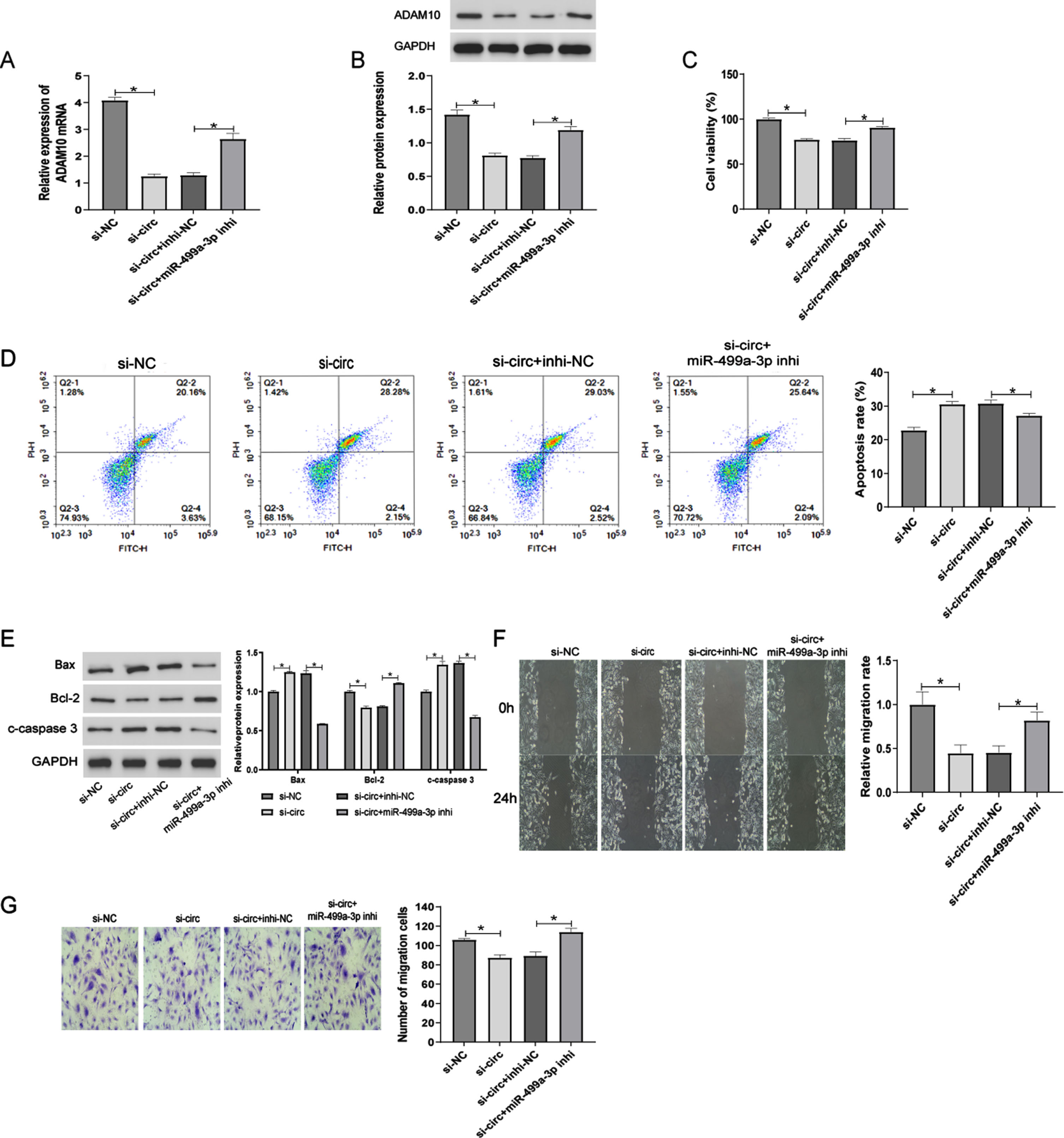
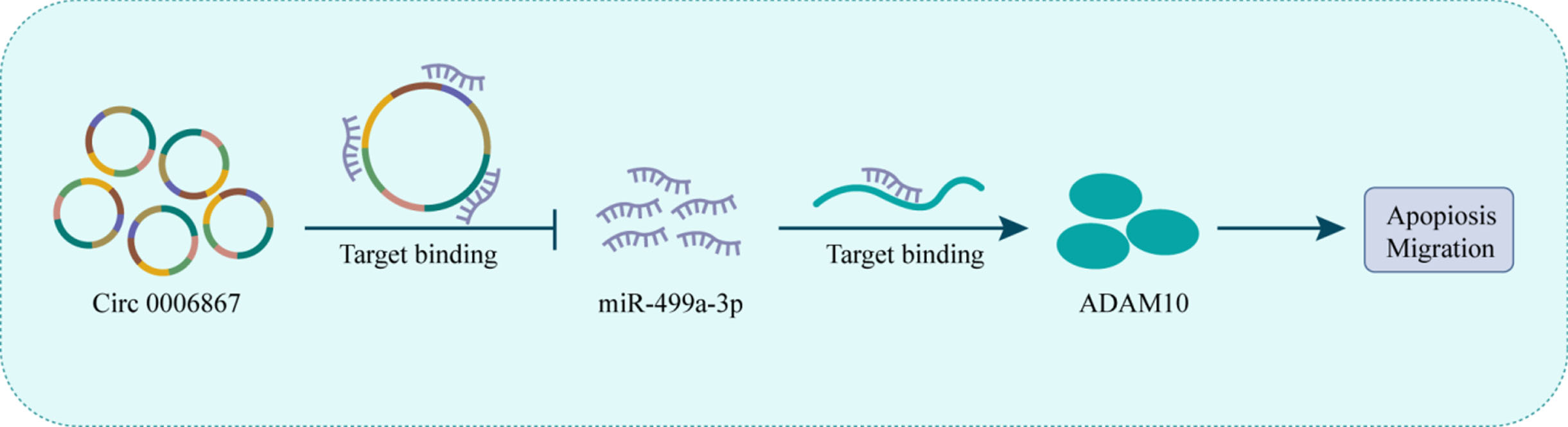
Fig. 6
Schematic representation of the the proposed mechanism of 00006867 in ox- LDL-induced endothelial cell injury. Circ_0006867 acts as a miR-499a-3p sponge to regulate the miR-499a-3p/ADAM10 pathway. Decreased circ_0006867 in ox-LDL-induced endothelial cell leads to the upregulation of miR-499a-3p, which downregulated ADAM10 expression, thereby increasing cell aptopsis and blocking cell migration.
4Discussion
Atherosclerosis is the common pathological basis of many cardiovascular diseases, such as CHD [23, 24]. Endothelial cell injury is considered to be the initial step in the progression of AS [25–27]. Therefore, identification of key molecules that regulate the proliferation, migration and apoptosis of endothelial cells is a critical step in the development of strategies for the prevention and treatment of AS. As one of the major risk factors for atherosclerosis, ox-LDL can increase reactive oxygen species production and the expression of leukocyte adhesion molecules, activate apoptotic pathway, and finally lead to endothelial cell dysfunction [28]. In this study, endothelial cell injury was stimulated by treatment with 100 mg/L ox-LDL for 24 h to simulate AS in vitro. We noted a significant downregulation of miR-499a-3p, which has been reported to be abnormally expressed in the serum of AS patients and to have a protect endothelial cells against apoptosis [12]. Zheng et al. found that lentivirus-mediated upregulation of miR-499a-3p could inhibit endothelial cell proliferation and migration [29]. In contrast to the findings of a previous study [12], we found that miR-499a-3p enhanced ox-LDL-induced endothelial cell apoptosis and inhibited cell migration.
Accumulating evidence demonstrates that circRNAs can serve as molecular sponges of miRNAs to regulate gene expression and thus, contribute to disease progression [30, 31]. In addition, circRNA-miRNA-mRNA regulatory networks are reportedly involved in the pathological process of AS [32, 33]. The biological functions of circRNAs were predicted by functional annotation of downstream target genes of miRNAs [34]. CircRSF1 was shown to regulate CCND2 expression by sponging miR-758, thus protecting against ox-LDL-induced endothelial cell injury in vitro [35]. Zhao et al. found that circ_USP36 knockdown reduced ox-LDL-mediated vascular smooth muscle cell injury by regulating the miR-182-5p/KLF5 axis [36]. Therefore, in the current study, we hypothesized that the regulatory mechanism of miR-499a-3p was mediated by circRNAs. Bioinformatics analysis revealed the presence of miR-499a-3p binding sites in circ_0006867. And silencing circ_00006867 inhibited the proliferation, migration and invasion of fibroblasts [37]. Subsequently, we characterized the existence of circ_0006867 and defined its expression trend in ox-LDL-induced endothelial cells. We showed that circ_0006867 negatively regulates miR-499a-3p and dual luciferase reporter assays further confirmed this binding relationship. While ADAM10 is a target gene of mir-499a-3p. ADAM10 is highly expressed in AS plaques, promotes endothelial cell proliferation and migration, and is related to plaque progression and neovascularization [38–40]. Additionally, Vorst et al found that endothelial ADAM10 has an atheroprotective effect, most likely by limiting ox-LDL-induced inflammation [41]. Further Rescue experiments revealed that circ_00006867 attenuated ox-LDL-induced endothelial cell apoptosis and promoted migration via the miR-499a-3p/ADAM10 axis. Thus, our study not only elucidates the regulatory role of miR-499a-3p in endothelial cells, but also implicates circ_0006867 as a potential intervention target for atherosclerosis.
However, some limitations of our study should be acknowledged. First, miR-499a-3p expression in endothelial cells requires validation in a larger cohort of AS patients. A retrospective analysis of patient follow-up data is needed to determine the relationship between miR-499a-3p and the diagnosis, treatment and prognosis of AS patients. We selected the target circRNA based only on the highest free energy and significant P-value, which excluded other circRNAs involved in this process. Therefore, a systematic screen of the entire transcriptome is required to fully elucidate the regulatory mechanism of miR-499a-3p. In addition, in vivo studies are required to further validate our findings and provide a reliable target for atherosclerosis intervention.
There is growing evidence that circRNAs regulate gene expression through sponging miRNAs and play major roles in the development of AS. Our findings enrich the knowledge of the mechanism by which circ_0006867 acted as a molecular sponge of miR-499a-3p and thus, affects ADAM10 expression.
In conclusion, this study clarifies a new mechanism of AS progression, which has important implications for better clinical treatment. Exploration of AS interventions based on circRNA-miRNA-mRNA regulatory networks is certainly a promising prospect.
Ethics approval and consent to participate
None.
Consent for publication
Not applicable.
Acknowledgments
None.
Funding
This work was supported by the Programs for Natural Science Foundation of Guangxi Province of China (Grant No. 2018GXNSFBA050068); Guangxi Key Laboratory of Precision Medicine in Cardio-cerebrovascular Diseases Control and Prevention (No.17-259-85); Guangxi Clinical Research Center for Cardio-cerebrovascular Diseases (No. AD17129014) and Guangxi Medical High-level Key Talents “139” Plan Project (No. G201901006).
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflict of interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; JGH and HLZ conducted the experiments, Ji-Ge Hong wrote the manuscript, ZYZ supervised the manuscript.
Supplementary material
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/CH-231895.
References
[1] | Wolf D , Ley K . Immunity and inflammation in atherosclerosis. Circ Res. (2019) ;124: (2):315–27. |
[2] | Kruger-Genge A , Blocki A , Franke RP , Jung F . Vascular endothelial cell biology: An update. Int J Mol Sci. (2019) ;20: (18). |
[3] | Anatol Kontush LC , Isabelle E-B , Patrice T , Robert S , Anne N-S , John Chapman M . Mildly oxidized LDL particle subspecies are distinct in their capacity to induce apoptosis in endothelial cells: Role of lipid hydroperoxides. FASEB J. (2003) ;17: (1):88–90. |
[4] | Krol J , Loedige I , Filipowicz W . The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. (2010) ;11: (9):597–610. |
[5] | Wu M , Wang H , Kong D , Shao J , Song C , Yang T , et al. miR-452-3p inhibited osteoblast differentiation by targeting Smad4. PeerJ. (2021) ;9: :e12228. |
[6] | de Yebenes VG , Briones AM , Martos-Folgado I , Mur SM , Oller J , Bilal F , et al. Aging-associated miR-217 aggravates atherosclerosis and promotes cardiovascular dysfunction. Arterioscler Thromb Vasc Biol. (2020) ;40: (10):2408–24. |
[7] | Li CT , Jiang X , He XQ , Li DY , Chen SH , Yao SX , et al. Plasma microRNAs as potential biomarkers in diagnosis of acute venous thromboembolism. Clin Hemorheol Microcirc. (2023) . |
[8] | Wang Z , Zhang J , Zhang S , Yan S , Wang Z , Wang C , et al. MiR30e and miR92a are related to atherosclerosis by targeting ABCA1. Mol Med Rep. (2019) ;19: (4):3298–304. |
[9] | Zhao L , Wang B , Sun L , Sun B , Li Y . Association of miR-192-5p with atherosclerosis and its effect on proliferation and migration of vascular smooth muscle cells. Mol Biotechnol. (2021) ;63: (12):1244–51. |
[10] | Zhang Y , Wang H , Xia Y . The expression of miR-211-5p in atherosclerosis and its influence on diagnosis and prognosis. BMC Cardiovasc Disord. (2021) ;21: (1):371. |
[11] | Su G , Sun G , Liu H , Shu L , Liang Z . Downregulation of miR-34a promotes endothelial cell growth and suppresses apoptosis in atherosclerosis by regulating Bcl-2. Heart Vessels. (2018) ;33: (10):1185–94. |
[12] | Xu Z , Han Y , Liu J , Jiang F , Hu H , Wang Y , et al. MiR-135b-5p and MiR-499a-3p promote cell proliferation and migration in atherosclerosis by directly targeting MEF2C. Scientific Reports. (2015) ;5: (1). |
[13] | Chen LL . The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. (2020) ;21: (8):475–90. |
[14] | Haddad G , Lorenzen JM . Biogenesis and function of circular RNAs in health and in disease. Frontiers in Pharmacology. (2019) ;10. |
[15] | Zhang P , Wang W , Li M . Role and mechanism of circular RNA circ_0050486 in regulating oxidized low-density lipoprotein-induced injury in endothelial cells. Clin Hemorheol Microcirc. (2022) ;82: (2):107–24. |
[16] | Liu F , Gao B , Wang Y . CircIRAK1 aggravates ox-LDL-induced endothelial cell injury in atherosclerosis via TRIM14 upregulation by binding to miR-330-5p1. Clin Hemorheol Microcirc. (2022) . doi: 10.3233/CH-221551. |
[17] | Zhu L , Zhao P , Meng X , Jin H , Tuo B . Circ_0091822 aggravates ox-LDL-induced endothelial cell injury through targeting the miR-661/RAB22A axis. Clin Hemorheol Microcirc. (2023) ;83: (1):47–59. |
[18] | Wei W , Tang M , Wang Q , Li X . Circ_HECW2 regulates ox-LDL-induced dysfunction of cardiovascular endothelial cells by miR-942-5p/TLR4 axis. Clin Hemorheol Microcirc. (2022) . doi: 10.3233/CH-221550. |
[19] | Shen S , Wu Y , Chen J , Xie Z , Huang K , Wang G , et al. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Annals of the Rheumatic Diseases. (2019) ;78: (6):826–36. |
[20] | Yu H , Pan Y , Dai M , Xu H , Li J . Circ_0003423 alleviates ox-LDL-induced human brain microvascular endothelial cell injury via the miR-589-5p/TET2 network. Neurochem Res. (2021) ;46: (11):2885–96. |
[21] | Zhang X , Lu J , Zhang Q , Luo Q , Liu B . CircRNA RSF1 regulated ox-LDL induced vascular endothelial cells proliferation, apoptosis and inflammation through modulating miR-135b-5p/HDAC1 axis in atherosclerosis. Biol Res. (2021) ;54: (1):11. |
[22] | Martínez Jaramillo C , Trujillo Vargas CM . LRBA in the endomembrane system. Colombia Médica. (2018) ;49: (3):236–43. |
[23] | Libby P , Theroux P . Pathophysiology of coronary artery disease. Circulation. (2005) ;111: (25):3481–8. |
[24] | Herrington W , Lacey B , Sherliker P , Armitage J , Lewington S . Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res. (2016) ;118: (4):535–46. |
[25] | Gimbrone MA Jr , Garcia-Cardena G . Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. (2016) ;118: (4):620–36. |
[26] | Rossi R , Nuzzo A , Olaru AI , Origliani G , Modena MG . Endothelial function affects early carotid atherosclerosis progression in hypertensive postmenopausal women. J Hypertens. (2011) ;29: (6):1136–44. |
[27] | Mudau M , Genis A , Lochner A , Strijdom H . Endothelial dysfunction: The early predictor of atherosclerosis. Cardiovasc J Afr. (2012) ;23: (4):222–31. |
[28] | Kattoor AJ , Kanuri SH , Mehta JL . Role of Ox-LDL and LOX-1 in atherogenesis. Curr Med Chem. (2019) ;26: (9):1693–700. |
[29] | Zheng H , Li J , Chen Y , Gong D , Wen J , Mai L , et al. Effect of lentivirus-mediated miR-499a-3p on human umbilical vein endothelial cells. BioMed Research International. (2020) ;2020: :1–13. |
[30] | Zheng Q , Bao C , Guo W , Li S , Chen J , Chen B , et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. (2016) ;7: :11215. |
[31] | Yuan S , Liang J , Zhang M , Zhu J , Pan R , Li H , et al. CircRNA_005647 inhibits expressions of fibrosis-related genes in mouse cardiac fibroblasts via sponging miR-27b-3p. Nan Fang Yi Ke Da Xue Xue Bao. (2019) ;39: (11):1312–9. |
[32] | Miao L , Yin R-X , Zhang Q-H , Liao P-J , Wang Y , Nie R-J , et al. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Scientific Reports. (2019) ;9: (1):18314. |
[33] | Wang L , Zheng Z , Feng X , Zang X , Ding W , Wu F , et al. circRNA/lncRNA-miRNA-mRNA network in oxidized, low-density, lipoprotein-induced foam cells. DNA Cell Biol. (2019) ;38: (12):1499–511. |
[34] | Panda AC . Circular RNAs act as miRNA sponges. Adv Exp Med Biol. (2018) ;1087: :67–79. |
[35] | Wei Z , Ran H , Yang C . CircRSF1 contributes to endothelial cell growth, migration and tube formation under ox-LDL stress through regulating miR-758/CCND2 axis. Life Sci. (2020) ;259: :118241. |
[36] | Zhao Q LY , Wang X , Zhang XJ . Circ_USP36_miR-182-5p_KLF5 axis regulates the_ox-LDL-induced injury in human umbilical_vein smooth muscle cells. Am J Transl Res. (2020) ;15: (12):7855–69. |
[37] | Pang Q , Lin X , Sun J , Hu J , Dai S , Shen Y , et al. Comprehensive analysis of circular RNA expression in ceRNA networks and identification of the effects of hsa_circ_0006867 in keloid dermal fibroblasts. Frontiers in Molecular Biosciences. (2022) ;9: :800122. |
[38] | Schulz B , Pruessmeyer J , Maretzky T , Ludwig A , Blobel CP , Saftig P , et al. ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circ Res. (2008) ;102: (10):1192–201. |
[39] | Donners MM , Wolfs IM , Olieslagers S , Mohammadi-Motahhari Z , Tchaikovski V , Heeneman S , et al. A disintegrin and metalloprotease 10 is a novel mediator of vascular endothelial growth factor-induced endothelial cell function in angiogenesis and is associated with atherosclerosis. Arterioscler Thromb Vasc Biol. (2010) ;30: (11):2188–95. |
[40] | Yang K , Lu L , Liu Y , Zhang Q , Pu LJ , Wang LJ , et al. Increase of ADAM10 level in coronary artery in-stent restenosis segments in diabetic minipigs: High ADAM10 expression promoting growth and migration in human vascular smooth muscle cells via Notch 1 and 3. PLoS One. (2013) ;8: (12):e83853. |
[41] | van der Vorst EPC , Maas SL , Theodorou K , Peters LJF , Jin H , Rademakers T , et al. Endothelial ADAM10 controls cellular response to oxLDL and its deficiency exacerbates atherosclerosis with intraplaque hemorrhage and neovascularization in mice. Front Cardiovasc Med. (2023) ;10: :974918. |




