Development of multiplex PCR systems for expression profiling of human cardiomyocytes induced to proliferate by lentivirus transduction of upcyte genes
Abstract
Severe heart diseases such as myocarditis and cardiomyopathy are often characterized by progressive damages of contractile heart tissue which ultimately can lead to terminal heart failure. There is a need for relevant in vitro cultures of human cardiomyocytes to study pathogenic processes and to perform pharmacological testing of new heart drugs. By using the upcyte/EPCC (enhanced primary cell culture) approach for direct multiplication of organ-specific cells, we established proliferating human cardiomyocyte cultures derived from atrial appendages. For qualitative cardiac expression profiling we established a comprehensive set of multiplex PCR assays, selected from a panel of 32 genes, to rapidly screen changes at the transcriptional level in human ventricular and atrial cardiomyocytes. Our multiplex PCR approach revealed some donor variability of native atrial heart tissue that need to be confirmed by further studies with more samples. Our initial studies further indicated that characteristic heart muscle cell markers such as MLC-2 a, MLC-2v, CHRM2, ADRB1, DES, EDRNB, C x40 and KCNA5 were down-regulated when isolated cardiomyocytes were taken into primary cell culture. Compared to native heart tissue, proliferating atrial cardiomyocytes lacked expression of those cardiac markers but still expressed MYCD, GATA-4, Cx43, SERCA2, BNP, Tbx5, EDNRA and ACTB. Surprisingly, atrium-derived cardiomyocytes started to express NFAc4 in passage three, and cardiomyocyte marker expressions of Cx43 and BNP were even increased over cultivation time.
In conclusion, our novel multiplex PCR assays should be useful for expression profiling of native heart tissues from patients with different disease conditions and for characterization of in vitro cardiomyocyte cultures.
Abbreviations
EPCC | enhanced primary cell cultures |
HE | hematoxylin – eosin |
qPCR | quantitative PCR |
1Introduction
Cardiovascular diseases still is the number one cause of death in industrialized countries. Chronic heart diseases such as cardiomyopathies can lead to progressive degeneration of contractile heart tissue and ultimately can cause cardiovascular death or heart failure-related disability [1]. To better understand molecular processes leading to myocardial diseases and to validate new cardiovascular drugs, relevant in vitro systems of human atrial and ventricular cardiomyocytes are required. However, use of primary human cardiomyocyte cultures is restricted due to the postmitotic state of this cell type [2–5]. Attempts for specific cardiomyocyte differentiation from pluripotent stem cells are still hampered by the fetal phenotype of obtained heart muscle cells [6, 7]. In addition, in vitro cell cultures differentiated from pluripotent stem cells might contain several distinct cell types including those that could cause teratoma formation in vivo [8–10]. We have been using the upcyte/EPCC (enhanced primary cell culture) approach for direct multiplication of organ-derived cell types as an alternative to stem cells. This has already been demonstrated for human hepatocytes to be a very powerful procedure [11–14].
Here we report on human atrial cardiomyocytes that were induced to proliferate by lentiviral transduction of upcyte/EPCC factors. To benchmark expression profiles of cardiomyocyte differentiation markers in atrial and ventricular tissue biopsies, we developed comprehensive, rapid and robust multiplex PCR protocols to monitor defined target genes at the transcriptional level. Applying these protocols to proliferating human cardiomyocytes allows to monitor their differentiation status during long-term cultivation and in vivo.
2Materials and methods
2.1Human biopsy material
Atrial and ventricular tissues were obtained from patients receiving cardiac surgery because of coronary artery or valve disease. All patients gave written informed consent. The study was approved by the ethic committee of the Medical Faculty of Technical University Dresden. Heart muscle tissue was processed as described before [15]. Ventricular tissues is designated as ‘ventricular’, atrial specimens are designated as B7, B10 and B11. Heart tissue samples were stored in DMEM supplemented with 10% FBS at 4°C until enzymatic dissociation.
2.2Cryosectioning of heart biopsy material
Heart tissues were embedded in pre-cooled Neg-50 freezing medium (Richard-Allan Scientific, Kalmazoo, USA) and frozen to – 50°C onto the appropriate sample plate. The sample was cut with cryomicrotome MicromHM 560 (Microm GmbH, Walldorf, Germany) into 9μm sections at –21°C. Sections were mounted onto SuperFrost®Plus glass slides (Menzel GmbH, Braunschweig, Germany), air dried for 20 h and stored at –20°C.
2.3Hematoxylin - eosin staining
Cryosections were fixed with pre-cooled 4% formaldehyde for 10 min at 4°C followed by treatment with pre-cooled methanol/acetone (1:1) for 10 min at –20°C. This leads to membrane permeabilisation and dehydration of the cross sections. To remove the fixation solution, slides were washed for 5 min with phosphate buffered saline (PBS). Slides were incubated for 4 min in Mayer’s Hematoxylin (Fremdling, Geberskirchen, Germany), washed twice by rinsing with distilled water (ddH2O) and for further 4 min with tap water. Sections were incubated for 1 h in 1% eosin (Bio Optica, Milano, Italy), then shortly washed by rinsing with ddH2O followed by fixation of cryosections with an alcohol series of 50%, 80%, 96%, and three times 100% ethanol, respectively. Sections were stored in xylol and mounted with biomount.
2.4Indirect immunofluorescence analysis
Cryosections were dried at room temperature for 20 min. Mounted tissue sections were surrounded by a fat pen before they were fixed in pre-cooled formaldehyde at 4°C for 10 min and in methanol/ acetone (1:1) at –20°C for 10 min. Glass slides were dried for 20 min at room temperature followed by washing with PBS for 3 min. Unspecific binding sites were blocked with 1% bovine serum albumin (BSA) in PBS in a humidified chamber at room temperature for 1 h. Cryosections were incubated with primary antibody diluted in 1% BSA in PBS at 37°C in a humidified chamber for 1 h (list of antibodies in Table 1.). Slides were washed 3 times in PBS for 5 min, and cryosections were incubated with the secondary antibody diluted with 1% BSA in PBS and 0.2μg/ml DAPI in a dark and humidified chamber at 37°C for 1 h. Slides were washed again three times in PBS for 5 min. To prevent fluorescence bleaching, cryosections were covered with fluorescent mounting medium (Dako, Hamburg, Germany). Slides were stored at 4°C in the dark until analysis with fluorescence microscope.
2.5Enzymatic dissociation of heart biopsies and differential adhesion of cardiomyocytes
Tissues were minced and washed three times with HBSS buffer (8 g/l NaCl, 0.4 g/l KCl, 0.06 g/l KH2PO4, 1 g/l glucose, 47.86 mg/l Na2HPO4, 0.35 g/l NaHCO3) with active shaking at 37°C for 4 min. Tissues were then incubated in 0.25% trypsin and 0.12 mg/ml collagenase 2 (Biochrom AG, Berlin, Germany) in HBSS buffer at 37°C for 10 min. Dissociated cells were collected in DMEM supplemented with 10% FBS (Biochrom AG, Berlin, Germany). This procedure was repeated six times. Collected cells were centrifuged for 5 min at 580 g, washed with HBSS buffer and centrifugation was repeated. Cells were resuspended in myocyte growth medium (PromoCell GmbH, Heidelberg, Germany) supplemented with 1% penicillin/streptomycin. After complete attachment, cells were incubated with 1×trypsin/EDTA (Biochrom AG, Berlin, Germany) for 9 min and supernatant was discarded. Trypsin-incubation was repeated one to three times to detach non-cardiomyocytes from culture flasks.
Biopsy derived cells were cultivated in myocyte growth medium supplemented with 5% FBS, 0.5 ng/ml epidermal growth factor, 2 ng/ml basic fibroblast growth factor, 5μg/ml insulin (PromoCell GmbH, Heidelberg, Germany) and 1% penicillin/streptomycin. A lentivirus vector system (Invitrogen, Karlsruhe, Germany) was used for transduction of proliferation-inducing genes (Upcyte genes) into cardiomyocytes as described recently for hepatocytes [12]. Sub-culturing of cells was performed with the Detach Kit 30 (PromoCell GmbH, Heidelberg, Germany). Briefly, cells were washed once with HBSS buffer. For detaching, cells were incubated with trypsin/EDTA solution at room temperature; trypsin was inactivated by the same volume of Trypsin Neutralization Solution.
2.6Microscopy
For double-immunofluorescence analysis, IX81 fluorescence microscope (Olympus, Tokyo, Japan) combined with the xenon burner MT20 (Olympus) was used. Pictures were taken with FView II camera (Olympus) and documented with Olympus cellR-imaging software.
Routine cell culture pictures were taken with the phase contrast microscope CKX41 (Olympus) and documented with Olympus cellF-imaging software.
2.7RNA isolation and cDNA synthesis
For total RNA extraction, the tissue (50 – 100 mg) was homogenized in 1 ml peqGOLD TriFASTtrademark (peqLab, Erlangen, Germany). Samples were stored at –80°C until mRNA purification. To ensure complete dissociation of nucleoprotein complexes, samples were thawed and left for 5 min at room temperature. For RNA isolation, 200μl chloroform were added. The sample was shaken vigorously for 15 sec, left at room temperature for 5 min and centrifuged at 12,000 g and 4°C for 5 min. The aqueous phase was transferred into a new tube, incubated with 0.5 ml isopropanol for 15 min on ice and centrifuged at 12,000 g and 4°C for 10 min. The supernatant was removed and the pellet was washed twice with 1 ml 75% ethanol. The RNA pellet was air-dried at room temperature and resuspended in 20μl pre-heated nuclease free water.
Preparation of total cellular RNA was performed according to the manufacturer’s protocol of the Gene MATRIX Universal RNA Purification Kit (Roboklon, Berlin, Germany), but the cells were lysated directly in the culture well. cDNA synthesis was performed with SuperScripttrademark II reverse transcriptase (ThermoFisher Scientific, Waltham, USA) according to manufacturer’s protocol using oligo(dT)20 primers (ThermoFisher Scientific). Since mRNA quality is crucial [16], we decided to check obtained cDNA by applying qPCR on representative target genes ADRB1, cTnT, GAPDH, SERCA2, SCN5A, MLC-2v and MLC-2a (Table 2). A qPCR reaction mixture included 250 nM of each dNTP, 0.25μM primer (sequences see Table 3), 20–50 ng template cDNA, 1×PCR buffer, 2.5 mM MgCl2, 0.75×Evagreen (Biotium, Inc., California, USA) and 1 U Taq polymerase (Ares Bioscience GmbH, Cologne, Germany). qPCR was performed in a iQ5 Thermo Cycler (Bio-Rad, Hercules, USA) for 40 cycles with 30 sec at 94°C, 35 sec at 60°C and 1 min 30 sec at 72°C. Raw data were exported as comma-separated values and processed using RKWard (v. 0.6.5) [17] as described [18–21]. For melting curve analysis dedicated functionality from the MBmca package [22] (v. 0.0.3-4) was used. The chipPCR package [19] (v. 0.0.8–10) was used for amplification curve pre-processing and Cq calculation by the second derivative maximum method.
2.8Multiplex PCRs for qualitative gene expression analysis
We established eight multiplex PCRs covering four to seven target gene transcripts each. Primers were either taken from published work or designed them de novo as indicated in Table 3. The defined target gene transcripts (HPRT1, MYCD, MLC-2a, Tbx5, DES, ACTC1, GATA-4, RPLP0, MLC-2v, CHRM2, ADRB1, EDNRB, Cx43, SERCA2, PLN, BNP, NFATc4, EDNRA, ACTB, ANP, KCNH2, cTnT, KCNA5, Cx40) cover biomarkers for ion channels, receptors, contractile apparatus, cardiac transcription factors, fibroblast biomarkers, and reference genes, respectively. Primers were tested on biopsy samples as described in [15]. Basic concepts of our multiplex RT-PCRs were (I) use of two reference genes (RPLP0, HPRT1) and (II) redundancy of selected targets in the multiplex RT-PCRs. By using this approach we aimed to reduce the risk of false negatives. Amplicon products of each target gene transcript were of different sizes, ranging from 89 to 334 bp, permitting their discrimination in agarose gel electrophoresis. The PCR reaction mixture included 0.032 U Uracil-DNA glycosylase (UDG) (ThermoFisher Scientific), 1 ng template cDNA, optimal primer concentration (Table 4), 0.5×Evagreen (Biotium), 1×PCR buffer and 1×Maxima Probe qPCR Master Mix (ThermoFisher Scientific). Multiplex PCRs were performed with iQ5 Thermo Cycler (Bio-Rad). The UDG treatment lasted 15 min at 50°C. UDG was inactivated for 10 min at 95°C. PCR cycles (40) were run according to the following conditions: 45 sec denaturation at 94°C, 120 sec annealing at 59.5°C,and 90 sec amplification at 68.5°C.
3Results
In this study we were interested to establish in vitro cell strains of human cardiomyocytes. Cell cultures of human cardiomyocytes might help to understand pathological processes leading to heart disease. However, adult human cardiomyocytes possess only a limited live span in vitro and do not proliferate. This dramatically limits their use as cell culture model. One aim of this study was to generate a viral vector which induces cardiomyocyte proliferation starting from native human cardiac tissue. We employed a lentivirus vector system to express so-called “upcyte/EPCC proliferation factors”. The same approach was already used successfully to obtain human proliferating primary-like hepatocytes [11–13]. Here we infected primary cardiomyocytes that were enriched from atrial and ventricular biopsies by six cycles of trypsin-collagenase tissue dissociation followed by differential adhesion onto cell culture surfaces. Within one week following lentiviral upcyte gene transduction, cardiomyocytes turned on to proliferate. We used heart biopsy material and cell strains derived thereof to establish novel sets of 32 PCR primers for single and multiplex expression profiling of cardiomyocyte differentiation markers.
3.1Immunohistochemistry as baseline for expression analysis of heart muscle differentiation markers
We first performed hematoxylin and eosin staining for routine histochemical analysis of biopsy materials. Figure 1A shows intensive staining of mono- and binucleated cardiomyocytes in both atrial and ventricular tissues. Those cells were surrounded by smaller cells which might be endothelial cells and cardiac fibroblasts. Altogether, ventricular tissue appeared to contain a tighter arrangement of cardiomyocytes compared with the tissue obtained from atrial appendages whose function still is not completely understood. As a baseline for heart muscle expression profiling, we next analyzed by immunohistochemistry typical structural and non-structural markers of cardiomyocytes, endothelial cells and fibroblasts. As demonstrated in Fig. 1B, both tissues were positive for connexin 43, the main gap junction protein of the heart, which is also expressed in other organs such as bone and brain [23, 24]. In addition, both tissues were positive for staining with antibodies against SERCA2 and myosin heavy chain 7 while even axial orientation of atrial muscle fibers is demonstrated by myosin staining. Non-cardiac protein vimentin, known to be expressed in fibroblasts, smooth muscle - and endothelial cells, was found in regions in proximity but distinct to mono- and binucleated cardiomyocytes. The same was found when cells were stained with an antibody against von Willebrand Factor (vWF), a protein expressed in endothelial cells to regulate blood coagulation. In Fig. 1B, vWF staining demonstrates a ventricular blood vessel.
By using double-immunofluorescence analysis of atrial tissue cyrosections (Fig. 1C), we found connexin 43 and myosin to be colocalized in cardiomyocytes, as expected. Double-staining of atrial cryosections with anti-myosin and anti-vWF antibodies revealed close vicinity of cardiomyocytes and endothelial cells.
3.2Enrichment of cardiomyocytes by differential adhesion
Heart material is naturally composed of different cell types. Cardiac fibroblasts are the most frequent cells in human heart [25] followed by cardiomyocytes, vascular endothelial and smooth muscle cells. The fact that cardiomyocytes tend to attach more tightly to cell culture surfaces than fibroblasts and endothelial cells allow for their enrichment by applying several cycles of trypsinization, a process called differential adhesion [26]. Heart muscle cell types can be discriminated by their morphology. Vascular endothelial cells showed a characteristic fried egg-like morphology with one central nucleus (Fig. 2A). Elongated cells with one central nucleus represent cardiac fibroblasts as shown in Fig. 2B. Small tissue complexes composed of cardiomyocyte clusters could be observed prior to their attachment onto culture flask surface. Phase contrast microscopy revealed in ventricular cardiomyocytes rectangular cell shapes with parallel organized filaments of the contractile apparatus (Fig. 2D) while those filaments appeared less well structured in material from atrial appendages (Fig. 2C).
Upon adherence to culture flask surfaces, cardiomyocytes changed their morphology into triangle like shapes with disorganized filaments. Cells seemed to increase their sizes; a fraction of cardiomyocytes still were binucleated (Fig. 2E&F). It is known that cardiomyocytes might contain two nuclei [27, 28]. Our preparations of attached human cardiomyocytes did not proliferate nor were they able to perform contractions. Following recombinant lentivirus infection, cells started to proliferate but did not further change their morphology (Fig. 2G-J).
3.3Multiplex PCR for cardiomyocyte expression profiling
To conventionally track cardiomyocyte marker expression during the time between usage of a heart muscle biopsy to establish a primary culture of cardiomyocytes over their proliferation induction for several passages, we established conditions for multiplex PCR of cDNAs. Those PCR products were analyzed as end-point reaction, i.e. a given marker is expressed or not without information of initial transcript quantities. In total, we defined 32 genes relevant for the monitoring of human cardiomyocyte differentiation. From this panel we selected 24 genes and combined them in multiplex PCRs. Selected primers were used for qPCR applications (Table 2). Depending on the expected PCR fragment size and the amplification efficiency for each transcriptional target, multiplex approaches were established starting with duplex, triplex and so on primer pairs until up to seven primer pairs could be combined. Multiplexed amplifications were validated to ensure primer pair specificity for a particular target sequence and minimal interactions among the target sequences/primer multiplex PCRs. This was accomplished by comparing the results of single-plex and multiplex PCRs, ensuring identical PCR products in each setup. To estimate the detection limit of our multiplex PCR, cDNA dilution experiments were performed (not shown).
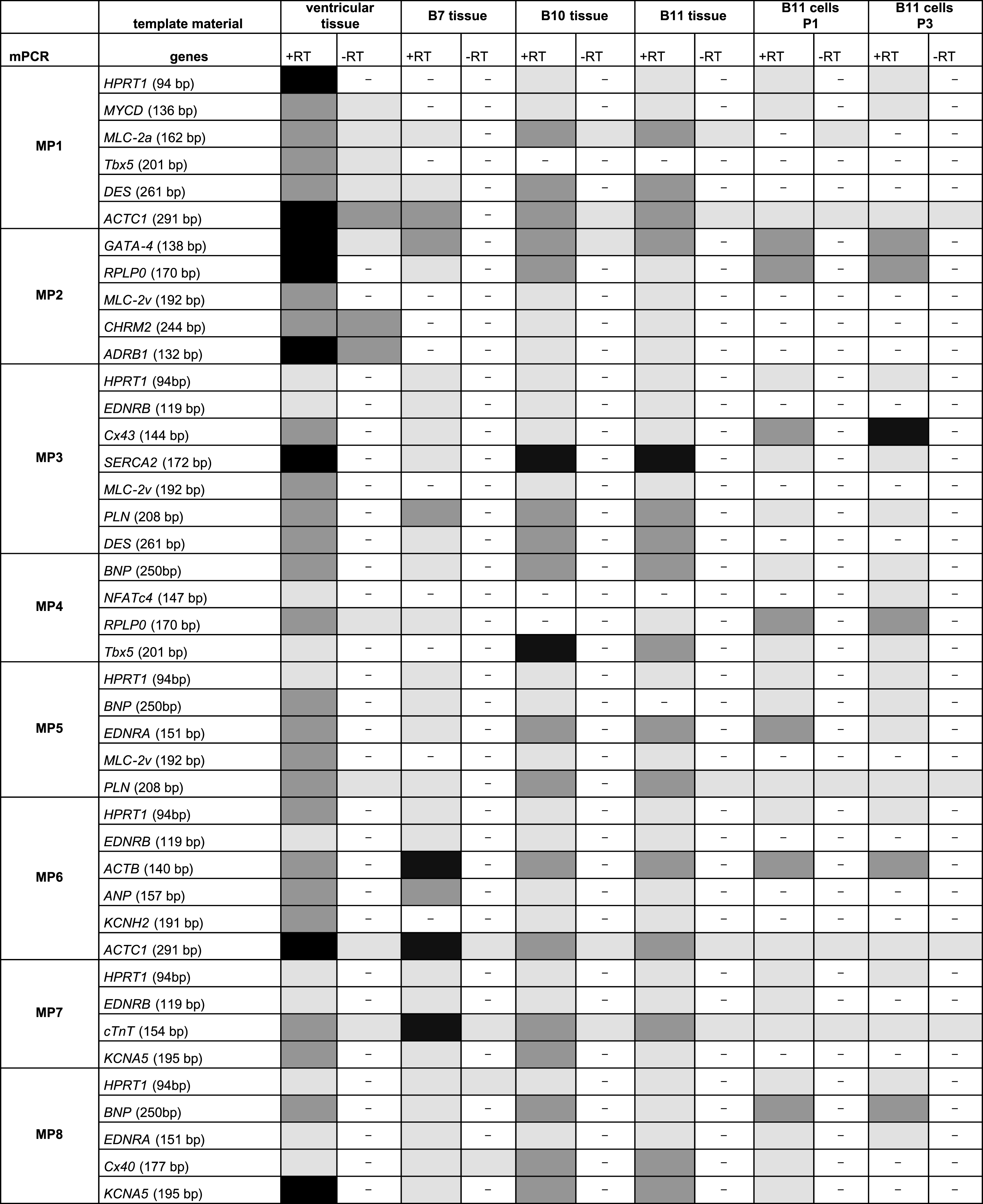
The final multiplex set consisted of eight primer pair combinations to be used for cardiomyocyte expression profiling (Fig. 3). Each multiplex approach includes a primer pair for a reference transcript that was either HPRT1 or RPLP0. Based on these eight multiplex approaches (Fig. 3, MP1 – MP8), native heart tissues from clinical biopsies were analyzed for cardiac marker expressions (Table 5). One ventricular tissue and three atrial tissues obtained from auricula atrii were included in this study. Multiplex PCR analysis of the one ventricular tissue revealed gene expressions for all tested cardiomyocytes markers. Strong amplicon signals were obtained for ACTC1, ADRB1, SERCA2 and KCNA5. Amplicon signal for EDNRB, NFATc4, EDNRA and Cx40 appeared somewhat lower as analyzed semi-quantitatively with agarose gel electrophoresis.
Compared to ventricular tissue, native tissues from atrial appendages apparently were negative for NFATc4, only weakly positive for Cx43, but strongly positive for Cx40 (Table 5). Interestingly, these three atrial tissues even showed differences in their expression profiles. Atrial tissue B7 revealed a lack of expression for cardiomyocyte markers MYCD, MLC-2v, CHRM2, ADRB1, Tbx5 and KCNH2, and only weak expression levels for SERCA2. On the other hand, this biopsy yielded strong signals for ACTB and cTnT. Compared to B7 tissue, atrial cardiomyocytes of tissues B10 and B11 were positive for all tested cardiomyocytes markers except NFATc4.
Thus, our multiplex PCR approach revealed some donor variability of native atrial heart tissue that need to be confirmed by analyzing more samples. Donor variability might be important for further studies on cardiomyocyte expression profiles of this tissue as well as for research applications of isolated atrial cardiomyocytes.
B11 tissue-derived cardiomyocytes that were subjected to proliferation induction by recombinant lentivirus infection were analyzed by the same multiplex PCR approach (Table 5). Our initial studies indicate that cardiomyocyte marker expressions are modified upon primary cell culturing and over three proliferative passages, a process which might take place due to dedifferentiation. Compared to the native atrial tissue, cultured B11 cardiomyocytes lacked expression of cardiac markers MLC-2a, MLC-2v, CHRM2, ADRB1, DES, EDRNB, Cx40 and KCNA5. However, proliferating cardiomyocytes still expressed MYCD, GATA-4, Cx43, SERCA2, BNP, Tbx5, EDNRA and ACTB. Surprisingly, B11-derived cardiomyocytes started to express NFAc4 in passage three, and cardiomyocyte marker expressions of Cx43 and BNP were even increased over cultivation time.
4Discussion
Physiologically relevant in vitro cultures of human cardiomyocytes are important tools to study molecular processes leading to myocardial damage. They are also required as models to study infection routes of myocarditis-inducing viruses such coxsackie virus B3 [29, 30], for validation of new cardiovascular drugs and for toxicity testing. We aimed to establish procedures for proliferation induction of primary human cardiomyoctes by using the upcyte/EPCC approach which already had been implemented for long-term cultures of primary-like human hepatocytes [11–13]. Furthermore, we wanted to establish fast screening protocols for cardiomyocyte expression profiling by transcriptomics based on multiplex PCR of cDNAs. In total, we validated 32 primer pairs; 24 primer pairs were used for multiplex PCR experiments shown in this study. We should note here that we aimed to include primers for vWF and fibroblast specific protein 1 (FSP1) but failed to find suitable primer pairs which do not show unspecific amplifications, primer dimers or double bands. Therefore, we excluded them from the current primer panel. Our approach can be used with laboratory standard PCR equipment. Compared to other approaches such as low-density microarrays, our multiplex PCRs should provide valuable information about gene expression profiles of heart cells at reasonable costs and hands-on-time. The redundant use of selected primers was useful to reduce false negatives which are a challenge in multiplex PCRs. Besides the application of primers in multiplex PCRs, we also used them in qPCRs for cDNA characterization. qPCR is widely acknowledged as the most sensitive method to quantify nucleic acids [31]. We think that our primer sets can be widely used for applications such as qPCR and digital PCR to investigate changes in gene expression and donor variability. However, method validation based on common guidelines for PCR assay optimization is crucial for new PCR applications[32].
We isolated adult human ventricular and atrial cardiomyocytes from clinical biopsies by using six cycles of enzymatic dissociation followed by differential cell adhesion. Heart surgery of the left atrium auricle (LAA) is often performed to avoid thrombus formation in patients with atrial fibrillation [33]. Since most patients subjected to cardiac surgery belong to the age group >60 years, quality and quantity of isolated primary cardiomyocytes might already be impaired by the patient’s clinical condition and due to general age-related processes affecting cardiomyocyte function. Indeed, it was shown that success rates of primary cardiomyocyte cultures depend on donor age [34]. Furthermore, both quality and quantity of individual biopsy material substantially influence the total yield of viable cardiomyocytes after enzymatic tissue dissociation. Others reported that cardiomyocytes constitute only 20% of all cells within the human myocardium [35]. Moreover, the partial oxygen pressure in tissues such as heart is at a range of 4–20 mm Hg (0.5–2.5 kPa) [36] while routine cell culture conditions involve exposure of cells to nearly atmospheric oxygen partial pressure of 159 mm Hg. Thus, due to increased oxidative stress during in vitro culturing, reactive oxygen species might accumulate and reduce the number of viable cells.
4.1Evidence for dedifferentiation processes in human atrial cardiomyocytes upon in vitro proliferation induction
It was shown that the presence of proliferating cardiac fibroblasts enhances dedifferentiation of atrial myocytes in vivo and in vitro. Cardiomyocytes were able to re-differentiate when fibroblast proliferation was blocked. To prevent enhanced dedifferentiation and overgrowth of cardiomyocyte cultures by cardiac fibroblast or vascular endothelial cells, cardiomyocytes need to be enriched. In this study we used a protocol of enzymatic tissue dissociation followed by differential adhesion onto cell cultures surfaces leading to successive enrichment of cardiomyocytes. This procedure is based on the fact that fibroblasts and endothelial cells adhere faster to cell culture surface than cardiomyocytes. Reversely, once cardiomyocytes are attached, their fraction can be further enriched by several trypsinizations removing fast detaching cells such as remaining fibroblasts and endothelial cells. Finally, we obtained populations of non-proliferating cells with typical bi-nucleated cardiomyocytes (Fig. 2E&F). After their infection with recombinant lentivirus for constitutive expression of proliferation-inducing genes (upcte/EPCC genes), quiescent cells started to proliferate and maintained expression of cardiomyocyte specific markers such as MYCD, GATA-4, Cx43, SercA2, BNP or Tbx5 for several passages (Table 5). On the other hand, proliferating atrial cardiomyocyte apparently lost expression of muscle differentiation markers such as MLC-2a, MLC-2v, CHRM2, ADRB1, DES, EDRNB, Cx40 and KCNA5. Dedifferentation can also be visualized by cluster analysis (Fig. 4). This figure demonstrates that atrial biopsy-derived B11 cardiomyocytes were shifted into another cluster group upon proliferation induction.
Although cardiomyocyte differentiation is realized by spatial expression pattern, some genes are expressed in the heart ubiquitously. Those genes are SERCA2 [38], troponin isoforms and α-actinin.
In adult human heart, β-MHC is restricted to the ventricle whereas α-MHC is predominantly expressed in the atria, but few ventricular myocardium fibers also contain α-MHC [39–41]. Similar to β-MHC expression, the regulatory MLC-2v expression is restricted to the ventricle already at week seven of development while MLC-1v ventricular restriction occurs later. MLC-2a expression was found at high levels in both atria and at low levels in the left ventricle [42] which might be related to α-MHC expression. During our study we detected variations of MLC isoform expression in atrium and ventricle.
To allow for electrical signal transmission, cells are coupled via gap junctions composed of six connexons. Two connexons make up a transmembrane channel between two cells. Three different connexin types are expressed in cardiac myocytes; these are Cx43, 40 and 45. While Cx43 is mainly expressed in the ventricular myocardium, atria cells express predominantly Cx40. During this study, Cx43 expression was detected both in native ventricular and atrial tissues. SA and AV node cells express only Cx45, and all three connexins are found in e.g. Purkinjie fibers (reviewed in: [43]). One unexpected finding in our study was that proliferating atrial cardiomyocytes increased expression of Cx43, the predominant ventricular connexin. Furthermore, we found expression of NFATc4 in ventricular tissue as well as in proliferating atrial cardiomyocytes of passage three, but not in earlier passages or in atrial tissue. Apparently, with human atrial myocytes this has not been observedbefore.
5Outlook
In this study we demonstrate successful cardiomyocyte enrichment and proliferation induction by using the upcyte/EPCC approach which is a technique for direct multiplication of primary cells. By contrast, many groups focus on cardiomyocyte differentiation from pluripotent stem cells such as embryonic stem cells (hESC) and induced pluripotent stem cells (iPS). Using hESCs, contractile cardiomyocytes can be obtained and enriched by several techniques those cells clearly show molecular, structural and functional properties of early-stage cardiomyocytes [44]. Their immature differentiation status is also reflected by the fact that they are actively proliferating unlike adult cardiomyocytes. In recent years, several laboratories established procedures for cardiomyocyte differentiation from iPS. It was shown that up to 80% of stem cells could be differentiated into heterogeneous populations of atrial, ventricular and cardiac conductive cells [45–48]. Furthermore, those cell populations might still contain undifferentiated iPS cells that could lead to teratoma formation in vivo. Nevertheless, it has been shown that fetal cardiomyocytes obtained from human tissues are able to survive upon transplantation into the adult heart while transplanted adult cardiomyocyte showed a reduced viability in vivo [49]. Therefore, a dedifferentiation process of adult cells, as is shown here, might even be helpful if such cells would be used for in vivo studies. However, it should also be feasible to avoid dedifferentiation processes if proliferation induction in human cardiomyocytes is made reversible. We are currently working on inducible promoters [50] to control for expression of upcyte/EPCC factors.
References
[1] | Maron BJ , Towbin JA , Thiene G , Antzelevitch C , Corrado D , Arnett D , et al. Contemporary definitions and classification of the cardiomyopathies: An American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation (2006) ;113: (14):1807–16. |
[2] | Zebrowski DC , Vergarajauregui S , Wu CC , Piatkowski T , Becker R , Leone M , et al. Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. Elife (2015) ;4. |
[3] | Mahmoud AI , Kocabas F , Muralidhar SA , Kimura W , Koura AS , Thet S , et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature (2013) ;497: (7448):249–53. |
[4] | Ikenishi A , Okayama H , Iwamoto N , Yoshitome S , Tane S , Nakamura K , et al. Cell cycle regulation in mouse heart during embryonic and postnatal stages. Dev Growth Differ (2012) ;54: (8):731–8. |
[5] | van Amerongen MJ , Engel FB . Features of cardiomyocyte proliferation and its potential for cardiac regeneration. J Cell Mol Med (2008) ;12: (6A):2233–44. |
[6] | Ribeiro AJ , Ang YS , Fu JD , Rivas RN , Mohamed TM , Higgs GC , et al. Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci U S A (2015) ;112: (41):12705–10. |
[7] | Veerman CC , Kosmidis G , Mummery CL , Casini S , Verkerk AO , Bellin M . Immaturity of human stem-cell-derived cardiomyocytes in culture: Fatal flaw or soluble problem? Stem Cells Dev (2015) ;24: (9):1035–52. |
[8] | Bulic-Jakus F , Katusic BA , Juric-Lekic G , Vlahovic M , Sincic N . Teratoma: From spontaneous tumors to the pluripotency/malignancy assay. Wiley Interdiscip Rev Dev Biol (2016) ;5: (2):186–209. |
[9] | Hultman I , Bjork L , Blomberg E , Sandstedt B , Ahrlund-Richter L . Experimental teratoma: At the crossroad of fetal- and onco-development. Semin Cancer Biol (2014) ;29: :75–9. |
[10] | Fujita J , Fukuda K . Future prospects for regenerated heart using induced pluripotent stem cells. J Pharmacol Sci (2014) ;125: (1):1–5. |
[11] | Burkard A , Dahn C , Heinz S , Zutavern A , Sonntag-Buck V , Maltman D , et al. Generation of proliferating human hepatocytes using upcyte((R)) technology: Characterisation and applications in induction and cytotoxicity assays. Xenobiotica (2012) ;42: (10):939–56. |
[12] | Herzog N , Hansen M , Miethbauer S , Schmidtke KU , Anderer U , Lupp A , et al. Primary-like human hepatocytes genetically engineered to obtain proliferation competence display hepatic differentiation characteristics in monolayer and organotypical spheroid cultures. Cell Biol Int (2016) ;40: (3):341–53. |
[13] | Levy G , Bomze D , Heinz S , Ramachandran SD , Noerenberg A , Cohen M , et al. Long-term culture and expansion of primary human hepatocytes. Nat Biotechnol (2015) ;33: (12):1264–71. |
[14] | Norenberg A , Heinz S , Scheller K , Hewitt NJ , Braspenning J , Ott M . Optimization of upcyte(R) human hepatocytes for the in vitro micronucleus assay. Mutat Res (2013) ;758: (1-2):69–79. |
[15] | Rödiger S , Schierack P , Bohm A , Nitschke J , Berger I , Frommel U , et al. A highly versatile microscope imaging technology platform for the multiplex real-time detection of biomolecules and autoimmune antibodies. Adv Biochem Eng Biotechnol (2013) ;133: :35–74. |
[16] | Karlsson O , Segerstrom L , Sjoback R , Nylander I , Boren M . qPCR based mRNA quality score show intact mRNA after heat stabilization. Biomol Detect Quantif (2016) ;7: :21–6. |
[17] | Rödiger S , Friedrichsmeier T , Kapat P , Michalke M . RKWard: A Comprehensive Graphical User Interface and Integrated Development Environment for Statistical Analysis with R J Stat Softw (2012) ;49: (9):1–34. |
[18] | Pabinger S , Rödiger S , Kriegner A , Vierlinger K , Weinhäusel A . A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol Detect Quantif (2014) ;1: (1):23–22. |
[19] | Rödiger S , Burdukiewicz M , Schierack P . chipPCR: An R package to pre-process raw data of amplification curves. Bioinformatics (2015) ;31: (17):2900–2. |
[20] | Rödiger S , Burdukiewicz M , Blagodatskikh KA , Schierack P . R as an Environment for the Reproducible Analysis of DNA Amplification Experiments. The R Journal (2015) ;7: (2):127–50. |
[21] | Spiess AN , Deutschmann C , Burdukiewicz M , Himmelreich R , Klat K , Schierack P , et al. Impact of smoothing on parameter estimation in quantitative DNA amplification experiments. Clin Chem (2015) ;61: (2):379–88. |
[22] | Rödiger S , Böhm A , Schimke I . Surface Melting Curve Analysis with R. The R Journal (2013) ;5: (2):37–53. |
[23] | Plotkin LI . Connexin 43 hemichannels and intracellular signaling in bone cells. Front Physiol (2014) ;5: :131. |
[24] | Salmina AB , Morgun AV , Kuvacheva NV , Lopatina OL , Komleva YK , Malinovskaya NA , et al. Establishment of neurogenic microenvironment in the neurovascular unit: The connexin 43 story. Rev Neurosci (2014) ;25: (1):97–111. |
[25] | Camelliti P , Borg TK , Kohl P . Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res (2005) ;65: (1):40–51. |
[26] | Tao J , Ma Y , Li X , Chen B , Yang Y , Ma X , et al. [An improved protocol for primary culture of cardiomyocyte from neonatal rat]. Zhonghua Xin Xue Guan Bing Za Zhi (2014) ;42: (1):53–6. |
[27] | Huang CF , Chen YC , Yeh HI , Chen SA . Mononucleated and binucleated cardiomyocytes in left atrium and pulmonary vein have different electrical activity and calcium dynamics. Prog Biophys Mol Biol (2012) ;108: (1-2):64–73. |
[28] | Shperling ID , Arakelian LA . The number and some other characteristics of isolated human ventricular cardiomyocytes under pathological conditions. Cor Vasa (1990) ;32: (4):327–34. |
[29] | Meyer RG , Meyer-Ficca ML , Kaiser H , Selinka HC , Kandolf R , Kupper JH . Plasmid-based generation of recombinant coxsackievirus B3 particles carrying capsid gene replacement replicons. Virus Res (2004) ;104: (1):17–26. |
[30] | Selinka HC , Wolde A , Pasch A , Klingel K , Schnorr JJ , Kupper JH , et al. Comparative analysis of two coxsackievirus B3 strains: Putative influence of virus-receptor interactions on pathogenesis. J Med Virol (2002) ;67: (2):224–33. |
[31] | Svec D , Tichopad A , Novosadova V , Pfaffl MW , Kubista M . How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif (2015) ;3: :9–16. |
[32] | Wadle S , Lehnert M , Rubenwolf S , Zengerle R , von SF . Real-time PCR probe optimization using design of experiments approach. Biomol Detect Quantif (2016) ;7: :1–8. |
[33] | Wilbring M , Jung F , Weber C , Matschke K , Knaut M . Reduced Incidence of Thromboembolic Events After Surgical Closure of Left Atrial Appendage in Patients With Atrial Fibrillation. Innovations (Phila) (2016) ;11: (1):24–30. |
[34] | Smith DA , Glover JL , Townsend LE , Maupin DE . A method for the harvest, culture, and characterization of human adult atrial myocardial cells: Correlation with age of donor. In Vitro Cell Dev Biol (1991) ;27A: (12):914–20. |
[35] | Rubart M , Field LJ . Cardiac regeneration: Repopulating the heart. Annu Rev Physiol (2006) ;68: :29–49. |
[36] | Sitkovsky M , Lukashev D . Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat Rev Immunol (2005) ;5: (9):712–21. |
[37] | Rucker-Martin C , Pecker F , Godreau D , Hatem SN . Dedifferentiation of atrial myocytes during atrial fibrillation: Role of fibroblast proliferation in vitro . Cardiovasc Res (2002) ;55: (1):38–52. |
[38] | Gaborit N , Le BS , Szuts V , Varro A , Escande D , Nattel S , et al. Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol (2007) ;582: (Pt 2):675–93. |
[39] | Everett AW . Isomyosin expression in human heart in early pre- and post-natal life. J Mol Cell Cardiol (1986) ;18: (6):607–15. |
[40] | Bouvagnet P , Leger J , Pons F , Dechesne C , Leger JJ . Fiber types and myosin types in human atrial and ventricular myocardium. An anatomical description. Circ Res (1984) ;55: (6):794–804. |
[41] | Gorza L , Mercadier JJ , Schwartz K , Thornell LE , Sartore S , Schiaffino S . Myosin types in the human heart. An immunofluorescence study of normal and hypertrophied atrial and ventricular myocardium. Circ Res (1984) ;54: (6):694–702. |
[42] | Chuva de Sousa Lopes SM , Hassink RJ , Feijen A , van Rooijen MA , Doevendans PA , Tertoolen L , et al. Patterning the heart, a template for human cardiomyocyte development. Dev Dyn (2006) ;235: (7):1994–2002. |
[43] | Severs NJ , Bruce AF , Dupont E , Rothery S . Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res (2008) ;80: (1):9–19. |
[44] | Mummery C , Ward-van OD , Doevendans P , Spijker R , van den Brink S , Hassink R , et al. Differentiation of human embryonic stem cells to cardiomyocytes: Role of coculture with visceral endoderm-like cells. Circulation (2003) ;107: (21):2733–40. |
[45] | Zhang J , Klos M , Wilson GF , Herman AM , Lian X , Raval KK , et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: The matrix sandwich method. Circ Res (2012) ;111: (9):1125–36. |
[46] | Lian X , Zhang J , Azarin SM , Zhu K , Hazeltine LB , Bao X , et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc (2013) ;8: (1):162–75. |
[47] | Xu H , Yi BA , Wu H , Bock C , Gu H , Lui KO , et al. Highly efficient derivation of ventricular cardiomyocytes from induced pluripotent stem cells with a distinct epigenetic signature. Cell Res (2012) ;22: (1):142–54. |
[48] | Weng Z , Kong CW , Ren L , Karakikes I , Geng L , He J , et al. A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells. Stem Cells Dev (2014) ;23: (14):1704–16. |
[49] | Reinecke H , Zhang M , Bartosek T , Murry CE . Survival, integration, and differentiation of cardiomyocyte grafts: A study in normal and injured rat hearts. Circulation (1999) ;100: (2):193–202. |
[50] | Meyer-Ficca ML , Meyer RG , Kaiser H , Brack AR , Kandolf R , Kupper JH . Comparative analysis of inducible expression systems in transient transfection studies. Anal Biochem (2004) ;334: (1):9–19. |
[51] | Vandesompele J , De PK , Pattyn F , Poppe B , Van RN , De PA , et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol (2002) ;3: (7):RESEARCH0034. |
[52] | Ball AJ , Levine F . Telomere-independent cellular senescence in human fetal cardiomyocytes. Aging Cell (2005) ;4: (1):21–30. |
[53] | Bearzi C , Rota M , Hosoda T , Tillmanns J , Nascimbene A , De AA , et al. Human cardiac stem cells. Proc Natl Acad Sci U S A (2007) ;104: (35):14068–73. |
Figures and Tables
Fig.1
Histological and immunohistochemical analyses of heart muscle tissues. Native atrial and ventricular tissues were cut into 9μm cryosections and fixed with formaldehyde and methanol/acetone as outlined in Materials and Methods. A) HE staining of cardiac tissue sections. Nuclei are stained blue by hematoxylin, and eosinophilic areas (proteins) are colored pink by eosin. Section was documented with the Olympus microscope CKX 41. *, accumulation of cells visualized by their nuclei. +, w/o cells. Scale bar: 500μm. B) Tissue sections were characterized by indirect immunofluorescence analysis for cardial proteins expression such as connexin 43 (Cx43), sarcoplasmic reticulum calcium (SERCA 2) ATPase, myosin and myosin heavy chain 7 (MYH7). Fibroblasts were identified with the anti-vimentin primary antibody. For endothelial cell detection, the anti-von Willebrand Factor (vWF) primary antibody was used. Immunofluorescence was documented with Olympus microscope IX81 equipped with the Olympus IXUCB filter and xenon burner MT20. Scale bar 100μm. C) Double-immunofluorescence analysis was performed for native atrial tissue. Colocalisation was shown for Cx43 (Cy3) with myosin (FITC) but not for vWF (Cy3) and myosin (FITC). Immunofluorescence was documented with Olympus microscope IX81. Scale bar: 25μm.
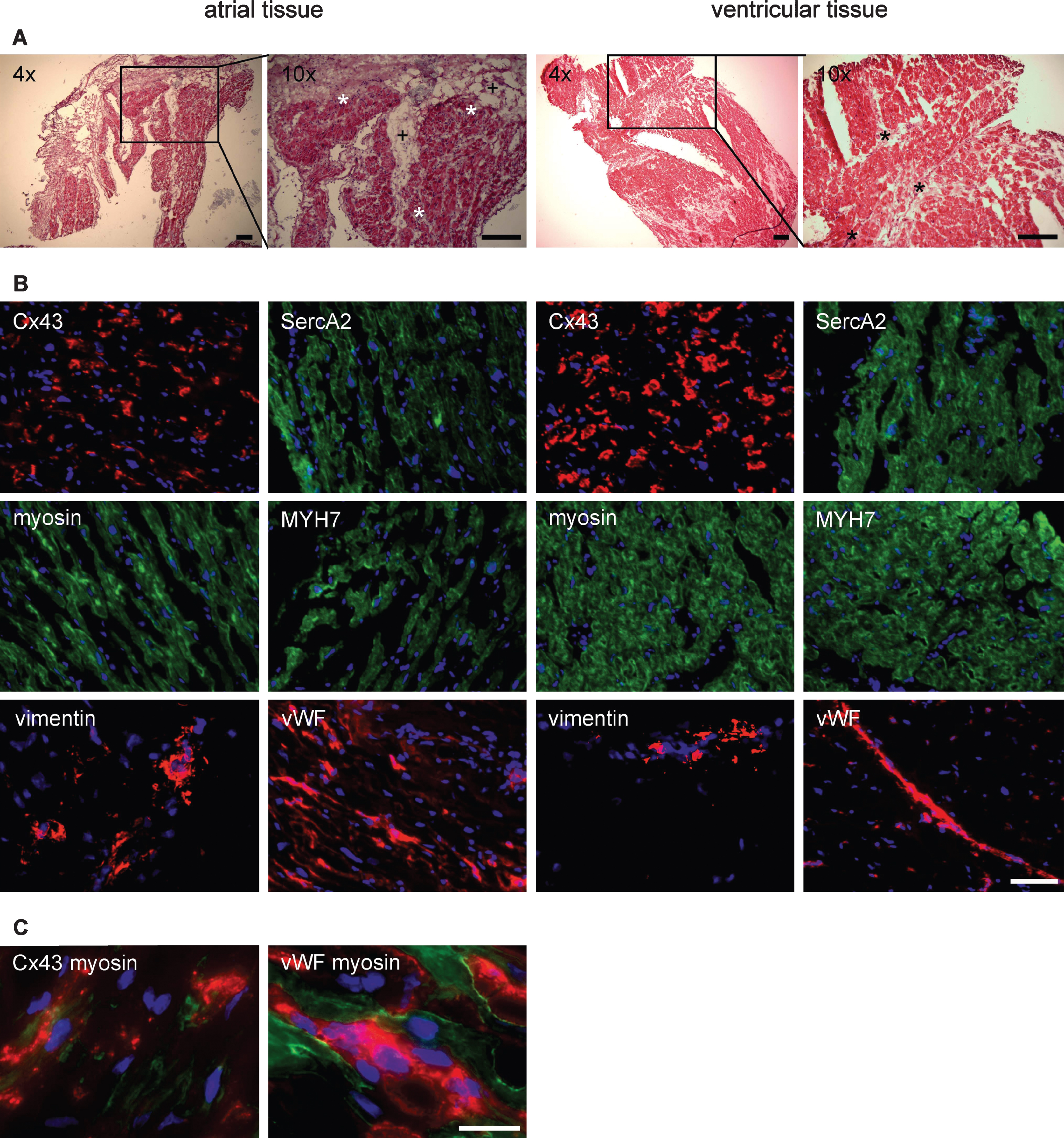
Fig.2
Phase contrast microscopy analysis of heart muscle cell types. Primary heart biopsies were dissociated and isolated cells were documented with phase contrast microscopy. A) Cells with endothelial cell-like morphology; B) cell with fibroblast-like morphology; C) atrial cardiomyocytes prior to attachment; D) ventricular cardiomyocytes prior to attachment; E) atrial cardiomyocytes six days after attachment; F), ventricular cardiomyocytes six days after attachment; G) proliferating atrial cardiomyocytes, passage 1; H) proliferating ventricular cardiomyocytes, passage 1; I) proliferating atrial cardiomyocytes in passage 8 and J) proliferating ventricular cardiomyocytes, passage 9. Pictures were taken with Olympus microscope CKX 41 using phase contrast. Scale bar: 50μm. Arrows, binucleated cardiomyocytes.
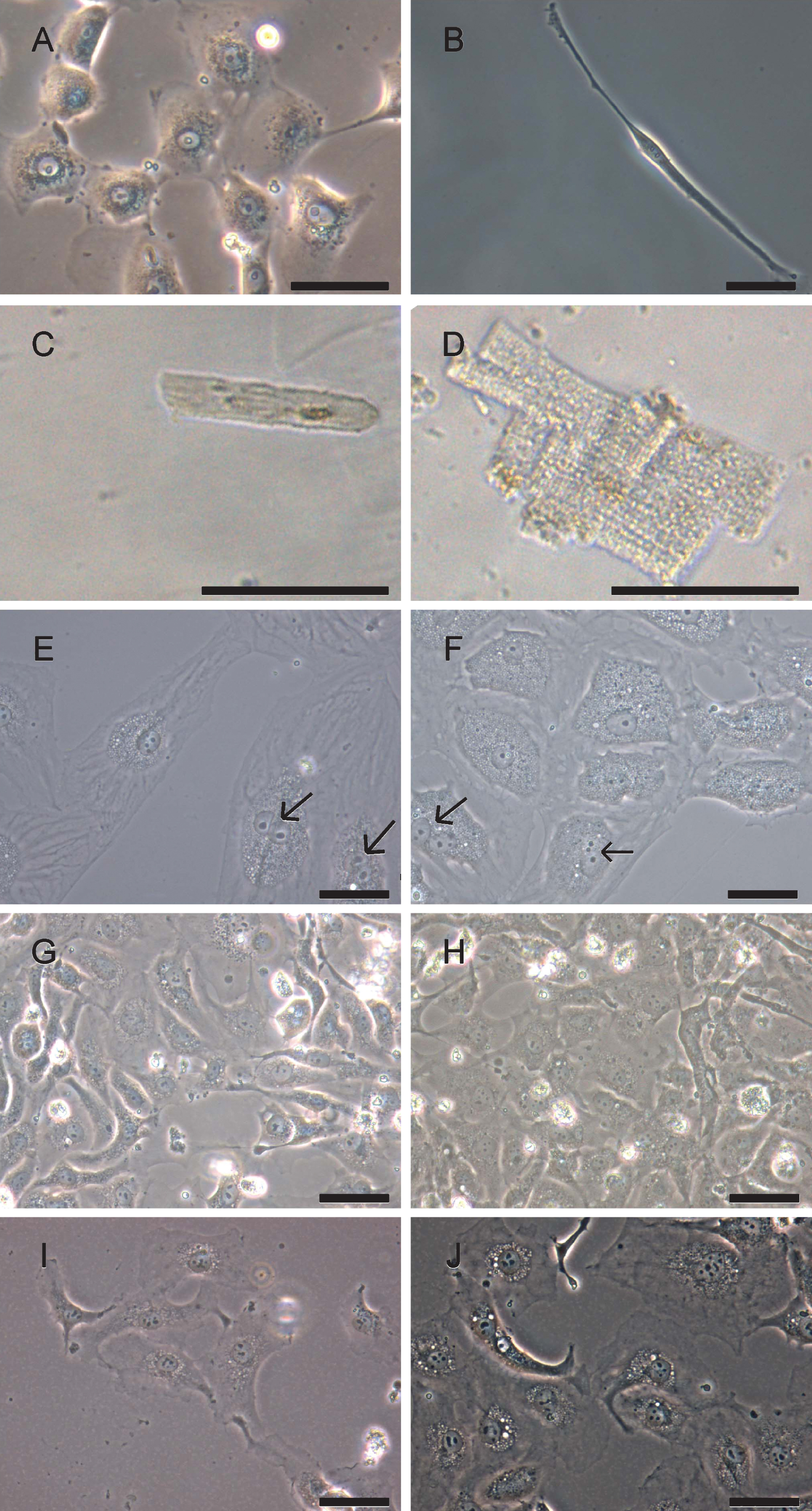
Fig.3
Agarose gel electrophoresis analysis of multiplex PCR on cDNA from ventricular heart muscle tissue. Total RNA of ventricular tissue was prepared, and mRNA was processed into cDNA by reverse transcriptase. Eight combinations of primer pairs that included cardiomyocyte-specific marker or reference gene transcripts (HPRT1 or RPLP0) were set for multiplex PCR. Multiplex PCR amplicons were compared to single loci PCR, respectively. A) Multiplex PCR (MP1) approach amplifying six transcript cDNAs and single transcripts with increasing fragment sizes: HPRT1, MYCD, MLC-2a, Tbx5, DES and ACTC1; B) Multiplex PCR (MP2) approach amplifying five transcript cDNAs and single transcripts with increasing fragment sizes: GATA-4, RPLP0, MLC-2v, CHRM2 and ADRB1; C) Multiplex PCR (MP3) approach amplifying seven transcript cDNAs and single transcripts with increasing fragment sizes: HPRT1, EDNRB, Cx43, SERCA2, MLC-2v, PLN and DES; D) Multiplex PCR (MP4) approach amplifying four transcript cDNAs and single transcripts with increasing fragment sizes: BNP, NFATc4, RPLP0 and Tbx5; E) Multiplex PCR (MP5) approach amplifying five transcript cDNAs and single transcripts with increasing fragment sizes: HPRT1, BNP, EDNRA, MLC-2v and PLN; F) Multiplex PCR (MP6) approach amplifying six transcript cDNAs and single transcripts with increasing fragment sizes: HPRT1, EDNRB, ACTB, ANP, KCNH2 and ACTA1; G) Multiplex PCR (MP7) approach amplifying four transcript cDNAs and single transcripts with increasing fragment sizes: HPRT1, EDNRB, cTnT and KCNA5; H), Multiplex PCR (MP8) approach amplifying five transcript cDNAs and single transcripts with increasing fragment sizes: HPRT1, BNP, EDNRA, Cx40 and KCNA5. Amplicons were compared to 50 bp marker.
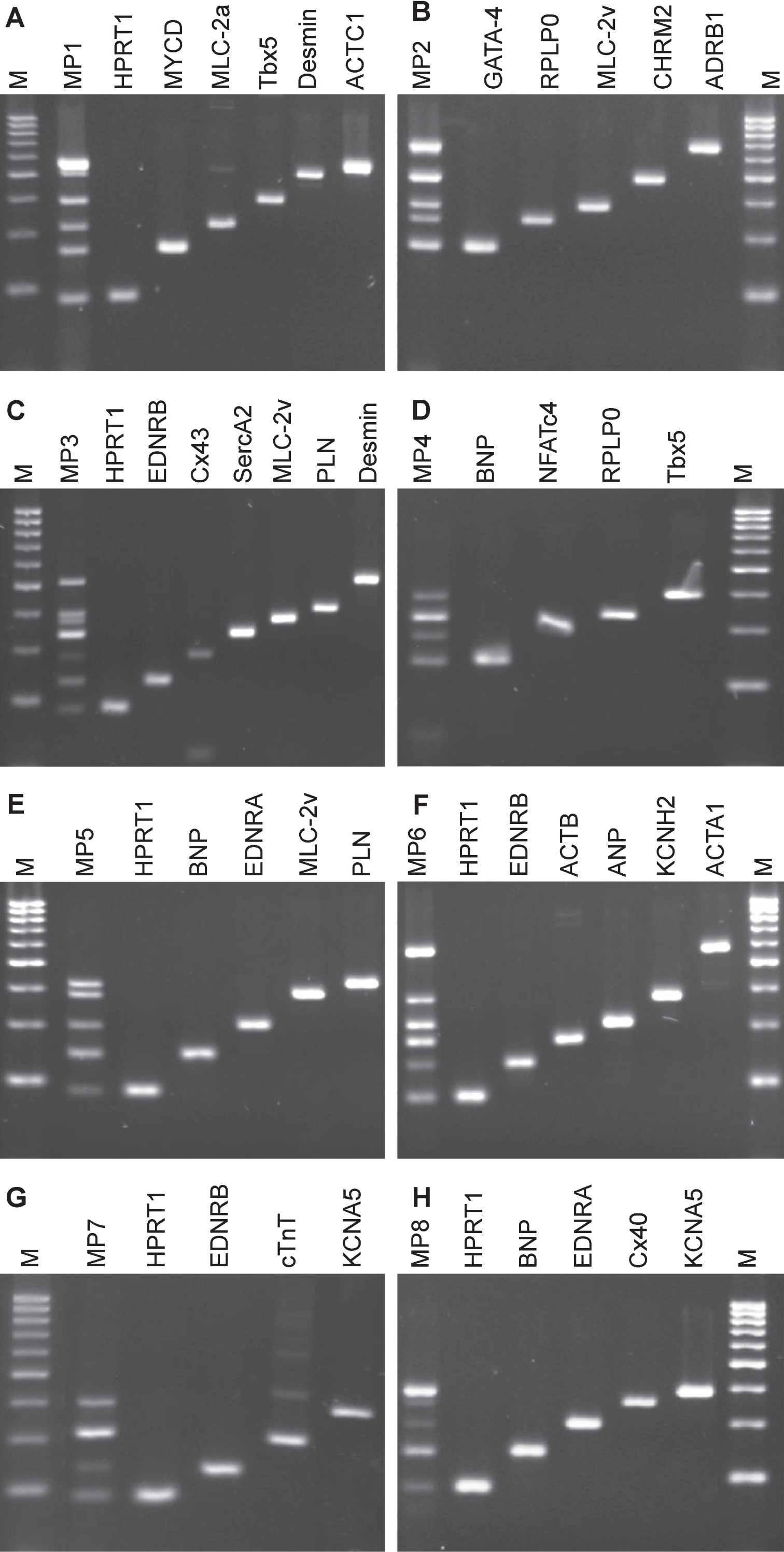
Fig.4
Cluster analysis of multiplex PCRs. cDNAs from human heart tissue (atrium: B7, B10, B11; ventricle) and lentivirus-infected atrial cells (B11P1, B11P3) were clustered based on 24 gene transcripts (HPRT1, MYCD, MLC-2a, Tbx5, DES, ACTC1, GATA-4, RPLP0, MLC-2v, CHRM2, ADRB1, EDNRB, Cx43, SERCA2, PLN, BNP, NFATc4, EDNRA, ACTB, ANP, KCNH2, cTnT, KCNA5, Cx40) encoded in eight multiplex PCRs. Amplification product patterns from agarose gels were transformed to dichotomous data (transcript absent = 0, transcript present = 1). Clusters were determined using the Manhattan Distance Measure and Hierarchical Cluster Analysis as described in [15]. All specimens of one cluster have similar gene expression patterns. Primary tissues B7, B10, B11 and ventricle form one cluster. Proliferating atrial cells (B11P1, B11P3) formed an independent cluster. Reversely transcribed (+RT) and non-reversely transcribed cDNAs (–RT) were analyzed in two distinct branches.
![Cluster analysis of multiplex PCRs. cDNAs from human heart tissue (atrium: B7, B10, B11; ventricle) and lentivirus-infected atrial cells (B11P1, B11P3) were clustered based on 24 gene transcripts (HPRT1, MYCD, MLC-2a, Tbx5, DES, ACTC1, GATA-4, RPLP0, MLC-2v, CHRM2, ADRB1, EDNRB, Cx43, SERCA2, PLN, BNP, NFATc4, EDNRA, ACTB, ANP, KCNH2, cTnT, KCNA5, Cx40) encoded in eight multiplex PCRs. Amplification product patterns from agarose gels were transformed to dichotomous data (transcript absent = 0, transcript present = 1). Clusters were determined using the Manhattan Distance Measure and Hierarchical Cluster Analysis as described in [15]. All specimens of one cluster have similar gene expression patterns. Primary tissues B7, B10, B11 and ventricle form one cluster. Proliferating atrial cells (B11P1, B11P3) formed an independent cluster. Reversely transcribed (+RT) and non-reversely transcribed cDNAs (–RT) were analyzed in two distinct branches.](https://content.iospress.com:443/media/jcb/2016/2-1/jcb-2-1-jcb15025/jcb-2-jcb15025-g004.jpg)
Table 1
List of antibodies used in this study
| antibody | species | dilution | provider |
| anti-connexin 43 | polyclonal, rabbit | 1:8,000 | Sigma-Aldrich, Missouri, USA |
| anti-MHY7 | monoclonal, mouse | 1:3,000 | Santa Cruz Biotechnology, INC., Texas, USA |
| anti-myosin, clone NOQ7.5.4D | monoclonal, mouse | 1:1,000 | Sigma-Aldrich, Missouri, USA |
| anti-SERCA2 | monoclonal, mouse | 1:500 | Thermo Fisher Scientific, Waltham, USA |
| anti-vimentin | monoclonal, mouse | 1:200 | Progen Biotechnik GmbH, Heidelberg, Germany |
| anti-von Willbrand Factor | polyclonal, rabbit | 1:800 | Sigma-Aldrich, Missouri, USA |
| Cy3 conjugated goat anti-rabbit IgG (H + L) | 1:200 | Dianova, Hamburg, Germany | |
| mouse anti-goat IgG-FITC | 1:100 | Santa Cruz Biotechnology, INC., Texas, USA |
Table 2
Characterization by qPCR of heart tissue-derived cDNA
| Gene | Cq (n = 3) | ΔCq |
| ADRB1 | 28.98±0.91 | 11.02±0.91 |
| cTnT | 22.71±0.08 | 17.29±0.08 |
| GAPDH | 22.85±0.75 | 11.98±2.9 |
| SERCA2 | 25.91±0.3 | 5.30±0.98 |
| SCN5A | 29.69±1.1 | 4.33±0.25 |
| MLC-2v | 31.31±0.26 | 4.75±5.6 |
| MLC-2a | 20.01±0.32 | 13.25±2.8 |
A reference gene (GAPDH) and six cardiomyocte markers were selected for a characterization of the cDNA (biopsy B11). The Cq values varied between 20 and 30. ΔCq (Cq - RT – Cq +RT) describes the Cq difference of the reverse transcribed sample to the non-reverese transcribed sample.
Table 3
Primers used in this study
| No | Target | gDNA | Function | Primer Sequence (5′ ⟶ 3′) | GC [% ] | Size [bp] TM [°C] | Source | |
| 1 | ACTA1 | CA | f: TATCGAGCACGGCATCATCA | 50 | 288 | # | ||
| actin, alpha, skeletal muscle | r: TAAATGGGCACGTTGTGGGT | 50 | ||||||
| 2 | ACTB | CA | f: CTGGAACGGTGAAGGTGACA | 55 | 140 | [51] | ||
| actin, beta | r: AAGGGACTTCCTGTAACAATGCA | 43.5 | 82.5±0.0 | |||||
| 3 | ACTC1 | CA | f: CTTCATTGGTATGGAATCTGCTG | 41.7 | 291 | # | ||
| actin, alpha, cardiac muscle 1 | r: TACTCTTGCTTGCTAATCCAC | 42.9 | 83.0±0.0 | |||||
| 4 | ADRB1 | + | RC | f: CCACAGAAGATGTTACTTGCAC | 45.5 | 334 | # | |
| adrenergic, beta-1-, receptor | 334 bp | r: ACACAGTAGTTAGTTGCACCA | 42.9 | |||||
| 5 | ADRB1 | + | RC | f: AGGGATTTCTACCTCACACTG | 47.6 | 133 | # | |
| adrenergic, beta-1-, receptor | 133 bp | r: ACAGAGTCACATGTCACAGAG | 47.6 | 79.2±0.0 | ||||
| 6 | ADRB2 | + | RC | f: GATTTCAGGATTGCCTTCCA | 45 | 233 | # | |
| adrenergic, beta-2-, receptor | 233 bp | r: GTACTACAATTCCTCCCTTGTG | 45.5 | 84.8±0.1 | ||||
| 7 | ANP, NPPA§ | HF | f: GACTCCTCTGATCGATCTGC | 55 | 157 | # | ||
| atrial natriuretic peptide A | r: GTTATCTTCAGTACCGGAAGC | 47.6 | 82.5±0.0 | |||||
| 8 | BMP2 | CD | f: ATGCTAGACCTGTATCGCAG | 50 | 123 | # | ||
| bone morphogenetic protein 2 | r: TTCCAAAGATTCTTCATGGTGG | 40.9 | ||||||
| 9 | BMP2 | CD | f: TTCCACCATGAAGAATCTTTGG | 40.9 | 181 | # | ||
| bone morphogenetic protein 2 | r: TTAATTCGGTGATGGAAACTGC | 40.9 | ||||||
| 10 | BMP2 | CD | f: CCACCATGAAGAATCTTTGGA | 42.9 | 271 | # | ||
| bone morphogenetic protein 2 | r: GCTTGCATTCTGATTCACCA | 45 | ||||||
| 11 | BNP, NPPA§ | + | HF | f: TCCTGCTCTTCTTGCATCTGG | 52.4 | 122 | # | |
| brain natriuretic peptide | 354 bp | r: TTTGCCCTGCAAATGGTTG | 47.4 | 89.0±0.0 | ||||
| 12 | CHRM2 | RC | f: GGATAAAGAAGGACAAGAAGG | 42.9 | 244 | # | ||
| cholinergic receptor, muscarinic 2 | r: GCTCATATTAGAGGCAACAGCA | 45.5 | 87.2±0.0 | |||||
| 13 | cTnI, TNNI3§ | CA | f: AAATCTAAGATCTCCGCCTCG | 47.6 | 274 | # | ||
| cardiac troponin I | r: TCAGATCTGCAATCTCCGTG | 50 | ||||||
| 14 | cTnT, TNNT2§ | + | CA | f: AGTGGGAAGAGGCAGACTGA | 55 | 154 | [52] | |
| cardiac troponin T | 697 bp | r: CGAACTTCTCTGCCTCCAAG | 55 | 84.2±0.0 | ||||
| 15 | Cx40, GJA5§ | GJ (atrial) | f: CCTCTTCATATTCCGTATGCT | 42.9 | 177 | # | ||
| gap junction protein, alpha 5, 40 kDa | r: GTGGAGACGAAGATGATCTG | 50 | 86.3±0.0 | |||||
| 16 | Cx43, GJA1§ | + | GJ (vent.) | f: CACATCAGGTGGACTGTTTCCTCT | 50 | 145 | [53] | |
| gap junction protein, alpha 1, 43kDa | r: TTAACCCGATCCTTAACGCCCTTG | 50 | 82.0±0.1 | |||||
| 17 | DES | + | IF | f: CCTTAAGAAAGTGCATGAAGAG | 40.9 | 262 | # | |
| Desmin | 581 bp | r: TCGGTATTCCATCATCTCCT | 45 | 87.2±0.0 | ||||
| 18 | EDNRA | RC | f: AATTGTTTCCAGTCATGCCTC | 42.9 | 151 | [15] | ||
| endothelin receptor type A | r: AGTTCATGCTGTCCTTATGG | 45 | 83.5±0.0 | |||||
| 19 | EDNRB | RC | f: TCTGTTGGTATTGGACTATATTGG | 37.5 | 119 | [15] | ||
| endothelin receptor type B | r: AGCATAAGCATGACTTAAAGCAG | 39.1 | 77.5±0.0 | |||||
| 20 | GAPDH | RG | f: TGCACCACCAACTGCTTAGC | 55 | 89 | [51] | ||
| Glyceraldehyde-3-phosphate dehydrogenase | r: GGCATGGACTGTGGTCATGAG | 57.1 | 82.6±0.5 | |||||
| 21 | GATA-4, GATA4§ | TF | f: CCAATCTCGATATGTTTGACGAC | 43.5 | 138 | # | ||
| GATA binding protein 4 | r: CGTTCATCTTGTGGTAGAGG | 50 | 85±0.0 | |||||
| 22 | HPRT1 | RG | f: GGCAGTATAATCCAAAGATGGTC | 43.5 | 95 | [15] | ||
| hypoxanthine phosphoribosyltransferase 1 | r: TTCAAATCCAACAAAGTCTGGC | 40.9 | 83.0±0.0 | |||||
| 23 | KCNA5 | + | IC | f: CGCAGAGTATTTGAAGTCAC | 45 | 195 | # | |
| potassium voltage-gated channel subfamily A member 5 | r: GGCTACAAATACATCCATCTC | 42.9 | 83.0±0.0 | |||||
| 24 | KCNH2 | IC | f: ATAGGCAAACCCTACAACAG | 45 | 191 | # | ||
| potassium voltage-gated channel subfamily H member 2 | r: AAGATGCTAGCATACATGAGG | 42.9 | 86.7±0.0 | |||||
| 25 | MLC-2a, MYL7§ | CA (atrial) | f: ACAAGGATGAGTTCAAGCAG | 45 | 162 | # | ||
| myosin, light chain 7, regulatory | r: CTCATTCCTCTTTCTCGTCTC | 47.6 | 85.6±0.0 | |||||
| 26 | MLC-2v, MYL2§ | CA (vent.) | f: ACAGGGATGGCTTCATTGAC | 50 | 192 | [52] | ||
| myosin, light chain 2, regulatory, cardiac, slow | r: ATGCGTTGAGAATGGTTTCC | 45 | 82.8±0.1 | |||||
| 27 | MYCD, MYOCD§ | CA | f: CCAAACTCTTCTTCAACGCC | 50 | 136 | # | ||
| myocardin | r: ATTCAGAGACCTTCAGATCATCC | 43.5 | 81.5±0.1 | |||||
| 28 | NFATc4, NFATC4§ | TF | f: CTGCTGAGGGATACAATAAGGT | 45.5 | 147 | # | ||
| nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 | r: TGATCTCACTCACTTCCTCC | 50 | 86.5±0.0 | |||||
| 29 | Nkx-2.5, NKX2-5§ | + | TF | f: CAGGTCAAGATCTGGTTCCA | 50 | 218 | # | |
| NK2 transcription factor related, locus 5 | r: TAGGCGTTATAACCGTAGGG | 50 | ||||||
| 30 | PLN | + | CA | f: GAGAACTGGTGGTTAATATGTG | 40.9 | 208 | # | |
| phospholamban | r: AC CTGCATTGGATGTTAGGCTG | 47.8 | ||||||
| 31 | RPLP0 | RG | f: AAATGTTTCATTGTGGGAGC | 40 | 170 | # | ||
| ribosomal protein, large, P0 | r: ATATGAGGCAGCAGTTTCTC | 45 | 86.5±0.0 | |||||
| 32 | SCN5A | IC | f: CATCTTTGGGTCTTTCTTCAC | 42.9 | 268 | # | ||
| sodium channel, voltage-gated, type V, alpha subunit | r: CACCATATTCAAGCAGATCAG | 42.9 | 85.0±0.0 | |||||
| 33 | SERCA2, ATP2A2§ | CA | f: GTCACTCCACTTCCTGATCC | 55 | 172 | [15] | ||
| ATPase, Ca2 + transporting, cardiac muscle, slow switch 2 | r: CTCCAGTATTGCAGGTTCCA | 50 | 85.1±0.1 | |||||
| 34 | Tbx5, TBX5§ | CD | f: GATAATAAATGGTCTGTGACGGG | 43.5 | 201 | # | ||
| T-box 5 | r: GTGTAATCTAGGCTGGTATTTGTG | 41.7 | 71.5±0.0 | |||||
| 35 | VIM | IF | f: CCCTTGACATTGAGATTGCC | 50 | 110 | [15] | ||
| Vimentin | r: CCAGATTAGTTTCCCTCAGGT | 47.6 |
If possible, primers were designed to cover overlapping intron-exon borders, fragment sizes for genomic DNA (gDNA) are indicated. Amplicon melting temperatures were aimed to exceed 75°C. +, amplification of genomic DNA; #, this study. The GC-content ranges between 37.5 and 7.1% (46.4±4.5%). RG, reference gene; TF, (cardiac) transcription factor; IC, ion channel; CA, contractile apparatus; RC, receptor; HF, heart failure; CD, cardiac differentiation; GJ, gap junction; IF, intermediate filament. TM melting temperature. All values are reported as median±median absolute deviation. §, Abbreviation according to the HUGO Gene Nomenclature Committee (HGNC).
Table 4
Final primer concentration
| Primer | MP1 | MP2 | MP3 | MP4 | MP5 | MP6 | MP7 | MP8 |
| pair | [μM] | [μM] | [μM] | [μM] | [μM] | [μM] | [μM] | [μM] |
| ACTB | 0.075 | |||||||
| ACTC1 | 0.375 | 0.25 | ||||||
| ADRB1 | 1 | |||||||
| ANP | 0.075 | |||||||
| BNP | 0.3 | 0.3 | 0.4 | |||||
| CHRM2 | 0.7 | |||||||
| cTnT | 0.15 | |||||||
| Cx40 | 0.8 | |||||||
| Cx43 | 0.125 | |||||||
| DES | 0.25 | 0.125 | ||||||
| EDNRA | 0.3 | 0.2 | ||||||
| EDNRB | 0.125 | 0.25 | 0.5 | |||||
| GATA-4 | 0.25 | |||||||
| HPRT1 | 0.3 | 0.075 | 0.15 | 0.15 | 0.15 | 0.15 | ||
| KCNA5 | 0.5 | 0.4 | ||||||
| KCNH2 | 0.5 | |||||||
| MLC-2a | 0.05 | |||||||
| MLC-2v | 0.0075 | 0.0375 | 0.075 | |||||
| MYCD | 0.5 | |||||||
| NFATc4 | 0.98 | |||||||
| PLN | 0.125 | 0.1 | ||||||
| RPLP0 | 0.0075 | 0.245 | ||||||
| SERCA2 | 0.1 | |||||||
| Tbx5 | 0.7 | 0.98 |
Table 5
Overview on results with multiplex PCR
 |




