InterpretME: A tool for interpretations of machine learning models over knowledge graphs
Abstract
In recent years, knowledge graphs (KGs) have been considered pyramids of interconnected data enriched with semantics for complex decision-making. The potential of KGs and the demand for interpretability of machine learning (ML) models in diverse domains (e.g., healthcare) have gained more attention. The lack of model transparency negatively impacts the understanding and, in consequence, interpretability of the predictions made by a model. Data-driven models should be empowered with the knowledge required to trace down their decisions and the transformations made to the input data to increase model transparency. In this paper, we propose InterpretME, a tool that using KGs, provides fine-grained representations of trained ML models. An ML model description includes data – (e.g., features’ definition and SHACL validation) and model-based characteristics (e.g., relevant features and interpretations of prediction probabilities and model decisions). InterpretME allows for defining a model’s features over data collected in various formats, e.g., RDF KGs, CSV, and JSON. InterpretME relies on the SHACL schema to validate integrity constraints over the input data. InterpretME traces the steps of data collection, curation, integration, and prediction; it documents the collected metadata in the InterpretME KG. InterpretME is published in GitHub11 and Zenodo22. The InterpretME framework includes a pipeline for enhancing the interpretability of ML models, the InterpretME KG, and an ontology to describe the main characteristics of trained ML models; a PyPI library of InterpretME is also provided33. Additionally, a live code44, and a video55 demonstrating InterpretME in several use cases are also available.
1.Introduction
Interpretability is the degree to which humans can understand the decisions made by computational frameworks. Specifically, in Artificial Intelligence (AI), the higher the interpretability of predictive models, the easier for humans to understand why a model makes certain decisions. The recent advancements and complexity of machine learning (ML) methods have demonstrated their success in forecasting complex problems (e.g., disease diagnosis or progression [29,31]). Unfortunately, they are often opaque and do not offer interpretations for their predictions, and it is not always possible to comprehend the results. Interpretable predictive models have rapidly become a relevant problem [5]. Nevertheless, although various tools aim to interpret the algorithmic decisions of ML models [27,34], they are incapable of capturing the knowledge required to translate a model’s insights into the application domain. On the contrary, KGs encode data and knowledge; they, together with domain ontologies (e.g., ML Schema [30]), represent building blocks for increasing the understanding of the behavior and effects of a predictive model.
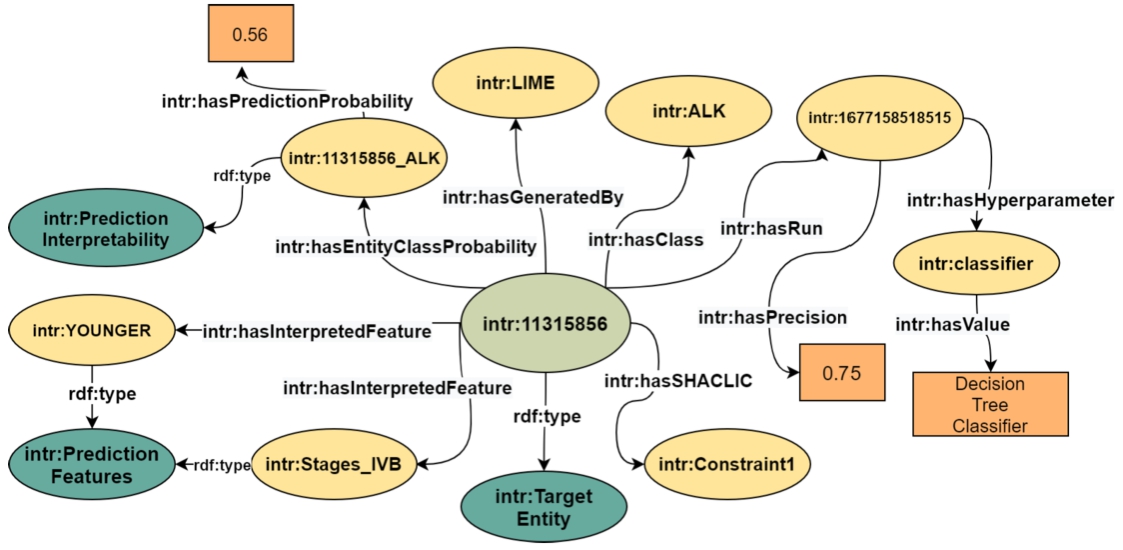
Fig. 1.
The InterpretME KG comprises the target entity highlighted in light green. This entity is characterized by several key features highlighted in yellow, including hyperparameters, LIME features, and SHACL integrity constarints. Literals are in orange and classes are in dark green.
Our Tool: We propose an analytical tool, named InterpretME, for tracing and explaining the predictive models built over data collected from KGs or from CSV, and JSON files. InterpretME is a stand-alone framework that works locally on individual systems, with many runs that can be integrated into a single InterpretME KG to compare interpretations. By specifying predictive model features, the InterpretME pipeline increases ML model interpretability. The InterpretME ontology combines concepts from existing ontologies (e.g., ML Schema [30], PROV-O66) to represent key properties of learned predictive models and their execution.
InterpretME resorts to SHACL for providing a meaningful description of a target entity (as shown in Fig. 1). The target entity is a specific node from the input KG or CSV, and JSON file that is of particular interest of a predictive model. Figure 1 depicts a target entity representing a lung cancer patient (intr:11315856) which is classified as an ALK positive. It is also described in terms of a Decision Tree Classifier with a run (intr:1677158518515) on certain features (e.g., intr:Stages_IVB, intr:YOUNGER). The classification has a precision of 0.75 and LIME [34] generated a local interpretation of the probability 0.56 for this classification.
InterpretME is designed to work with both KGs (accessible via Web interfaces, e.g., SPARQL endpoints), and datasets in various formats (e.g., CSV, and JSON). InterpretME models – as factual statements – everything learned while training an ML model and its interpretation; these statements compose the InterpretME KG. To retrieve data for a predictive pipeline, the user must provide either a SPARQL endpoint or the path to a dataset. If the input data is retrieved from KGs, InterpretME can validate constraints expressed in SHACL, align the InterpretME KG to the input KGs, and execute federated queries. On the other hand, if the input data is collected from datasets in CSV, and JSON formats, a user can also explore interpretations with the InterpretME KG. The latest version of InterpretME is customized for supervised ML models (e.g., decision trees and random forests) and interpretable tools (e.g., LIME). InterpretME is publicly available as a resource in GitHub1 and Zenodo2; a PyPI library is also available3. We empirically evaluate InterpretME in real-world KGs regarding execution time, interpretation quality, and traceability of target entities. The observed results reveal the key role of Semantic Web technologies in the interpretation of the outcomes of predictive models. As a result, InterpretME assists ML model users by allowing for exploring – using SPARQL queries – metadata represented as factual statements in the InterpretME KG.
The rest of the paper is structured as follows: Section 2 describes the main concepts and motivates our work. Section 3 defines the InterpretME architecture, while Section 4 reports the results of our experimental studies. Section 5 presents the main characteristics of InterpretME as a resource. Section 6 discusses the state of the art, and our conclusions and future work are outlined in Section 7.
2.Preliminaries and motivation
2.1.Main concepts
2.1.1.Predictive modeling frameworks
Fig. 2.
Predictive modeling conceptualization by De Bie et al. [5]. It comprises four stages: Data Engineering, Data Exploration, Model Building, and Exploitation. They offer three forms of automation: Mechanization, Composition, and Assistance.
![Predictive modeling conceptualization by De Bie et al. [5]. It comprises four stages: Data Engineering, Data Exploration, Model Building, and Exploitation. They offer three forms of automation: Mechanization, Composition, and Assistance.](https://content.iospress.com:443/media/sw-prepress/sw--1--1-sw233511/sw--1-sw233511-g002.jpg)
Predictive Modeling comprises methods to forecast future outcomes based on data encoding what has happened in the past (e.g., historical data) and what is currently happening (e.g., current data). Predictive modeling resorts to a variety of models and algorithms (e.g., random forest, gradient boosting model or decision trees) to solve predictive problems (e.g., classification or outlier detection) [32]. Despite the large spectrum of mature models and well-defined problems, predictive modeling is a complex task that requires user expertise in the domain context and modeling. De Bie et al. [5] conceptualizes predictive modeling in four stages, as depicted in Fig. 2. a) Data Engineering comprises the tasks of acquiring, organizing, curating, and preparing data for prediction modeling. b) Data Exploration includes the understanding of an application domain, and the interpretation of missing values, integrity constraints, and data structures. c) Model Building involves the selection of the predictive model (e.g., machine or deep learning), target function, hyperparameters, and relevant features. d) Exploitation covers the transformation of the predictions into the decisions on the application domain. Predictive modeling complexity and the mandatory requisite of knowledgeable users motivate automated techniques for facilitating the implementation of the stages in Fig. 2.
De Bie et al. [5] identify three forms of automation: i) Mechanization is used in well-known tasks that can be performed algorithmically without human intervention. Functions and modules, for example, can be used to perform tasks such as clustering algorithms and standardizing table values. Packages for performing such operations are available as a library that can be easily integrated into any project. ii) Composition is performed on a series of tasks that are implemented as workflows in high-level programming languages. For example, algorithm selection, hyperparameter optimization, model selection, and so on fall under the stage of Model Building, which is covered in Composition. iii) Assistance implemented to enhance interpretability and user efficiency by presenting visualizations, prediction scores, and explanatory model decisions. This includes monitoring what humans do during a learning process so that an automated assistant can identify inappropriate choices and make useful recommendations.
Current developments in automation have mainly impacted the stages of Model Building and Exploitation. In particular, Automated Machine Learning (AutoML) systems (e.g., AutoML77 and AutoWeka [24]) successfully implement mechanization, and can automatize the processes of model building and feature selection, and the optimization of the hyperparameters and target functions. Furthermore, interactive tools like RapidMiner,88 the Local Interpretable Model-agnostic Explanations (LIME) framework [34], and SHapely Additive exPlanations (SHAP) [27] provide assistance by reporting explanations of a model’s results. Specifically, LIME relies on local surrogate models to explain the prediction of each entity of a test dataset. It computes – for the tested entities – the feature contribution for each target class and the prediction probability. In general, LIME is independent of the original algorithm given to the model and works locally to provide explanations for the prediction relative to each instance. LIME tries to fit the local model using sample data points that are similar to the instance being explained.
2.1.2.Knowledge graph frameworks
The Semantic Web [4] aims at humans and machines working cooperatively in data exchange. Technologies capable of encoding semantics have been defined to achieve this goal. One of these technologies is the Resource Description Framework (RDF) [26], i.e., the W3C standard for publishing and exchanging data over the web. An RDF graph
SPARQL [33] is the W3C recommendation language to query RDF graphs. SPARQL 1.1 [17] provides the SERVICE clause and allows for specifying a federated query over various RDF graphs accessible via SPARQL endpoints. However, usually, SPARQL endpoints prohibit using these clauses. Federated query engines offer the possibility to answer federated queries without the need to know against which endpoint a subquery will be executed [1].
The Shapes Constraint Language (SHACL) [23] is the W3C recommendation language to define integrity constraints over RDF data. SHACL integrity constraints are expressed in RDF and modeled as a network of shapes, called shape schema. A shape consists of i) a definition of the target; usually an RDF class or set of nodes, and ii) a set of constraints that are imposed over the instances of the target. Constraints can also link shapes to each other, hence, a shape network. An RDF graph’s entity that matches the target definition of a shape, satisfies the shape if it validates all the constraints of the shape; the problem is, in general, intractable [7]. However, algorithms have been proposed and implemented to validate tractable SHACL fragments [6,12] efficiently.
Definition 2.1
Definition 2.1(InterpretME KG).
An InterpretME KG is a heterogeneous graph where each node and edge is assigned to a type, i.e., InterpretME KG = (V, E, L, T), where: i) V represents the set of nodes, where each node
2.2.Motivating example
The motivation for our work originates from the lack of automated assistance despite the great potential of integrating knowledge graphs with predictive modeling frameworks. Tracing and explaining the predictive models built over data collected from the KGs in order to provide assistance is the main goal of our tool InterpretME. An InterpretME KG comprises the vocabulary shown in Fig. 3 and is further detailed in Section 5. Even though the state-of-the-art successfully developed automated machine learning systems, through mechanization and composition, pipelines for predictive modeling are unable to generate human- and machine-readable decisions to assist users and enhance their efficiency. LIME creates human-readable interpretations for a particular predicting task. They cannot deliver factual statements about interpretations of the target entity. They also fail to take into account the target entity’s properties in the predictive model and the input KG.
Fig. 3.
Vocabulary. Figure 3a shows the properties of the classes used to describe the machine learning models. Figure 3b and Fig. 3c depicts the extension of ML schema used to describe the predictive model characteristics and also incorporates the SHACL validation results. The required properties and classes are utilize for presenting the traced knowledge of the predictive models in the InterpretME KG.
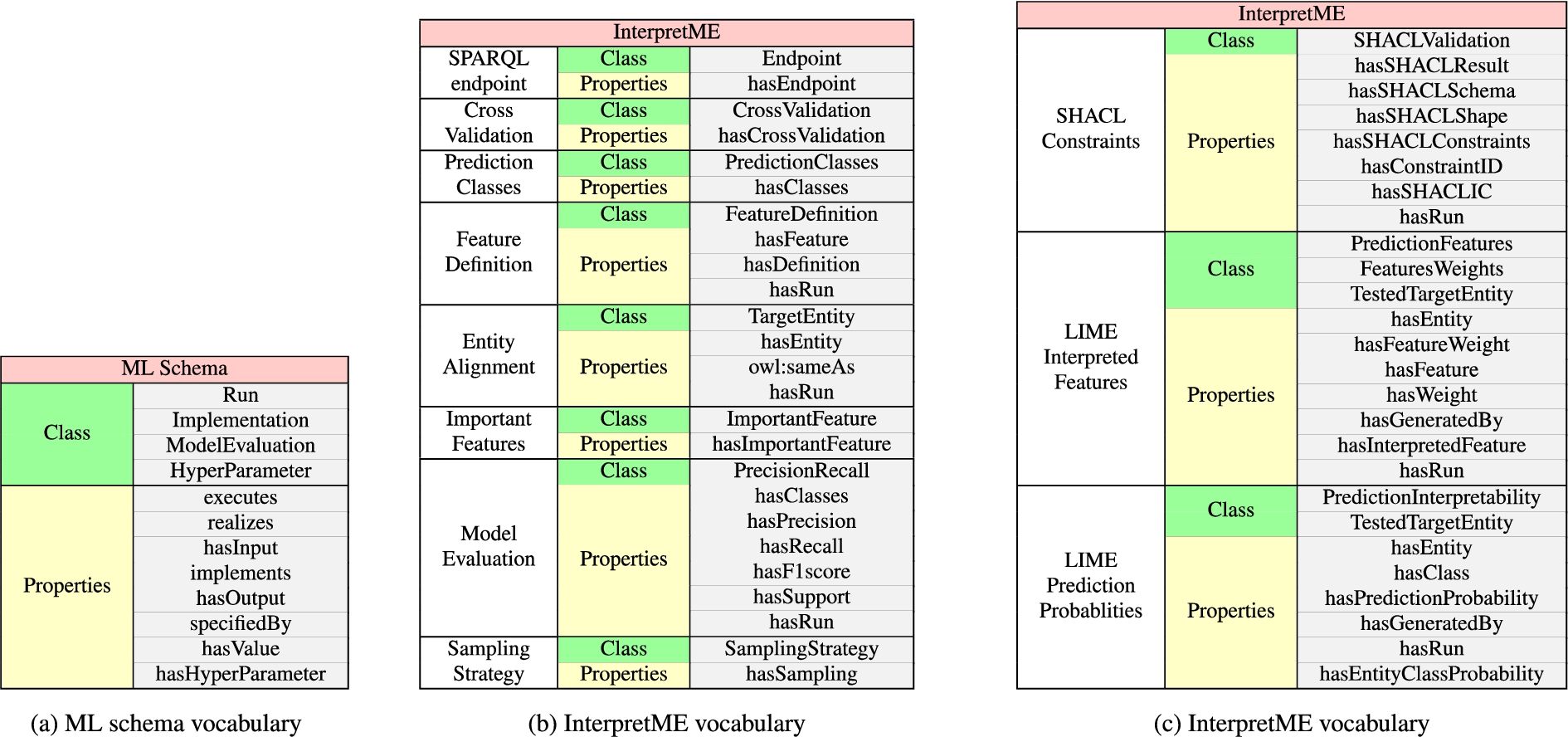
Fig. 4.
Lung Cancer KG Example. Figure 4a depicts the motivation of a pipeline that integrates knowledge graphs and data-driven frameworks, i.e., AutoML, Decision Trees, LIME. SPARQL queries are utilized to extract the data from KG, the classification task is to predict whether a patient is positive for biomarker and Fig. 4b illustrates the questions reported by domain experts regarding the issue in interpreting data-driven frameworks such as LIME, that still lack in terms of interpretability and traceability.
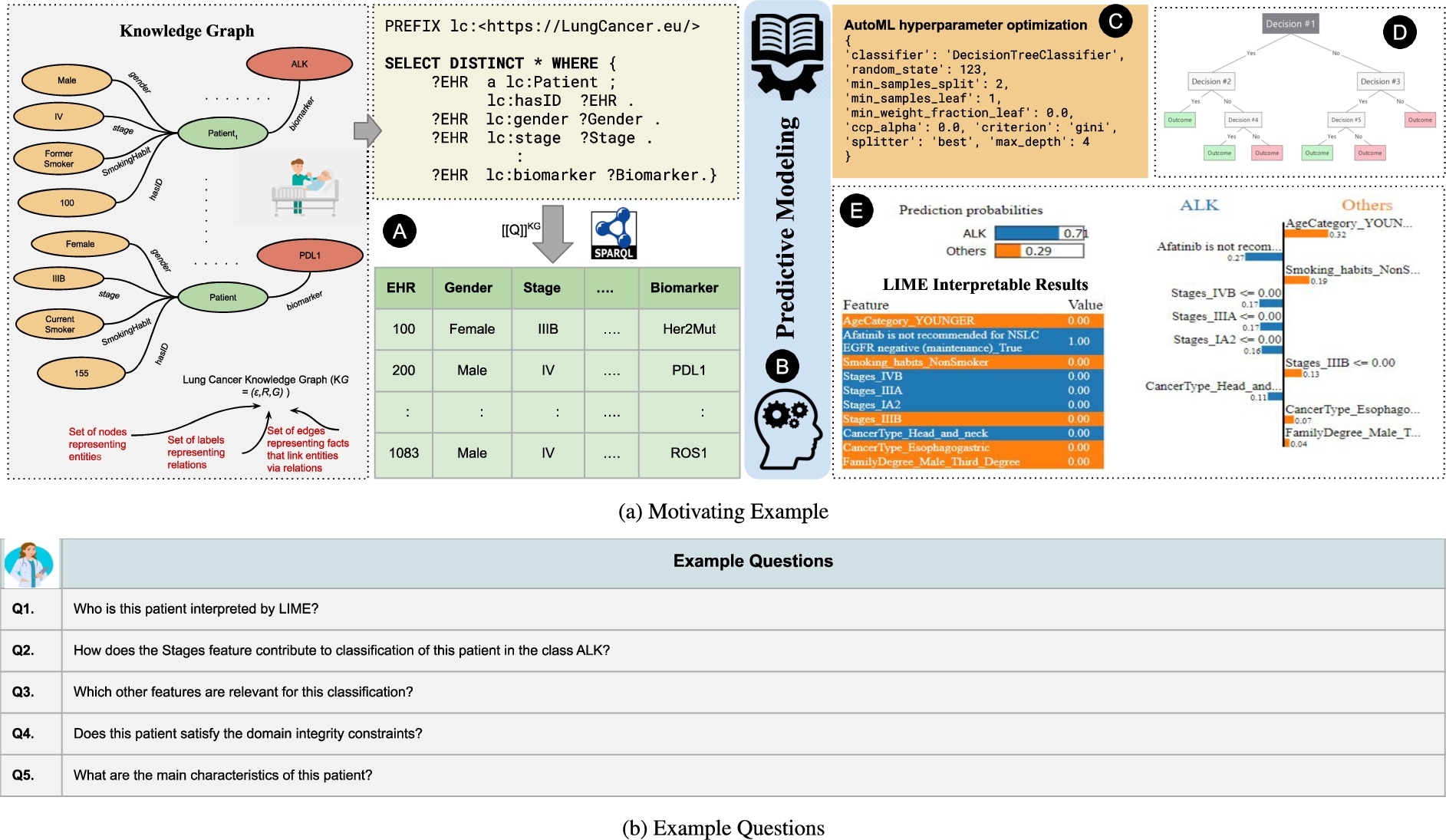
Figure 4a depicts a predictive modeling pipeline, where an automated machine learning system (e.g., AutoML) is utilized for model and feature selection, and hyperparameters’ optimization. Moreover, interpretable tools (e.g., LIME) provides interpretable results. Figure 4a illustrates the pipeline  ; an input dataset
; an input dataset  is collected from an RDF knowledge graph that integrates data about lung cancer patients. The SPARQL query is used to extract features from the KG, describing the main characteristics of a lung cancer patient, i.e., patient identifier (a.k.a. EHR is electronic health record of a patient), Gender, Age, cancer stage (a.k.a. CancerStage), smoking habits (a.k.a. SmokingHabit), and lung cancer Biomarkers. Additionally, the dataset comprises information about the cancer types of the relatives of the lung cancer patients; this information is maintained in the features CancerType and FamilialDegree. The predictive task is a binary classification to predict if a patient will be positive for the biomarker ALK or by any other biomarker. AutoML performs model selection and hyperparameter optimization
is collected from an RDF knowledge graph that integrates data about lung cancer patients. The SPARQL query is used to extract features from the KG, describing the main characteristics of a lung cancer patient, i.e., patient identifier (a.k.a. EHR is electronic health record of a patient), Gender, Age, cancer stage (a.k.a. CancerStage), smoking habits (a.k.a. SmokingHabit), and lung cancer Biomarkers. Additionally, the dataset comprises information about the cancer types of the relatives of the lung cancer patients; this information is maintained in the features CancerType and FamilialDegree. The predictive task is a binary classification to predict if a patient will be positive for the biomarker ALK or by any other biomarker. AutoML performs model selection and hyperparameter optimization  ; based on AutoML recommendations, the random forests and decision tree models are selected to implement the classification problem. Further, the interpretable surrogate tools LIME and Decision trees are utilized to provide local interpretations of each patient in the test dataset. Decision trees yield the relevant features which contribute to the model outcomes
; based on AutoML recommendations, the random forests and decision tree models are selected to implement the classification problem. Further, the interpretable surrogate tools LIME and Decision trees are utilized to provide local interpretations of each patient in the test dataset. Decision trees yield the relevant features which contribute to the model outcomes  .
.  depicts an exemplar entity where LIME determines a prediction probability of 0.7 to belong to the target class ALK (i.e., Class 0), otherwise, 0.3 target class Others (i.e., Class 1). LIME also identifies the top 10 relevant features for the target entity and assigns weights. These outcomes allow for understanding the quality of the implemented pipeline. Nevertheless, when they are reported to oncologists, many questions may still arise Fig. 4b: Q1) Who is this patient interpreted by LIME? Q2) How does the Stages feature contribute to the classification of this patient in the target class ALK? Q3) Which other features are relevant for this classification? Q4) Does this patient satisfy the domain integrity constraints? Q5) What are the main characteristics of this patient? The oncologists gave us the feedback that InterpretME facilitates their understanding of the reasons for the classification. Notably, the users comprehended better the answers to the queries over the InterpretME KG and the Lung Cancer KG than simply the answers of LIME. Although, the user would have been able to interpret the results produced by LIME, he/she would need to trace back these results to the original data properties to know if, for example, the reported patient violates the domain constraints or not. For instance, a domain constraint, ‘If a patient receives a drug for the treatment, then he or she is cured’. InterpretME combines local explanations with knowledge graphs and SHACL constraints to ensure traceability. In order to support our justification, a user case study was also carried out; it will be discussed in Section 4.5. The assessment outcomes suggest that using KGs to document ML pipelines contributes to a better understanding of the predictions suggested by an ML model, as well as the interpretations provided by LIME.
depicts an exemplar entity where LIME determines a prediction probability of 0.7 to belong to the target class ALK (i.e., Class 0), otherwise, 0.3 target class Others (i.e., Class 1). LIME also identifies the top 10 relevant features for the target entity and assigns weights. These outcomes allow for understanding the quality of the implemented pipeline. Nevertheless, when they are reported to oncologists, many questions may still arise Fig. 4b: Q1) Who is this patient interpreted by LIME? Q2) How does the Stages feature contribute to the classification of this patient in the target class ALK? Q3) Which other features are relevant for this classification? Q4) Does this patient satisfy the domain integrity constraints? Q5) What are the main characteristics of this patient? The oncologists gave us the feedback that InterpretME facilitates their understanding of the reasons for the classification. Notably, the users comprehended better the answers to the queries over the InterpretME KG and the Lung Cancer KG than simply the answers of LIME. Although, the user would have been able to interpret the results produced by LIME, he/she would need to trace back these results to the original data properties to know if, for example, the reported patient violates the domain constraints or not. For instance, a domain constraint, ‘If a patient receives a drug for the treatment, then he or she is cured’. InterpretME combines local explanations with knowledge graphs and SHACL constraints to ensure traceability. In order to support our justification, a user case study was also carried out; it will be discussed in Section 4.5. The assessment outcomes suggest that using KGs to document ML pipelines contributes to a better understanding of the predictions suggested by an ML model, as well as the interpretations provided by LIME.
3.InterpretME: An interpretable pipeline for predictive modeling over knowledge graphs
3.1.Tracing metadata in machine learning pipelines
We aim to collect metadata during the different stages of a predictive model pipeline. Figure 5 shows, in a pictorial view, the characteristics of the data collected towards the improvement of automation Assistance; this figure is based on the conceptualization of predictive modeling proposed by De Bie et al. [5] reported in Fig. 2. In the Data Engineering stage, InterpretME captures metadata from the input KG (e.g., features’ definition, endpoint, and target classes) and records the SHACL constraints, used for data validation. During Model Building, suggestions from AutoML systems can be considered. InterpretME traces the optimized hyperparameters and estimated features’ relevancy, and records the model performance metric outcomes (e.g., precision) for a particular run. SHACL validation reports are also included in the InterpretME KG. InterpretME also exploits decision trees and visualizes the validation report to enhance exploration. Lastly, for Exploitation, InterpretME aligns target entities with entities in KGs, where the model’s features are defined. These facts allow for i) tracing back the main properties of target entities in the input KG; ii) understanding interpretability of models’ results (e.g., LIME prediction probabilities, interpreted features, and associated weights); and iii) checking SHACL validation reports. InterpretME identifies uniquely all the collected metadata. As a result, tracing the collected metadata enhances explanations about an ML model behavior.
3.2.The InterpretME architecture
Figure 6 depicts the InterpretME architecture. InterpretME is a tool for fine-grained representations of the main characteristics of a trained predictive modeling framework. The architecture of InterpretME deals with training the predictive models and collecting information generated as output of predictive models (i.e., model accuracy, list of important features, prediction probabilities, and classified classes for each instance). InterpretME provides assistance (Fig. 5) to the user by tracing metadata to generate instances of the InterpretME KG. Federated queries are obtained on top of the InterpretME KG and the input KG to trace back, and answers questions in Fig. 4b. InterpretME takes a JSON file as input (i.e., endpoints of KGs, features’ definition, target definition, SHACL constraints, sampling strategy, and class definition)99; a SPARQL query is generated based on the feature definition given by the user and the query is used to retrieve the application domain data from the input KGs. Users perform the step of providing a SPARQL endpoint or path to a dataset and its corresponding feature definition in the input JSON file. InterpretME performs additional steps automatically, such as extracting data based on the query, running predictive models, saving prediction results, and so on. The structure of the query enables entities in the input KGs to be aligned to the identifiers of the entities in the ML models’ datasets. These alignments facilitate the SHACL validation and federated query processing over the InterpretME KG and the input KGs.
 InterpretME evaluates the SHACL constraints over the nodes of the input KGs and outputs a validation report per constraint and target entity. These results indicate whether an entity validates or invalidates the constraints defined by the user. The validation reports states the validity of data used by the predictive models1010. Thus, SHACL constraints are a crucial part of the InterpretME KG. Their evaluation identifies if a particular entity is violating the integrity constraints, where True represents that the particular entity is valid, inversely represented by False.
InterpretME evaluates the SHACL constraints over the nodes of the input KGs and outputs a validation report per constraint and target entity. These results indicate whether an entity validates or invalidates the constraints defined by the user. The validation reports states the validity of data used by the predictive models1010. Thus, SHACL constraints are a crucial part of the InterpretME KG. Their evaluation identifies if a particular entity is violating the integrity constraints, where True represents that the particular entity is valid, inversely represented by False.
 Preprocessing includes transforming data collected from the input KGs or from CSV, and JSON files into a form that can be used in the training of a predictive model. Many machine learning models cannot handle categorical values directly, InterpretME generates one hot-encodings using the Python library sklearn to transform the model’s categorical features into binary features that can be further used for predictive models1111; it makes training data more expressive, and can be easily re-scaled. Also, the target class can be defined in InterpretME. Sampling strategies (e.g., under-sampling or over-sampling) are also utilized to reduce data imbalance so that the machine learning algorithms can perform better. Many machine learning algorithms, like decision trees, random forests, and neural networks resort to class distribution in the training dataset to compute the probability of instances in each class when the model will be used to make predictions. Model configuration (during Preprocessing) can be done based on automated systems’ selections (e.g., AutoML/OpenML) or based on a user’s preferences. Automated tools (e.g., Optuna [2]) can be used to generate optimized hyperparameters for predictive models.
Preprocessing includes transforming data collected from the input KGs or from CSV, and JSON files into a form that can be used in the training of a predictive model. Many machine learning models cannot handle categorical values directly, InterpretME generates one hot-encodings using the Python library sklearn to transform the model’s categorical features into binary features that can be further used for predictive models1111; it makes training data more expressive, and can be easily re-scaled. Also, the target class can be defined in InterpretME. Sampling strategies (e.g., under-sampling or over-sampling) are also utilized to reduce data imbalance so that the machine learning algorithms can perform better. Many machine learning algorithms, like decision trees, random forests, and neural networks resort to class distribution in the training dataset to compute the probability of instances in each class when the model will be used to make predictions. Model configuration (during Preprocessing) can be done based on automated systems’ selections (e.g., AutoML/OpenML) or based on a user’s preferences. Automated tools (e.g., Optuna [2]) can be used to generate optimized hyperparameters for predictive models.
Fig. 6.
The InterpretME Architecture. The input of InterpretME is either Knowledge graphs or datasets in formats, i.e., CSV, and JSON. In Mechanization layer, SHACL validation, data preprocessing, and AutoML are utilized. The Composition layer performs the predictive task, and the interpretable tools like Decision trees and LIME are implemented for understanding the predictions. In Assistance layer, InterpretME generates an InterpretME KG with the traced metadata of the trained predictive model, to provide users with more enhanced and reliable interpretations. Generating and Exploiting the InterpretME KG involves RML mappings and Federated Query Processing.
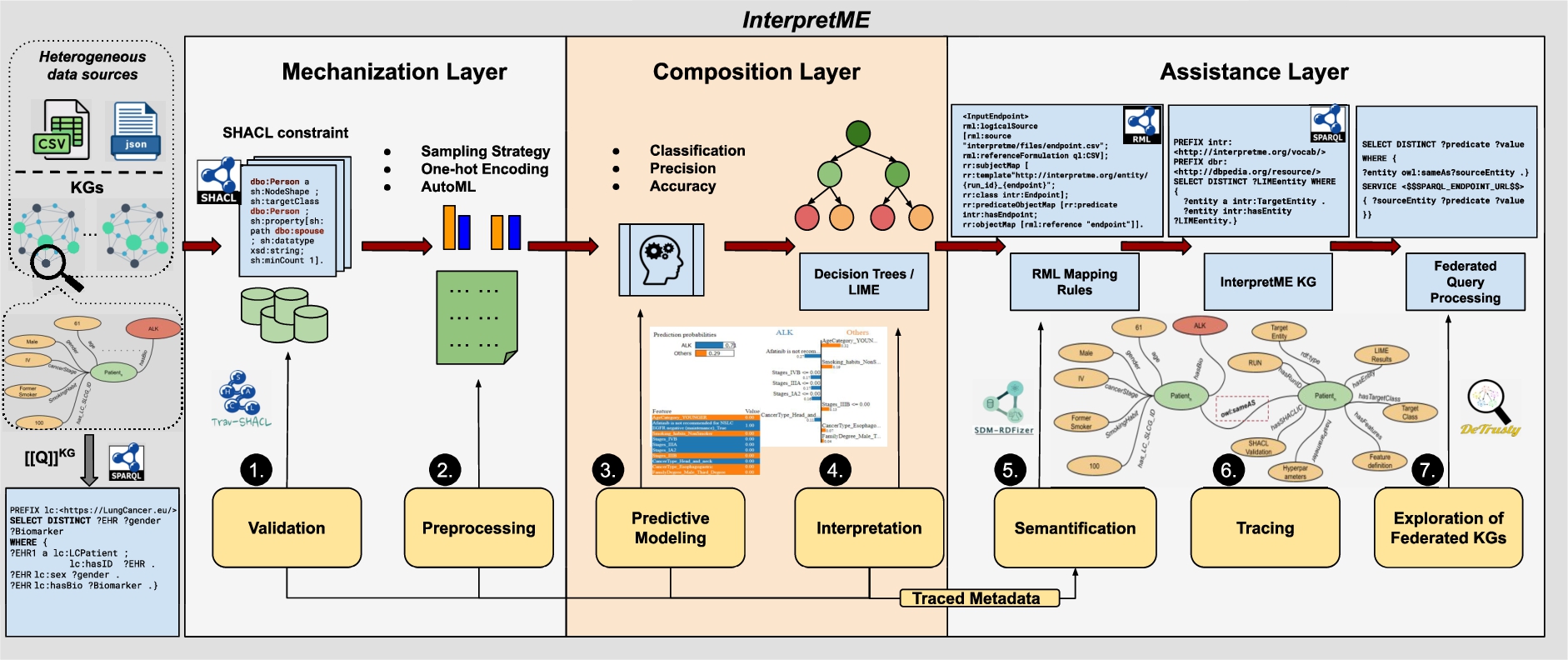
 Predictive Modeling. An automated model can also perform stratified shuffle split cross-validation with random forest, and identify the relevant features; they are used to train a decision tree classifier to predict and visualize the outcomes of the predictive models, and report on precision and accuracy1212.
Predictive Modeling. An automated model can also perform stratified shuffle split cross-validation with random forest, and identify the relevant features; they are used to train a decision tree classifier to predict and visualize the outcomes of the predictive models, and report on precision and accuracy1212.
 Interpretation is utilized to enhance the understanding of the trained predictive model. Also, a visualization of the decision tree can be obtained with associated constraints. This module provides more insights of a validated/invalidated feature, and interpretable surrogate tools like LIME [34] (as in Fig. 4a) yields the most relevant features and local explanations list. They reflect the contribution of each feature to the prediction.
Interpretation is utilized to enhance the understanding of the trained predictive model. Also, a visualization of the decision tree can be obtained with associated constraints. This module provides more insights of a validated/invalidated feature, and interpretable surrogate tools like LIME [34] (as in Fig. 4a) yields the most relevant features and local explanations list. They reflect the contribution of each feature to the prediction.
 Semantification is a module of InterpretME, where mapping rules are defined with the RDF Mapping Language (RML) [8] using the data collected from the predictive models. RML expresses – using triples maps – the conversion of raw data, e.g., JSON, CSV, and XML, into RDF KGs. RML states correspondences between traced metadata and factual statements in the InterpretME KG. Listing 3.3 demonstrates an example of an RML triples map where, the
Semantification is a module of InterpretME, where mapping rules are defined with the RDF Mapping Language (RML) [8] using the data collected from the predictive models. RML expresses – using triples maps – the conversion of raw data, e.g., JSON, CSV, and XML, into RDF KGs. RML states correspondences between traced metadata and factual statements in the InterpretME KG. Listing 3.3 demonstrates an example of an RML triples map where, the
 During Tracing, SPARQL queries can be executed to explore the InterpretME results of a particular target entity. Users can identify patterns and correlations between the given features and also perform statistical analysis of a particular target entity. The InterpretME KG provides clarification and eases the interpretation of the model’s prediction of a particular entity aligned with the SHACL validation results.
During Tracing, SPARQL queries can be executed to explore the InterpretME results of a particular target entity. Users can identify patterns and correlations between the given features and also perform statistical analysis of a particular target entity. The InterpretME KG provides clarification and eases the interpretation of the model’s prediction of a particular entity aligned with the SHACL validation results.
 Exploration of Federated KGs. Query processing allows for querying the InterpretME KG and the input KGs (if there is any). A user is involved in executing federated SPARQL queries to gain more insights from the predictive model’s decision. Mostly results of interpretable tools are hard to interpret and understand the characteristics of target entity’s behavior in predictive models. As a result, the advantage of comparing the predictive model’s characteristics in the InterpretME KG to the characteristics of a specific entity in the input KGs is obtained by using query processing. Thus, a user can trace back via query federation to interpret the results for a specific target entity with input characteristics aligned with SHACL validation results. Exemplar queries are shown in GitHub1313.
Exploration of Federated KGs. Query processing allows for querying the InterpretME KG and the input KGs (if there is any). A user is involved in executing federated SPARQL queries to gain more insights from the predictive model’s decision. Mostly results of interpretable tools are hard to interpret and understand the characteristics of target entity’s behavior in predictive models. As a result, the advantage of comparing the predictive model’s characteristics in the InterpretME KG to the characteristics of a specific entity in the input KGs is obtained by using query processing. Thus, a user can trace back via query federation to interpret the results for a specific target entity with input characteristics aligned with SHACL validation results. Exemplar queries are shown in GitHub1313.
3.3.Running example
To exemplify different components of the InterpretME architecture, we have used the following running example (Fig. 7). In our GitHub repository, we provide a comparable example with the French Royalty KG1414. Because of privacy issues, the Lung Cancer KG cannot be published, but this example allows for reproducing our results.
 The Lung Cancer KG integrates the features’ and class target definitions about lung cancer patients; their constraints are defined in terms of SHACL. Features’ definitions are classified into independent and dependent variables. Independent variables are features that the user selects for analyzing the model’s prediction. On the other hand, a dependent variable is a feature under investigation that is expected to change in response to changes in the independent variable. They are used later in the predictive modeling pipeline and are defined as follows:
The Lung Cancer KG integrates the features’ and class target definitions about lung cancer patients; their constraints are defined in terms of SHACL. Features’ definitions are classified into independent and dependent variables. Independent variables are features that the user selects for analyzing the model’s prediction. On the other hand, a dependent variable is a feature under investigation that is expected to change in response to changes in the independent variable. They are used later in the predictive modeling pipeline and are defined as follows:

where EHR is the electronic health record of a patient, the first part states the feature name (i.e., Gender), and the later part (i.e., <https://LungCancer.eu/vocab/sex>) describes the feature in the KG. Figure 7 shows the traced input configuration, i.e., features’ definitions, sampling strategies, class definitions, number of important features, SPARQL endpoint, etc. that are represented in the form of RDF triples, later used for the InterpetME KG. In the current running example Fig. 7, the target class (Biomarker) is imbalanced (i.e., target class ALK has 872 instances of patients while target class Others has 432 instances of patients). Therefore, the under-sampling technique1515 is selected for this use case to tackle the problem of imbalance.
Fig. 7.
Running example demonstrates the InterpretME internal architecture over the Lung cancer KG. At each stage, all that InterpretME traces about the features’ definition, SHACL validation, and the predictive model characteristics are shown as an RDF graph. InterpretME integrates all these traced metadata to generate the InterpretME KG.
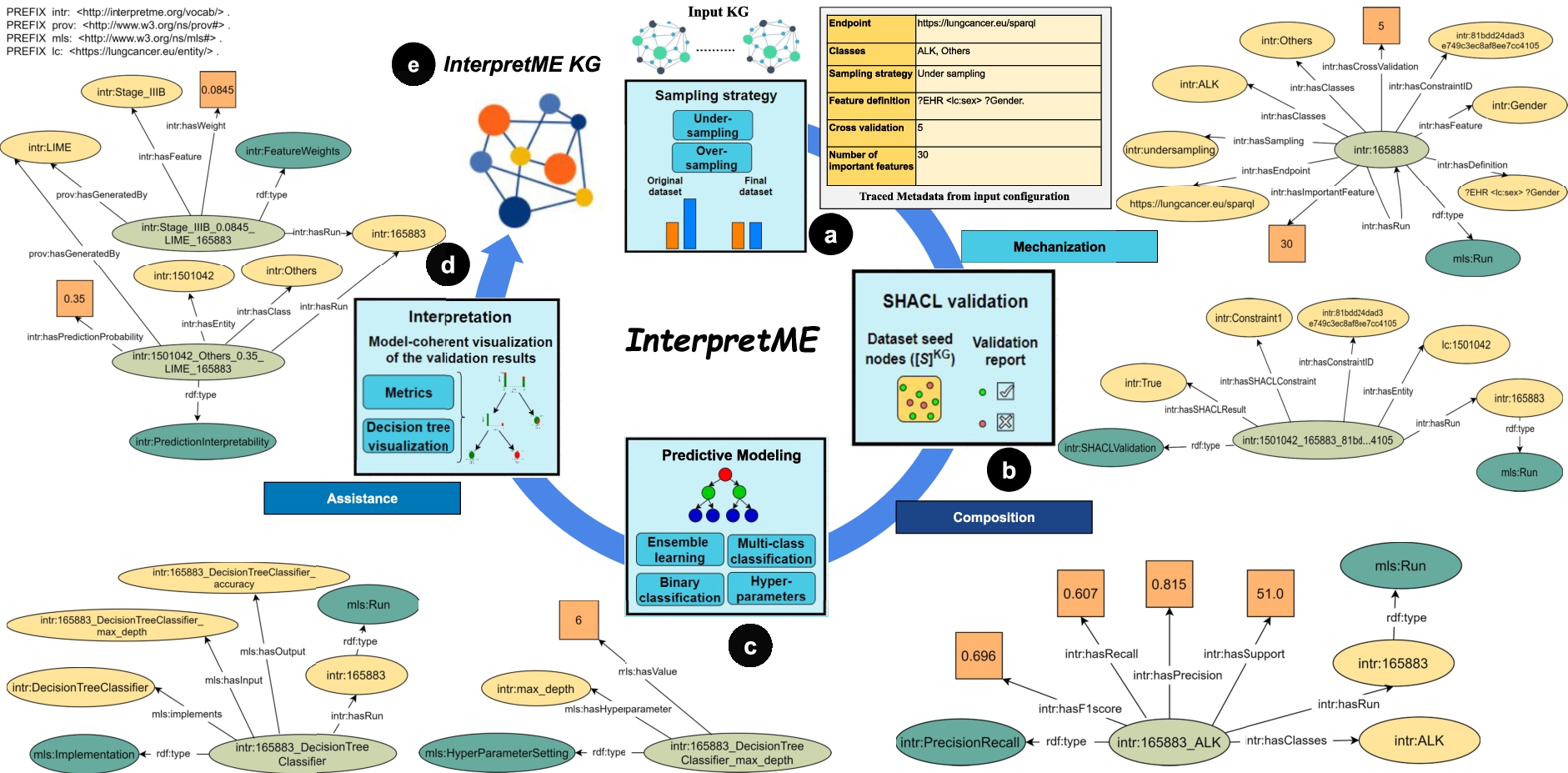
 Considering our running example in Fig. 7, the SHACL shapes represent integrity constraints. A SHACL schema defines integrity constraints, used by InterpretME for validating results and for uncovering their impact on a model’s decisions. Here, the “constraint”: “Afatinib is not recommended for NSCLC EGFR negative (hasDrug)” is applied over the extracted data from the input KG. The above constraint states a medical protocol that Afatinib is a drug that is not recommended for a patient being EGFR negative. Figure 7 depicts the traced metadata about SHACL validation, i.e., the patient lc: 1501042 with EGFR positive satisfies the constraint, an entity alignment is performed to trace the original entity of the input KG with SHACL validation results.
Considering our running example in Fig. 7, the SHACL shapes represent integrity constraints. A SHACL schema defines integrity constraints, used by InterpretME for validating results and for uncovering their impact on a model’s decisions. Here, the “constraint”: “Afatinib is not recommended for NSCLC EGFR negative (hasDrug)” is applied over the extracted data from the input KG. The above constraint states a medical protocol that Afatinib is a drug that is not recommended for a patient being EGFR negative. Figure 7 depicts the traced metadata about SHACL validation, i.e., the patient lc: 1501042 with EGFR positive satisfies the constraint, an entity alignment is performed to trace the original entity of the input KG with SHACL validation results.
 The preprocessed and sampled data with optimized hyperparameters is then fed to the automated tools for model building (e.g., ensemble learning) to perform the predictive task. Here, the automated model can also perform stratified shuffle split cross-validation with models like Random forest, AdaBoost classifier, Gradient boost classifier, and identify the relevant features; they are used to train a decision tree classifier to predict and visualize the outcomes12. InterpretME stores the metadata about the evaluation of the model for a particular run.
The preprocessed and sampled data with optimized hyperparameters is then fed to the automated tools for model building (e.g., ensemble learning) to perform the predictive task. Here, the automated model can also perform stratified shuffle split cross-validation with models like Random forest, AdaBoost classifier, Gradient boost classifier, and identify the relevant features; they are used to train a decision tree classifier to predict and visualize the outcomes12. InterpretME stores the metadata about the evaluation of the model for a particular run.
 The trained predictive model in
The trained predictive model in  can be visualized by means of Decision trees; they can be visualized with SHACL constraints to observe which subtree validates or invalidates the protocol1616. To understand the predictive model’s outcomes, InterpretME resorts to interpretable tools, e.g., LIME, to provide a local explanation of each entity. LIME provides the prediction probability of the target class ALK (i.e., Class 0) as 0.65 and target class Others (i.e., Class 1) as 0.35. For example, Fig. 7 demonstrates that the target entity intr:1501042 has a prediction probability of 0.35 for class Others (i.e., Class 1) interpreted by LIME. The relevant features list is often used to illustrate which features may cause a change in the prediction of the trained model. LIME results are also represented in the InterpretME KG; a user can utilize SPARQL queries to perform statistical analyses over these factual statements.
can be visualized by means of Decision trees; they can be visualized with SHACL constraints to observe which subtree validates or invalidates the protocol1616. To understand the predictive model’s outcomes, InterpretME resorts to interpretable tools, e.g., LIME, to provide a local explanation of each entity. LIME provides the prediction probability of the target class ALK (i.e., Class 0) as 0.65 and target class Others (i.e., Class 1) as 0.35. For example, Fig. 7 demonstrates that the target entity intr:1501042 has a prediction probability of 0.35 for class Others (i.e., Class 1) interpreted by LIME. The relevant features list is often used to illustrate which features may cause a change in the prediction of the trained model. LIME results are also represented in the InterpretME KG; a user can utilize SPARQL queries to perform statistical analyses over these factual statements.
 Traced metadata are semantified using RML. RML mapping rules specify the role of transforming collected metadata into RDF triples for the InterpretME KG1717. The unified schema of InterpretME represents the meaning of data in the InterpretME KG1818. The following RML syntax is used to define one of the mapping rules:
Traced metadata are semantified using RML. RML mapping rules specify the role of transforming collected metadata into RDF triples for the InterpretME KG1717. The unified schema of InterpretME represents the meaning of data in the InterpretME KG1818. The following RML syntax is used to define one of the mapping rules:

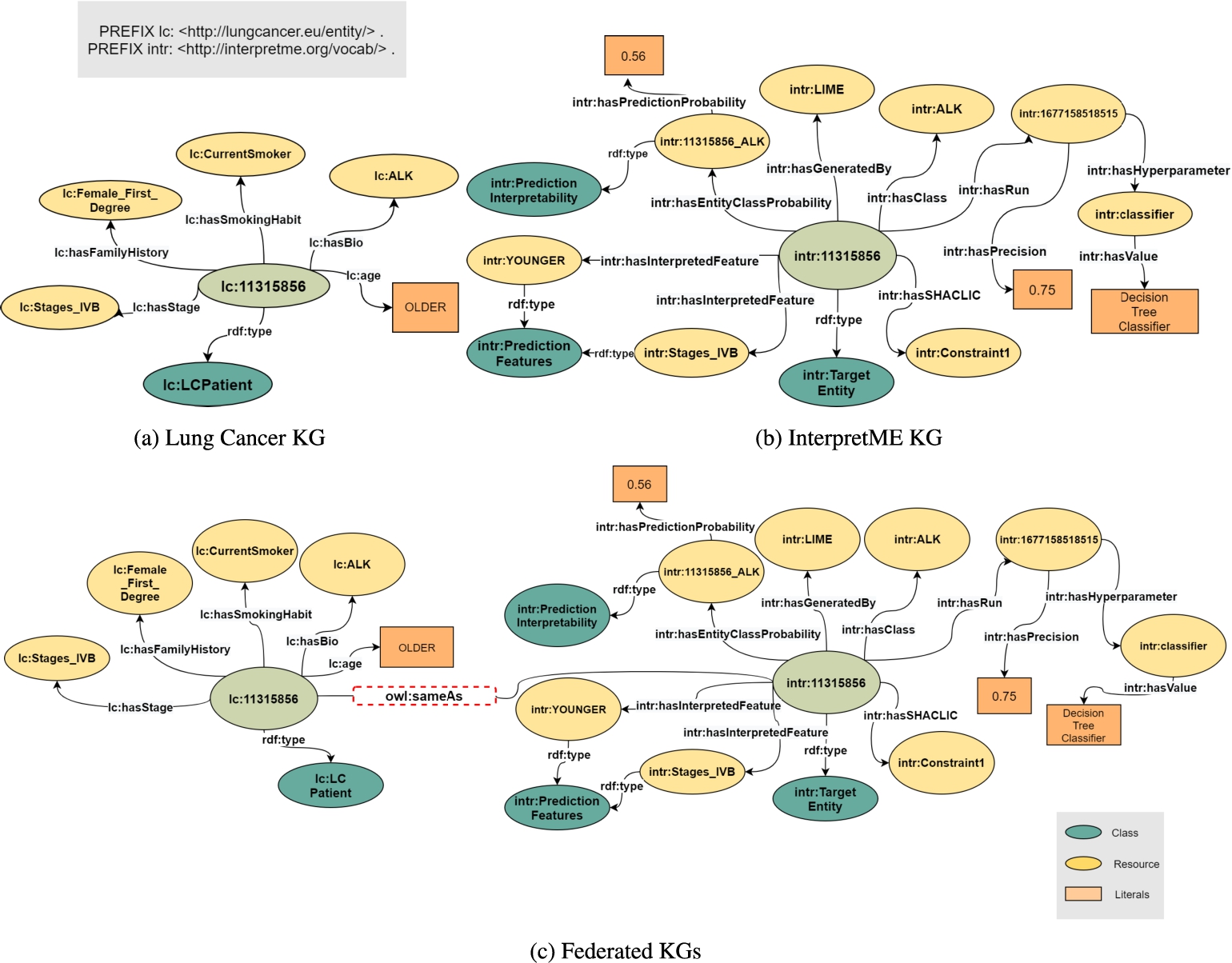
SDM-RDFizer [21] integrates entities collected during the training of a ML model into the InterpretME KG as factual statements. RML provides a declarative definition of the classes and properties of the InterpretME ontology and ML schema to the collected entities. As a result, the InterpretME KG comprises machine and human statements that document the trained ML behavior. The generated RDF data is uploaded to an instance of Virtuoso. Entities in the InterpretME KG and the input KGs are aligned to ensure traceability. InterpretME can accept input from multiple KGs and therefore the process of entity alignment as shown in Fig. 8 plays a vital role. The federation of KGs requires the identification of a specific entity, as well as the source of the entity. As a result, the URI of an entity in the input KGs differs from the URI in the InterpretME KG. The InterpretME federated query engine allows for tracing back the ML models’ results, i.e., main properties of target entities, LIME prediction probabilities, and SHACL validation reports. InterpretME has the advantage of assessing target entities not only based on ML models but also based on the entity’s properties. Appendix includes some queries to be executed over the SPARQL endpoint of InterpretME; their answers provide insights about the behavior of the trained ML models. Additionally, a short tutorial is available to demonstrate the use of InterpretME14.
Fig. 8.
Entity Alignment. Figure 8a shows a target entity lc:11315856 from the Input KG with the features, i.e., age, gender, smoking habit, and biomarker. Figure 8b illustrates the InterpretME KG of a target entity (intr:11315856) with the traced metadata from the predictive model. Figure 8c demonstrates the entity from the Lung cancer KG is aligned with the target entity from the InterpretME KG via owl:sameAs. The out-degree distribution of lc:11315856 is 6 in the input KG, 11 in the InterpretME KG, and goes to 17 in the federated KGs.
4.Empirical evaluation
We evaluate InterpretME with the goal of answering the following research questions: RQ1) What is the impact of integrating predictive modeling frameworks with KGs to enhance interpretability? RQ2) Can InterpretME trace decisions made by predictive models? RQ3) To which extent does the InterpretME KG satisfy standard quality criteria? RQ4) How much is the observed overhead at each stage of InterpretME? The experimental settings utilized to evaluate InterpretME are as follows:
Benchmarks. Table 1 presents our KGs in numbers. The French Royalty KG [16] is fully curated. For each person in the KG, we added the class dbo:Person, the number of children, the number of predecessors, and triple-related counts like the number of distinct predicates. Furthermore, we added the ground truth for the relationship hasSpouse. Forecasting whether a member of the French Royalty has a spouse is our predictive task. We created ten SHACL constraints of which some are inspired by the logical rules presented in [16], e.g., if I have a child, this child has another parent which is not me. The other constraints follow a similar pattern, e.g., if a person has two different parents, the parents are likely to be married. We use a private KG from the biomedical domain – The Lung Cancer KG [40] – which integrates lung cancer patients mentioned as in Section 2. This KG includes information describing the main characteristics of a lung cancer patient. The prediction task is a binary classification to predict the biomarker of the patient, which can be ALK or others. The SHACL constraints used are the medical protocols that recommend when treatments should be prescribed according to a patient’s biomarkers; we defined four different SHACL constraints stating that EGFR negative patients should not take Afatinib or Gefitinib.
Table 1
Statistics about input KG and InterpretME KG
| Knowledge graph | #triples | #entities | #predicates | #objects | #triples /#entities | |
| French Royalty | Input KG | 31,599 | 3,439 | 133 | 4,390 | 9.18 |
| InterpretME KG | 245,981 | 30,914 | 152 | 35,622 | 7.96 | |
| Lung Cancer | Input KG | 77,466,343 | 21,420,170 | 357 | 15,053,863 | 2.30 |
| InterpretME KG | 62,393 | 8,877 | 152 | 11,546 | 7.03 | |
Machine Learning Models. InterpretME resorts to ensemble learning techniques to solve the previously described prediction tasks. Random forests create multiple decision trees by bootstrapping datasets created from the KGs, and randomly selecting a subset of variables at each step of the decision tree. A random forest generates the list of relevant features iteratively with a stratified shuffle split cross-validator (5 fold-validation with training 70% and testing 30%). A decision tree uses this list to train the predictive model. The generated classification decision tree is easy to understand due to its visualization of the decision process. Accuracy without the random-forest stratified shuffle split is 69% in the Lung Cancer KG predictive task and 95% in the French Royalty KG, but with our approach, accuracy is increased to 77.50% and 96.17%, respectively.
Metrics. Execution Time: The time elapsed at each component of the pipeline is reported. The reported times correspond to absolute wall-clock system time in seconds as reported by Python’s time.time() method. The average execution time of five runs is reported. The experiments are run on a machine equipped with an Intel® Core(TM)i7- 10850H at 2.71 GHz and 16 GiB RAM.
Fig. 9.
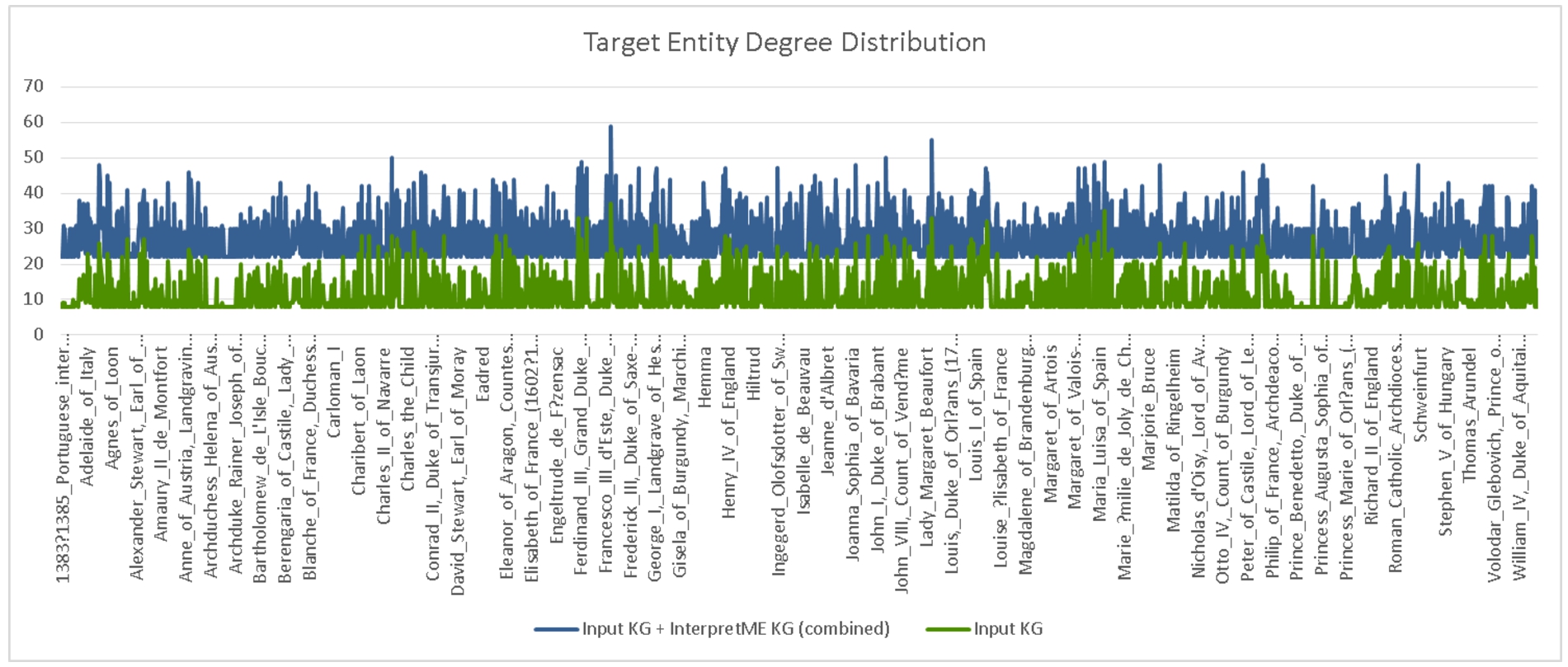
Degree distributions of the entity targets in the French royalty KG. The distribution in blue depicts the degrees of the target entities semantically enhanced with InterpretME, while green shows their original degree distribution.
4.1.Degree distribution of the InterpretME KG
In graph theory and complex networks, an entity’s degree (or the number of neighbors ignoring edges’ direction) is defined as the number of relations with other entities. Increasing the degree of an entity expresses a richer, more accurate view of the particular entity. Better context means better interpretability, not just volume. For instance, in the predictive task described in the running example Section 3.3, the predictive model states the decision of a particular target entity; it is unclear how these decisions are generated with only knowing the node of a patient and its attributes.While InterpretME tries to increase more contextual edges of a patient node via annotating the contextual information and behavior about the patient (e.g., Validation report) in the pipeline. In a nutshell, increasing the degree of a node with more context makes the interpretation not only human-understandable but also machine-readable. InterpretME traces the entities of the target classes (e.g., ALK) defined as dependent variables via SPARQL queries over the input KGs. In the InterpretME KG, these entities are described in terms of metadata collected by InterpretME components (e.g., hasInterpretedFeature, hasFeatureWeight, hasEntityClassProbability, hasPredictionProbability, hasSHACLResult, hasSHACLConstraint). For instance, Fig. 8 shows entity alignment. Given a predictive task over the Lung Cancer KG, we know the main characteristics of a target entity lc:11315856 (i.e., LCPatient, age, smoking habit, biomarker) in the input KG. Additionally, InterpretME traces both the characteristics of a predictive model (i.e., prediction, accuracy, relevant features, feature contribution, hyperparameter’s, model, target class) and the knowledge about the entity satisfying the domain constraint. The entity in the input KG (lc:11315856) is aligned with the target entity via owl:sameAs. InterpretME applied an extension of the Leibniz’s Inference Law [14], which states that if X owl:sameAs Y, then
Figure 9 depicts the degree distribution results of the French Royalty KG; the average number of neighbors is WithInterpretME: 27.19 (with a standard deviation of 6.13) and 11.39 (with a standard deviation of 5.06) WithoutInterpretME. InterpretME increases the number of RDF triples that describe a target entity; a user can query these triples to explore the predictive model decisions. The InterpretME KG properties are retrieved. These values of degree distribution quantify the information gained for each target entity in terms of all that InterpretME traces. The execution of queries 1 and 41919 over the InterpretME KG retrieves the values of these properties for intr:Louis_XIV. As a result of tracing the machine learning pipeline, the number of properties of intr:Louis_XIV (i.e., outdegree) goes from 32 in the French Royalty KG to 55, and thus, the entity’s degree is increased. Based on these outcomes, we can answer the RQ1), and InterpretME enhances the interpretability of a target entity dbr:Louis_XIV and provide the user more contextual insights into the prediction task.
4.2.Traceability
We evaluate InterpretME in terms of the traceability of a target entity. Table 2 reports on the average number of answers to the type of questions presented in our motivating example (Section 2). InterpretME efficiently traces the target entity and provides the user with additional information about the prediction probability of the entity. Also, it helps users to uncover relevant features of an entity that contribute to the prediction, with assigned weight distribution for the top-10 features. The federated query engine, DeTrusty [37], evaluates SPARQL queries to retrieve data from the original KG, the InterpretME KG, or both. Instances in the InterpretME KG are linked to the entity in the original KG via owl:sameAs. The KGs are accessible via SPARQL endpoints. In Table 2, the questions presented in Section 2 are expressed as SPARQL queries over the two KGs. For the French Royalty KG, InterpretME models LIME interpretations in terms of 10 RDF triples (on average), and the contribution of a given feature is documented with 2 RDF triples on average for the binary classification task. Moreover, target entities satisfy the 10 domain integrity constraints, and the average number of main characteristics of the target entities in the French Royalty KG is 12.24. The average number of answers in the Lung Cancer KG is higher because a lung cancer patient is described in more detail than a person in the French Royalty KG. Based on these outcomes, we can positively answer RQ2) because InterpretME can translate back to the original KGs, the decisions made by the predictive models.
Table 2
Average number of answers per target entity to questions from Fig. 4a
| ID | Question | Avg answers in | |
| Lung cancer KG | French royalty KG | ||
| Q1 | Which is the target entity interpreted by LIME? | 20.57 | 10.00 |
| Q2 | How does feature contribute to the classification of this entity? | 2.05 | 2.00 |
| Q3 | Which other features are relevant for this classification? | 11.65 | 4.47 |
| Q4 | Does this target entity satisfy the domain integrity constraints? | 4.00 | 10.00 |
| Q5 | What are the main characteristics of the target entity? | 46.28 | 12.24 |
Table 3
Evaluation metrics
| Metric | Value | Explanation | Formula (in [11]) |
| Accuracy | Section 2.2.1 | ||
| Synt. validity of RDF doc | 1 | RDF files are valid | |
| Synt. validity of literals | 1 | Literals have a label that defines a range of possible values | |
| Semant. validity of triples | 1 | Literals are collected from input or can be measured | |
| Trustworthiness | Section 2.2.2 | ||
| KG level | 0.25 | Automated data curation from structured data sources | |
| Statement level | 0.5 | Provenance is traced | |
| Unknown/empty values | 0 | Empty values are not indicated | |
| Relevancy | Section 2.3.1 | ||
| Ranking of statements | 0 | Ranking is not useful in this context | |
| Ease of Understanding | Section 2.4.1 | ||
| Description of resources | 1 | Resources have label and comment | |
| Labels in multiple lang | 0 | Labels are only available in English | |
| RDF serialization | 1 | Serialization in Turtle | |
| Self-describing URIs | 0.5 | Partial use of self-describing URIs | |
| Interlinking | Section 2.5.3 | ||
| Interlinking via owl:sameAs | 1 | Use of owl:sameAs for linking the InterpretME KG to the input KG | |
4.3.The quality of the InterpretME knowledge graph in numbers
This section answers RQ3) and reports the results of the evaluation of the InterpretME KG in terms of the quality metrics proposed by Färber et al. [11]. Four data quality categories are considered: i) Intrinsic category is independent of the use case context; ii) Contextual category depends on the application context of a data consumer; iii) The form of how information is available is quantified by the representational category; and iv) Accessibility category determines how data can be accessed. Table 3 depicts all the metrics by category from Färber et al. [11] to check the quality of the InterpretME KG. They help the user to better estimate the pros and cons of InterpretME. Färber et al. [11] propose extended quality parameters and categories (i.e, trustworthiness and interlinking). The scores for quality metrics (Table 3) are assigned manually, following the scores’ definition by Färber et al. [11]. Since, the InterpretME KG covers a new domain of structured data, the eloquence of several suggested criteria [11] is still relatively low. For example, the metric used in the representational category, such as Labels in multiple languages shown in Table 3 can be enhanced from 0 to 1 with the inclusion of other languages. However, we are optimistic that these values will be increased once real-world applications begin to use InterpretME.
Fig. 10.
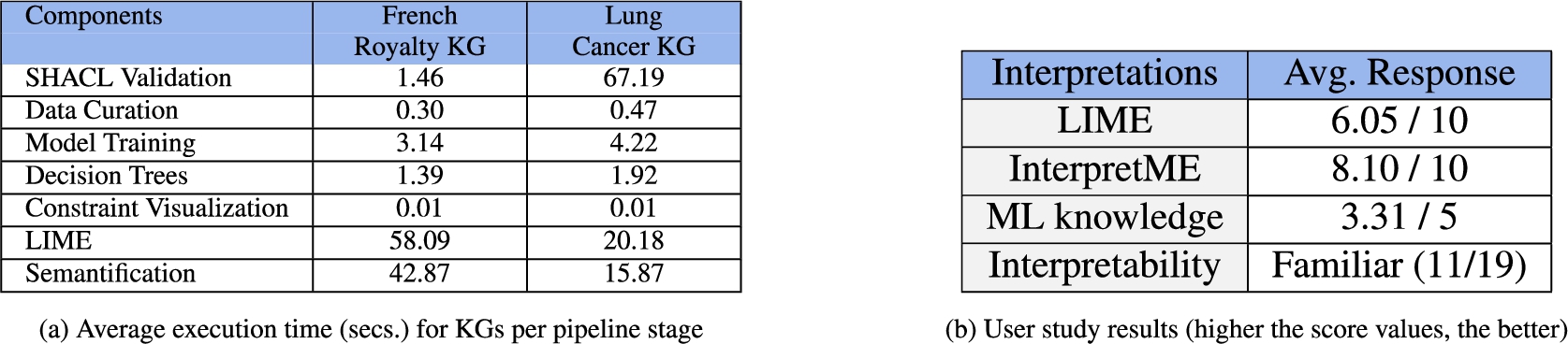
Evaluation. Figure 10a presents impact of execution time in seconds at each stage of InterpretME pipeline. Figure 10b reports on a user study conducted with 19 participants (ages ranging from 20 to 45); on average the participants were familiar with the concept of Interpretability, and experience level of ML is 3.31. Participants rated LIME interpretation 6.05, while InterpretME was rated with 8.10.
4.4.Execution time
This section describes Fig. 10a and aims at answering RQ4), i.e., the overhead caused by each of the components of InterpretME. The analysis of the execution time reveals the impact of different parameters on the performance of the InterpretME steps. We have calculated the average execution time based on the two use cases, French Royalty and Lung Cancer KGs by taking the average of five runs per use case. The French Royalty KG comprises
The average execution time of the first component, i.e., the SHACL validation, is 1.46 and 67.19 seconds, respectively. The second component – Preprocessing – where data is extracted and preprocessed to be given to the machine learning model, takes on average 0.30 and 0.47 seconds in the two use cases, respectively. The next component is training the model (Predictive Modeling); the average execution time is 3.14 and 4.22 seconds for the French Royalty KG and the Lung Cancer KG, respectively. Further, Interpretation component, where the decision trees for the model are generated, takes an average 1.39 and 1.92 seconds, respectively. For French Royalty, the number of target entities is slightly higher compared to the Lung Cancer KG. Thus, using LIME to generate the interpretable results requires 58.09 and 20.18 seconds in the use cases, respectively. Lastly, the Semantification of the traced metadata takes 42.87 and 15.87 seconds for the French Royalty KG and the Lung Cancer KG, respectively.
The reported results are consistent with the fact stated by Figuera et al. [12] that the SHACL validation is mainly impacted by the KG size and number of SHACL constraints. The validation of the Lung Cancer constraints takes much longer than for the French Royalty, since the Lung Cancer KG is much larger and the constraints used are more complex, e.g., we are using the concept of linked shapes for the Lung Cancer KG. As a result, the ML pipeline execution time is also impacted, as well as the generation of the InterpretME KG instances.
4.5.User study
We report the results of a user study2020 (Fig. 10b) about interpretability in InterpretME. We aim to assess how each participant understands the term interpretability. Further, we conduct a user experience evaluation of the framework based on usability criteria. The goal is to answer the following questions: i) Is it beneficial to integrate knowledge graphs for interpretability? This question is intended to determine the impact of knowledge graphs in the interpretability of a target entity. ii) How clear is the notion of the interpretation from LIME on an increasing scale from 1–10? This question seeks to examine the interpretations provided by LIME. iii) How clear is the notion of the interpretation from InterpretME on an increasing scale from 1–10? This question aims to assess the conceptual and practical understanding of the InterpretME KG. iv) Do we have a positive or negative attitude towards the tool? This question indicates participants’ attitudes toward InterpretME in terms of usability, reliability, and trustability.
During a lecture of data management, we shared the overall 12 questions to 19 participants that included 7 master students from computer science, 5 researchers from academia, 3 software developers, 2 medical students, and 2 Ph.D. students. The vast majority of our participants (
In the (i) question, most participants believed that KGs play a significant role in the interpretability of ML models and comply with the fact mentioned in [38]. Participants were asked to assess LIME interpretations before creating the InterpretME KG. Most participants who were familiar with ML found it difficult and computational. The (ii) and (iii) question focuses on the benefit of the interpretation given by the InterpretME KG compared to LIME. Almost all of the participants (
5.InterpretME as a tool
InterpretME is publicly available as a Python library on PyPI3. While the core of the pipeline is entirely new, InterpretME reuses available tools from the community. InterpretME pipeline() receives a JSON file as a configuration input from the user to extract all the features’ definition, SPARQL endpoints or Datasets, SHACL constraints, target classes, and sampling strategies. Features are defined in the form of independent and dependent variables, provided to the InterpretME pipeline to perform the prediction tasks. For the SHACL validation, InterpretME relies on Trav-SHACL [12] since it is capable of validating a SHACL schema against a SPARQL endpoint and scales better compared to other approaches with the same capability. InterpretME provides the Preprocessing component for data curation, optimize hyperparameters, and sampling strategy. Predictive Modeling provides binary and multi-class classification with ensemble learning techniques. Also, cross-validation and classification reports are obtained for a particular predictive model. Currently, InterpretME uses LIME [34] to create model interpretations. RML [8] is used to define the process of integrating the traced metadata into the InterpretME KG; it is semantified using SDM-RDFizer [21] (version 4.5.5 [20]), an efficient RML-compliant engine for KG creation. The RDF data is uploaded into an instance of Virtuoso 7.20.3233. The following pipeline() command executes the whole pipeline; It includes the extraction of data and metadata from the input data, validating SHACL constraints, preprocessing the data, running predictive models, semantifying the results, and populating the InterpretME KG:

The InterpretME ontology extends ML Schema [10], a vocabulary to deal with machine learning algorithms. ML Schema can be used to describe different algorithms, implementation, model evaluation, and the input and output considered by the algorithms; it also represents relationships between ML algorithms and their executions. InterpretME reuses 12 concepts in the mappings from ML Schema as shown in the Fig. 3a. They include classes mls:Run, mls:Implementation, mls:ModelEvaluation, and mls:HyperParameterSetting, and the relations mls:hasInput, mls:hasOutput, and mls:hasHyperParameter. The InterpretME ontology also include classes and properties to represent ML algorithm results with LIME interpretations (e.g., intr:PredictionInterpretability and intr:hasPredictionProbability), and the results of validating SHACL schemas over the input KG (e.g., intr:SHACLValidation, intr:hasSHACLResult, and intr:hasSHACLConstraint); the description of these classes and properties are depicted in Fig. 3b and Fig. 3c. Following the FAIR principles [15], the InterpretME ontology is publicly available on an instance of VoCoL2424; it enables the management, understanding, and exploration of the ontology.

The data from the InterpretME KG – and from the input KGs – can be queried using the federated query engine DeTrusty (version 0.12.3 [37]). DeTrusty is based on MULDER [9], i.e., it uses semantic source descriptions during decomposition and planning. Rohde [36] describes the vision of incorporating the SHACL validation result into SPARQL query answers by executing the validation during query processing. While we are not providing an engine fulfilling this vision, a similar outcome can be achieved when querying the original KG together with the InterpretME KG since the SHACL validation result is part of it. InterpretME is a stand-alone framework that works locally on individual systems, with multiple runs that can be combined into a single InterpretME KG to compare interpretations. InterpretME comprises the following resources: i) the InterpretME pipeline enhancing interpretability of machine learning models; ii) the InterpretME KG describing the predictive model characteristics; and iii) the InterpretME ontology specifying the vocabulary for describing the main characteristics of trained predictive models. InterpretME is utilized on top of the RDF KGs of the following projects: 1) CLARIFY2525 to define machine learning models to predict biomarkers of a lung cancer patient based on demographic features (e.g., age and gender), smoking habits, and relatives with cancer; 2) ImProVIT2626 to develop models to predict the impact of the immune system and demographic features into the response of Hepatitis B and Influenza vaccines; and 3) P4-LUCAT2727 to implement predictive models to forecast the relapse after surgery or the disease progression in advanced stages.
6.Related work
Tools for supporting interpretability Artificial intelligence and machine learning have gained global prominence across numerous domains, resulting in a growing demand for automated machine learning frameworks with assistance in both research and various sectors. De Bie et al. [5] discuss the challenges to achieve automated machine learning and highlight that existing tools contribute to mechanization and composition automatization, lacking support in assistance. InterpretME also aims at bridging this gap, and resorts to Semantic Web technologies to enhance users’ assistance. Lundberg et al. [27] propose an interpretation framework called SHAP based on coalition game theory (Shapely values). SHAP provides feature contributions for each individual instance, global explanations, and feature importance. Ribeiro et al. [34] present LIME, an approach for local surrogate models, which are used to explain individual predictions of a pipeline. Local surrogate models are trained to approximate the predictions of models locally, instead of training a surrogate model globally. As a result, LIME generates human-friendly explanations for target entities. However, these explanations are not machine-readable and cannot be translated into the domain application. InterpretME overcomes these limitations, and provides fine-grained representations of local interpretations which are linked to the target entities in the domain application KGs.
Semantic web technologies in machine learning Semantic Web technologies like ontologies are used to improve the accuracy of predictive models. Ristoski et al. [35] provide a comprehensive survey of the use of Semantic Web Technologies in data mining and knowledge discovery and highlights the potential benefits and challenges of the need for more efficient algorithms and tools in practice. Kulmanov et al. [25] study the role of ontologies in semantic similarity and machine learning, and present ontology embeddings as background knowledge. Further, ontologies can constrain the output of a machine learning model, i.e., making the model consistent with the axioms of the ontology. Min et al. [28] improve the performance of predictive models by using ontological adjustments, i.e., using the hierarchy of an ontology to move samples of rare classes into the next broader concept. On average, Min et al. [28] reduce the area under the receiver operating curve (AROC) by 9.0% in predicting the effectiveness of antidepressants in patients with rare conditions. Haug et al. [18] propose the combination of a large enterprise data warehouse with medical knowledge from a disease-oriented ontology. This combination allows for automating the generation of computable diagnostic models, which aim at supporting researchers in generating and evaluating tools for real-time clinical diagnosis. Similarly, InterpretME resorts to Semantic Web technologies to document ML pipelines and enhances not only accuracy but also improves interpretability.
7.Conclusions and future work
InterpretME enhances predictive models by incorporating metadata throughout the predictive modeling pipeline. Leveraging concepts from the ML Schema and employing a cutting-edge SHACL engine for efficient constraint validation, the InterpretME KG ensures accurate and insightful results. Our empirical observations have found that InterpretME facilitates the description of predictive model insights using factual statements and establishes links to application domain KGs, thereby bridging the gap between data meaning and predictive modeling [13].
InterpretME supports random forests, decision trees, and the LIME interpretable model in its current version. However, our future plans involve integrating additional models and tools. Additionally, we aim to establish connections between InterpretME and automated ML systems and causal KGs [22]. These enhancements will further enrich domain understanding and expand the portfolio of Semantic Web tools available for data analytics.
Notes
25 EU H2020 Funded project https://www.clarify2020.eu/.
26 German Funded project https://www.tib.eu/en/research-development/project-overview/project-summary/improvit.
27 EraMed project https://p4-lucat.eu/.
Acknowledgements
This work has been supported by the project TrustKG- Transforming Data in Trustable Insights with grant P99/2020, the EU H2020 RIA project CLARIFY (GA No. 875160) and the EraMed project P4-LUCAT (GA No. 53000015).
Appendices
Appendix
AppendixSPARQL queries to represent statistical queries over the InterpretME KG
Listing 1.
SPARQL query to retrieve target entity and the associated feature contributions generated by LIME

Listing 2.
SPARQL query to retrieve target class and the probability ordered in decreasing order

These queries can be executed over the SPARQL endpoint of the InterpretME KG2828 to gain some insights into the behavior of the trained predictive model. Listing 1 presents a SPARQL query that collects the information about the entities that define the class intr:TargetEntity. The projected attributes include the entity from the input KG that is classified into a specific target class and the interpretations provided by LIME which include interpreted features and their feature contributions. Based on a particular interpreted feature we can analyze the feature contribution from the highest to lowest value.
Listing 2 depicts a SPARQL query that retrieves the count of the target entities with respect to the target class that entity is classified into and the prediction probability generated by LIME.
Listing 3 performs a SPARQL query that performs the statistical analysis of probability, i.e., MAX, MIN, AVG about the classification of target entities in a specific target class.
Listing 3.
SPARQL query to retrieve the MAX and MIN and AVG of the prediction probability generated by LIME for the target entity

References
[1] | M. Acosta, M. Vidal, T. Lampo, J. Castillo and E. Ruckhaus, ANAPSID: An adaptive query processing engine for SPARQL endpoints, in: The Semantic Web – ISWC 2011, (2011) . |
[2] | T. Akiba, S. Sano, T. Yanase, T. Ohta and M. Koyama, Optuna: A next-generation hyperparameter optimization framework, (2019) . doi:10.48550/ARXIV.1907.10902. |
[3] | M. Arenas, C. Gutierrez and J. Pérez, Foundations of RDF databases, in: Reasoning Web. Semantic Technologies for Information Systems. Reasoning Web 2009, Lecture Notes in Computer Science, Vol. 5689: , Springer, Berlin, Heidelberg, (2008) . doi:10.1007/978-3-642-03754-2_4. |
[4] | T. Berners-Lee, J. Hendler and O. Lassila, The semantic web: A new form of web content that is meaningful to computers will unleash a revolution of new possibilities, (2001) . http://www.sciam.com/article.cfm?articleID=00048144-10D2-1C70-84A9809EC588EF21. |
[5] | T.D. Bie, L.D. Raedt, J. Hernández-Orallo, H.H. Hoos, P. Smyth and C.K.I. Williams, Automating data science, Commun. ACM 65: (3) ((2022) ), 76–87. doi:10.1145/3495256. |
[6] | J. Corman, F. Florenzano, J.L. Reutter and O. Savković, Validating SHACL constraints over a SPARQL endpoint, in: The Semantic Web – ISWC 2019, Springer, Cham, (2019) . doi:10.1007/978-3-030-30793-6_9. |
[7] | J. Corman, J.L. Reutter and O. Savković, Semantics and validation of recursive SHACL, in: The Semantic Web – ISWC 2018, Springer, Cham, (2018) . doi:10.1007/978-3-030-00671-6_19. |
[8] | A. Dimou, M. Vander Sande, P. Colpaert, R. Verborgh, E. Mannens and R. Van de Walle, RML: A generic language for integrated RDF mappings of heterogeneous data, in: 7th Workshop on Linked Data on the Web, (2014) . |
[9] | K.M. Endris, M. Galkin, I. Lytra, M.N. Mami, M.-E. Vidal and S. Auer, Querying interlinked data by bridging RDF molecule templates, in: Transactions on Large-Scale Data- and Knowledge-Centered Systems XXXIX, Vol. 11310: , Springer, Berlin, Heidelberg, (2018) . doi:10.1007/978-3-662-58415-6_1. |
[10] | D. Esteves, A. Ławrynowicz, P. Panov, L. Soldatova, T. Soru and J. Vanschoren, ML schema core specification, (2016) . http://www.w3.org/2016/10/mls/. |
[11] | M. Färber, F. Bartscherer, C. Menne and A. Rettinger, Linked data quality of DBpedia, Freebase, OpenCyc, Wikidata, and YAGO, Semantic Web Journal ((2017) ). doi:10.3233/SW-170275. |
[12] | M. Figuera, P.D. Rohde and M.-E. Vidal, Trav-SHACL: Efficiently validating networks of SHACL constraints, in: The Web Conference, ACM, New York, NY, USA, (2021) . doi:10.1145/3442381.3449877. |
[13] | T. Gebru, J. Morgenstern, B. Vecchione, J.W. Vaughan, H.M. Wallach, H. Daumé III and K. Crawford, Datasheets for datasets, Commun. ACM 64: (12) ((2021) ), 86–92. doi:10.1145/3458723. |
[14] | D. Gries and F.B. Schneider, A Logical Approach to Discrete Math, Texts and Monographs in Computer Science, (1993) . doi:10.1007/978-1-4757-3837-7. |
[15] | P. Groth and M. Dumontier, Introduction – FAIR data, systems and analysis, Data Science 3: (1) ((2020) ), 1–2. doi:10.3233/DS-200029. |
[16] | N. Halliwell, F. Gandon and F. Lecue, User scored evaluation of non-unique explanations for relational graph convolutional network link prediction on knowledge graphs, in: K-CAP ’21: Proceedings of the 11th Knowledge Capture Conference, ACM, New York, NY, USA, (2021) . doi:10.1145/3460210.3493557. |
[17] | S. Harris and A. Seaborne, SPARQL 1.1 query language, (2013) . https://www.w3.org/TR/2013/REC-sparql11-query-20130321/. |
[18] | P. Haug, J. Holmen, X. Wu, K. Mynam, M. Ebert and J. Ferraro, Ontology-based tools to expedite predictive model construction, AMIA Summits on Translational Science Proceedings 2014 ((2014) ), https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4419766/pdf/1859946.pdf. |
[19] | A. Hogan, E. Blomqvist, M. Cochez, C. D’amato, G.D. Melo, C. Gutierrez, S. Kirrane, J.E.L. Gayo, R. Navigli, S. Neumaier et al., Knowledge graphs, ACM Computing Surveys 54(4) ((2021) ). doi:10.1145/3447772. |
[20] | E. Iglesias, S. Jozashoori and D. Chaves-Fraga, SDM-RDFizer v4.4.2, (2022) . doi:10.5281/zenodo.6511933. |
[21] | E. Iglesias, S. Jozashoori, D. Chaves-Fraga, D. Collarana and M.-E. Vidal, SDM-RDFizer: An RML interpreter for the efficient creation of RDF knowledge graphs, in: CIKM ’20: Proceedings of the 29th ACM International Conference on Information & Knowledge Management, ACM, New York, NY, USA, (2020) . doi:10.1145/3340531.3412881. |
[22] | U. Jaimini and A.P. Sheth, CausalKG: Causal knowledge graph explainability using interventional and counterfactual reasoning, IEEE Internet Comput. 26(1) ((2022) ). doi:10.1109/MIC.2021.3133551. |
[23] | H. Knublauch and D. Kontokostas, Shapes constraint language (SHACL), (2017) . https://www.w3.org/TR/2017/REC-shacl-20170720/. |
[24] | L. Kotthoff, C. Thornton, H.H. Hoos, F. Hutter and K. Leyton-Brown, Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA, J. Mach. Learn. Res. 18: ((2017) ), 25:1–25:5. https://www.cs.ubc.ca/labs/algorithms/Projects/autoweka/papers/16-599.pdf. |
[25] | M. Kulmanov, F.Z. Smaili, X. Gao and R. Hoehndorf, Semantic similarity and machine learning with ontologies, Briefings in Bioinformatics 22(4) ((2020) ). doi:10.1093/bib/bbaa199. |
[26] | O. Lassila and R.R. Swick, Resource description framework (RDF) model and syntax specification, (1999) . https://www.w3.org/TR/1999/REC-rdf-syntax-19990222/. |
[27] | S.M. Lundberg and S.-I. Lee, A unified approach to interpreting model predictions, in: NIPS ’17: Proceedings of the 31st International Conference on Neural Information Processing Systems, Curran Associates Inc., Red Hook, NY, USA, (2017) . |
[28] | H. Min, F. Alemi, C.A. Hane and V.S. Nori, Improving the accuracy of predictive models for outcomes of antidepressants by using an ontological adjustment approach, Applied Sciences 12(3) ((2022) ). doi:10.3390/app12031479. |
[29] | M.A. Myszczynska, P.N. Ojamies, A.M.B. Lacoste, D. Neil, A. Saffari, R. Mead, G.M. Hautbergue, J.D. Holbrook and L. Ferraiuolo, Applications of machine learning to diagnosis and treatment of neurodegenerative diseases, Nat Rev Neurol 16: ((2020) ), 440–456. doi:10.1038/s41582-020-0377-8. |
[30] | P.I. Nakagawa, L.F. Pires, J.L.R. Moreira, L.O. Bonino da Silva Santos and F. Bukhsh, Semantic description of explainable machine learning workflows for improving trust, Applied Sciences 11(22) ((2021) ). doi:10.3390/app112210804. |
[31] | M.F. Pinto, H. Oliveira, S. Batista, L. Cruz, M. Pinto, I. Correia, P. Martins and C. Teixeira, Prediction of disease progression and outcomes in multiple sclerosis with machine learning, Sci Rep 10 ((2020) ). doi:10.1038/s41598-020-78212-6. |
[32] | R. Prasad, L. Joseph and R.C. Deo, Modeling and forecasting renewable energy resources for sustainable power generation: Basic concepts and predictive model results, in: Translating the Paris Agreement into Action in the Pacific, Springer, Cham, (2020) . |
[33] | E. Prud’hommeaux and A. Seaborne, SPARQL query language for RDF, (2008) . https://www.w3.org/TR/2008/REC-rdf-sparql-query-20080115/. |
[34] | M.T. Ribeiro, S. Singh and C. Guestrin, “Why should I trust you?”: Explaining the predictions of any classifier, in: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, ACM, New York, NY, USA, (2016) . doi:10.1145/2939672.2939778. |
[35] | P. Ristoski and H. Paulheim, Semantic web in data mining and knowledge discovery: A comprehensive survey, J. Web Semant. ((2016) ). doi:10.1016/j.websem.2016.01.001. |
[36] | P.D. Rohde, SHACL constraint validation during SPARQL query processing, in: Proceedings of the VLDB 2021 PhD Workshop, CEUR-WS.org, Aachen, Germany, (2021) . http://ceur-ws.org/Vol-2971/paper05.pdf. |
[37] | P.D. Rohde, DeTrusty v0.12.3, (2023) . doi:10.5281/zenodo.8095810. |
[38] | I. Tiddi and S. Schlobach, Knowledge graphs as tools for explainable machine learning: A survey, Artif. Intell. ((2022) ). doi:10.1016/j.artint.2021.103627. |
[39] | M. van Bekkum, M. de Boer, F. van Harmelen, A. Meyer-Vitali and A. ten Teije, Modular design patterns for hybrid learning and reasoning systems, Appl. Intell. 51: (9) ((2021) ), 6528–6546. doi:10.1007/s10489-021-02394-3. |
[40] | M. Vidal, K.M. Endris, S. Jazashoori, A. Sakor and A. Rivas, Transforming heterogeneous data into knowledge for personalized treatments – A use case, Datenbank-Spektrum 19: (2) ((2019) ), 95–106. doi:10.1007/s13222-019-00312-z. |

![Tracing Metadata depicts what all InterpretME traces (e.g., model characteristics) the predictive model characteristics at each forms of mechanization, i.e., Mechanization, Composition, Assistance. Based on Figure proposed by De Bie et al. [5] presented in Fig. 2.](https://content.iospress.com:443/media/sw-prepress/sw--1--1-sw233511/sw--1-sw233511-g005.jpg)



