Blue Brain Nexus: An open, secure, scalable system for knowledge graph management and data-driven science
Abstract
Modern data-driven science often consists of iterative cycles of data discovery, acquisition, preparation, analysis, model building and validation leading to knowledge discovery as well as dissemination at scale. The unique challenges of building and simulating the whole rodent brain in the Swiss EPFL Blue Brain Project (BBP) required a solution to managing large-scale highly heterogeneous data, and tracking their provenance to ensure quality, reproducibility and attribution throughout these iterative cycles. Here, we describe Blue Brain Nexus (BBN), an ecosystem of open source, domain agnostic, scalable, extensible data and knowledge graph management systems built by BBP to address these challenges. BBN builds on open standards and interoperable semantic web technologies to enable the creation and management of secure RDF-based knowledge graphs validated by W3C SHACL. BBN supports a spectrum of (meta)data modeling and representation formats including JSON and JSON-LD as well as more formally specified SHACL-based schemas enabling domain model-driven runtime API. With its streaming event-based architecture, BBN supports asynchronous building and maintenance of multiple extensible indices to ensure high performance search capabilities and enable analytics. We present four use cases and applications of BBN to large-scale data integration and dissemination challenges in computational modeling, neuroscience, psychiatry and open linked data.
1.Introduction
The growing complexity of many scientific domains has led to the emergence of cross-disciplinary teams and methods for performing scientific investigations and producing new scientific knowledge [10]. A “digital data deluge” [21,25] has emerged thanks to the capability of researchers and engineers to collect, store and share large datasets at unprecedented rates from experimental measurements, sensor networks [2,30] and computer simulations of complex systems including high energy physics [19] and the rodent brain [34]. This data deluge, along with increasingly available compute power, has led to new approaches in managing and integrating diverse, large-scale data and in data-driven methods in science such as machine learning [32] and simulation [15] enabling fast iteration of experimentation and knowledge discovery cycles.
In neuroscience, the Swiss brain research initiative Blue Brain Project (BBP or Blue Brain)11 has pioneered data-driven modeling and supercomputer-based numerical simulations methods to build accurate and biologically detailed digital reconstructions and simulations of the rodent brain [16,33,34]. This approach is challenging as it involves cross-disciplinary teams of scientists and engineers performing a series of complex operations:
– Acquiring heterogeneous, sparse, multi-modal experimental data (e.g. neuron morphologies, electrophysiology recordings, ion channel recordings and parameters from literature), generated by internal or external sources and at different levels of organisation of the brain (i.e. sub-cellular, cellular, circuit, brain region and whole brain);
– Preparing the sparse data through curation, integration (e.g. within a brain atlas giving a spatial context), features extraction (e.g. neuron morphometrics and electrophysiological features), densification (e.g using different inference techniques and algorithms to reconstruct missing data) and validation;
– Using the densified data to generate large biophysically detailed computational models (such as single cell, circuit or brain region models) then used to perform simulations from which predictions can be made and new knowledge can be acquired and thereafter shared with the research community.
Tracking the provenance of the neuroscience data as it gets generated, analyzed and integrated in computational models, is essential for attribution, quality assessment, and reproducibility. Blue Brain developed Blue Brain Nexus (BBN or Nexus)22 to address these challenges.
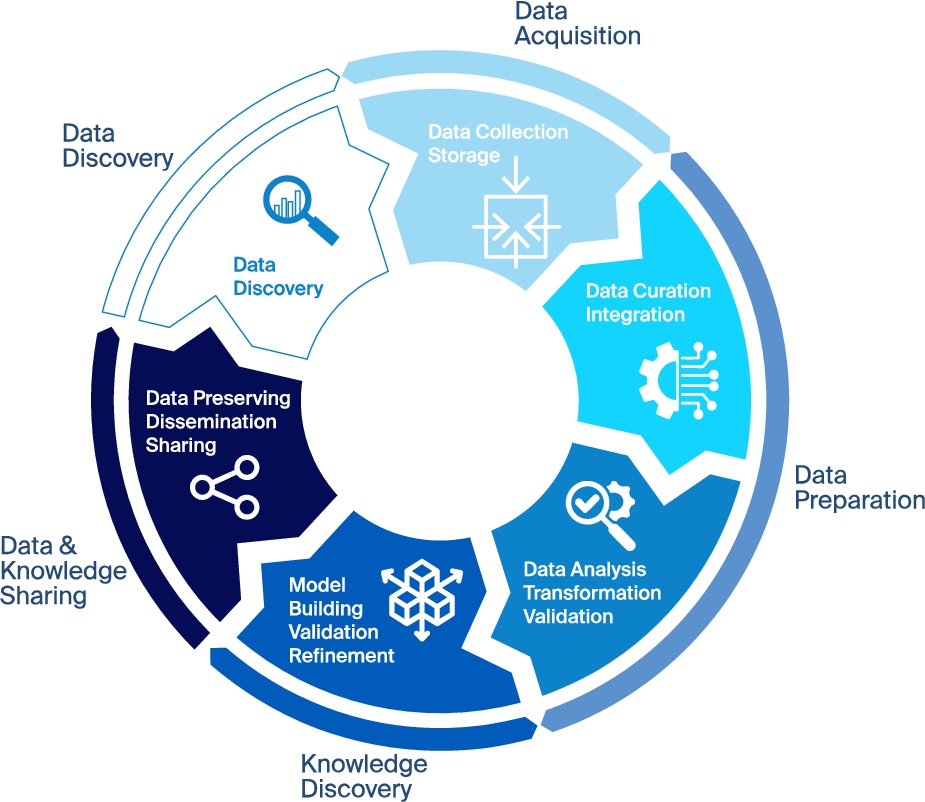
Blue Brain’s data-driven and simulation approach follows an iterative scientific investigation cycle described in high level terms in Fig. 1 from data discovery, acquisition and preparation to knowledge discovery and dissemination. With each iteration, knowledge may be gained, potentially changing how data are related and how they should be interpreted and validated. Furthermore this cycle was also found to occur in data mining [17], big data [13], machine learning communities [41] and in more and more scientific and engineering fields [36].
The challenges around managing the data and knowledge produced alongside the data-driven science cycle have grown significantly with the scale of data and the collaborative and accelerated nature of the iterations.
Fig. 1.
A typical data-driven science iterative cycle involves the following five steps: a) data discovery when searching for available data for an investigation; b) data acquisition when collecting and storing data; c) data preparation encompassing data curation, integration, transformation, analysis and validation; d) knowledge discovery related to building, refining and validating models from data; and e) data and knowledge sharing and dissemination.
This paper presents Blue Brain Nexus (BBN), an integrated ecosystem of domain agnostic data and knowledge graph management systems and tools open sourced by the Swiss brain research initiative Blue Brain. BBN supports the iterative cycle of data-driven science depicted in Fig. 1 and addresses its challenges by going beyond metadata cataloging and by using an open, scalable, extensible, standards-based and interoperable technology stack. The rest of the paper is structured as follows. Section 2 introduces the challenges raised alongside the data-driven science iterative cycle as well as knowledge graphs and related work using them to address the aforementioned challenges. Section 3 gives an overview of BBN components as well as architecture. BBN components’ design principles, features, extension points, performance benchmarks are presented in Section 4 and Section 5. BBN production deployment at Blue Brain and its impact in supporting biologically detailed simulation of the rodent brain is detailed in Section 6. Three adoptions of BBN are presented in Section 7 demonstrating BBN production deployment and usage across different organisations and use cases. Finally, the lessons learned in using BBN in production settings as well as future directions are presented and discussed for conclusion in Section 8.
2.Motivation and background
2.1.Data-driven science challenges
Building, deploying and maintaining a data and knowledge management system to enable the data-driven science iterative cycle raises a number of organisational and technical challenges.
Data discovery challenges: Data across one or many organisations can be scattered across multiple systems and repositories without connections. These siloed data are often hard to find and access because of a lack of rich and searchable metadata [9,38]. This situation leads to multiple sources of truth for data causing researchers and engineers to discover and reuse potentially inconsistent data across different applications and workflows. Data silos can also lead to missed opportunities for scientific investigations due to the lack of visibility on available data and a duplication of effort across different research groups – or worse, within a single one. A platform for data and knowledge management should allow organisations to unify or bridge data silos by enabling not only flexible and incremental data linking but also by exposing such linked data through APIs and user interfaces for access and consumption.
Data acquisition challenges: Data can be collected from a variety of sources, including internal environments, where the data collection process may be well-controlled, as well as external sources, where it may be less controlled. In both cases, the volume and variety of data raise transport, storage and preservation challenges. While using increasingly cheap hard drives to store large volume of data is no longer a challenge [19], adapting to data scale evolution and preserving highly variable data generation contexts are more challenging. Indeed, collected data can be generated from different processes, experiments or simulations and may involve different organisations, subjects, protocols, contributors, storage systems and APIs. As a result, collected data often come in different sizes, quality levels, formats (e.g. tabular, relational and binary) and structures (e.g. unstructured, semi-structured and structured).
As dynamic and evolving activities, scientific investigations may also acquire data at different rates, possibly leading to variability of their significance and meaning depending on the scientific context [4]. The ability to adapt to the scale and complexity of newly collected data and to robustly track their provenance (who, when, where, how, why) is key to assessing data quality as well as the trustworthiness and reliability of a given data source.
Data preparation challenges: Data preparation often involves curation, validation, integration, and transformation of diverse data. It also encompasses data analysis as well as potentially extracting features from them and is often related to data wrangling [18]. For example, curating data may relate to handling missing or duplicated values as well as data fusion while integration is more about combining them with other data. Transformation involves normalizing, reshaping and mapping data representation to common formats suitable for targeted analysis. An example of transformation would be to represent data with optimized feature vectors, a widely preferred data shape to perform data classification in machine learning or data mining communities.
Making sure curated and transformed data as well as the features extracted from them meet expected quality is key for data-driven methods. For example, detecting training data issues early on and ensuring features’ presence and shape are critical steps towards a good quality model in machine learning [40,41].
The ability to validate data is therefore important to detect anomalies that may be introduced during the data acquisition and preparation steps. For example, validation may involve ensuring expected outputs of a given experiment in terms of value ranges or ensuring consistent and common data shape and format across an organisation. As such, data validation capability is a key requirement for maintaining research data quality as it evolves and increasing its overall utility for downstream tasks and pipelines.
Knowledge discovery challenges: The resulting prepared data with optimized representations can then be used to discover non-trivial patterns, relations or new classes of data but also to build models using a variety of techniques including classification, regression and optimization. The accuracy and utility of these techniques can be greatly improved by combining multiple datasets across many sources, thus increasing the coverage of a given subject of study. Examples of sources can consist of reference databases or knowledge bases whether specialized such as UniProt [45] for protein sequence and functional information, community-driven like Wikipedia33 or expert-based representing domain knowledge often in the form of ontologies such as the Gene Ontology [6].
As models are validated and refined, new insights emerge, resulting often in new classes of data, new relationships between data or new interpretation and perspective on a given subject of study. For example, new neuron types can be discovered from single cell transcriptomic data, leading to new classification of neurons which have then enabled new and specific annotation and searches for neuron morphological and electrophysiological data. Tracking the data and the modeling processes from which a specific piece of knowledge (e.g. a new class of data) was derived is challenging. Machine learning models, for example, are usually trained on a specific set of data, validated on a different one and involve many different parameters. Recording such provenance is crucial to reproduce the model building process and potentially refine it as new data and knowledge are acquired and prepared. Consistently managing data, metadata, schemas and ontologies together and making the sources of data that drive the evolution of a knowledge framework explicit can facilitate the critical review, assessment and credit assignment for new discoveries.
Data and knowledge sharing challenges: Publishing and sharing research data and methods are fundamental parts of the scientific process and the basis of the dissemination of scientific results. Furthermore, they are important drivers to foster collaboration by enabling data reuse and supporting research results’ reproducibility. It is therefore not a surprise to witness the emergence of a tremendous number of publicly accessible research data repositories on the web encompassing various scientific domains. Those repositories are increasingly promoted by publishers as well as research funding agencies.
Supporting research data sharing and publishing comes, however, with a set of challenges summarized in the FAIR (Findable, Accessible, Interoperable, Reusable) guiding principles for scientific data management [49]. Persistent and globally unique data identifiers – along with a consistent versioning scheme as well as open and interoperable APIs – are key capabilities to make data findable and accessible [35]. High quality metadata – defined by the authors of [20] as the usage of controlled and standard terms for both metadata fields and values to describe, for example, data provenance – was identified to result, when missing, in issues for data and research results’ reproducibility [7] and reusability [38]. Furthermore authors in [31] identified eight groups of dataset reusability features all related to documenting the context in which the data was generated. Addressing these issues requires the set of scientific and technical activities, protocols and contributors involved in the data generation to be described and shared as metadata (e.g. data about data). Such metadata should be represented using generic, expressive and machine-readable formats to cope with cross-disciplinary scientific domains, on the one hand, and enable data and metadata interoperability for people and software agents, on the other hand.
Without rich and high quality metadata, research data repositories – even those with open and accessible APIs – can become a source of a “new tragedy of the commons” [8,38]. In this situation, a shared repository may lose its overall utility and quality over time because of the collective action of researchers caring individually only about sharing their own data without providing provenance metadata.
Genericity and extensibility challenges: Cross-disciplinary and multi-modal scientific investigations require scalable data storage and flexible metadata representation to adapt to new research directions and results. As research activities evolve, use cases and needs vary greatly across different domains and groups. Research data management platforms should provide flexible and expressive metadata representation languages and formats to support different domains, modalities and scale. A “one-size-fits-all” issue arises inevitably if it is not possible to customize the system’s behavior and extend its feature set. Extensibility points are therefore key for a research data and knowledge management platform.
Production-level deployment and security challenges: Building, deploying and operating a data and knowledge management platform in production come with their own set of challenges with respect to scalability, availability, reliability and data integrity. They come also with security requirements to allow an organisation to control data and metadata access from data collection and generation to publishing and sharing.
Humans in the loop: Managing, finding, accessing and monitoring data and knowledge throughout the entire lifecycle require programmatic clients as well as web interfaces targeting expert and non-expert users as well as developers. The goal is to allow scientists and developers to incrementally prototype and manage a knowledge graph but also to build applications on top of it without having to necessarily understand the system’s technical internal mechanism.
2.2.Knowledge graphs for research data management
Addressing data-driven science challenges necessitates at its core enabling different targeted users – particularly scientists, data and knowledge engineers – to discover, manage, link and reuse heterogeneous and dynamic data as well as knowledge from different sources and describe their context in the form of high quality and complex metadata. Supporting these capabilities is a key use case for knowledge graphs leveraging RDF (Resource Description Framework) as a data representation model and implementing linked data principles [25] as well as semantic web technologies [42].
Knowledge graphs have recently seen rapid adoption as a key solution to support several industry-specific applications [39] and research data management use cases including: dataset integration and search at web scale [11], research data integration [29,47] and scholarly data publication [5,23]. Thanks to the increasingly growing fields of knowledge graph embedding [43,48] and graph to text conversion [1], knowledge graphs are also used in the context of recommender systems, question answering and natural language processing (NLP) tasks.
In a knowledge graph, a domain of interest is modeled as a set of entities each representing a piece of data, described individually and potentially linked to each other using a set of metadata and relationships [39]. Entities along with their metadata and relationships form a graph of data and represent factual knowledge about the domain of interest [27]. Thanks to the open, generic, standard and flexible nature of RDF and its associated query language SPARQL,44 RDF-based knowledge graphs can support managing and accessing heterogeneous data from different domains while enabling interoperable metadata. When using the expressive and machine-readable W3C SHACL55 (Shapes Constraint Language) validation language, users define and enforce complex constraints within schemas for high quality metadata.
Many general purpose RDF-based knowledge graph platforms are open sourced and made available as managed services such as the metaphactory platform [22] and LinkedDataHub66 or as downloadable artefacts such as metreeca77 to be deployed and used within organisations on premises. Some such as KGKT [28] are libraries with usage from a terminal or Jupyter notebooks while others such as Wikibase88 are for creating and maintaining knowledge bases from structured data. Authors in [37] surveyed tools and systems for semantic data integration. While such platforms support secured authoring, management and access of RDF-based knowledge graphs, most of them focus primarily on metadata. Data and knowledge engineers need to complement them with data storage and management capabilities. Jointly managing data, metadata and schemas within the same framework is key for keeping consistency between them, enabling high quality metadata and increasing the data longevity.
3.Blue Brain Nexus overview
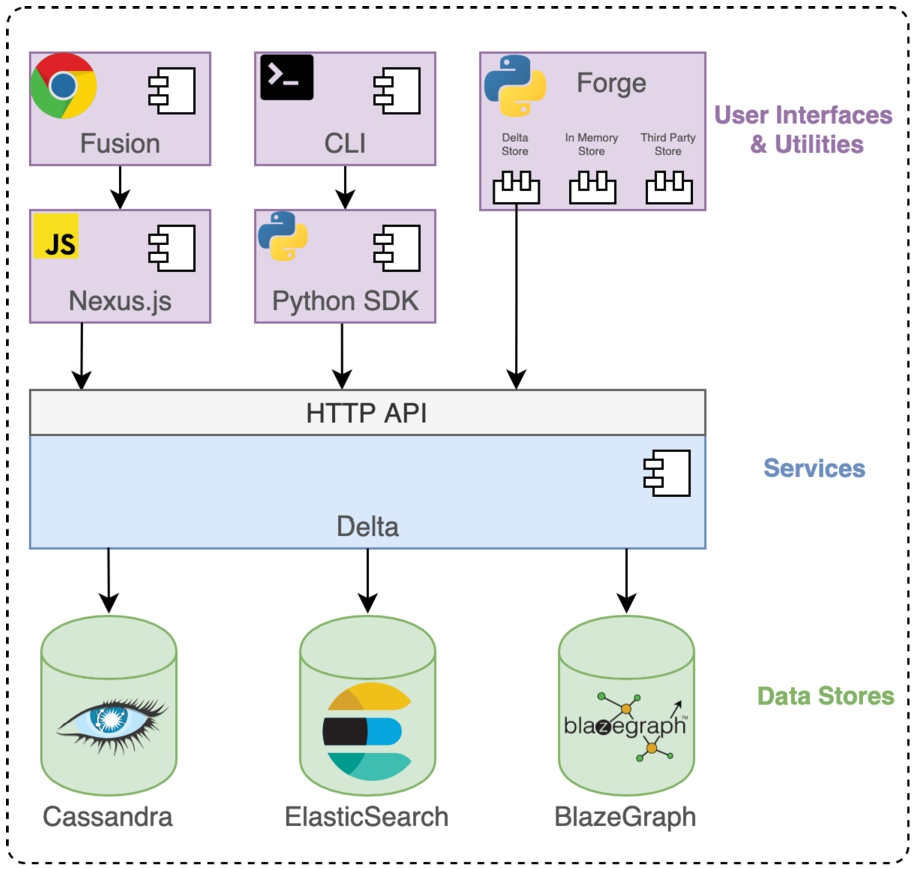
BBN is an ecosystem of integrated software components targeting different users and supporting them alongside the data-driven science iterative cycle. The ecosystem is made of three main components including: i) Nexus Delta, a set of services targeting developers for managing data and knowledge graph lifecycle; ii) Nexus Fusion, a web-based user interface enabling non-expert users to store, view, query, access and share (meta)data as well as expert users to manage knowledge graphs; and finally iii) Nexus Forge, a Python user interface enabling data and knowledge engineers to build knowledge graphs from various data sources and formats using data mappings, transformations and validations. An overview of the ecosystem is presented in Fig. 2.
Fig. 2.
An overview of Blue Brain Nexus ecosystem components: Nexus Delta uses off-the-shelf products as data stores (Cassandra, Elasticsearch, Blazegraph) and exposes as HTTP(s)-based API and to clients core functions and services such as Identity and Access Management as well as knowledge graph management and administration. Nexus Forge, a Python framework, supports a variety of data and knowledge engineering activities for building a knowledge graph. A Javascript library (Nexus.js) supporting a web application (Nexus Fusion). A Python library (Nexus SDK) and a command line interface (Nexus CLI). The entire stack can be deployed in container orchestration systems (e.g. OpenShift, Kubernetes), cloud providers (e.g. Amazon, Google) or on premises.
3.1.Services
Nexus Delta99 is a secure and scalable system carrying out core functions supporting the data-driven science iterative cycle and exposing them through a uniform API. It targets developers who want to minimize the cost of integrating and interfacing with a knowledge graph system as part of their existing infrastructure. Developers can thus build applications allowing users to securely create, manage and validate knowledge graphs. This component is based on a distributed and event streaming architecture, uniquely combining under a single API: a scalable data store using Cassandra,1010 a flexible RDF1111-based graph store using Blazegraph1212 and a powerful full-text search engine using Elasticsearch.1313 Nexus Delta enables high-level, declarative and expressive metadata representations and validation while still being able to turn them into a developer-friendly and secured RESTful API.
Nexus Delta is highly interoperable with existing data infrastructures by adopting open standards for authentication and authorization as well as for (meta)data storage, representation and access. Thanks to configurable data projections and automated full-text and graph-based indexing capabilities, it enables data and knowledge engineers to build custom views to share and access data. Support for distributed data with configurable storage backends enables the exploitation of existing cloud services and the integration with high-performance compute environments while maintaining secure data management.
3.2.User interfaces
Nexus Fusion1414 is a fully customizable, resource-centric front-end for Nexus Delta. It leverages Nexus.js, a JavaScript library that facilitates interfacing with Nexus Delta and building react.js based web applications. Nexus Fusion primarily targets non-expert users by lowering entry barriers to search, navigate and access a knowledge graph but also targets data and knowledge engineers enabling them to perform administrative tasks such as custom data views creation and query editing.
Nexus Forge1515 is a domain-agnostic and extensible Python framework enabling data scientists and knowledge engineers to build knowledge graphs from various sources and data formats by supporting the definition and execution of declarative data transformations and mappings, metadata validation based on the W3C SHACL recommendation and the storage and access of the resulting graph in-memory or in Nexus Delta.
3.3.Utilities
The Nexus Fusion and Nexus Forge components reuse a Javascript library (Nexus.js1616) and a Python Software Development Kit (Nexus Python SDK1717) respectively. These utilities provide developers with programming language specific wrappers around the Nexus Delta API to simplify the development of new applications. A command line interface (Nexus CLI1818) makes use of the Nexus Python SDK to provide developers with basic administrative access to Nexus Delta from a terminal.
In BBN, a simple knowledge graph creation scenario starts with the configuration of a secured project within an organisation just like in Github. By default, a project and all its content are only accessible to its creator with the ability to grant access to other clients and users. For example a project’s configuration can be the default storage media to be used to store actual data (e.g. local file system, Amazon S3 compatible object storage, remote POSIX file-system) or how the data should be indexed and viewed. Then the data (e.g an image in PNG format, a PDF, ZIP or CSV document) can be uploaded in the project and stored in the project’s configured storage media using one of the BBN user interfaces or utilities. The data can be described and linked with other data through metadata which can correspond to a file name, description or release date but also to a link to the file creator, license and other derived files. Validation applies to metadata and occurs before they get stored in the Nexus Delta’s primary store. Once the metadata are stored, the indexing processes controlled by the configured views are notified of the new metadata. The corresponding indices (i.e. in Elasticsearch and in Blazegraph) are then updated with the new metadata. The validation, storage and index update sequences occur for every metadata update in a project.
4.Nexus Delta
Nexus Delta supports a range of functions allowing secure storage, management, validation and connection of data from any domain in a knowledge graph. The system also allows users to create managed views (backed by Elasticsearch or Blazegraph) for searching all or a subset of a knowledge graph, shielding data and knowledge engineers from the complex tasks required to build and manage custom Elasticsearch indices or Blazegraph namespaces.
4.1.System design
Nexus Delta exposes to clients its core functions and services through a RESTful1919 API over hypertext transfer protocol (HTTP). It can be deployed in a clustered configuration to be able to scale horizontally on demand while being able to handle correctly the functions that require coordination such as change propagation to the managed views. The system uses Akka Cluster2020 for a decentralized, fault-tolerant, peer-to-peer based cluster membership following the gossip protocol2121 to randomly spread the cluster state as shown in Fig. 3. Active nodes in a cluster communicate on demand over the Transmission Control Protocol (TCP) or User Datagram Protocol (UDP) using a custom protocol provided by Akka Remoting2222 to achieve the necessary distribution of load and synchronization.
Apache Cassandra was chosen as an eventually consistent2323 persistent store such that the system:
– can store large amount of data via horizontal scaling;
– can handle arbitrary spikes in throughput for both read and write operations under low latency;
– favours availability over global strong consistency while retaining the ability to scale to arbitrary sizes.
Consistency is guaranteed at the level of a single resource (aggregate) and control is given to clients for executing coordinated changes across multiple resources like for example: uploading a file and creating a description of the uploaded file as a separate resource. The anatomy of Nexus Delta is depicted in Fig. 4.
Fig. 3.
Nexus Delta can be deployed in a decentralized clustered configuration where the nodes randomly spread the cluster membership via the gossip protocol. Akka Remoting over TCP or UDP is used to achieve distribution of load, consistency and synchronization.
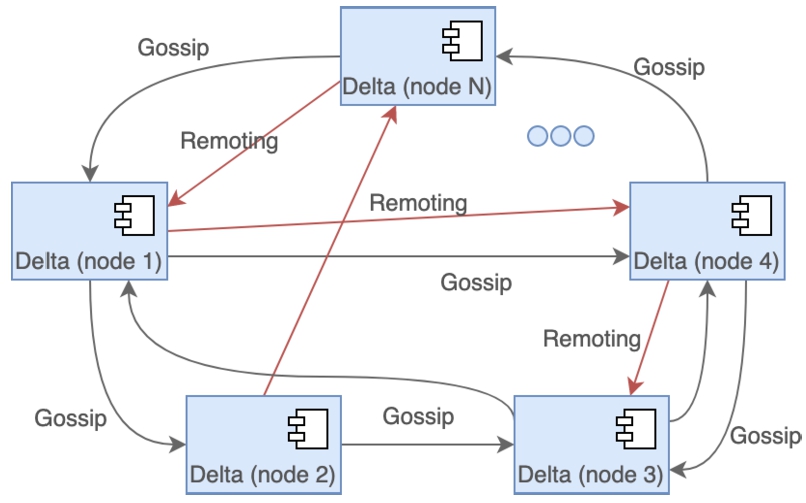
Fig. 4.
Anatomy of a Nexus Delta service. Each service has separate read and write models driven by specialized stores and asynchronous processes that project the data from the write model (the primary store) to an efficient read model backed by separate stores (e.g. Elasticsearch, Blazegraph). Asynchronous communication is realized by consuming the event log stream of an upstream service.
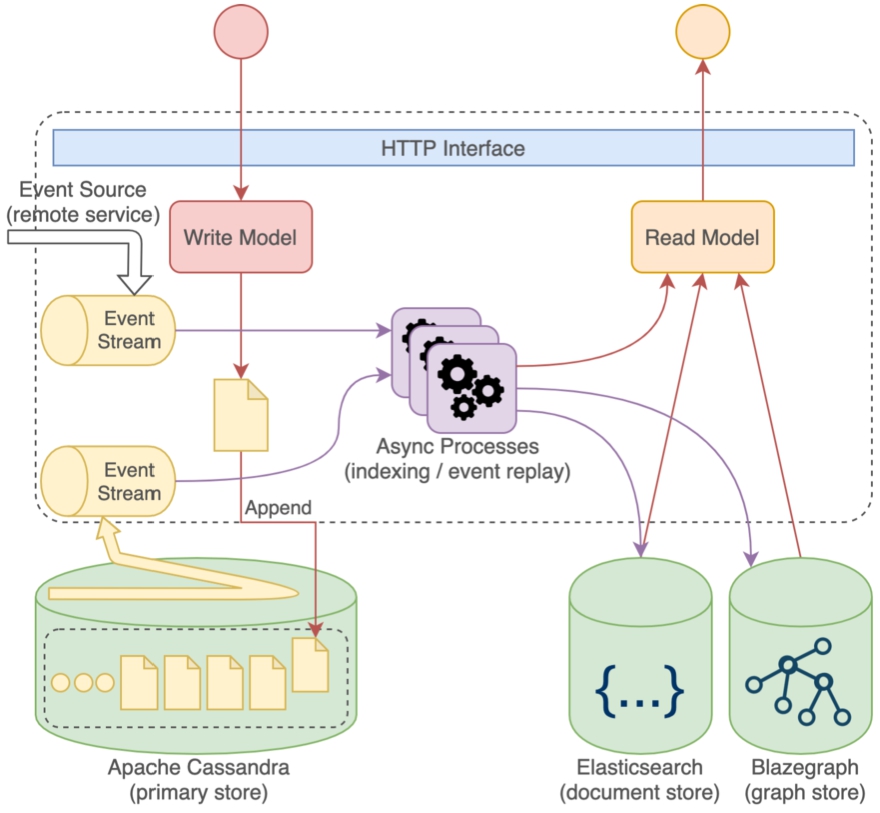
There is a clear separation between the read (indices) and write model (the primary store) of the system, following the command query responsibility segregation (CQRS) 2424 pattern. This allows for independent scaling of the read and write parts of the system, independent evolution of the models and the use of the appropriate internal (components) or external (stores) for efficient data access.
Request handlers translate requests pertaining to the write model into commands. Commands represent client intent for changing the application state (e.g. CreateResource). Before being evaluated, each command is validated for access, consistency and coherence with respect to the global application state. Evaluating a command produces an event that is appended to the global application event log. Requests pertaining to the read model are translated into queries that are being executed against the appropriate store. Direct resource current and past state queries are executed against the primary store (Apache Cassandra), SPARQL2525 queries are being executed against the triple store (Blazegraph) and general filtering, full-text search and any other document oriented specific queries are executed against the document store (Elasticsearch). Additionally, each service exposes its internal event log in a stable document format via Server Sent Events (SSE).2626
Nexus Delta uses an event sourcing persistence model where all changes to the global state of the system are recorded as a sequence of events. This event log is used for computing the current application state, reconstructing past states and coping with retroactive changes. Each resource in the system has its own virtual event log as a subset of the global event log as shown in Fig. 5. The implication of the chosen persistence model is that the system uses its database as an append-only store. Updates generate new events that are added to the log and data are never physically removed from the system, but rather marked as obsolete.
Fig. 5.
Data are recorded into the Nexus Delta system as an append only event log. Each resource state is derived from an implicit subset of the global event log.
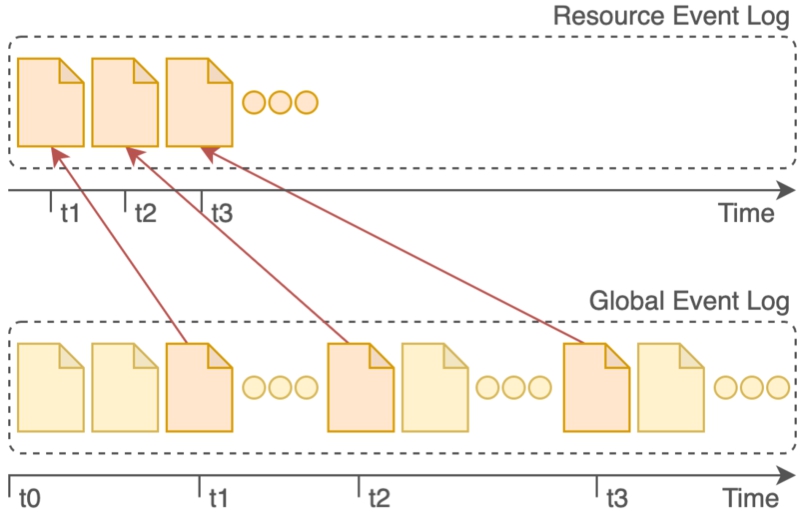
Nexus Delta ensures that additions to the primary store are synchronized with the systems used solely for querying purposes. The synchronization is performed as part of asynchronous processes (projections) that replay the event log, generate the necessary model and update each of the target systems. These processes keep track of their progress such that they can be resumed in case of global system failures, network partitions, cluster resizing or simple restarts (e.g. software updates). The processes are also capable of retrying individual updates with exponential backoff when the target systems become unavailable.
The asynchronous nature of the data indexing process provides some advantageous global system characteristics:
– Partial degradation instead of unavailability of the system when one of the dependent systems becomes unavailable. For example, if the Elasticsearch cluster becomes unavailable, only queries that target this store will be affected; the rest of the system continues to function normally.
– Natural bulkheading for arbitrary spikes in write operations. The throughput for ingesting data into the system does not affect the query performance.
– Optimal speed for propagating changes from the primary store to the target system (e.g. the speed of propagating changes to Elasticsearch is not affected by possible slower writes to Blazegraph).
– Indices can be rebuilt independently in an automated fashion when required (i.e. if a target system experiences data loss due to a hardware failure) by restarting the respective process.
– The read (query) part is eventually consistent; writes to the system propagate with a slight delay to the respective target systems.
4.2.Resource-centric RESTful API
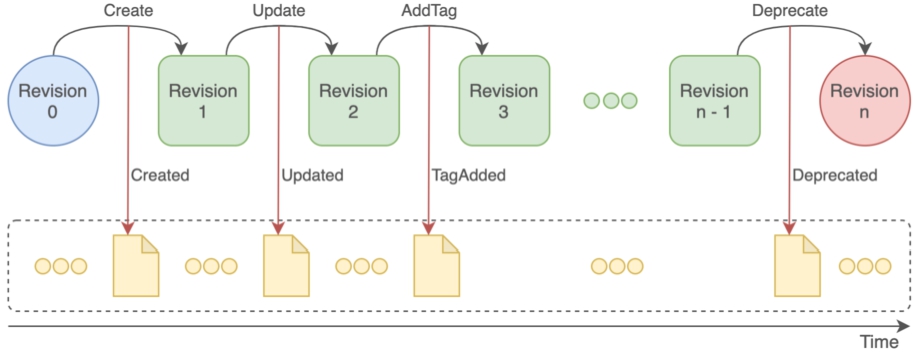
Nexus Delta is built following the REpresentational State Transfer (REST) architectural style where the client-system interaction is performed via access and manipulation of resources using stateless operations. Both system specific data (i.e a view configuration) and client data (e.g. files, metadata, schemas) are represented as resources with a lifecycle, as depicted in Fig. 6. A resource lifecycle is represented as an ordered series of state transitions recorded as events in the global event log. The state transitions correspond to resource creation, update, tag and deprecation and each of them yields a new revision. The current state (last revision) of a resource is computed by replaying all events pertaining to that resource in the order they occur.
Fig. 6.
An overview of the resource lifecycle in Nexus Delta and how state changes are recorded as resource events in the global event log.
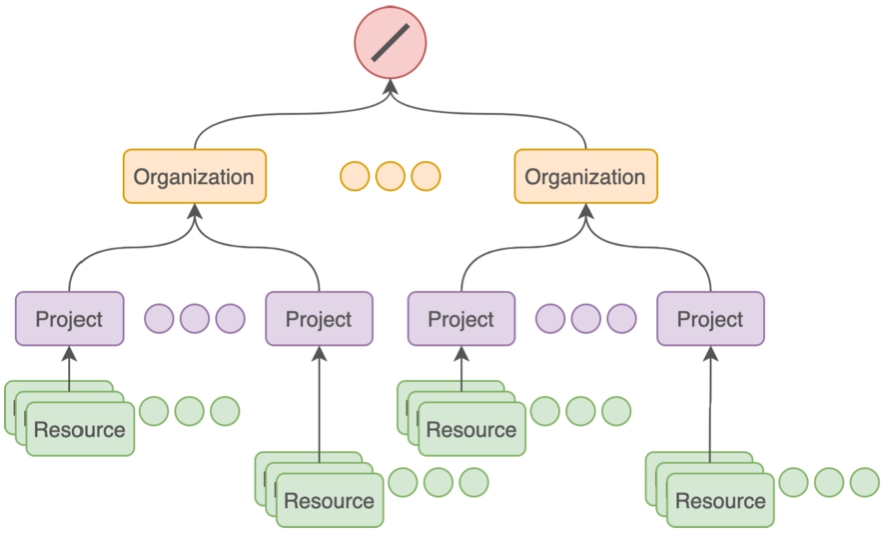
Resources are anchored to projects and organisations as shown in Fig. 7. Organisations and projects provide data boundaries allowing for logical isolation, grouping, varied configuration and access control policies.
Fig. 7.
Logical grouping of resources within projects and organisations in Nexus Delta.
The system uses a uniform positional addressing scheme determined by the resource type, scope, schema and identifier as shown in Fig. 8. This scheme allows for operating on a single resource or collections of resources using HTTP methods to express intent such as creation, update or listing.
Fig. 8.
Resource addressing and identification scheme in Nexus Delta. A project resource can define a collection of aliases and prefix mappings to shorten resource identifiers used in the addressing scheme (e.g. morphologies is an alias to “https://neuroshapes.org/morphology” and can be used as a segment for identification, either as a schema resource or the collection of resources constrained by it).

The resource_type segment represents a filter on the system supported resource types. The most general type is resources but additional ones are listed in Table 1.
Table 1
Supported resource types in Nexus Delta along with their descriptions
| Resource type | Description |
| realms | Resources that represent accepted Identity Providers (IdP) and their configurations. |
| identities | Read-only singleton resource providing information about an authenticated client (e.g. a user, a group). |
| permissions | Singleton resource defining the collection of supported permissions. |
| acls | Resources defining a three way mappings between identities, permissions and a target scope (global, organisation or project) |
| organisations | First level logical grouping of projects and resources managed in them. |
| projects | Second level logical grouping of resources. |
| schemas | Resources defining a collection of constraints (using W3C SHACL schemas) other resources can be validated against. |
| resolvers | Configuration resources for identifier-based resolution (i.e locating and fetching) of other resources (e.g JSON-LD context, schemas and ontologies using owl:imports). |
| views | Configuration resources controlling the metadata indexing processes and querying options. |
| storages | Configuration resources for defining file storage media. |
| files | Resources that represent user provided binary. |
| archives | Ephemeral resources for bulk data and metadata download as tarball archive file. |
| resources | A generic resource type representing user defined metadata and optionally constrained by schemas. |
The org_id and project_id segments represent a filter on the resource’s organisation and project scope. The schema_id represents a SHACL schema identifier allowing the system to validate resources upon creation and update (through POST or PUT) operations but also to select (through GET) all resources that conform to the constraints defined by the schema. Finally, the resource_id selects a unique resource within the parent scope. Resources of type views expose additional sub-resources corresponding to specific endpoints (e.g. documents/_search for an ElasticsearchView and graph/sparql for a SparqlView) for data search and access but expose also control sub-resources like documents/statistics or graph/offset.
All resources are unambiguously and uniquely identified by HTTP based internationalized resource identifiers (IRIs) within a given project. Clients can reuse existing identifiers (e.g. DOI2727 for publications or ORCID2828 for researchers) or let the system generate them. As using documented and management-details-independent identifiers is key to avoid common data access and collision issues [35], Nexus Delta-generated identifiers are independent of the system deployment address including the organisation, project and schema under which the resources are managed as shown in Fig. 8. Besides, using an HTTP-based IRI as part of another HTTP path is tedious and error prone because of the IRI’s length and the need to encode it. Consequently, the system enables resource identifiers to be shortened and compacted by means of aliases and prefix mappings defined in the project’s configuration.
All resource types except files have a JSON-LD2929 representation format. Files have a dual representation accessible through content negotiation: a JSON-LD representation for metadata such as file name and size and a binary representation for the actual file content. Additionally, both resources and files metadata can be viewed in N-Triples and DOT formats.
4.3.Authentication and authorization
The authentication and authorization functions are performed using four resource types: realms, permissions, acls and identities. Nexus Delta supports OpenID Connect,3030 OAuth 2.03131 and JSON Web Tokens (JWT)3232 allowing for a seamless integration with identity providers that follow the same open protocols. Several off-the-shelf products implement these protocols on top of the Lightweight Directory Access Protocol (LDAP), allowing members of an institution to authenticate to a Nexus Delta deployment using the same institution’s identity management system.
All HTTP requests accept an Authorization header with a JWT Bearer token as value when consuming the RESTful API. The general authorization flow is executed as follows:
– a provided token is validated against the identity providers defined through realms resources (issuer, signature, expiry, not before);
– the caller information (subject and group claims) is extracted and its identities (anonymous, authenticated with a provider, user and group membership) collected for further authorization;
– the collected identities are then compared to the configured access control lists to verify that the caller is authorized to perform the intended action such as listing resources in a given project or performing a file upload.
A detailed description of the authentication and authorization API along with examples are available as part of Nexus Delta documentation3333
4.4.Data validation
Knowledge graphs evolve over time as new data, schemas and ontologies get added and updated. The added data often come out of extract transform load (ETL) pipelines reading and transforming from various trusted or untrusted sources. Ensuring that those pipelines produce the expected outputs is key to maintain the knowledge graph quality as it evolves and to detect ETL pipelines’ anomalies. To enable data validation, Nexus Delta uses the W3C SHACL recommendation for defining, exchanging and enforcing constraints on metadata represented as RDF graphs. The validation using SHACL involves two types of resources:
– schema: defines in a shapes graph the constraints the metadata should conform to. A schema can define one or many shapes and is also serialized and exchanged in JSON-LD;
– metadata resource: the metadata RDF graph to be validated against the shapes of a given schema, also serialized and exchanged in JSON-LD.
The W3C SHACL recommendation only defines SHACL shapes and ways to logically combine them using Boolean operators (AND, OR, NOT, XONE). But shapes are almost never developed alone in production settings. It is therefore useful to be able to:
– reuse an already defined shape for modularity purposes: there is a need of an import mechanism telling a SHACL processor where to lookup, fetch and reuse an already managed shape. This is done through classic owl:imports mechanism that Nexus Delta resolvers exploit for SHACL schemas.
– include managed ontologies as part of the validation: it is key to be able to leverage class hierarchies during validation by injecting in the schema transitive closure of rdfs:subClassOf relations typically defined in ontologies.
– group and identify a collection of shapes in order to document and manage them (e.g. Create, Read, Update, Deprecate).
In Nexus Delta, a schema is a resource of type Schema and enables the above capabilities by using the following JSON-LD syntax. The corresponding JSON-LD context3434 is omitted in the example for simplicity.

Table 2
Schema main properties
| Key | Description | URI |
| @id | The identifier of the schema | |
| @type | The type of the schema. By default it is nxv:Schema | http://www.w3.org/1999/02/22-rdf-syntax-ns#ype |
| imports | Collection of schemas and ontologies identifiers to bring in the scope of the current schema when validating | http://www.w3.org/2002/07/owl#mports |
| shapes | The collection of SHACL shapes defined within the current schema | reverse of http://www.w3.org/2000/01/rdf-schema#isDefinedBy |
Table 2 details the main properties of a resource of type Schema.
Given a shapes graph (a schema) and a metadata graph (a resource) as inputs, the SHACL processor used in Nexus Delta starts by selecting the part of the metadata graph to focus on and then validates whether that part conforms to the shapes graph or not. The TopQuadrant SHACL API3535 written in Java and based on Apache Jena3636 is used as SHACL validator.
4.5.Data storage
Nexus Delta supports the storage of arbitrary binary data through files resources. As with other resource types, files are immutable once uploaded to the system and future updates represent new revisions. Compared to other resources that are represented as JSON, files differ in terms of size, materialization, handling during transport and how integrity is ensured. The files may have the following properties:
– The file size may be larger than the allocated process memory; to accommodate large file sizes the system uses a back-pressured streaming approach for both uploads and downloads.
– During upload the system computes a SHA-256 hash for the file and stores it as metadata along with the observed file size and the client provided media type. Clients can compare the resulting value of the SHA-256 hash with the expected value to assert the integrity of the operation (upload / download).
– The system automatically records two resource representations: the binary representation as submitted by the client and a JSON-LD representation for the provided and computed metadata. Content negotiation is used to discriminate between the two representations; the client can provide either the Accept: application/ld+json header to express the intent for retrieving the file metadata (the JSON-LD representation) or the Accept: */* header to express the intent of retrieving the binary representation.
– There are situations where the use of HTTP(S) for registering a file in the system is not feasible due to exceptional sizes (e.g. an image stack of a three-dimensional mouse brain tissue block with a size of 10 terabytes) or when using a shared clustered storage medium and the data already exist on the target storage but the system is not keeping track of it. In these situations a file can be directly materialized by making a reference to the existing file instead of performing a possible inefficient file upload.
Nexus Delta’s file specific API along with examples are available as part of its documentation.3737
The system supports multiple storage media for files. By default it uses a storage backed by the filesystem where Nexus Delta is running (either as a local drive or an arbitrary mount). The storage media supported for a project are controlled by resources of type Storage with more specific sub-types:
– DiskStorage – local filesystem mount.
– S3Storage – Amazon S3 compatible object storage.
– RemoteDiskStorage – remote filesystem mount via a HTTP(S) based integration service; this storage type relies on a remote HTTP service that exposes basic file operations on an underlying POSIX file-system. This is useful in organisations that are running a distributed network storage (e.g. Ceph, Gluster, GPFS, Lustre) that cannot be mounted directly on the system where Nexus Delta runs because of security or geographic location considerations.
4.6.Data access and sharing through views
In Nexus Delta, all or a subset of a project’s resources can be selected and projected to specific views exposing endpoints for search, aggregation, statistics and navigation. The processes that control these projections are defined by resources of types views. While focusing on process automation, security and control, Nexus Delta uses off-the-shelf open source products to provide two types of views: a document oriented view through Elasticsearch and a graph view through SPARQL 1.1 endpoints with Blazegraph. This built-in view support occurs per project and the created views inherit the project’s ACLs. Clients can thus select data and quickly put up endpoints for sharing and disseminating them while controlling their access.
Views define and configure criteria to select resources (e.g. by type, by schema or at a specific revisions using tags) and how they should be indexed (e.g. specific metadata to index). Two types of views are created per project by default upon project creation: an ElasticsearchView and a SparqlView. Additional ones can be created based on clients and user requirements. Table 3 details the different configuration options within an ElasticsearchView and a SparqlView while the documentation3838 details views specific API.
Table 3
The different configuration options within an ElasticsearchView and a SparqlView
| Option | Description | View type |
| resourceType | Only resources of the listed types will be selected and indexed. | ElasticsearchView, SparqlView |
| resourceSchemas | Only resources conformant to the listed schemas will be selected and indexed. | ElasticsearchView, SparqlView |
| resourceTag | Only resources with the provided tags will be selected and indexed at the state corresponding to the tags. | ElasticsearchView, SparqlView |
| includeMetadata | A boolean flag indicating whether to index (true) or not (false) Blue Brain Nexus added metadata (e.g. revision, createdAt, …). | ElasticsearchView, SparqlView |
| includeDeprecated | A boolean indicating whether to index (true) or not (false) deprecated resources. | ElasticsearchView, SparqlView |
| mapping | A JSON object corresponding to an Elasticsearch mapping document. It configure how Elasticsearch should index the resource. | ElasticsearchView |
| sourceAsText | A boolean indicating whether to index (true) or not (false) the JSON-LD payload of a resource as a string in Elasticsearch. | ElasticsearchView |
The indexing processes controlled by the views are executed incrementally by replaying the event log for each individual project and applying the necessary changes in the respective target system. As resources get created, updated and deprecated, each view incrementally applies the changes to maintain the indices up-to-date. The progress of indexing data is persisted to survive system restarts and presented to clients and users through the REST API and the Nexus Fusion interface for monitoring. If a view configuration is updated, the progress is reset and the process starts over with the new configuration.
Each view exposes its query endpoint as a sub-resource. A _search endpoint is exposed in the case of an ElasticsearchView while a sparql endpoint is exposed for a SparqlView. Queries to a view endpoint are first checked for authorization before being dispatched to the target system.
When querying, projects’ boundaries can be crossed by means of aggregate views of type AggregateElasticsearchView3939 and AggregateSparqlView4040 avoiding thus silos for resources managed in the system. They combine multiple views from one and/or many projects allowing the access of resources across projects. An aggregate view describes to which projects and views submitted queries are to be dispatched. Crossing projects’ boundaries implies additional authorization checks as the client needs to have access to the selected projects and views in order to run the submitted query.
The system also supports automated, incremental, multi-project and cross-deployment metadata aggregation, transformation and projection through CompositeViews which combine the capabilities of both Elasticsearch and Blazegraph.
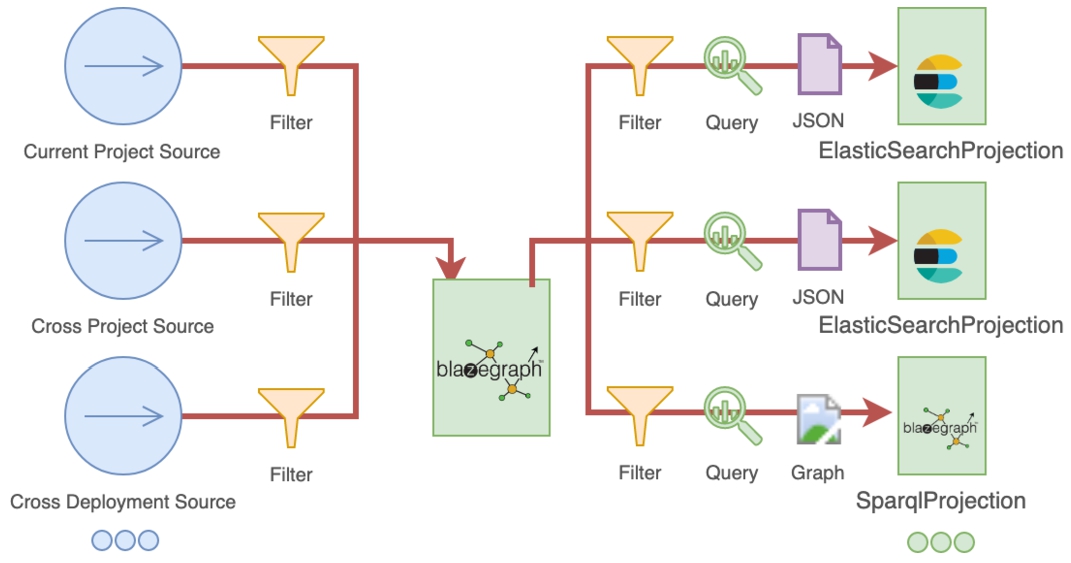
CompositeViews may have multiple sources of metadata (multiple projects in the current Nexus Delta cluster and/or projects in a different Nexus Delta deployment) and may project data to multiple specialized indices at the same time. The execution flow depicted in Fig. 9 is incremental as with the other view types where:
– metadata is aggregated from all the sources in an isolated Blazegraph namespace in a ordered series of changes (events in the Nexus Delta log). Filtering criteria can be applied to metadata sources to discard changes to resources that are not relevant;
– for every change a set of SPARQL CONSTRUCT queries are executed on the accumulated metadata to collect sets of triples (one query per index). Unlike ElasticsearchViews and SparqlViews that are resource centric, the queries executed to collect triples in CompositeViews can span any number of resources;
– the collected triples are then indexed in the target systems. In the case of SparqlProjections the sets of triples are bounded in named graphs and stored as is. In the case of ElasticsearchProjections a context is applied to sets of triples to frame them in JSON-LD documents that are finally stored.
Fig. 9.
The execution flow of CompositeViews indexing processes. Multiple sources are used to aggregate metadata in an isolated Blazegraph namespace. Queries are executed for each change to collect relevant sets of triples that are further stored in SPARQL based indices or transformed to JSON-LD and then stored in Elasticsearch indices. Filters can be applied on sources or projections to discard irrelevant information.
In terms of querying capabilities, CompositeViews allow querying the aggregation namespace, all indices of the same type (Elasticsearch or SPARQL) at once or individual indices. Due to the non-deterministic ordering of changes collected by a CompositeView (collection of changes may execute at different rates or sources may be unavailable) queries that do not target a single resource may return different results, depending on the order of the collected changes. CompositeViews accommodate a healing mechanism where the index process can be restarted in place at fixed intervals. Additionally, clients can choose to restart indexing for a single source, a single projection or the entire process.
4.7.Extension points
Supporting new requirements and use cases may require extending Nexus Delta capabilities. This could involve using a different system or application to organize and search data differently. By exposing the event log over HTTP(S) using Server-Sent Events, the system allows clients to replicate the internal streaming approach and the push-based asynchronous communication for resource projections during views creation. These clients can therefore maintain third party indices supporting potentially different use cases and system behavior as shown in Fig. 10.
Fig. 10.
Clients and services can subscribe to the event log stream, query the knowledge graph for additional information, generate derivations to be registered back into the system or maintain third-party indices.
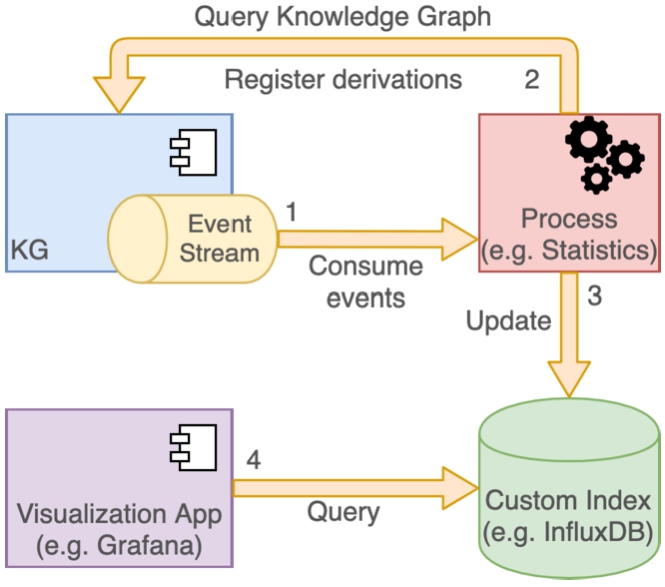
A very simple example of a system extension would be to compute and visualize statistics of the data in Nexus Delta (e.g. data size and count per type per project) and their evolution over time. For tracking metrics over time, an obvious choice for an index would be a time series database, like InfluxDB.4141 A third-party tool could:
– subscribe to the event log stream;
– collect the required information for each changed resource by querying the knowledge graph; a SPARQL SELECT query would allow the collection of the data size as well as the types and deprecation status of the resource for which the event was emitted;
– record a new entry in the time series database using the creation date as the metric timestamp, the data size as the metric value and the types and originating project as metric tags.
Since the subscription to the event log stream is continuous, as new data are added to the system, the third-party tool will automatically propagate the information in the time series database.
4.8.Synthetic benchmarks
Nexus Delta has been tested to verify that the implementation achieves the design goals with respect to a set of key non-functional requirements:
– horizontal scalability of the system;
– low latency resource ingestion and access;
– partial degradation of function when dependent systems (e.g. Elasticsearch and Blazegraph) become unavailable.
The tests have been executed for Nexus Delta version 1.4.2 in a reference environment made of a Kubernetes cluster hosted in Amazon Web Services (AWS). A load injector was configured with 8 vCPUs, 32 GiB RAM as a separate instance in the same virtual private cloud (VPC). The resource allocation and deployment configuration within the Kubernetes cluster was as follows:
– 1–12 node Nexus Delta cluster (8 vCPU, 8 GiB HEAP);
– 1–12 node Cassandra cluster (version 3.11.9 – 3.5 vCPU, 12 GiB HEAP, 250 GiB EBS storage of type GP3 with 10,000 provisioned input/output operations per second (iops));
– 1 node Blazegraph (version 2.1.5 – 2 vCPU, 4 GiB HEAP);
– 1 node Elasticsearch (version 7.4.0 – 4 vCPU, 8 GiB HEAP).
Prior to the test execution, indexing was disabled and a reasonable amount of data was injected into the system to ensure that it behaved well under a typical volume; specifically 120 million resources were created across 27 projects using an exponential distribution. The equivalent number of triples (22 for each resource plus 11 system metadata) was approximately: 4 billion. The tests covered read and write operations and excluded queries to the third-party indices (Elasticsearch and Blazegraph) as the goal was not to benchmark these systems.
The results are presented in Figs 11 and 12.
Fig. 11.
Throughput of read/write operations scales up with the size of the Nexus Delta and Cassandra clusters.
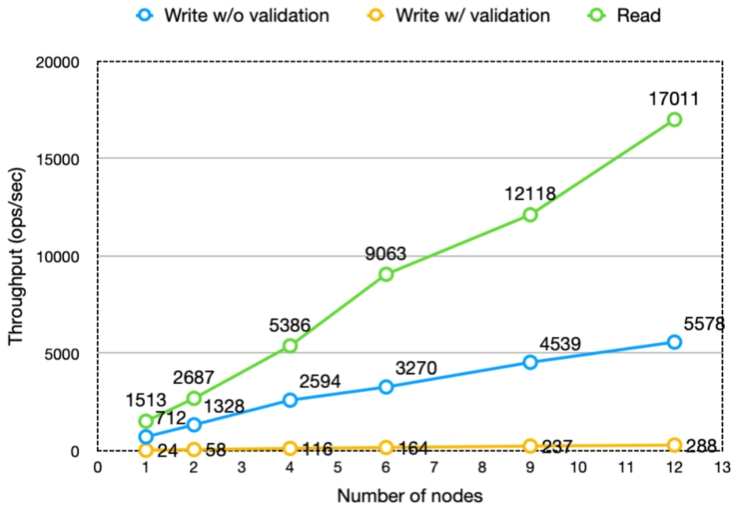
Fig. 12.
Latency of the requests (response times shown for the 50th and 95th percentile) is maintained in the same range as the size of the Nexus Delta and Cassandra clusters is scaled up.
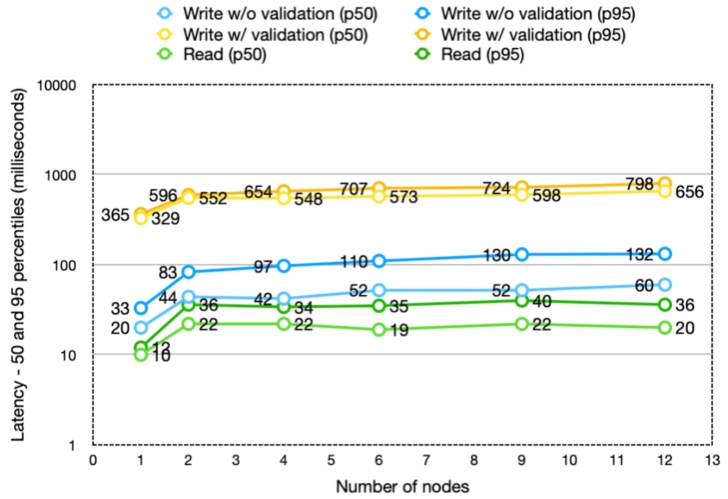
The results showed that the throughput scales almost linearly with the number of Nexus Delta nodes while maintaining the latencies in the same range. There is a significant difference in throughput between write requests with validation and those without. The reason is that validation is a very heavy function compared to the other functions of the system.
In most cases the increase in latency is minimal as adding nodes to the cluster increases the necessary data exchange between nodes when handling requests. The chance for the required data to be handled by the node that accepts each request decreases from 100% (single node), 16% (six-node cluster) to 8% (twelve-node cluster). If executing the request implies interaction with multiple resources (e.g. in the case of creating a resource with validation where the schema has import definitions) the chances drop close to 1%.
Taking parts of the system offline showed that the system continued to function with reduced availability as follows:
– when taking a Cassandra node offline, the system continued to function under expected conditions – full function;
– when taking a Delta node offline, the system continued to function under expected conditions – full function;
– when taking the Elasticsearch node offline, the system paused the indexing process, resource listing operations and direct Elasticsearch queries became unavailable; the system recovered immediately after bringing the node back online;
– When taking the Blazegraph node offline, the system paused the indexing process and direct Blazegraph SPARQL queries became unavailable; the system recovered immediately after bringing the node back online.
5.User interfaces
5.1.Nexus Fusion
Nexus Fusion is the web interface that enables users to interactively use Nexus Delta. Users can easily login and securely access organisations, projects and resources according to their permissions. In a given project, users can upload new data, create new resources and easily search for specific resources by their type or schema. Each listed resource can be selected and viewed thanks to a customizable page. Furthermore, Nexus Fusion enables data and knowledge engineers to interact with project views, more specifically, listing existing views, creating new views, monitoring views’ indexing progress and querying both SparqlView and ElasticsearchView.
The Nexus Fusion interface has been developed to be resource-centric and to enable users to search and discover managed resources. This has been made possible by enabling the presentation of resources catalogued in a knowledge graph using Studios. A Studio is a user configurable component where a collection of resources, resulting from a SPARQL query run against a SparqlView can be presented as a named Dashboard. In order to offer additional structure to a set of Studios, Dashboards can be grouped in named Workspaces. Finally, the set of available Studios can be discovered by users based on their permissions. Nexus Fusion provides a framework where resource-specific plugins can be developed, integrated and configured by software engineers to extend the set of BBN features to cope with specific requirements or present specific resources in a customized way.
Fig. 13.
A Studio in Nexus Fusion has workspaces (e.g. the “Neuron electrophysiology” workspace in (a)), which in turn have dashboards (e.g. the “All” dashboard in (a) listing all neuron electrophysiology recordings). A dashboard represents custom SPARQL query results displayed in an easy to read tabular format. Each entry in the table is a Nexus Delta resource. By clicking on a resource, users will be loading and viewing the plugins that have been configured for that type of resource (b).
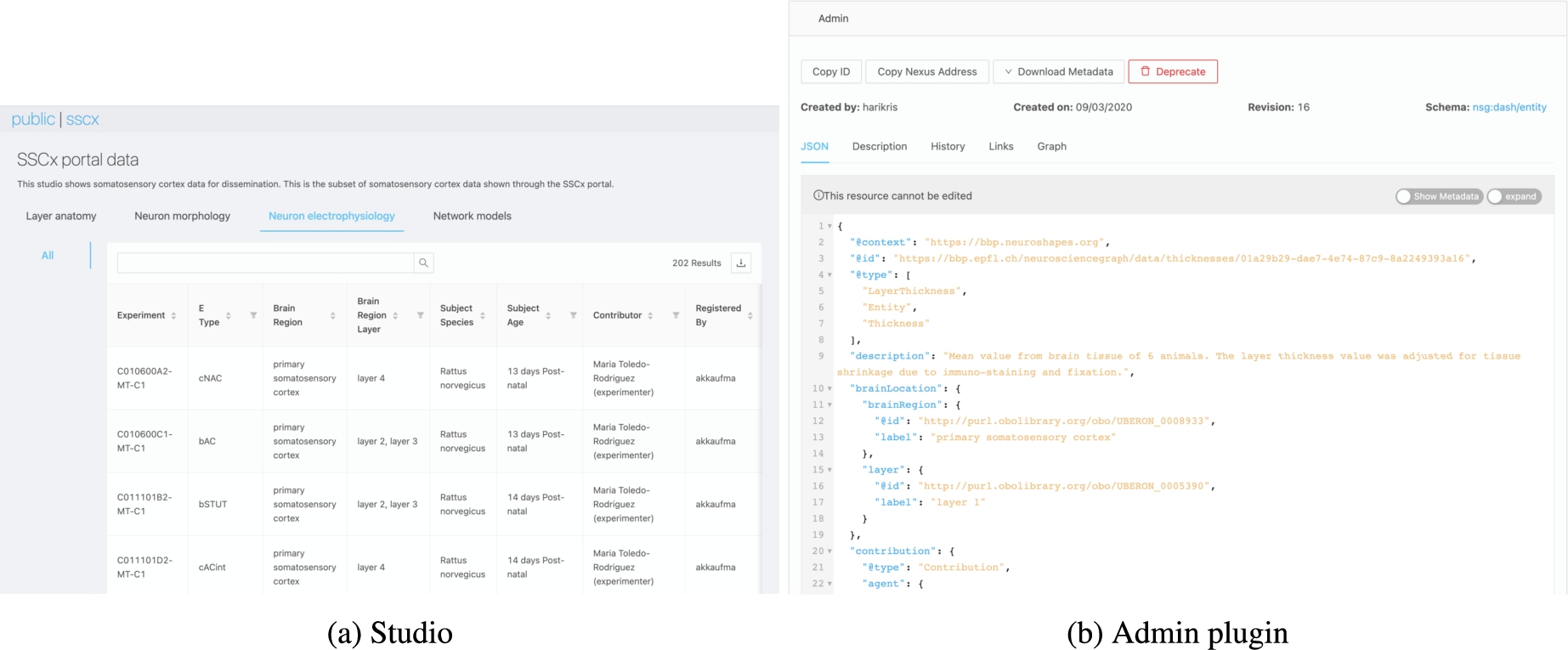
Fig. 14.
Plugins have been developed at Blue Brain Project for specific types of resources. Scientists can interactively explore 3D neuron morphologies through a plugin that reads and presents morphology data (a). Another plugin enables scientists to browse all static electrophysiology recordings for a specific neuron (b), with the ability to toggle to an interactive plot of the electrophysiology recordings.
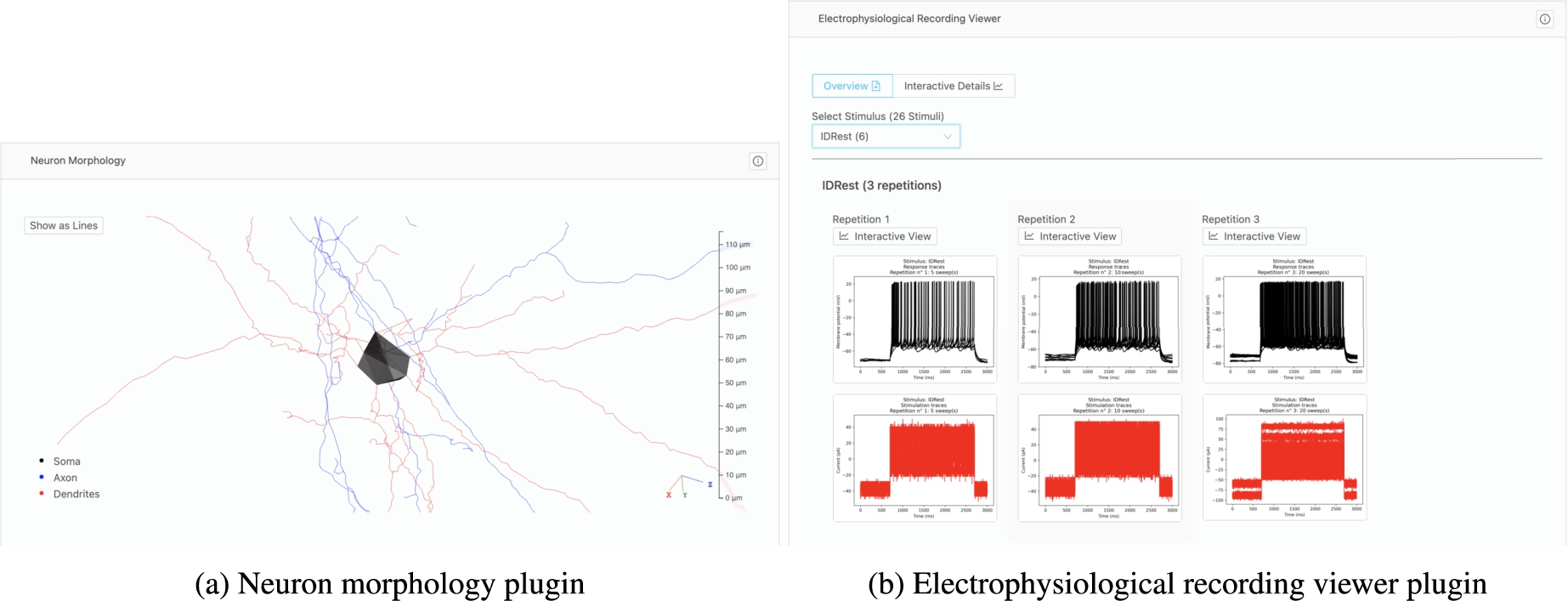
Thanks to its flexible and customizable nature, Nexus Fusion is extensively used within Blue Brain as many Studios have been configured as shown in Fig. 13, for example, to enable BBP scientists to search and access experimental data and models for reuse. Many plugins have been developed with BBP scientists to meaningfully present specific scientific datasets. Figure 14, for example, shows a neuron morphology 3D shape (a) and an interactive neuron electrophysiology viewer plugin (b). All these capabilities combined address the identified Data discovery and Humans in the loop challenges related to searching, navigating and accessing linked data for expert users.
5.2.Nexus Forge
Building knowledge graphs from various sources and data formats often involves data scientists and knowledge engineers to develop pipelines, whether automatised or prototyped in Jupyter notebooks, performing tasks to read, shape and map input data to structures that conform to schemas and ontologies modeling a targeted domain. With Nexus Forge, users can leverage a high level and simple Python interface4343 to:
– Load and access W3C SHACL specifications for specifying and enforcing (meta)data constraints. This capability addresses the data preparation challenges.
– Define, execute and share declarative mappings to transform data from different sources and formats to a targeted format and structure potentially conformant to defined data models. Mappings are predefined rules that encode the logic on how to transform data from a specific source so that they conform to a specific schema. A dictionary mapping language is supported.
– Validate the transformed data addressing data preparation challenges by making sure transformed (meta)data meet expected shape and quality as defined in schemas.
– Store the resulting transformed and validated data in stores such as Nexus Delta.
– Search and download (meta)data from the resulting knowledge graph.
6.Seminal use case: supporting biologically detailed simulation of the rodent brain at Blue Brain
Fig. 15.
The Blue Brain Project leverages Blue Brain Nexus to integrate a broad variety of heterogeneous data across multiple data centers. Furthermore, scientists and engineers at Blue Brain leverage the (meta)data integrated to power multiple scientific applications.
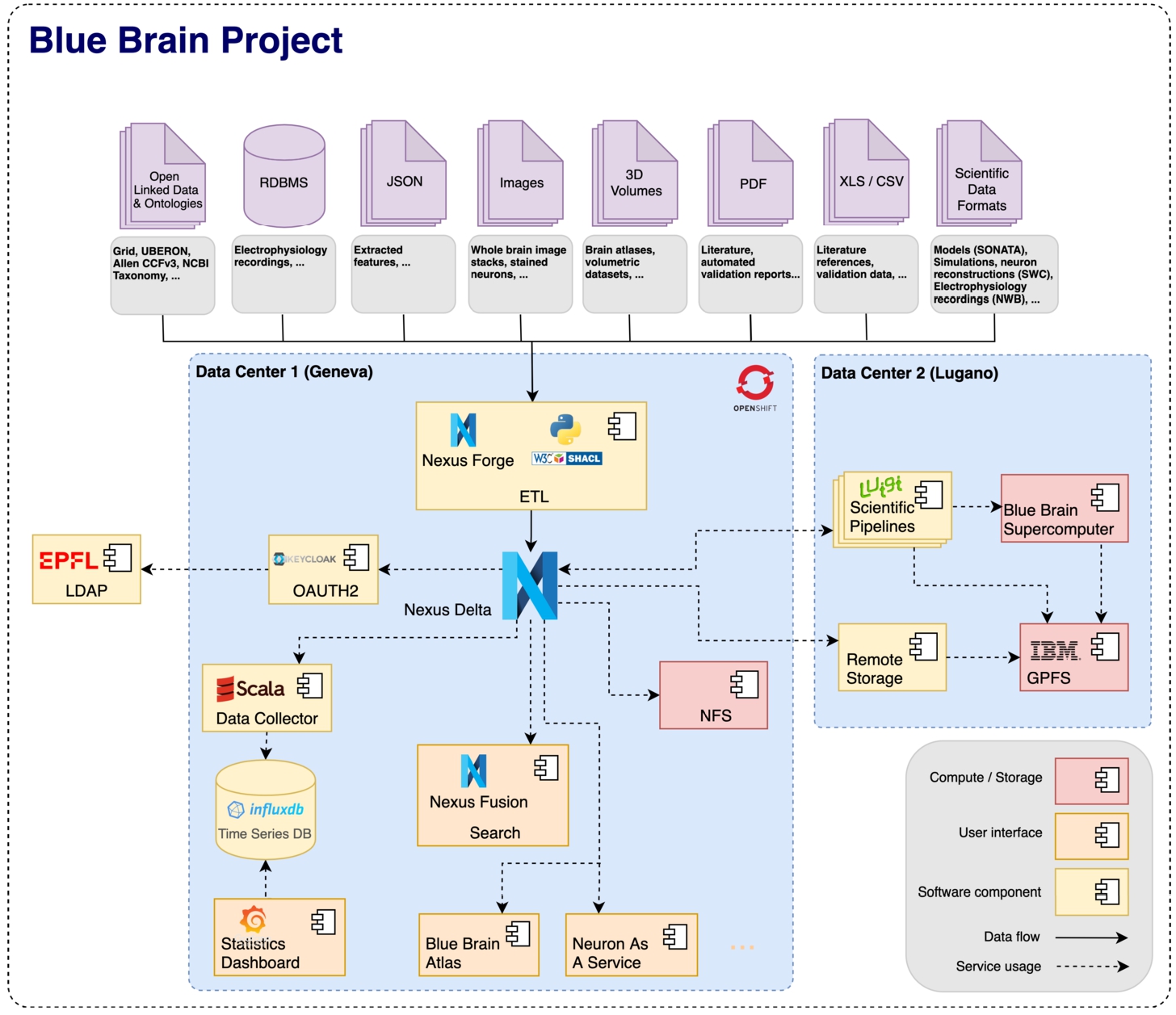
The original and main driving use case for designing and developing BBN is to support biologically detailed simulation of the rodent brain at Blue Brain.
At Blue Brain, BBN is deployed on premises and on a container cluster manager such as kubernetes which interfaces for authentication with the organisation’s LDAP system. It interoperates with the organisation’s parallel file system (GPFS) for read/write operations with direct connectivity to the Blue Brain supercomputer.4545 BBN is used by different applications and workflows targeting different users as shown in Fig. 15.
BBN allows Blue Brain’s scientists as well as data and knowledge engineers to perform the following tasks:
– Manage neuroscience data while tracking their provenance: the data are acquired from different sources both internal and external. Examples of data include neuron morphologies, neuron electrophysiological recordings, parameters from literature or brain atlases. Acquired data are first curated with metadata conformant to SHACL schemas such as the ones from Neuroshapes4646 including the Minimal Information about a Neuroscience DataSet (MINDS) [26] giving information about the subject from which the data were generated, the protocol used, the data type, the license, the data distribution (where to download the actual data) and brain location corresponding to a brain region name and/or precise coordinates within a brain atlas. Provenance of experimental data, models and simulation results is tracked using W3C PROV-O.4747
– Search and reuse curated data to derive new data and build models. Data and models can be searched using different metadata including types, species, brain regions, cell types, contributing laboratories, and protocols.
– Build custom web applications enabling visualization of neuroscience data such as neuron morphologies, electrophysiology recordings, and brain atlases.
Table 1 in the supplemental material details how BBN supports key data and knowledge management challenges at BBP.
7.Adoption of Blue Brain Nexus
BBN has already been deployed to address various use cases alongside the data-driven science iterative cycle in neuroscience, psychiatry and open linked data.
7.1.Knowledge graph for neuroscience data at the Human Brain Project
The Human Brain Project (HBP)4848 is a European flagship project with a ten-year horizon, aiming to understand the human brain and to translate neuroscience knowledge into medicine and technology [3]. Accordingly, it aims to build a collaborative information and communications technology-based scientific research infrastructure to allow a transdisciplinary community of researchers from over 130 institutions across Europe to share data and knowledge in the field of neuroscience, computing and brain-related medicine [50].
To enable thousands of scientists to find and publish diverse data across brain scales and modalities, the HBP has developed the EBRAINS Knowledge Graph platform4949 which uses BBN as shown in Fig. 16.
Fig. 16.
Within the EBRAINS Knowledge Graph (version 2), BBN is used to integrate (meta)data before being further indexed into purpose-built services in order to drive distinct user applications.
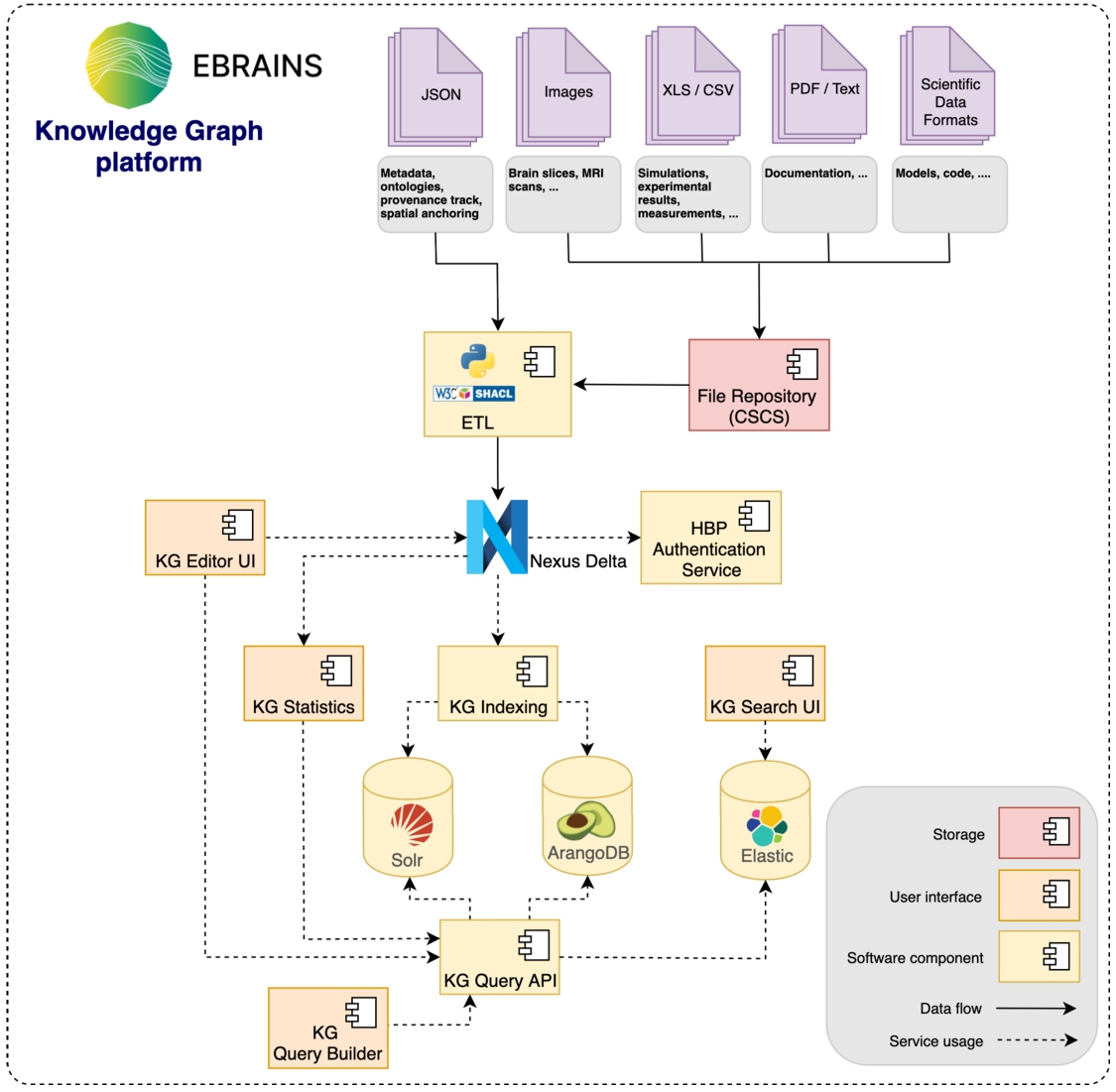
The HBP has developed four applications on top of the knowledge graph (KG) to drive different use cases.
KG Editor: This application is primarily used by the HBP Knowledge Graph team to integrate data into the EBRAINS Knowledge Graph. It allows users to create entities in the knowledge graph, edit and publish their metadata for public consumption.
KG Search:5050 This application enables researchers to find data that has been shared in the EBRAINS Knowledge Graph. It provides the following functionalities: data navigation by type (datasets, person, species), direct data download, and finding of artefacts related to a person or institution, with public or restricted access.
KG Query API: This application is intended for developers who want to integrate their own tools with the EBRAINS Knowledge Graph. It features a graphical interface that allows users to design their query visually. Most HBP applications consume data through this API.
KG Statistics: This application enables users to inspect visually the content of the EBRAINS Knowledge Graph. This application also allows users to configure access rights and monitor the platform deployment.
Table 2 in the supplemental material details the key EBRAINS Knowledge Graph platform challenges addressed by BBN.
7.2.Integrated clinical and research data management at the Krembil Centre for Neuroinformatics
The Krembil Centre for Neuroinformatics (KCNI or Krembil Centre) is an interdisciplinary, computationally focused research institute, located within the largest mental health hospital in Canada, the Centre for Addiction and Mental Health (CAMH) in Toronto. A core mandate of the Krembil Centre is to advance clinical and translational research by supporting advanced data management and analytics. KCNI supports a centralized source data management system, based on the Ontario Brain Institutes Brain-CODE platform [46], incorporating research-domain specific databases for neuroimaging (XNAT5151), omics/molecular data (LabKey5252) and assessments (REDCap [24]). CAMH is a Healthcare Information and Management Systems Society (HIMSS) Stage 7 hospital, with a fully implemented electronic medical record system, (provided by CERNER5353) that is accessible through a structured data warehouse. These clinical records are of significant value for clinical operations, physician decision support, as well as clinical and basic research. However, the standard commercial medical record implementation is tuned for acute rather than longitudinal behavioral health, and does not incorporate models of the care pathway. The Krembil Centre has deployed BBN in two distinct environments as shown in Fig. 17.
Fig. 17.
The Krembil Centre utilizes two distinct instances of BBN to facilitate the secure organisation of clinical records and research study data independently, employing a shared standardized schema to facilitate bi-directional data integration for translational clinical-research.
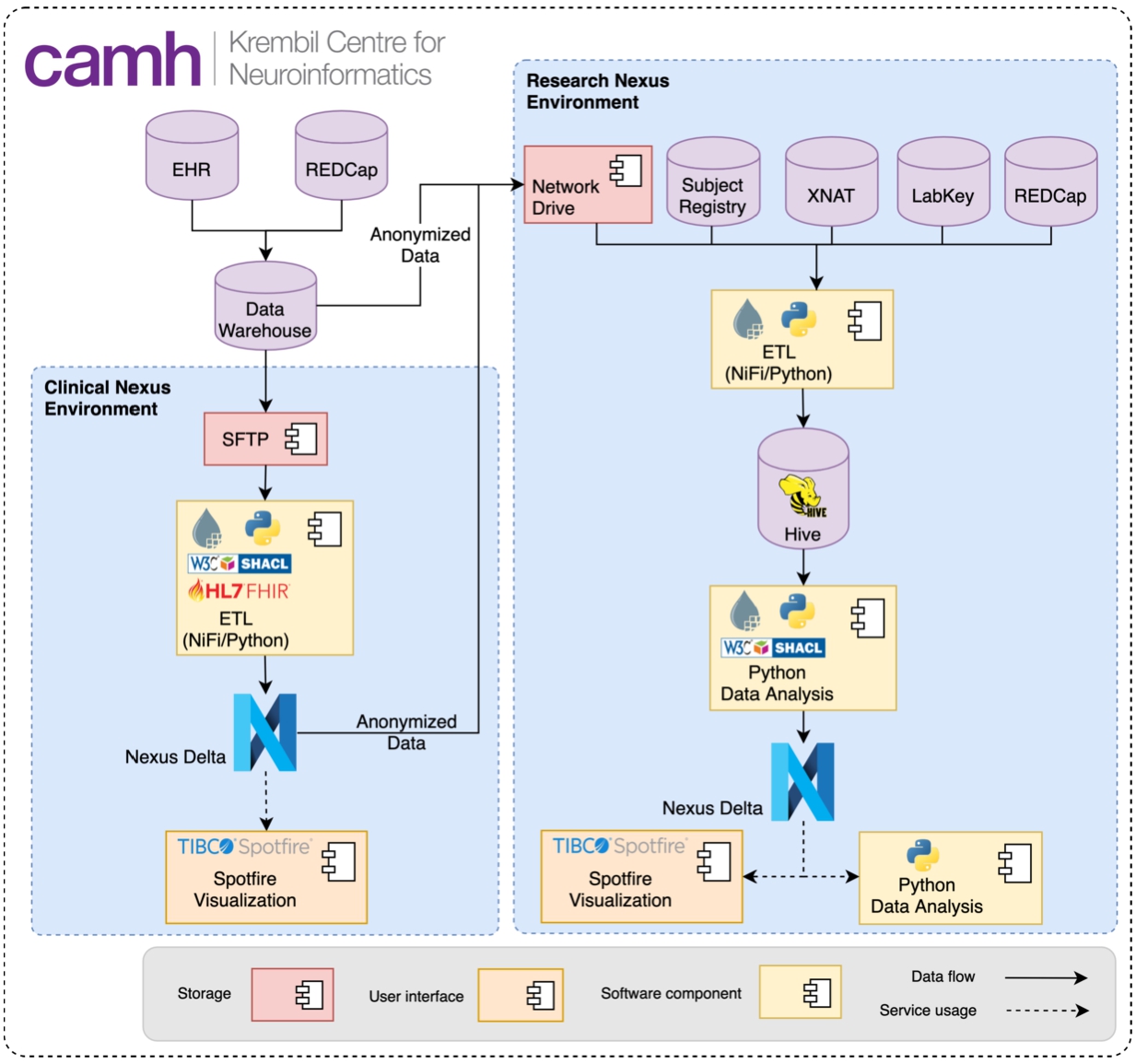
In a dedicated clinical environment, the CERNER schema was mapped to the interoperability standard Fast Healthcare Interoperability Resources (FHIR)5454 and made available for use by clinical operations and enable physician feedback and decision support. A clinical instance of REDCap for self-assessment data collection is integrated with FHIR via the MEDRED ontology [12]. Research data are similarly modelled with schemas associated with domain-specific requirements, including the Neuroimaging Data Model (NIDM5555) to represent data in a flexible semantic framework. BBN provides the core underlying semantic interoperability layer to facilitate data integration both across research studies and between clinical and research instances, leveraging common schemas and query functionalities. This capability allows for complex queries across data domains and structural hierarchies of the brain, integrating real-time clinical record information towards a rich knowledge commons to apply to accelerate discovery and care.
Electronic medical records (EMRs) are typically deployed as transactional systems designed for managing acute medical care and supporting billing requirements. This framework is not ideally constructed to handle the longitudinal needs of behavioral and mental health care, wherein patients require multiple, often recurring, encounters towards treatment and outcome management. Furthermore, it is essential for the practice of measurement-based and data-driven care to accurately present integrated care pathways to enact standardization and determine both adherence and diverging patient trajectories. The clinical instance of BBN implemented at the CAMH, allows the connection of disparate EMR data structures into an integrative holistic view of a patient. Through the adoption of a semantic graph framework, it is possible to more accurately model the patient as a persistent entity, the structure of modern behavioral health care delivery and their relative relationships.
BBN further supports iteration of data models, essential to the continuous process as pathways are modified in response to research discovery and refinement in clinical practice. This allows for a rich and dynamic care model that evolves over time. Additional benefits arise down-stream through the capabilities of semantic queries that can traverse the complex and interlinked graph of the patient population, to draw out key information and inferences.
Table 3 in the supplemental material summarizes the key challenges addressed by the deployment of BBN within the Krembil Centre.
7.3.Research Data Connectome Project
Fig. 18.
The Research Data Connectome uses BBN to implement a linked data pipelines allowing the project to extract, transform, validate, store and connect in a knowledge graph research data from different data providers’ repositories. The data can then be shared through BBN or through dedicated Web-based Services.
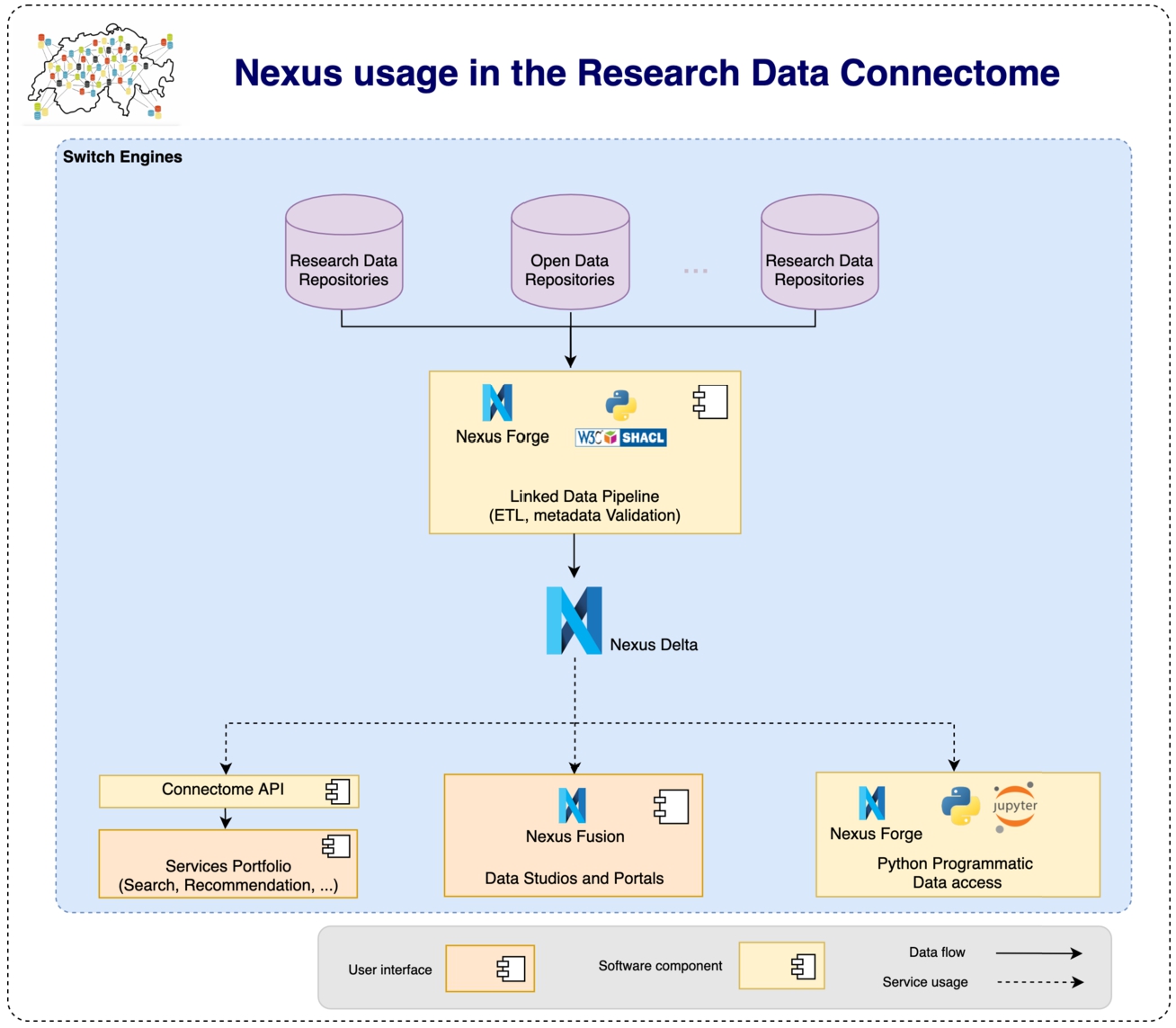
The Research Data Connectome project originated from SWITCH5656 innovation activities aiming at providing Switzerland researchers and universities with key infrastructure components, resources and services to achieve FAIR (Findable, Accessible, Interoperable, Reusable) research data and knowledge discovery as well as dissemination. The project main goal is to connect different type of data from various providers including research repositories across Switzerland and enable Switzerland’s researchers to discover them for reuse that could enable collaboration opportunities across different domains. To achieve such goal, SWITCH brought together many partners from different disciplines including data providers (including FORS,5757 DaSCH5858), research laboratories (including eXascale Infolab,5959,6060 SATW,6161 BCUL,6262 SARI6363 and Blue Brain6464) as well as individual researchers to design, implement and deploy a knowledge graph building and management pipeline [14] made of a set of resources, systems and services encompassing a:
– set of common SHACL schemas and ontology (RESCS Ontology6565) that made the knowledge graph6666 schema allowing the project to build applications and services while being shielded from the variety of (meta)data format and structures from upstream repositories;
– set of Guidelines and standards for research data quality;6767
– data extraction, transformation, validation and load pipeline6868 that normalise, structure, map and validate data from repositories to the knowledge graph schema;
– secure and scalable knowledge graph management system infrastructure to store, version and provide access to the resulting mapped data;
– set of end-user targeted services to discover, reuse and share stored data and knowledge.
The BBN ecosystem was chosen and deployed during the first pilot phase of the Research Data Connectome project after a review [14] of alternative open source solutions. Figure 18 shows how BBN is deployed at SWITCH.
Table 4 in supplemental material gives more details about the key challenges faced by the Research Data Connectome project and the BBN capabilities that address them.
7.4.Adoption discussion
BBN can be deployed, ran and managed locally, on premises or in the cloud. Thanks to this packaging scheme as Docker images, its deployment within BBP, KCNI, HBP and Switch infrastructures was greatly accelerated and improved. Across all four production deployments, BBN has proven to integrate well with different systems thanks to its interoperability capability based on open standards. Indeed, relying on standard authorization and authentication services, including OpenID Connect, OAuth 2.0 and JSON Web Tokens (JWT), enabled seamless integration with the four organisations’ identity management systems. Using W3C SHACL for validation, shaping data and schemas using RDF serialized as JSON-LD, and serving them using a REST architecture allowed easy integration and communication with third-party systems. The genericity and expressiveness of BBN domain modeling format ranging from JSON to W3C SHACL and its scalability addresses a large spectrum of complex data representation needs and use cases.
The availability of BBN Delta’s entire events journal over a secured HTTP based SSE interface is also another integration point. It enables other tools and systems to securely and transparently access, react and adapt to events occurring in the knowledge graph. Those tools can therefore monitor the knowledge graph and trigger subsequent jobs such as logging or indexing data in third-party systems. Furthermore, SSE allows developers to add additional features to BBN by building custom applications leveraging the knowledge graph data throughout their lifecycle.
BBN has been used primarily as a complement for existing tools and systems, bringing new features and capabilities that were missing. Examples of such features include validation of data and metadata to improve ETL pipelines’ output quality, serving as a metadata interoperability layer, support for comprehensive search capabilities ranging from document and faceted search to semantic and graph based queries through SPARQL 1.1. Data, metadata, schemas, ontologies can now be managed and kept in sync together. Developers across the four deployments of BBN also built custom applications to extend its feature set and adapt its behaviour. The developer friendly RDF JSON-LD serialization and the REST interface played an important role in on-boarding developers, enabling them to work with a data exchange format they are already familiar with. Furthermore, BBN internalizes the boilerplate code necessary to build, maintain and synchronize custom views from the data stored in the Cassandra primary store saving developers precious time. BBN enables a comprehensive implementation of the FAIR guiding principles for scientific data management. Tables 5 and 6 in the supplemental material detail BBN implementation of the FAIR principles.
Based on feedback from BBN production deployments across 3 adopters and use cases, a set of limitations and further improvements of BBN can be identified. While users of BBN can currently programmatically create knowledge graphs from different sources and formats using the python framework Nexus Forge, a web-based interface would allow users that are not data and knowledge engineers to also perform such complex tasks with a certain degree of automation. Publishing and disseminating built knowledge graphs or a subset of them either publicly or to a specific group of users can be done through Nexus Fusion by creating web-based data views generated from SPARQL or Elasticsearch queries. However writing such queries is challenging for non-expert users who are not always fully aware of how the data is structured or who just don’t know about those querying languages. Addressing this challenge would require enabling users to create data views directly from the results of simpler and user-friendly querying modalities such as keyword, natural language or faceted search. Furthermore the web-based data views could be automatically assigned with a DOI identifier. High quality knowledge graphs are valuable data sources for data science and machine learning pipelines which in return can greatly help in building (e.g NLP), populating (e.g. link prediction, node classification) and analysing (e.g. GNN: Graph Neural Network) them. Integration with widely used machine learning and graph analytics tools is a requirement to enable data scientists and machine learning engineers to benefit from and contribute to knowledge graphs. The data-driven science cycle involves numerous steps involving many users performing complex tasks. Bringing user collaboration features through the Nexus Fusion web interface is needed to accelerate iterations across this cycle when using BBN.
8.Conclusion and future directions
Data-driven science often involves an iterative knowledge discovery cycle which is, at its core, a collaborative journey. It involves multi-disciplinary teams of scientists and engineers working on collecting, analyzing, linking and classifying data, from which new knowledge is generated, shared and disseminated. We presented Blue Brain Nexus (BBN), an open source and scalable data and knowledge graph management system providing foundation to support this cycle.
BBN has thus far been deployed in production to address four different use cases alongside the data-driven science iterative cycle in the fields of computational modeling, neuroscience, psychiatry and linked open data. Many valuable lessons have been learned. First, these four production deployments have shown that a knowledge graph should first be considered part of existing infrastructures within which it can be easily deployed and operationalized. Secondly a knowledge graph should also be integrated in the ecosystem of tools and systems it complements and interacts with. It should support use cases’ scale, complexity, and evolution. Finally, the four use cases show that rapid onboarding of developers and users and the ease at which they can prototype and build custom front-end and backend applications on top of the knowledge graph are crucial to demonstrate added value.
BBN is actively and continuously being further developed and maintained by Blue Brain as a key technology and complementary approach to classical neuroinformatics in its data-driven approach and long term scientific goal and workflow for organzing brain tissue data and models [44]. BBN technology stack based on open, interoperable and domain agnostic standards supporting domain specific extension, combined with the three production deployments by adopters from large long term organisations where potential developers and contributors community can arise, shows BBN maturity, genericity, ability to support new use cases and evolution of existing ones as well as sustainability moving forward. Furthermore, we are actively seeking to expand the engagement of adopters in contributing to the ongoing development and to sustaining BBN.
Thanks to the lessons learned across the diverse use cases, new developments and improvements are considered as part of BBN roadmap including providing various users: i) with advanced web-based knowledge graph authoring features; ii) with web-based data views creation (from the results of simpler and user-friendly query modalities) and publishing as portal with support of DOIs assignment; iii) with web-based collaboration tools across all steps and actors of the data-driven science cycle; iv) with a plugin architecture for Nexus Delta to support developers with more modularity and domain specific extension features; v) with integration with machine learning widely used tools and pipelines.
Code, user and developer resources availability
BBN is an open source project with Apache 26969 as license. It is freely available on Github7070 and a sandbox7171 deployment is available as a playground. It comes with a set of developer oriented documentation such as APIs, installation and deployments instructions as well as user oriented documentation made of high-level and hands-on tutorials.7272,7373 BBN architecture and features described in this paper refers to the version 1.4 whose release notes are available in https://bluebrainnexus.io/docs/releases.
Author contributions
– Conceptualization / Design: MF.S., B.R., S.K. and S.H. conceived of the presented idea.
– Software design and development:
∗ Nexus Delta: MF.S., B.R., D.M., H.G., W.W.
∗ Nexus Fusion and Javascript SDK: M.D., J.M., K.P., I.L., J.L., D.N.M., N.S., AU.
∗ Nexus Forge: P.-A.F., AG.R., A.-K.K., MF.S.
∗ Nexus Python SDK: J.L., P.-A.F., S.K., MF.S., B.R., D.M., H.G., W.W., A.-K.K., H.L., S.H.
– Writing (original draft preparation): MF.S., B.R., S.K. and S.H. prepared the original draft of the manuscript and contributed to the final version of the manuscript.
– Writing (editing text contribution): MF.S., B.R., S.K., D.R., S.H., AU.
– Writing (review): MF.S., B.R., S.K., D.M., H.G., W.W., M.D., J.M., K.P., A.-K.K., H.L., J.L., P.-A.F., S.J., D.R., S.H., AU., AG.R., C.L.
– Validation / Benchmark: B.R. conceived of the benchmark design and setup. B.R., D.M., H.G., W.W., C.L. implemented, ran the benchmark and reported the results.
– Visualization / Illustrations:
– Supervision: H.M., S.H. and S.K. provided oversight and leadership responsibility for the presented idea and its implementation.
– Other contributions: H.M., A.-K.K., H.L., J.L., P.-A.F., S.J., D.N., N.S., provided use cases as subject matter experts as well as critical reviews helping drive the implementation of Blue Brain Nexus.
Funding
This study was supported by funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne, from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology.
Funding has been provided in part for Nexus Forge from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 (Human Brain Project SGA2).
Conflict of interest statement
The authors declare no competing financial interests.
Notes
Acknowledgements
We are grateful to Karin Holm from BBP project’s Operations team (Science Writing/Publications) and for Felix Schürmann the BBP’s Computing Division Director for reviewing this paper and for providing us with valuable feedback and comments. We also thank Jean-Denis Courcol, Genrich Ivaska, Pavlo Getta and Benoît Coste from the BBP’s Neuroscientific Software Engineering team for providing us with valuable use cases and user feedback. Many thanks to Olli Salo Antero, Mike Kenyon and Joshua Akers from the BBP’s Core Services team for providing the infrastructure to run BBN at BBP. We thank Jan Bjaalie, Oliver Schmid, David Kunzmann and Jeff Muller from the Human Brain Project for the useful discussions and for the useful and valuable use cases. We also thank Sebastian Sigloch from the Research Data Connectome, Adeel Ansari and Nikola Bogetic from KCNI for the fruitful discussions and for the useful and valuable use cases. We thank Patrycja Lurie, Jayakrishnan Nair, Nabil Alibou and Eugenia Oshurko, BBP’s Data and Knowledge Engineering team members and Simon Dumas BBP’s NeuroInformatics Software Engineering team member, for their valuable feedback. Many thanks to Michael Martin Vincent, Kate Mullins Elizabeth and Mathieu Chambon from the BBP’s Communications team for the support in creating illustrations for this paper.
References
[1] | O. Agarwal, H. Ge, S. Shakeri and R. Al-Rfou, Knowledge graph based synthetic corpus generation for knowledge-enhanced language model pre-training, in: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Association for Computational Linguistics, Online, (2021) , pp. 3554–3565. https://www.aclweb.org/anthology/2021.naacl-main.278. doi:10.18653/v1/2021.naacl-main.278. |
[2] | D. Alahakoon and X. Yu, Smart electricity meter data intelligence for future energy systems: A survey, IEEE Transactions on Industrial Informatics 12: (1) ((2016) ), 425–436. doi:10.1109/TII.2015.2414355. |
[3] | K. Amunts, A.C. Knoll, T. Lippert, C.M.A. Pennartz, P. Ryvlin, A. Destexhe, V.K. Jirsa, E. D’Angelo and J.G. Bjaalie, The Human Brain Project – Synergy between neuroscience, computing, informatics, and brain-inspired technologies, PLOS Biology 17: (7) ((2019) ), e3000344. doi:10.1371/journal.pbio.3000344. |
[4] | J. Andreu, C. Poon, R.D. Merrifield, S. Wong and G.-Z. Yang, Big data for health, IEEE Journal of Biomedical and Health Informatics 19: ((2015) ). doi:10.1109/JBHI.2015.2450362. |
[5] | A. Aryani and J. Wang, Research graph: Building a distributed graph of scholarly works using research data switchboard, 2017. https://monash.figshare.com/articles/Research_Graph_Building_a_Distributed_Graph_of_Scholarly_Works_using_Research_Data_Switchboard/4742413. doi:10.4225/03/58c696655af8a. |
[6] | M. Ashburner, C.A. Ball, J.A. Blake, D. Botstein, H. Butler, J.M. Cherry, A.P. Davis, K. Dolinski, S.S. Dwight, J.T. Eppig, M.A. Harris, D.P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J.C. Matese, J.E. Richardson, M. Ringwald, G.M. Rubin and G. Sherlock, Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium, Nature Genetics 25: (1) ((2000) ), 25–29. doi:10.1038/75556. |
[7] | M. Baker, 1,500 scientists lift the lid on reproducibility, Nature News 533: (7604) ((2016) ), 452. http://www.nature.com/news/1-500-scientists-lift-the-lid-on-reproducibility-1.19970. doi:10.1038/533452a. |
[8] | J.R. Bambauer, Tragedy of the data commons, SSRN Scholarly Paper, ID 1789749, Social Science Research Network, Rochester, NY, 2011. https://papers.ssrn.com/abstract=1789749. doi:10.2139/ssrn.1789749. |
[9] | M. Boeckhout, G.A. Zielhuis and A.L. Bredenoord, The FAIR guiding principles for data stewardship: Fair enough?, European Journal of Human Genetics 26: (7) ((2018) ), 931–936. doi:10.1038/s41431-018-0160-0. |
[10] | K. Börner, N. Contractor, H.J. Falk-Krzesinski, S.M. Fiore, K.L. Hall, J. Keyton, B. Spring, D. Stokols, W. Trochim and B. Uzzi, A multi-level systems perspective for the science of team science, Science translational medicine 2: (49) ((2010) ), 49cm24. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3527819/. doi:10.1126/scitranslmed.3001399. |
[11] | D. Brickley, M. Burgess and N. Noy, Google dataset search: Building a search engine for datasets in an open web ecosystem, in: The World Wide Web Conference, WWW ’19, Event-Place: San Francisco, CA, USA, ACM, New York, NY, USA, (2019) , pp. 1365–1375. ISBN 978-1-4503-6674-8. doi:10.1145/3308558.3313685. |
[12] | J.-P. Calbimonte, F. Dubosson, R. Hilfiker, A. Cotting and M. Schumacher, The MedRed ontology for representing clinical data acquisition metadata, in: The Semantic Web – ISWC 2017, C. d’Amato, M. Fernandez, V. Tamma, F. Lecue, P. Cudré-Mauroux, J. Sequeda, C. Lange and J. Heflin, eds, Lecture Notes in Computer Science, Springer International Publishing, (2017) , pp. 38–47. ISBN 978-3-319-68204-4. doi:10.1007/978-3-319-68204-4_4. |
[13] | P. Ceravolo, A. Azzini, M. Angelini, T. Catarci, P. Cudré-Mauroux, E. Damiani, A. Mazak, M. van Keulen, M. Jarrar, G. Santucci, K. Sattler, M. Scannapieco, M. Wimmer, R. Wrembel and F.A. Zaraket, Big data semantics, J. Data Semantics 7: (2) ((2018) ), 65–85. https://exascale.info/assets/pdf/ceravolo2018jods.pdf. doi:10.1007/s13740-018-0086-2. |
[14] | P. Cudré-Mauroux and eXascale Infolab, Design considerations on SWITCH’s connectome vision, 2020, https://www.semanticscholar.org/paper/6c11bcd3f048d27826d9aee6403c2ef723d8a3c6. |
[15] | G.T. Einevoll, A. Destexhe, M. Diesmann, S. Grün, V. Jirsa, M. de Kamps, M. Migliore, T.V. Ness, H.E. Plesser and F. Schürmann, The Scientific Case for Brain Simulations, Neuron 102: (4) ((2019) ), 735–744. http://www.sciencedirect.com/science/article/pii/S0896627319302909. doi:10.1016/j.neuron.2019.03.027. |
[16] | X. Fan and H. Markram, A brief history of simulation neuroscience, Frontiers in Neuroinformatics 13: ((2019) ). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6513977/. doi:10.3389/fninf.2019.00032. |
[17] | U.M. Fayyad, G. Piatetsky-Shapiro and P. Smyth, From data mining to knowledge discovery: An overview, in: Advances in Knowledge Discovery and Data Mining, U.M. Fayyad, G. Piatetsky-Shapiro, P. Smyth and R. Uthurusamy, eds, American Association for Artificial Intelligence, (1996) , pp. 1–34. ISBN 978-0-262-56097-9. http://dl.acm.org/citation.cfm?id=257938.257942. |
[18] | T. Furche, G. Gottlob, L. Libkin, G. Orsi and N.W. Paton, Data wrangling for big data: Challenges and opportunities, in: EDBT, (2016) . doi:10.5441/002/edbt.2016.44. |
[19] | E. Gibney, LHC plans for open data future, Nature 503: (7477) ((2013) ), 447. https://www.nature.com/articles/503447a. doi:10.1038/503447a. |
[20] | R.S. Gonçalves and M.A. Musen, The variable quality of metadata about biological samples used in biomedical experiments, Scientific Data 6: (1) ((2019) ), 1–15, https://www.nature.com/articles/sdata201921. doi:10.1038/sdata.2019.21. |
[21] | J. Gray, D.T. Liu, M. Nieto-Santisteban, A. Szalay, D.J. DeWitt and G. Heber, Scientific data management in the coming decade, SIGMOD Rec. 34: (4) ((2005) ), 34–41. doi:10.1145/1107499.1107503. |
[22] | P. Haase, D.M. Herzig, A. Kozlov, A. Nikolov and J. Trame, metaphactory: A platform for knowledge graph management, Semantic Web 10: (6) ((2019) ), 1109–1125. doi:10.3233/SW-190360. |
[23] | T. Hammond, M. Pasin and E. Theodoridis, Data integration and disintegration: Managing Springer Nature SciGraph with SHACL and OWL, in: Proceedings of the ISWC 2017 Posters & Demonstrations and Industry Tracks Co-Located with 16th International Semantic Web Conference (ISWC 2017), Vienna, Austria, October 23rd – to – 25th, 2017, N. Nikitina, D. Song, A. Fokoue and P. Haase, eds, CEUR Workshop Proceedings, Vol. 1963: , CEUR-WS.org, (2017) . http://ceur-ws.org/Vol-1963/paper493.pdf. |
[24] | P.A. Harris, R. Taylor, B.L. Minor, V. Elliott, M. Fernandez, L. O’Neal, L. McLeod, G. Delacqua, F. Delacqua, J. Kirby and S.N. Duda, The REDCap consortium: Building an international community of software platform partners, Journal of Biomedical Informatics 95: ((2019) ), 103208. http://www.sciencedirect.com/science/article/pii/S1532046419301261. doi:10.1016/j.jbi.2019.103208. |
[25] | T. Heath and C. Bizer, Linked Data: Evolving the Web into a Global Data Space, Synthesis Lectures on the Semantic Web: Theory and Technology, Morgan & Claypool. ISBN 978-1-60845-431-0. https://ieeexplore.ieee.org/document/6812934. doi:10.2200/S00334ED1V01Y201102WBE001. |
[26] | S.L. Hill, How do we know what we know? Discovering neuroscience data sets through minimal metadata, Nature Reviews Neuroscience 17: (12) ((2016) ), 735–736. https://www.nature.com/articles/nrn.2016.134. doi:10.1038/nrn.2016.134. |
[27] | A. Hogan, E. Blomqvist, M. Cochez, C. D’amato, G.D. Melo, C. Gutierrez, S. Kirrane, J.E.L. Gayo, R. Navigli, S. Neumaier, A.-C.N. Ngomo, A. Polleres, S. Rashid, A. Rula, L. Schmelzeisen, J. Sequeda, S. Staab and A. Zimmermann, Knowledge graphs, 2021, p. 257. https://hal-emse.ccsd.cnrs.fr/emse-03440299. doi:10.2200/S01125ED1V01Y202109DSK022. |
[28] | F. Ilievski, D. Garijo, H. Chalupsky, N.T. Divvala, Y. Yao, C. Rogers, R. Li, J. Liu, A. Singh, D. Schwabe and P. Szekely, KGTK: A toolkit for large knowledge graph manipulation and analysis, in: The Semantic Web – ISWC 2020, J.Z. Pan, V. Tamma, C. d’Amato, K. Janowicz, B. Fu, A. Polleres, O. Seneviratne and L. Kagal, eds, Springer International Publishing, Cham, (2020) , pp. 278–293. ISBN 978-3-030-62466-8. doi:10.1007/978-3-030-62466-8_18. |
[29] | M.Y. Jaradeh, A. Oelen, K.E. Farfar, M. Prinz, J. D’Souza, G. Kismihók, M. Stocker and S. Auer, Open research knowledge graph: Next generation infrastructure for semantic scholarly knowledge, 2019. arXiv:1901.10816 [cs]. doi:10.48550/arXiv.1901.10816. |
[30] | R. Kitchin, The real-time city? Big data and smart urbanism, GeoJournal 79: (1) ((2014) ), 1–14. doi:10.1007/s10708-013-9516-8. |
[31] | L. Koesten, P. Vougiouklis, E. Simperl and P. Groth, Dataset reuse: Toward translating principles to practice, Patterns 1: (8) ((2020) ), 100136. https://www.sciencedirect.com/science/article/pii/S2666389920301847. doi:10.1016/j.patter.2020.100136. |
[32] | Y. LeCun, Y. Bengio and G. Hinton, Deep learning, Nature 521: (7553) ((2015) ), 436–444. https://www.nature.com/articles/nature14539. doi:10.1038/nature14539. |
[33] | H. Markram, The Blue Brain Project, Nature Reviews Neuroscience 7: (2) ((2006) ), 153–160. https://www.nature.com/articles/nrn1848. doi:10.1038/nrn1848. |
[34] | H. Markram, E. Muller, S. Ramaswamy, M.W. Reimann, M. Abdellah, C.A. Sanchez, A. Ailamaki, L. Alonso-Nanclares, N. Antille, S. Arsever, G.A.A. Kahou, T.K. Berger, A. Bilgili, N. Buncic, A. Chalimourda, G. Chindemi, J.-D. Courcol, F. Delalondre, V. Delattre, S. Druckmann, R. Dumusc, J. Dynes, S. Eilemann, E. Gal, M.E. Gevaert, J.-P. Ghobril, A. Gidon, J.W. Graham, A. Gupta, V. Haenel, E. Hay, T. Heinis, J.B. Hernando, M. Hines, L. Kanari, D. Keller, J. Kenyon, G. Khazen, Y. Kim, J.G. King, Z. Kisvarday, P. Kumbhar, S. Lasserre, J.-V. Le Bé, B.R.C. Magalhães, A. Merchán-Pérez, J. Meystre, B.R. Morrice, J. Muller, A. Muñoz-Céspedes, S. Muralidhar, K. Muthurasa, D. Nachbaur, T.H. Newton, M. Nolte, A. Ovcharenko, J. Palacios, L. Pastor, R. Perin, R. Ranjan, I. Riachi, J.-R. Rodríguez, J.L. Riquelme, C. Rössert, K. Sfyrakis, Y. Shi, J.C. Shillcock, G. Silberberg, R. Silva, F. Tauheed, M. Telefont, M. Toledo-Rodriguez, T. Tränkler, W. Van Geit, J.V. Díaz, R. Walker, Y. Wang, S.M. Zaninetta, J. DeFelipe, S.L. Hill, I. Segev and F. Schürmann, Reconstruction and simulation of neocortical microcircuitry, Cell 163: (2) ((2015) ), 456–492. https://www.cell.com/cell/abstract/S0092-8674(15)01191-5. doi:10.1016/j.cell.2015.09.029. |
[35] | J.A. McMurry, N. Juty, N. Blomberg, T. Burdett, T. Conlin, N. Conte, M. Courtot, J. Deck, M. Dumontier, D.K. Fellows, A. Gonzalez-Beltran, P. Gormanns, J. Grethe, J. Hastings, J.-K. Hériché, H. Hermjakob, J.C. Ison, R.C. Jimenez, S. Jupp, J. Kunze, C. Laibe, N.L. Novère, J. Malone, M.J. Martin, J.R. McEntyre, C. Morris, J. Muilu, W. Müller, P. Rocca-Serra, S.-A. Sansone, M. Sariyar, J.L. Snoep, S. Soiland-Reyes, N.J. Stanford, N. Swainston, N. Washington, A.R. Williams, S.M. Wimalaratne, L.M. Winfree, K. Wolstencroft, C. Goble, C.J. Mungall, M.A. Haendel and H. Parkinson, Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data, PLOS Biology 15: (6) ((2017) ), e2001414. doi:10.1371/journal.pbio.2001414. |
[36] | F.J. Montáns, F. Chinesta, R. Gómez-Bombarelli and J.N. Kutz, Data-driven modeling and learning in science and engineering, Comptes Rendus Mécanique 347: (11) ((2019) ), 845–855. http://www.sciencedirect.com/science/article/pii/S1631072119301809. doi:10.1016/j.crme.2019.11.009. |
[37] | M. Mountantonakis and Y. Tzitzikas, Large-scale semantic integration of linked data: A survey, ACM Computing Surveys 52: ((2019) ), 1–40. doi:10.1145/3345551. |
[38] | M. Musen, We face a new “Tragedy of the Commons.” The remedy is better metadata, CBIIT, 2017. https://datascience.cancer.gov/news-events/blog/we-face-new-tragedy-commons-remedy-better-metadata. |
[39] | N. Noy, Y. Gao, A. Jain, A. Narayanan, A. Patterson and J. Taylor, Industry-scale knowledge graphs: Lessons and challenges, Queue 17: (2) ((2019) ), 20:48–20:75. doi:10.1145/3329781.3332266. |
[40] | N. Polyzotis, S. Roy, S.E. Whang and M. Zinkevich, Data management challenges in production machine learning, in: Proceedings of the 2017 ACM International Conference on Management of Data, SIGMOD ’17, Event-Place: Chicago, Illinois, USA, ACM, (2017) , pp. 1723–1726. ISBN 978-1-4503-4197-4. doi:10.1145/3035918.3054782. |
[41] | N. Polyzotis, S. Roy, S.E. Whang and M. Zinkevich, Data lifecycle challenges in production machine learning: A survey, SIGMOD Rec. 47: (2) ((2018) ), 17–28. doi:10.1145/3299887.3299891. |
[42] | P. Ristoski and H. Paulheim, Semantic web in data mining and knowledge discovery: A comprehensive survey, Journal of Web Semantics 36: ((2016) ), 1–22, http://www.sciencedirect.com/science/article/pii/S1570826816000020. doi:10.1016/j.websem.2016.01.001. |
[43] | M.R. Saeed, C. Chelmis and V.K. Prasanna, Extracting entity-specific substructures for RDF graph embeddings, Semantic Web ((2019) ), 1–22. doi:10.3233/SW-190359. |
[44] | F. Schürmann, J.-D. Courcol and S. Ramaswamy, Computational concepts for reconstructing and simulating brain tissue, in: Computational Modelling of the Brain: Modelling Approaches to Cells, Circuits and Networks, M. Giugliano, M. Negrello and D. Linaro, eds, Springer International Publishing, Cham, (2022) , pp. 237–259. ISBN 978-3-030-89439-9. doi:10.1007/978-3-030-89439-9_10. |
[45] | UniProt Consortium, UniProt: A worldwide hub of protein knowledge, Nucleic Acids Research 47: (D1) ((2018) ), D506–D515. doi:10.1093/nar/gky1049. |
[46] | A.L. Vaccarino, M. Dharsee, S. Strother, D. Aldridge, S.R. Arnott, B. Behan, C. Dafnas, F. Dong, K. Edgecombe, R. El-Badrawi, K. El-Emam, T. Gee, S.G. Evans, M. Javadi, F. Jeanson, S. Lefaivre, K. Lutz, F.C. MacPhee, J. Mikkelsen, T. Mikkelsen, N. Mirotchnick, T. Schmah, C.M. Studzinski, D.T. Stuss, E. Theriault and K.R. Evans, Brain-CODE: A secure neuroinformatics platform for management, federation, sharing and analysis of multi-dimensional neuroscience data, Frontiers in Neuroinformatics 12: ((2018) ). doi:10.3389/fninf.2018.00028. |
[47] | L. Vogt, R. Baum, P. Bhatty, C. Köhler, S. Meid, B. Quast and P. Grobe, SOCCOMAS: A FAIR web content management system that uses knowledge graphs and that is based on semantic programming, Database 2019: ((2019) ), baz067. doi:10.1093/database/baz067. |
[48] | Q. Wang, Z. Mao, B. Wang and L. Guo, Knowledge graph embedding: A survey of approaches and applications, IEEE Transactions on Knowledge and Data Engineering 29: (12) ((2017) ), 2724–2743. doi:10.1109/TKDE.2017.2754499. |
[49] | M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L.B. da Silva Santos, P.E. Bourne, J. Bouwman, A.J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C.T. Evelo, R. Finkers, A. Gonzalez-Beltran, A.J.G. Gray, P. Groth, C. Goble, J.S. Grethe, J. Heringa, P.A.C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S.J. Lusher, M.E. Martone, A. Mons, A.L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M.A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, The FAIR Guiding Principles for scientific data management and stewardship, Scientific Data 3: ((2016) ), 160018. doi:10.1038/sdata.2016.18. |
[50] | L. Zehl, S. Zafarnia, S. Köhnen, K. Andersson, M. Markovic, E. Legouée, C.H. Blixhavn, H. Kleven, X. Gui, P. Chervakov, O. Schmid, S.D. Bell, T. Gillespie, M.B. Abrams, A.P. Davison, J.C. Muller, T.B. Leergaard, K. Amunts, J.G. Bjaalie and T. Dickscheid, Integrating neuroscientific data into a unified database – From individual experiments to a standardized metadata collection using the Human Brain Project Neuroinformatics Platform, 2018. http://abstracts.g-node.org/abstracts/465d8b99-9137-4f14-9eec-3cec2bdfcf5c. doi:10.12751/NNCN.BC2018.0236. |




