A review of monoclonal antibodies in COVID-19: Role in immunotherapy, vaccine development and viral detection
Abstract
The harmful COVID-19 pandemic caused by the SARS-CoV-2 coronavirus imposes the scientific community to develop or find conventional curative drugs, protective vaccines, or passive immune strategies rapidly and efficiently. Passive immunity is based on recovering hyper-immune plasma from convalescent patients, or monoclonal antibodies with elevated titer of neutralizing antibodies with high antiviral activity, that have potential for both treatment and prevention. In this review, we focused on researching the potentiality of monoclonal antibodies for the prevention and treatment of COVID-19 infection. Our research review includes antibody-based immunotherapy, using human monoclonal antibodies targeting SARS-CoV-2 viral protein regions, specifically the spike protein regions, and using hyper-immune plasma from convalescent COVID-19 patients, in which monoclonal antibodies act as immunotherapy for the cytokine storm syndrome associated with the COVID-19 infection. In addition, we will demonstrate the role of the monoclonal antibodies in the development of candidate vaccines for SARS-CoV-2. Moreover, the recent progress of the diagnostic mouse monoclonal antibodies’ role will be highlighted, as an accurate and rapid diagnostic assay, in the antigen detection of SARS-CoV-2. In brief, the monoclonal antibodies are the potential counter measures that may control SARS-CoV-2, which causes COVID-19 disease, through immunotherapy and vaccine development, as well as viral detection.
1.The definition and production of monoclonal antibodies
Monoclonal antibodies are antibodies generated mainly from B cells-lymphocytes. These cells cannot propagate in cell culture for a long period of time. So, there are several methods used to run and produce them in stable cell lines, specifically using B cells-based protocols. If the source of these cells are mouse lymphocytes, the achieved murine monoclonal antibodies are used mainly for immunodiagnostic applications. In the case of using cells from human origin, particularly from convalescent patients, as patients with COVID-19, the produced human monoclonal antibodies are considered therapeutic antibodies targeting the SARS-CoV-2 [1].
The classic method for the production of the mouse monoclonal antibodies is based on the cell fusion of the splenic cells of immunized mice (for examples, immunization with SARS-CoV-2 proteins) and the mouse myeloma cell lines, using fusing media, such as Polyethylene glycol (PEG), then cell cloning and sub-cloning for the final production of the specific mouse monoclonal antibodies. The generated mouse monoclonal antibodies are mainly used for immunodiagnosis [2, 3]. In contrast, the generation of potential therapeutic recombinant human mAb includes the cloning of cDNA’s, encoding the variable regions of the heavy and light chains into expression plasmids, containing the human IgG1 heavy chain and Ig kappa light chain constant regions, respectively. Both plasmids contain the interleukin-2 signal sequence to enable efficient secretion of the recombinant antibodies. The recombinant human antibodies are then produced in HEK-293T cells after transfection with sets of the IgG1 heavy and light chain expression plasmids and purified using protein-A affinity chromatography [4].
The need to treat the emergent SARS-CoV-2 that caused the current fling in health problem worldwide, directed the focus to the development of monoclonal antibody-based passive immunotherapy, in order to deliver a rapid response. Although, several investigations and primary data have been published on specific neutralizing antibodies in the preclinical phase [4, 5], no monoclonal antibodies have yet been available on the market. The increase understanding of MERS-CoV and SARS-CoV in recent years has stimulated the research community to make significant advancement in the immunotherapy design of COVID-19 in a short period of time by applying the current anti-viral treatment which presented hopeful data for MERS and SARS [6].
The virus has a genome with almost 80% similarity to the previously described SARS-CoV and 96% identity to the BatCoV RaTG13 of bats [7], which include the genome codes for 3 surface proteins: the membrane, the envelope, and the spike proteins. Both, the membrane, and the envelope play a role in the viral assembly and its release. The virus-host cell receptor interaction is mediated through the trimetric spike protein (S protein), which binds to the angiotensin-converting enzyme 2 (ACE2) of the human respiratory cells [8, 9] (Fig. 1, created with BioRender.com). Studies show that the S protein cleavage by proteolytic enzymes determine the zoonotic nature of the virus and whether, or not, the virus can cross species. For example, the Ugandan bat strain, MERS-like CoV is found to have the ability to bind to the human cells but fails to mediate infectivity [10]. However, SARS-CoV-2 protein sequence shows a unique Furin cleavage site (RRAR, Fig. 1) which has not yet been identified whether it is cleaved or not by the human Furin endoprotease. This site has been created by an insertion of 12 nucleotides in the cleavage site of the hosts’ proteases, which normally cleave the S protein of Coronaviruses on S1 and S2. This insertion led to the acquisition of 3 O-linked glycosylation [11]. It is worth mentioning that the Bat CoV (RaTG13) isolated from Yunnan, China, does not have the Furin cleavage sequence [12]. We may propose that the Furin cleavage site that appeared in the SARS-CoV-2, might have been acquired by the recombination with other viruses; a process that may have taken weeks or occurred within the last few years.
Figure 1.
Structural diagram of Spike region of SARS-CoV-2. Figure is created with BioRender.com.
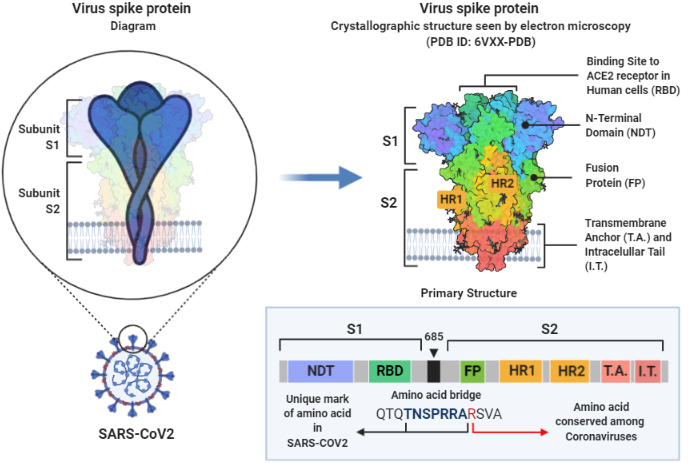
Although, the structural similarity between SARS-CoV-2 and the SARS-CoV is high (94% after excluding the receptor binding motif), unfortunately several receptor binding domain (RBD) directed monoclonal antibodies (S230, 80R, CR3014 and m396) fail to cross react, at 1
Figure 2.
Diagrammatic procedures of classic hybridoma Technology for production of monoclonal antibodies.
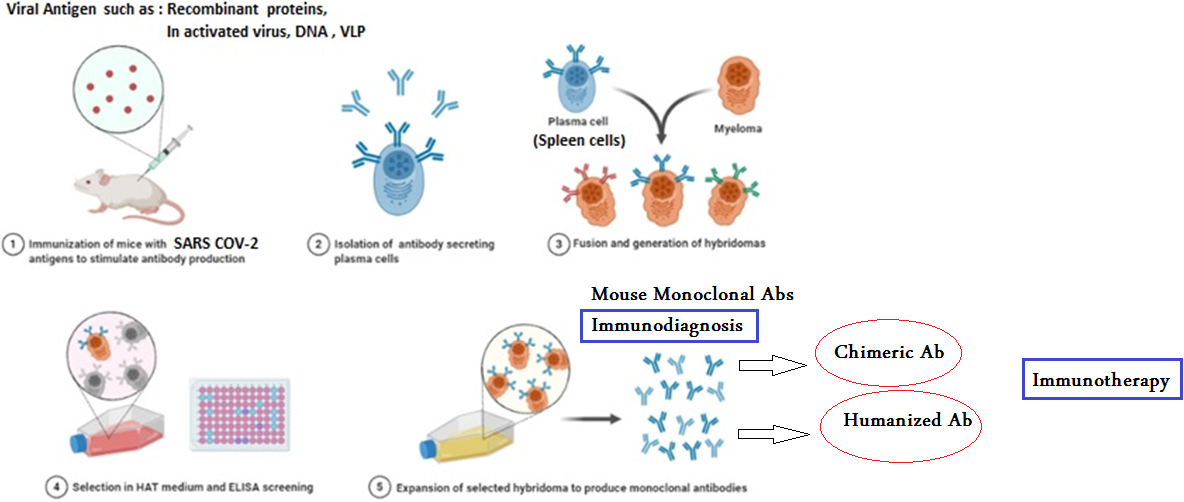
Since the hybridoma technique (Fig. 2) was introduced, in 1975 (Fig. 2, created with BioRender.com) as an effective approach to have enough quantities of pure mAbs; therapeutic trials have been conducted and continue to neutralize viral surface epitopes, as in SARS-CoV [15] and MERS [16]. These principles of hybridoma technique (Fig. 2) are, now, used to generate human monoclonal antibodies to neutralize the SARS-CoV-2.
The potential, different, approaches for using the hybridoma technique are:
1. Using the classic hybridoma technology for the production of mouse monoclonal antibodies against the specific SARS surface epitopes from the spike protein; then use the specific hybrid cell line that produces the specific mouse monoclonal antibodies for the extraction of RNA and performing humanization and chimeric human monoclonal antibodies. This model was achieved by Wang et al. [4] to generate the human monoclonal antibody 47D11. The antibody showed its neutralizing capacity against common epitopes from SARS-CoV and SARS-CoV-2 viruses in Vero cell culture.
2. The use of peripheral blood mononuclear cells (PBMCs), from convalescent patients with COVID-19, as a source of B-cell lymphocytes. From the peripheral blood mononuclear cells (PMBCs), CD22+B cells are separated by Magnetic Activated Cell Sorting (MACS) then immortalized by EBV [17, 18]. In April 17th 2020, the clinical trials identifier number NCT04354766 was assigned to a clinical trial that uses generated monoclonal antibodies from immortalized B cells, isolated from convalescent patients to neutralize SARS-CoV-2 [19].
3. The use of PBMCs, from convalescent COVID-19 patients, for the separation of CD27+B cells and using the cell fusion procedure with human cell lines. These hypothesis and approaches were carried out by Puligedda et al. [20] and produced mAbs that have potent neutralizing activity against poliovirus.
4. Using phage display technology to screen native human semisynthetic phage library, (Tomlinson I library) for the receptor binding domain (RBD) of SARS-CoV-2. In COVID-19 patients [21], 3 antibody formats (scFv, scFv-Fc and IgG1) have been identified and exhibited high binding specificity to the RBD of the trimetric spike glycoprotein. The scFv antibody format demonstrates specific binding to an epitope that partially overlap the angiotensin-converting enzyme 2 (ACE2)-interacting sites on the CoV-2 RBD. The IgG1 format binds to the cells expressing membrane-bound SARS-CoV-2 S-protein, while the scFv-Fc format shows specific binding to the RBD, in docking studies, through the use of Flow cytometry assay and ELISA tests, respectively. Another phage display protocol uses one native llama VHH and one synthetic humanized llama VHH libraries [22]. This protocol uses the next generation of antibody therapy approach described by Wang et al. [4]. The approach is based on the generation of bi- or tri-specific antibodies which may target different antigens at the same time to block the S/ACE2 interaction-site. This method would improve the restrictions related to the polyclonal and monoclonal combination of immunotherapies. The developed multi-specific antibodies show enhanced affinity, avidity and improve S/ACE2 blocking by using an in-silico approach that also fuses VHHs to Fc domains. Moreover, the developed bi-specific antibody demonstrates effective S/ACE2 hindering which is better than the separate monoclonal VHH-Fcs. The antibodies 80R, CR3014, CR3022, M396 and B1 are all identified by phage display method and they all target the S protein and block the interaction of S1 region with the ACE2 receptor, except for the B1 antibody which targets the S2 domain and blocks the interaction of S2 with the ACE2 receptor.
2.The potential use of the antibody-based immunotherapy for COVID-19
2.1Hyper immune- convalescent plasma for the treatment of COVID-19 infection
Hyper immune plasma and intravenous immunoglobulin infusions are safe and applicable treatments for a broad range of human infections. During outbreaks and the need to get immediate and effective measures, the use of convalescent serum, or plasma, or complete blood continues to grow and should be used under the WHO guidance. Recovering plasma was used in the 2014 Ebola outbreak, which prevailed in West Africa, for its better-quality of survival, relative to the standard therapy [23]. However, the extensive immune deregulation is a major cause for the severity and harmful manifestations of COVID-19, including the cytokine storm syndrome, which is an overexcited immune-inflammatory reaction to an infection, resulting in acute tissues and organs injury [24]. The timing of the therapy with convalescent patients’ plasma is likely to be precarious; if given at the wrong time, to a COVID-19 patient, the immune-plasma could be ineffective, or even inspire/increase a cytokine storm syndrome. Casadevall and Pirofski [25] underline the dangers of the passive usage of convalescent immunoglobulin, which falls into two categories, the serum disease, and the antibody-dependent enhancement of infection. The serum disease is concomitant with the contamination of other blood pathogens, whereas the antibody dependent enhancement theoretically could create antibodies to one strain of coronavirus, and increase the infection to another coronavirus strain [25]. Moreover, Nguyen et al. [26] reports that the restricted usage of the Intravenous Immunoglobulin (IVIG) includes variable specific neutralizing antibody titer(s) against novel pathogens. The use of IVIG in COVID-19 patients may cause immunoglobulins to bind to the Fc
In COVID-19 patients, the convalescent plasma can reverse the inflammatory processes and inhibit the consequences of viral infection. The proposed mechanisms of inhibition included the presence of IgM/IgG neutralizing antibodies which have the capability to inhibit the viral entry to the host cells and activate the hosts’ immune system through antibody dependent cellular immunity [27]. Acosta-Ampudia et al. [28], shows that the plasma of recovered COVID-19 patients contains autoantibodies, cytokines and metabolites different in composition from those of the severe symptomatic cases. This profile is responsible for an early and transient hyper immunity known as the cytokine storm, in patients with severe symptomatic infection. This convalescent plasma, at day 28, may increase lymphocytes (T and B cells) post transfusion and reduce rations of IL-6/IFN-
The use of convalescent plasma has produced promising results for the treatment of the COVID-19 disease, in 2 studies reviewed by Shen et al. [29], and Duan et al. [30]. Despite the promising data of these studies, the use of convalescent plasma has the following concerns:
(1) Immunoglobulins are not 100% homologous between individuals.
(2) The viral load in patients is expected to be higher than the Immunoglobulins transferred to them which mean that the patients will need more than one dose.
(3) The transferred Igs may give negative feedback to the patient’s immune system which may result in the reduction of the Igs that are naturally produced by the patient’s immune system.
(4) The possible infusion reactions occurring, between the produced antibodies and the transferred antibodies that could cause immune complexes, may lead to kidney inflammation, or other consequences.
(5) The viral latency.
(6) The subtypes of immunoglobulins between donor and recipient.
(7) There is not enough information about the dominant IgA in the mucosal tissue.
All of these limitations for using convalescent plasma of COVID-19 patients, along with its possibility to aggravate the disease, discourage the use of intravenous immunoglobulins (IVIG) [26].
However, Joyner et al. [31], shows that the infusion of convalescent plasma presents only 1.2% overall serious adverse events, 4 hours after transfusion, in 5000 patients ages ranging from 18–97. Their experience provides no signal of toxicity beyond what is expected from using plasma in severely ill patients. Additionally, given the deadly nature of COVID-19 and the large count of critically ill patients, with multiple comorbidities included in their analyses, the mortality rate does not appear to be excessive. Several clinical trials have been lunched and assigned clinical trial numbers, which include phases 1 to 3 and are listed in Esmaeilzadeh and Elahi [19]. Accordingly, the previously mentioned limitations would pave the way to consider monoclonal antibodies for the neutralization of SARS-CoV-2 that can focus on specific target epitopes on viral particles and pose a potential protection and immunotherapy for COVID-19 [32].
Barnes et al. [33] shows that the plasma IgGs isolated and purified from convalescent plasma, of patients recovered from COVID-19, are more potent neutralizers. It targets not only the RBD of the s-protein epitopes but also epitopes of S1A (in the NTD from amino acid 12 to 306) outside the RBD, as opposed to their plasma Fabs counterparts. The epitope mapping of the RBD and S1A of the s-protein that uses the Negative-Stain Electron Microscopy Polyclonal Epitope Mapping (nsEMPEM) and Single-Particle Cryo-Electron Microscopy proved that these sites are not affected by the mutations, commonly occurring in the SARS-CoV-2 isolates. This may pave the way for antibody therapies or vaccine development.
2.2Therapeutic monoclonal antibodies targeting SARS-CoV-2
Pharmaceutical companies are in favor to produce monoclonal antibodies as a treatment for COVID-19 including Celltrion, AstraZeneca and Regeneron. The first human study to treat COVID-19 is, using LY-CoV555 neutralizing mAb against the spike protein, and is currently in progress, in phase II trial and is carried out by the pharmaceutical company “Lilly”. The neutralizing antibody LY-CoV555 with a dose of 2800 mg, appeared to accelerate the natural decline in viral load over time, whereas the other doses (700 mg, or 7000 mg) have not [34]. Recently, Bamlanivimab and Casirivimab-Imdevimab are novel virus-neutralizing monoclonal antibodies authorized to treat mild to moderate COVID-19 infected outpatients who are at a risk for progression to a severe level of infection. Also, early treatment may show efficacy in reducing progression in severely infected cases [32]. However, the U.S. Food and Drug Administration issued an emergency use authorization (EUA) for Casirivimab and Imdevimab (Regeneron Pharmaceuticals Inc, https://www.regeneron.com/casirivimab-imdevimab) to be administered together for the treatment of mild to moderate COVID-19 infection in adults and pediatric patients (https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19).
Figure 3.
Neutralizing monoclonal antibodies and selection of neutralizing viral epitope as candidate vaccine. Figure is created with BioRender.com.
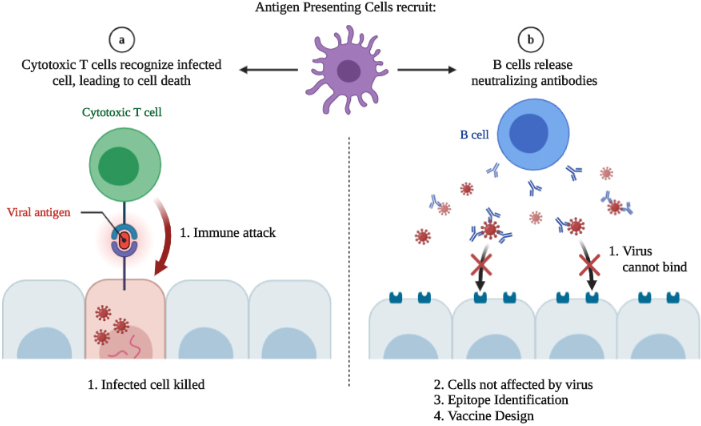
2.3Monoclonal antibodies against SARS-CoV-2 epitopes, as a candidate for COVID-19 vaccine
The neutralizing therapeutic antibodies and vaccine development, which are the potential counter measures controlling SARS-CoV-2 that cause COVID-19 disease, reports that the high dose vs. low dose viral infection of SARS-CoV-2 may affect the lymphocytic immune reactions. The high dose viral exposure belongs to the cases of health-care workers and individuals, who have currently sick or have asymptomatic family members spreading the virus. The low dose viral infection may lead to the stimulation response of the T and B cell lymphocytes, as well as the production of neutralizing antibodies and rapid viral clearance (spontaneous clearance). Whereas the high dose viral infection could cause the increase of the severity of the disease and late viral clearance. This may be due to lymphopenia leading to the ineffectiveness of the T and B cell, then the initiation of the cytokine storm and tissue damage [35]. So generally, the low viral dose is a better candidate for the vaccine; especially if it stimulates cellular immunity and humoral immunity, particularly concerning the production of the neutralizing antibodies. These neutralizing antibodies have the capability to bind to the viral particles, eliminating any free viral particle that can infect the target cells. In other words, these neutralizing antibodies binds with the Angiotensin Converting receptor 2 (ACE2) preventing the binding of the virus particles with the target cells, which in-turn can prevent intracellular replication and viral assembly, as well as viral transmission from cell to cell. So, if we have these neutralizing antibodies based on the neutralizing assays, we can determine the neutralizing epitope and use it as a candidate vaccine (Fig. 3b).
Both human monoclonal antibodies, from hybridoma technology, and plasma of convalescent COVID-19 patients are encouraging immunological strategies for the development of immunotherapy for COVID-19 infection. Recently, Begum and Ray [36] suggested that using B cell lymphocytes producing antibody isolates from convalescent SARS-CoV-2 individuals, for the generation of human B cell hybridomas, and a mixture of human mAbs, for multiple immunogenic targets of SARS-CoV-2 spike protein, might deliver an antibody mix for a long-term treatment therapy with a convalescent focus. Neutralizing monoclonal antibodies bond exactly on the neutralizing epitopes, on the spike protein of SARS-CoV-2, as opposed to the widespread variety of antibodies in plasma. Some plasma antibodies will be neutralizing ones, but many others are non-neutralizing (off-target effects), potentially causing tissue injury. For example, a non-neutralizing antibody binding to a non-spike protein viral antigen could result in a damaging antibody reactions (type I-III) and antibody dependent disease development. These conflicting properties are less likely to occur with the use of a single or combined monoclonal antibodies, which are presently in the initial clinical trials phase [37]. Interestingly, the availability of both non-neutralizing and neutralizing monoclonal antibodies generated in this study will enable the assessment of the S protein immunogens, for the presentation of neutralizing and non-neutralizing epitopes, and inspire effective candidates for vaccine design. The design of an immunogen that improves the quality of mAbs, elicited by natural infection, may well appear as an essential target for vaccine research.
Figure 4.
Diagrammatic potential of the promising monoclonal antibodies against spike epitopes of SARS-CoV-2.
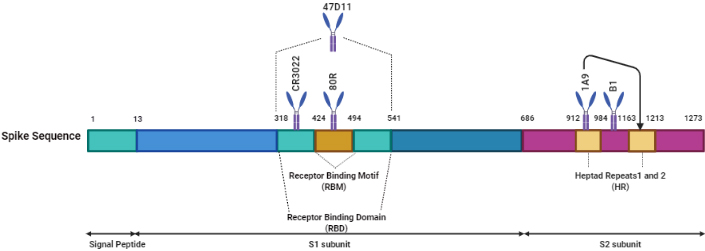
Scientists have issued papers of specific therapeutic human monoclonal antibodies (hmAbs) (B38, H4, 47D11) targeting SARS-CoV-2 with the prospect that these antibodies are effective. Heavy and light-chains of both B38 and H4 hmAbs which are generated in Nicotiana benthamiana plant leaves show specific binding to the SARS-CoV-2 RBD and in-vitro efficient neutralization capacity [38]. However, ACE2 is an endogenous membrane protein that assists in the infection of SARS-CoV-2. During COVID-19 infection, the extracellular peptidase domain of ACE2, blocks the receptor binding domain (RBD), of the spike protein of SARS-CoV-2. The sole focus of scientists is to produce mAbs targeting the S protein and RBD. Several studies, in vivo, demonstrates the high neutralizing potency of the antibodies targeting these epitopes against SARS-CoV-2 [39, 40, 41]. Other work was conducted to isolate mAbs from memory B cells that were sorted from PBMCs of convalescent patients, who recovered from infection [42].
Separation of B cells from convalescent COVID-19 patients, the collection of B cells that are efficient for generating the needed neutralizing antibodies and the cloning of the neutralizing monoclonal antibodies are labor-intensive tasks. Chi and his colleagues [42] were able to prove that antibodies raised against epitopes, other than RBD, would neutralize the virus. They identified a neutralizing mAb named 4A8 which recognizes an epitope of the N-Terminal Domain (NTD) on the spike glycoprotein. This antibody shows the capacity to neutralize the SARS-CoV-2 virus either in authentic or pseudo-forms. However, the 4A8 antibody fails to recognize the RBD. Almost all generated mAbs, that are targeting the S protein epitope (s), are in the clinical trials, either single, such as, the REGN10933, LY3819253, CT-P59, BRII-198, or combined, such as, LY3819253+LY3832479, REGN10933+REGN10987, AZD8895+AZD1061 [43]. Other mAbs are with the nucleotide analog that blocks the viral RNA polymerase named Remdesivir, such as, the LY3819253.
Another challenge in the development of mAbs is the possible mutations in the S glycoprotein of the SARS-CoV-2 virus. These mutations would affect the neutralizing capacity of an antibody leading to the escape of the variant. Korber et al. [44], identified 14 mutation points in the S protein targeted by all mAbs in the clinical trials. All these mutations are rare, or of low frequency except for one mutation; D614G (outside the RBD) which affects the stabilization of the interprotomer that is found in 3577 counts globally and may suggest a role in human-to human viral transmission. Now, this variant constitutes over 97% of the isolates globally [45] and may cause a conformational shift in the S protein due to the interruption occurring between S1 and S2 contacts. In 9 countries, different counts of mutations are discovered, such as 8 in Europe and 65 in the USA. Interestingly, Yurkovetskiy et al. [45] tested human mAb with the D614G mutation and concluded that it could not affect the antibodies targeting the RBD to cause the blockage of the ACE2 binding with the RBD. During infection, the host proteases cleave the S protein on the S1 (N-terminal) and the S2 (C-terminal) subunits. The S1 contains the signal peptide NTD and RBD, whereas the S2 subunit contains the heptad repeats (HR) 1 and 2, which are necessary membrane components. These 2 repeats have been fused and nominated, also as potential candidates, to block viral fusion based on the data of 1A9 monoclonal antibody against SARS-CoV HR2 domain [46].
The hypothesis of using this antibody against SARS-CoV-2 is based on the fact that the S2 protein is highly conserved. Figure 4, created with BioRender.com, shows the potential promise of the monoclonal antibodies against the spike epitopes of SARS-CoV-2.
Table 1
The cytokine storm members and available monoclonal antibodies included during COVID-19 infection
| Cytokine | Available mAb/antagonist | Adverse effects of using the antibody | Clinical trials | Reference | ||
|---|---|---|---|---|---|---|
| Name | Action/results | Name | Action/results | |||
| IL-6 | Destroys the alveoli membrane/hemorrhage in lungs then fibrosis | 1-Tocilizumab (mAb) 2-Sarilumab (mAb) 3-Clazakizumab (mAb) | Binds then block the action of IL-6. No direct action on the virus/Inhibits the destructive effect of the virus. | 1-Increase in hepatic enzymes 2-Skin allergy 3-Infection by opportunistic fungi | Phase III | [46, 48, 49, 50] |
| IL-1 | Produced by macrophages, induces fever/respiratory fibrosis | Canakinumab (mAb) | Blocks IL-1 | Not identified | – | [19] |
| IL-17 | Produced by lymphoid cells, activates T-helper 17 cells, activates secretion of proinflammatory mediators and infiltration of neutrophils/lung tissue damage | No available | – | Not identified | Not yet | [51] |
| IFN- | Produced by CD | Emapalumab (mAb) | Blocks IFN- | Not identified | Phase II/III | [52] |
| TNF- | One of the first cytokines produced during viral infection, induces differentiation od dendritic cells | XPro1595 (soluble protein used in Alzheimer’s disease) | Inhibits the TNF- | Not identified | Phase II | [53] |
3.Monoclonal antibodies in cytokine storm control
In 2019, patients with coronavirus disease, had a huge number of inflammatory immune cells, such as T cells and macrophages, which were stimulated and infiltrated into the respiratory tissues in response to the generated chemokines and cytokines released by the infected cells. In a low, or mild COVID-19 infection, SARS-CoV-2 would be cleared by these cells, whereas in severe infection, major cytokines, such as, interleukin-6 (IL-6), IL-1
4.The role of monoclonal antibodies in SARS-CoV-2 immunodetection
While molecular biology diagnosis has been widely used in the diagnosis of COVID-19, it cannot be used to evaluate the progress of COVID-19 and cannot be used to identify old infection and humoral immunity [54, 55, 56, 57]. The limitations of these diagnostic techniques have developed increasing demand for rapid and simpler tests that are immunity based. For example, RT-PCR, while very reliable and accurate, is only capable of identifying the presence of viral RNA and cannot determine the state or progression of the infection at a given time, in an infected patient. In contrast, immunological tests have several advantages (rapid, sensitive, specific, safety reagents, qualitative or quantitative determination) [58]. These tests measure immunological responses to the SARS-CoV-2, allowing for the differentiation between old and recently infected cases. They play an important role in COVID-19 epidemiology, vaccine development, and immune response [59, 60]. They are also tools that use components of the immune response, as whole parts of the test [57], in which the quantitation of the analyte depends on the reaction of an antigen (analyte) and an antibody [58]. The enzyme-based immunoassays detect the antigen with specifically manufactured monoclonal antibodies [54]. This antigen may be found in body fluids (Oropharyngeal swab, saliva, sputum, blood, urine, stool) [59]. Recently, the immunological test was used for the identification of SARS-CoV-2 proteins, using monoclonal antibodies (Fig. 5) (created with BioRender.com). Multiple forms of N protein or S protein (S1, S2) are used as targets of SARS-CoV-2 monoclonal antibodies [61, 62, 63, 64]. The monoclonal antibodies against SARS-CoV-2 nucleocapsid proteins that have been produced, might form the basis for a future rapid antigen detection test [65, 66].
Figure 5.
Immunodetection of SARS-CoV-2 based on specific monoclonal antibodies using ELISA or lateral flow assay.
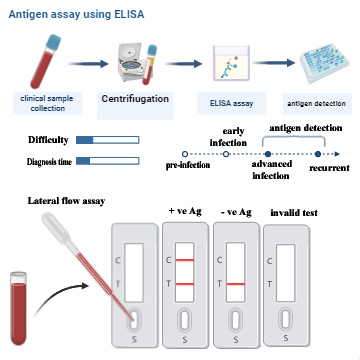
These days, there are many immunoassay techniques that are approved or still pending approval, where their versatility and creativity could increase the different ways, with which one could detect pathogens in a clinical sample study. Thus, great efforts have been made to develop tests for the rapid detection of SARS-CoV-2 antigens, which target viral antigen in biological samples, such as nasopharyngeal secretions. Several rapid antigen tests have been proposed [67, 68], and now is considered, the most used tool for SARS-CoV-2 antigens detection. Recently Weitzel et al. [69], reported that rapid antigen detection has the potential to serve as an alternative diagnostic method, especially as a screening tool for patients with high viral loads during early and infective stages of infection [69].
Porte et al [70] evaluated a novel rapid antigen detection test (RDT) SARS-CoV-2, in transport medium, of nasopharyngeal (NP) and oropharyngeal (OP) swabs from suspected COVID-19 cases. They used the fluorescence immunochromatographic SARS-CoV-2 antigen test (Bioeasy Biotechnology Co., Shenzhen, China) for diagnostic accuracy, instead of Real Time (RT) PCR. The sensitivity and specificity of the RDT assay averages around 93.9% and 100%, respectively. The antigen Detection (AD) tests are based on specific monoclonal antibodies, which are more rapid, less laborious and less expensive alternatives [71]. Recently, Favresse et al. [71] compared the clinical performance of 5 AD tests, including 4 rapid AD (RAD) tests (biotical, Panbio, Healgen, and Roche) and 1 automated AD test (VITROS). The result of this study demonstrates that the performance of the RAD tests is more modest and allows us to identify RT-PCR positive patients with higher viral loads. The sensitivity ranges from 93.1% to 96.6%, meaning that some samples with high viral loads were missed. The VITROS automated assay shows a 100% for both sensitivity and specificity. So Favresse et al. [71] concluded that compared to RAD tests, the VITROS assay is fully aligned with the RT-PCR, which may provide the identification of SARS-CoV-2 contagious patients with a faster, easier, and cheaper approach.
5.Concluding remarks and future perspective
Along the past three decades, the use of monoclonal antibodies, in the treatment of various diseases, proved to be effective and easily delivered. These molecules have several advantages to be used against the SARS-CoV-2 virus specifically; probably due to the fact that they come from convalescent patients with COVID-19 disease. Till today, more than 70 monoclonal antibodies are in either preclinical or clinical pipeline programs for development, which may offer a potential rapid manufacturing to face the pandemic. It is well known that vaccines may provide long term protection after few weeks of application. Monoclonal antibodies would be a complement to the vaccines especially during the pandemic, as they offer immediate protection upon administration and lasts for weeks or months. This will, also, help in the treatment of infected patients and may prevent young and elderly people from being infected. However, it is important to study the safety, manufacturing capacities, production time, price and availability of these products (monoclonal antibodies, vaccines, etc) in all countries.
Compliance with ethics requirements
Our article does not contain any studies with human or animal subjects.
Availability of data and material
Not applicable.
Funding
This review article was partially supported by Science, Technology and Innovation Funding Authority (STIFA) Emergency Targeted Program of COVID-19, Egypt, Project ID: 43703 granted to Yasser E. Shahein and the Corona call, Ideation Fund, Academy of Scientific Research and Technology; Project ID: 7012 to Mohamed M. Omran
Authors’ contributions
AT, YS, MO, MA, KA wrote the draft and AT, YS and MO finalized it, while AR performed the editing. All authors read and approved the final version of the manuscript.
Conflict of interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
References
[1] | J.P. Henderson, What are protective antibody responses to pandemic SARS-CoV-2? J. Clin. Invest 130: (12) ((2020) ), 6232–6234. |
[2] | A. Tabll, A.T. Abbas, S. El-Kafrawy and A. Wahid, Monoclonal antibodies: principles and applications of immmunodiagnosis and immunotherapy for hepatitis C virus, World J. Hepatol 7: (22) ((2015) ), 2369–2383. |
[3] | C.A. Abdel Malak, T.H. Abelhafez, A.A. Tabll, M.M. Mashaly, R. El Shenawy, Y.S. El-Abd, M.H. Shaker and M.K. El-Awady, Neutralizing activity and safety of human monoclonal antibodies against hepatitis C virus, Hum Antibodies 26: (3) ((2017) ), 127–134. |
[4] | C. Wang, W. Li, D. Drabek, N.M.A. Okba, R. van Haperen, A.D.M.E. Osterhaus et al., A human monoclonal antibody blocking SARS-CoV-2 infection, Nat. Commun 11: (1) ((2020) ), 2251. |
[5] | B. Ju, Q. Zhang, J. Ge, R. Wang, J. Sun, X. Ge, J. Yu and S. Shan, Human neutralizing antibodies elicited by SARS-CoV-2 infection, Nature 584: (7819) ((2020) ), 115–119. |
[6] | B. Shanmugaraj, K. Siriwattananon, K. Wangkanont and W. Phoolcharoen, Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19), Asian Pac. J. Allergy Immunol 38: (1) ((2020) ), 10–18. |
[7] | Y. Yan, W.I. Shin, Y.X. Pang, Y. Meng and J. Lai, The first 75 days of novel coronavirus (SARS-CoV-2) outbreak: recent advances, prevention, and treatment, Int. J. Environ. Res. Public Health 17: (7) ((2020) ), 2323. |
[8] | P. Gollapalli, B.S. Sharath, H. Rimac, P. Patil, S.K. Nalilu, S. Kandagalla and P. Shetty, Pathway enrichment analysis of virus-host interactome and prioritization of novel compounds targeting the spike glycoprotein receptor binding domain-human angiotensin-converting enzyme 2 interface to combat SARS-CoV-2, J. Biomol. Struct. Dyn ((2020) ), 1–14. doi: 10.1080/07391102.2020.1841681. |
[9] | G. Simmons, P. Zmora, S. Gierer, A. Heurich and S. Pöhlmann, Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research, Antiviral Res 100: (3) ((2013) ), 605–614. |
[10] | S.J. Anthony, K. Gilardi, V.D. Menachery, T. Goldstein, B. Ssebide, R. Mbabazi, I. Navarrete-Macias, E. Liang, H. Wells, A. Hicks, A. Petrosov, D.K. Byarugaba, K. Debbink, K.H. Dinnon, T. Scobey, S.H. Randell, B.L. Yount, M. Cranfield, C.K. Johnson, R.S. Baric, W.I. Lipkin and J.A. Mazet, Further evidence for bats as the evolutionary source of middle east respiratory syndrome coronavirus, mBio 8: (2) ((2017) ), e00373-17. |
[11] | K.G. Andersen, A. Rambaut, W.I. Lipkin, E.C. Holmes and R.F. Garry, The proximal origin of SARS-CoV-2, Nat. Med 26: (4) ((2020) ), 450–452. |
[12] | D. Wrapp and N. Wang, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science 367: (6483) ((2020) ), 1260–1263. |
[13] | X. Tian, C. Li, A. Huang, S. Xia, S. Lu and Z. Shi, Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody, Emerg. Microbes Infect 9: (1) ((2020) ), 382–385. |
[14] | J. Shang, G. Ye, K. Shi, Y. Wan, C. Luo, H. Aihara, Q. Geng, A. Auerbach and F. Li, Structural basis of receptor recognition by SARS-CoV-2, Nature 581: (7807) ((2020) ), 221–224. |
[15] | B. Rockx, D. Corti, E. Donaldson, T. Sheahan, K. Stadler, A. Lanzavecchia and R. Baric, Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge, J. Virol 82: (7) ((2008) ), 3220–3235. |
[16] | J. Goo, Y. Jeong, Y.S. Park, E. Yang, D.I. Jung, S. Rho, U. Park, H. Sung, P. Park, J. Choi, S. Seo, N. Cho, H. Lee, J. Lee, J. Kim and M. Song, Characterization of novel monoclonal antibodies against MERS-coronavirus spike protein, Virus Res 278: ((2020) ), 197863. |
[17] | J. Fraussen, K. Vrolix, P. Martinez-Martinez, M. Losen, E. Meulemans, M.H. De Baets, P. Stinissen and V. Somers, A novel method for making human monoclonal antibodies, J. Autoimmun 35: (2) ((2010) ), 130–134. |
[18] | A. Tabll, Y. El Abd, N.G. El Din, R. El Shenawy, T.H. Abdelhafez, M. El Awady, H. El-Mohamady and S. Viazov, Establishment of human clones producing neutralizing human monoclonal antibodies to the envelope E1/E2 protein of hepatitis C virus by EBV immortalization of immune CD22+ B cells, Hum. Antibodies 22: (3–4) ((2013) ), 55–65. |
[19] | A. Esmaeilzadeh and R. Elahi, Immunobiology and immunotherapy of COVID-19: a clinically updated overview, J. Cell. Physiol 236: (4) ((2021) ), 2519–2543. |
[20] | R.D. Puligedda, V. Vigdorovich, D. Kouiavskaia, C.D. Kattala, J.Y. Zhao, F.H. Al-Saleem, K. Chumakov, D.N. Sather and S.K. Dessain, Human IgA monoclonal antibodies that neutralize poliovirus, produced by hybridomas and recombinant expression, antibodies (Basel, Switzerland), 9: (1) ((2020) ), 5. |
[21] | H.A. Parray, A.K. Chiranjivi, S. Asthana, N. Yadav, T. Shrivastava, S. Mani, C. Sharma and P. Vishwakarma, Identification of an anti-SARS-CoV-2 receptor-binding domain-directed human monoclonal antibody from a naïve semisynthetic library, J. Biol. Chem 295: (36) ((2020) ), 12814–12821. |
[22] | J. Dong, B. Huang, Z. Jia, B. Wang and S. Gallolu Kankanamalage, Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity, Emerg. Microbes Infect 9: (1) ((2020) ), 1034–1036. |
[23] | F. Sahr, R. Ansumana, T.A. Massaquoi, B.R. Idriss, F.R. Sesay, J.M. Lamin et al., Evaluation of convalescent whole blood for treating ebola virus disease in freetown, sierra leone, J. Infect 74: (3) ((2017) ), 302–309. |
[24] | R.J. Jose and A. Manuel, COVID-19 cytokine storm: the interplay between inflammation and coagulation, Lancet Respir. Med 8: (6) ((2020) ), e46–e47. |
[25] | A. Casadevall and L.A. Pirofski, The convalescent sera option for containing COVID-19, J. Clin. Invest 130: (4) ((2020) ), 1545–1548. |
[26] | A.A. Nguyen, S.B. Habiballah, C.D. Platt, R.S. Geha, J.S. Chou and D.R. McDonald, Immunoglobulins in the treatment of COVID-19 infection: proceed with caution!, Clin. Immunol. (Orlando, Fla) 216: ((2020) ), 108459. |
[27] | M. Rojas, Y. Rodríguez, D.M. Monsalve, Y. Acosta-Ampudia, B. Camacho, J.E. Gallo et al., Convalescent plasma in Covid-19: possible mechanisms of action, Autoimmun. Rev 19: (7) ((2020) ), 102554. |
[28] | Y. Acosta-Ampudia, D.M. Monsalve, M. Rojas, Y. Rodríguez, J.E. Gallo, J.C. Salazar-Uribe, M.J. Santander, M.P. Cala, W. Zapata, M.I. Zapata, R. Manrique, J.M. Pardo-Oviedo, B. Camacho, C. Ramírez-Santana and J.M. Anaya, CP-COVID-19 group, COVID-19 convalescent plasma composition and immunological effects in severe patients, J. Autoimmun 118: ((2021) ), 102598. |
[29] | C. Shen, Z. Wang, F. Zhao, Y. Yang, J. Li, J. Yuan et al., Treatment of 5 critically III patients with COVID-19 with convalescent plasma, JAMA 323: (16) ((2020) ), 1582–1589. |
[30] | K. Duan, B. Liu, C. Li and H. Zhang, Effectiveness of convalescent plasma therapy in severe COVID-19 patients, Proc. Natl. Acad. Sci. USA 117: (17) ((2020) ), 9490–9496. |
[31] | M.J. Joyner, K.A. Bruno, S.A. Klassen, K.L. Kunze, P.W. Johnson, E.R. Lesser, C.C. Wiggins, J.W. Senefeld, A.M. Klompas, D.O. Hodge, J.R.A. Shepherd, R.F. Rea, E.R. Whelan, A.J. Clayburn, M.R. Spiegel, S.E. Baker, K.F. Larson, J.G. Ripoll, K.J. Andersen, M.R. Buras, M.N.P. Vogt, V. Herasevich, J.J. Dennis, R.J. Regimbal, P.R. Bauer, J.E. Blair, C.M. van Buskirk, J.L. Winters, J.R. Stubbs, N. van Helmond, B.P. Butterfield, M.A. Sexton, J.C. Diaz Soto, N.S. Paneth, N.C. Verdun, P. Marks, A. Casadevall, D. Fairweather, R.E. Carter and R.S. Wright, Safety update: COVID-19 convalescent plasma in 20,000 hospitalized patients, Mayo Clinic Proc 95: (9) ((2020) ), 1888–1897. |
[32] | A.M. Pallotta, C. Kim, S.M. Gordon and A. Kim, Monoclonal antibodies for treating COVID-19, Cleveland Clin. J. Med ((2021) ). doi: 10.3949/ccjm.88a.ccc074. |
[33] | C.O. Barnes, A.P. West, Jr., K.E. Huey-Tubman, M.A.G. Hoffmann, N.G. Sharaf, P.R. Hoffman et al., Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies, Cell 182: (4) ((2020) ), 828–842.e816. |
[34] | P. Chen, A. Nirula, B. Heller, R.L. Gottlieb, J. Boscia, J. Morris, G. Huhn, J. Cardona, B. Mocherla, V. Stosor, I. Shawa, A.C. Adams, J. van Naarden, K.L. Custer, L. Shen, M. Durante, G. Oakley, A.E. Schade, J. Sabo, D.R. Patel, P. Klekotka and D.M. Skovronsky, BLAZE-1 investigators, SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19, N. Engl. J. Med 384: (3) ((2021) ), 229–237. |
[35] | G.D. Sempowski, K.O. Saunders, P. Acharya, K.J. Wiehe and B.F. Haynes, Pandemic preparedness: developing vaccines and therapeutic antibodies for COVID-19, Cell 181: (7) ((2020) ), 1458–1463. |
[36] | F. Begum and U. Ray, Polymonoclonal (not polyclonal) antibodies derived from convalescent human B cell hybridomas might be a better therapeutic option than single target monoclonal antibodies, ACS Pharmacol. Transl. Sci 3: (4) ((2020) ), 786–787. |
[37] | Z. Ku, X. Xie, E. Davidson, X. Ye, H. Su, V.D. Menachery, Y. Li, Z. Yuan, X. Zhang, A.E. Muruato, A.G.I. Escuer, B. Tyrell, K. Doolan, B.J. Doranz, D. Wrapp, P.F. Bates, J.S. McLellan, S.R. Weiss, N. Zhang, P.Y. Shi and Z. An, Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape, Nat. Commun 12: (1) ((2021) ), 469. |
[38] | B. Shanmugaraj, K. Rattanapisit, S. Manopwisedjaroen, A. Thitithanyanont and W. Phoolcharoen, Monoclonal antibodies B38 and H4 produced in Nicotiana benthamiana neutralize SARS-CoV-2 in vitro, Front. Plant Sci 11: ((2020) ), 589995. |
[39] | T.F. Rogers, F. Zhao, D. Huang, N. Beutler, A. Burns, W.T. He, O. Limbo, C. Smith, G. Song, J. Woehl, L. Yang, R.K. Abbott, S. Callaghan, E. Garcia, J. Hurtado, M. Parren, L. Peng, S. Ramirez, J. Ricketts, M.J. Ricciardi, S.A. Rawlings, N.C. Wu, M. Yuan, D.M. Smith, D. Nemazee, J.R. Teijaro, J.E. Voss, I.A. Wilson, R. Andrabi, B. Briney, E. Landais, D. Sok, J.G. Jardine and D.R. Burton, Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model, Science 369: (6506) ((2020) ), 956–963. |
[40] | S.J. Zost, P. Gilchuk, J.B. Case, E. Binshtein, R.E. Chen and J.P. Nkolola, Potently neutralizing and protective human antibodies against SARS-CoV-2, Nature 584: (7821) ((2020) ), 443–449. |
[41] | W.B. Alsoussi, J.S. Turner, J.B. Case, H. Zhao, A.J. Schmitz, J.Q. Zhou, R.E. Chen, T. Lei, A.A. Rizk, K.M. McIntire, E.S. Winkler, J.M. Fox, N.M. Kafai, L.B. Thackray, A.O. Hassan, F. Amanat, F. Krammer, C.T. Watson, S.H. Kleinstein, D.H. Fremont, M.S. Diamond and A.H. Ellebedy, A potently neutralizing antibody protects mice against SARS-CoV-2 infection, J. Immunol 205: (4) ((2020) ), 915–922. |
[42] | X. Chi, R. Yan, J. Zhang, G. Zhang, Y. Zhang, M. Hao, Z. Zhang, P. Fan, Y. Dong, Y. Yang, Z. Chen, Y. Guo, J. Zhang, Y. Li, X. Song, Y. Chen, L. Xia, L. Fu, L. Hou, J. Xu, C. Yu, J. Li, Q. Zhou and W. Chen, A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2, Science 369: (6504) ((2020) ), 650–655. |
[43] | L. Yang, W. Liu, X. Yu, M. Wu, J.M. Reichert and M. Ho, COVID-19 antibody therapeutics tracker: A Global online database of antibody therapeutics for the prevention and treatment of COVID-19, Antibody Ther 3: (3) ((2020) ), 205–212. |
[44] | B. Korber, W.M. Fischer, S. Gnanakaran, H. Yoon, J. Theiler, W. Abfalterer, N. Hengartner, E.E. Giorgi, T. Bhattacharya, B. Foley, K.M. Hastie, M.D. Parker, D.G. Partridge, C.M. Evans, T.M. Freeman, T.I. de Silva, Sheffield COVID-19 Genomics Group, C. McDanal, L.G. Perez, H. Tang, A. Moon-Walker, S.P. Whelan, C.C. LaBranche, E.O. Saphire and D.C. Montefiori, Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus, Cell 182: (4) ((2020) ), 812–827.e819. |
[45] | L. Yurkovetskiy, X. Wang, K.E. Pascal, C. Tomkins-Tinch, T.P. Nyalile, Y. Wang, A. Baum, W.E. Diehl, A. Dauphin, C. Carbone, K. Veinotte, S.B. Egri, S.F. Schaffner, J.E. Lemieux, J.B. Munro, A. Rafique, A. Barve, P.C. Sabeti, C.A. Kyratsous, N.V. Dudkina, K. Shen and J. Luban, Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant, Cell 183: (3) ((2020) ), 739–751.e738. |
[46] | Z. Zheng, V.M. Monteil, S. Maurer-Stroh, C.W. Yew, C. Leong, N.K. Mohd-Ismail, S. Cheyyatraivendran Arularasu, V.T.K. Chow, R.T.P. Lin, A. Mirazimi, W. Hong and Y.J. Tan, Monoclonal antibodies for the S2 subunit of spike of SARS-CoV-1 cross-react with the newly-emerged SARS-CoV-2, Euro Surveill 25: (28) ((2020) ), 2000291. |
[47] | T. Tanaka, M. Narazaki and T. Kishimoto, Immunotherapeutic implications of IL-6 blockade for cytokine storm, Immunotherapy 8: (8) ((2016) ), 959–970. |
[48] | P. Mehta, D.F. McAuley, M. Brown, E. Sanchez, R.S. Tattersall and J.J. Manson, COVID-19: consider cytokine storm syndromes and immunosuppression, Lancet (London, England) 395: (10229) ((2020) ), 1033–1034. |
[49] | M. Cascella, I. Mauro, E. de Blasio, A. Crispo, A. del Gaudio, S. Bimonte, A. Cuomo and P.A. Ascierto, Rapid and impressive response to a combined treatment with single-dose tocilizumab and NIV in a patient with COVID-19 Pneumonia/ARDS, Medicina (Kaunas) 56: (8) ((2020) ), 377. |
[50] | X. Xu, M. Han, T. Li, W. Sun, D. Wang, B. Fu, Y. Zhou, X. Zheng, Y. Yang, X. Li, X. Zhang, A. Pan and H. Wei, Effective treatment of severe COVID-19 patients with tocilizumab, Proc. Natl. Acad. Sci. USA 117: (20) ((2020) ), 10970–10975. |
[51] | A. Carugno, D.M. Gambini, F. Raponi, P. Vezzoli, E. Robustelli Test, M.E.G. Arosio, A. Callegaro and P. Sena, Coronavirus disease 2019 (COVID-19) rash in a psoriatic patient treated with Secukinumab: Is there a role for Interleukin 17? Dermatol. Ther 33: (6) ((2020) ), e14011. |
[52] | L. Chen, H. Deng, H. Cui, J. Fang, Z. Zuo, J. Deng, Y. Li, X. Wang and L. Zhao, Inflammatory responses and inflammation-associated diseases in organs, Oncotarget 9: (6) ((2018) ), 7204–7218. |
[53] | J. Price, J.M. Lord and P. Harrison, Inflammaging and platelet hyperreactivity: a new therapeutic target? J. Thromb. Haemost 18: (1) ((2020) ), 3–5. |
[54] | K. Khalaf, N. Papp, J.T. Chou, D. Hana, A. Mackiewicz and M. Kaczmarek, SARS-CoV-2: pathogenesis, and advancements in diagnostics and treatment, Front. Immunol 11: ((2020) ), 570927. |
[55] | V. Roy, S. Fischinger, C. Atyeo, M. Slein, C. Loos, A. Balazs, C. Luedemann, M.G. Astudillo, D. Yang, D.R. Wesemann, R. Charles, A.J. Lafrate, J. Feldman, B. Hauser, T. Caradonna, T.E. Miller, M.R. Murali, L. Baden, E. Nilles, E. Ryan, D. Lauffenburger, W.G. Beltran and G. Alter, SARS-CoV-2-specific ELISA development, J Immunol. Methods 484-485 ((2020) ), 112832. |
[56] | M.P. Motley, E. Bennett-Guerrero, B.C. Fries and E.D. Spitzer, Review of viral testing (polymerase chain reaction) and antibody/serology testing for severe acute respiratory syndrome-coronavirus-2 for the intensivist, Crit. Care Explor 2: (6) ((2020) ), e0154. |
[57] | X. Cheng, G. Chen and W.R. Rodriguez, Micro- and nanotechnology for viral detection, Anal. Bioanal. Chem 393: (2) ((2009) ), 487–501. |
[58] | J. He, P. Hu, Y. Gao, S. Zheng, C. Xu, R. Liu, L. Fang, R. Li, C. Han, J. An, J. Dong, G. Deng, L. Sun and Y. Lv, Comparison and application of different immunoassay methods for the detection of SARS-CoV-2, J. Med. Virol 92: (11) ((2020) ), 2777–2784. |
[59] | D.T.H. Moura, T.R. McCarty, I.B. Ribeiro, M.P. Funari, P.V.A.G. Oliveira, A.A. Miranda Neto, E.S.D. Monte Júnior, F. Tustumi, W.M. Bernardo, E.G.H. Moura and C.C. Thompson, Diagnostic characteristics of serological-based COVID-19 testing: a systematic review and meta-analysis, Clinics (Sao Paulo) 75: ((2020) ), e2212. |
[60] | L.J. Carter, L.V. Garner, J.W. Smoot, Y. Li, Q. Zhou, C.J. Saveson, J.M. Sasso, A.C. Gregg, D.J. Soares, T.R. Beskid, S.R. Jervey and C. Liu, Assay techniques and test development for COVID-19 diagnosis, ACS Cent, Sci 6: (5) ((2020) ), 591–605. |
[61] | N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G.F. Gao and W. Tan, China novel coronavirus investigating and research team, a novel coronavirus from patients with pneumonia in China, 2019, N. Engl. J. Med 382: (8) ((2020) ), 727–733. |
[62] | R.W. Peeling, C.J. Wedderburn, P.J. Garcia, D. Boeras, N. Fongwen, J. Nkengasong, A. Sall, A. Tanuri and D.L. Heymann, Serology testing in the COVID-19 pandemic response, Lancet Infect. Dis 20: (9) ((2020) ), E245–E249. |
[63] | Y. Hirotsu, M. Maejima, M. Shibusawa, Y. Nagakubo, K. Hosaka, K. Amemiya, H. Sueki, M. Hayakawa, H. Mochizuki, T. Tsutsui, Y. Kakizaki, Y. Miyashita, S. Yagi, S. Kojima and M. Omata, Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients, Int. J. Infect. Dis 99: ((2020) ), 397–402. |
[64] | D. Lin, L. Liu, M. Zhang, Y. Hu, Q. Yang, J. Guo, Y. Dai, Y. Xu, Y. Cai, X. Chen, K. Huang and Z. Zhang, Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak, Eur. J. Clin. Microbiol. Infect. Dis 39: (12) ((2020) ), 2271–2277. |
[65] | L. Huang, S. Tian, W. Zhao, K. Liu, X. Ma and J. Guo, Multiplexed detection of biomarkers in lateral-flow immunoassays, Analyst 145: (8) ((2020) ), 2828–2840. |
[66] | C. Sheridan, Fast, portable tests come online to curb coronavirus pandemic, Nat. Biotechnol 38: (5) ((2020) ), 515–518. |
[67] | A. La Marca, M. Capuzzo, T. Paglia, L. Roli, T. Trenti and S.M. Nelson, Testing for SARS-CoV-2 (COVID-19): a systematic review and clinical guide to molecular and serological in-vitro diagnostic assays, Reprod. Biomed. Online 41: (3) ((2020) ), 483–499. |
[68] | Y.W. Tang, J.E. Schmitz, D.H. Persing and C.W. Stratton, Laboratory diagnosis of COVID-19: current issues and challenges, J. Clin. Microbiol 58: (6) ((2020) ), e00512–20. |
[69] | T. Weitzel, P. Legarraga, M. Iruretagoyena, G. Pizarro, V. Vollrath, R. Araos, J.M. Munita and L. Porte, Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium, Travel. Med. Infect. Dis 39: ((2021) ), 101942. |
[70] | L. Porte, P. Legarraga, V. Vollrath, X. Aguilera, J.M. Munita, R. Araos, G. Pizarro, P. Vial, M. Iruretagoyena, S. Dittrich and T. Weitzel, Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples, Int. J. Infect. Dis 99: ((2020) ), 328–333. |
[71] | J. Favresse, C. Gillot, M. Oliveira, J. Cadrobbi, M. Elsen, C. Eucher, K. Laffineur, C. Rosseels, S. van Eeckhoudt, J.B. Nicolas, L. Morimont, J.M. Dogné and J. Douxfils, Head-to-Head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection, J. Clin. Med 10: (2) ((2021) ), 265. |




