Erratum to: SETD2 Regulates the Methylation of Translation Elongation Factor eEF1A1 in Clear Cell Renal Cell Carcinoma
[Kidney Cancer, 6 (2022), 179–193, DOI 10.3233/KCA-220009] https://content.iospress.com/articles/kidney-cancer/kca220009
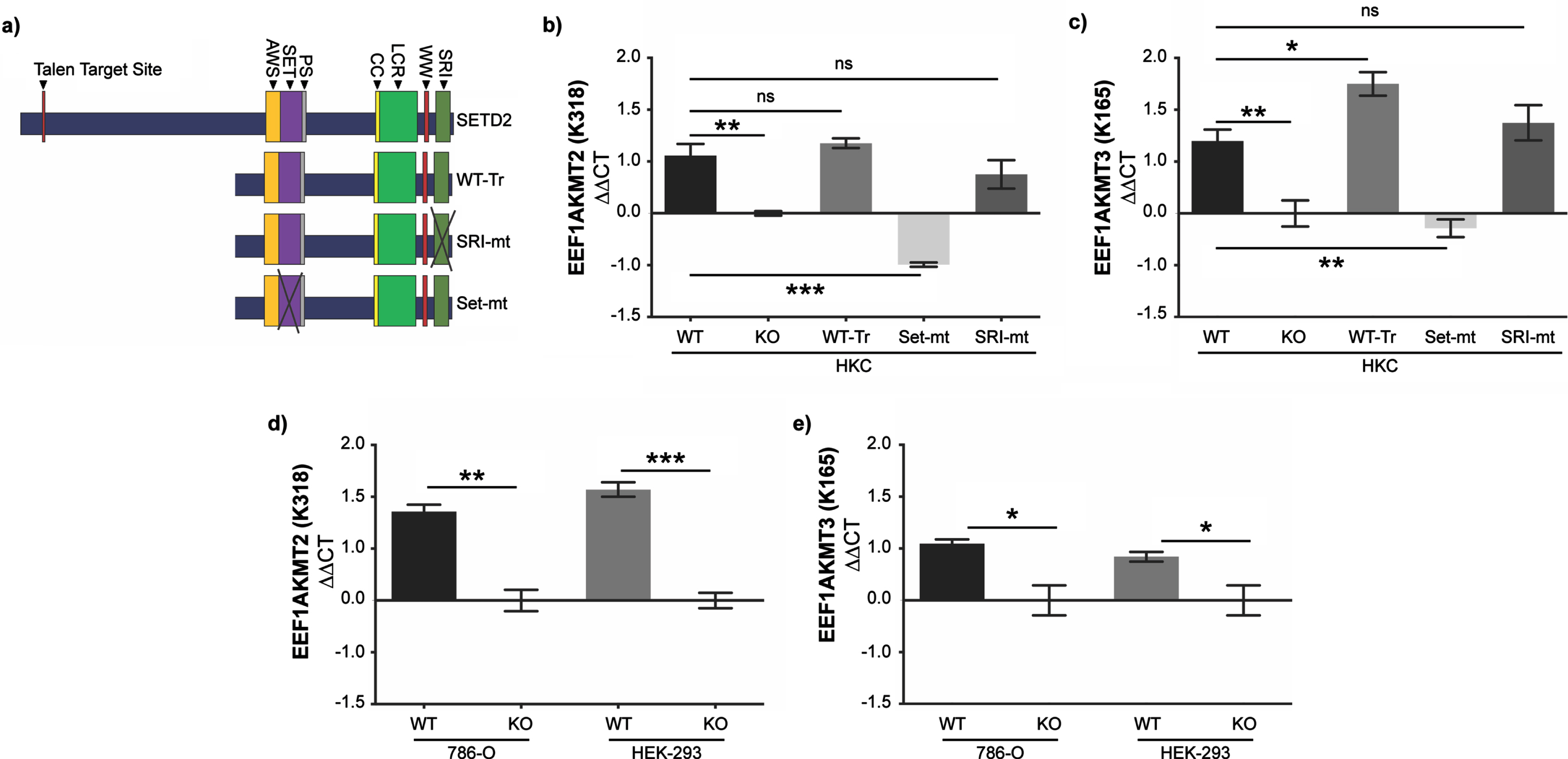
On page 185, the order of the panels in Figure 3 in the published version did not correlate with the figure legend. An updated figure is included below so that it correlates accurately with the figure legend. In addition, an updated Results paragraph is included below that accurately correlates with this reorganized figure.
None of the data changed. Rather, the order of the panels was adjusted. The error had no bearings on the interpretation of the results, nor did it influence the conclusions of this work.
Decreased eEF1A1 Methyltransferase Expression in SETD2-KO
While we have robust data that trimethylation of K165 and monomethylation of K318 on eEF1A1 decreases in SETD2-KO cells, neither lysine matches a known consensus sequence for SETD2-mediated lysine-methylation (i.e., KxP or KxxG) (10, 20, 21). Furthermore, lysine methyltransferases responsible for these PTMs have previously been identified: EEF1AKMT3 methylates K165 and EEF1AKMT2 methylates K318 (39– 41). Therefore, we sought to evaluate whether the expression of these enzymes was dependent on SETD2 protein and, if so, gain insight into the mechanism of this regulation. To perform this analysis, we utilized WT and SETD2-KO HKC cells as well as SETD2-KO HKC cells with various SETD2 constructs and mutations (Fig. 3 a). These cell lines were designed by Hacker et al. and include a truncated but functional SETD2 gene (WT-Tr), a SET-domain mutant (SET-mt), and an SRI-domain mutant (SRI-mt) (24). To briefly summarize, SETD2-KO HKC cell lines were generated using TAL effector nucleases (TALENs) targeting exon 3 of SETD2 (24). Individual clones of TALEN-treated cells were isolated and deletion confirmed by demonstrating loss of H3K36me3 on immunocytochemistry and verifying inactivation of both alleles of SETD2 via Sanger sequencing. We validated SETD2 status in HKC cell lines by western blot (Sup Fig 1). Subsequently, a truncated wild-type FLAG-tagged form of SETD2 (amino acids 1323– 2564; WT-Tr), which includes all known functional domains, was exogenously expressed in the SETD2-KO cells (full length SETD2 is very large thus making it difficult to express full length SETD2) (24). The SET-mt (R1625 C) and an SRI-mt (R2510 H) were generated from truncated SETD2. Using RT-qPCR (see Materials and Methods for details), we observe that loss of SETD2 expression was associated with decreased expression of EEF1AKMT2 (Fig. 3 b) and EEF1AKMT3 (Fig. 3c). Interestingly, the WT-Tr and SRI-mt constructs recover EEF1AKMT2/3 expression to wildtype levels, whereas the SET-mt does not in HKC cells (Fig. 3 b-c). SETD2-dependent expression of EEF1AKMT2/3 was validated in the human ccRCC 786-O cells as well as HEK-293T cells (human embryonic kidney cells) (using similar methods as described above for generated SETD2-KO cells) (Fig. 3d-e). These data suggest that SET domain function of SETD2 is necessary for the expression of EEF1AKTM2/3 in human kidney and kidney cancer cells.




