Novel neural network architecture using sharpened cosine similarity for robust classification of Covid-19, pneumonia and tuberculosis diseases from X-rays
Abstract
COVID-19 is a rapidly proliferating transmissible virus that substantially impacts the world population. Consequently, there is an increasing demand for fast testing, diagnosis, and treatment. However, there is a growing need for quick testing, diagnosis, and treatment. In order to treat infected individuals, stop the spread of the disease, and cure severe pneumonia, early covid-19 detection is crucial. Along with covid-19, various pneumonia etiologies, including tuberculosis, provide additional difficulties for the medical system. In this study, covid-19, pneumonia, tuberculosis, and other specific diseases are categorized using Sharpened Cosine Similarity Network (SCS-Net) rather than dot products in neural networks. In order to benchmark the SCS-Net, the model’s performance is evaluated on binary class (covid-19 and normal), and four-class (tuberculosis, covid-19, pneumonia, and normal) based X-ray images. The proposed SCS-Net for distinguishing various lung disorders has been successfully validated. In multiclass classification, the proposed SCS-Net succeeded with an accuracy of 94.05% and a Cohen’s kappa score of 90.70%; in binary class, it achieved an accuracy of 96.67% and its Cohen’s kappa score of 93.70%. According to our investigation, SCS in deep neural networks significantly lowers the test error with lower divergence. SCS significantly increases classification accuracy in neural networks and speeds up training.
1Introduction
A novel coronavirus (COVID-19), brought on by the SARS infection and immediately spread to other countries, was discovered in China toward the end of 2019. The symptoms of the covid-19 virus include fever, hypotension, vomiting, headache, and a feeling of chest tightness [1]. The World Health Organization (WHO) estimates that 187 million people worldwide were affected by this virus, increasing human mortality [2]. Unfortunately, because of the significant growth of covid-19 patients, health centers may become overwhelmed, resulting in a physician and radiology shortage. Chest imaging technology is needed to diagnose covid-19 pneumonia and severe lung damage, differentiate both viral and bacterial pneumonia and address other respiratory problems including Tuberculosis (TB) [3]. On the other hand, researchers are working on a covid-19 vaccine to prevent the virus from pre-foliating. The first approach is to develop a new way to help people who are sick and sanitation requirements to keep others from getting sick. Technological breakthroughs have been achieved by introducing mobile applications to monitor and track human contact [4]. The reverse transcription-polymerase chain reaction (RT-PCR) is used to reveal covid-19 in people’s early stages of the virus. However, a more severe issue is that RT-PCR is expensive and needs complex equipment and technical expertise [5]. Physicians are looking to the modalities report to address the RT-PCR problem. With this modality, one can discern bilateral patchy and confluent patterns, nodular opacities, ground glass and consolidative opacities, and opacities in the ground glass.
The imaging modalities utilized to find covid-19 include Computed tomography (CT) [6], X-ray [7], and ultrasound. CT scans give excellent resolution but expose patients to more radiation than X-rays and require expensive equipment. X-ray images are more readily available to a larger population due to low-cost X-ray imaging facilities. Due to the daily increase in cases, manual evaluation of modalities would be time-consuming, and radiologists would have difficulty diagnosing covid-19. Clinical and AI researchers have emphasized automating the identification of pneumonia or other lung disorders in the recent pandemic years, particularly in their early detection and identification. Computer-aided diagnostic (CAD) systems are more effective and efficient for diagnosing medical imaging [8]. The CAD is a second opinion from a doctor, especially in cases where the diagnosis is difficult to determine with the human eye. Recent research on CAD systems using artificial intelligence technologies has been successful [9]. Convolutional Neural Networks (CNN) automatically extract discriminate features from medical images that are used to predict outcomes. The CNN architectures in the CAD systems allow better classification of medical images providing large datasets [10]. CNN, Deep Belief Network [11], Capsule Network [12], Generative Adversarial Network (GAN) [13], and other DL technologies [14] have been reported for medical image diagnosis. X-rays will be used to evaluate the feasibility of early automated identification and categorization between healthy patients, COVID-19, pneumonia, and tuberculosis. We have developed an SCS model to assess whether or not a patient is healthy or has lung syndrome (pneumonia, TB, and normal). The proposed SCS-Net has demonstrated outstanding performance in classifying multiclass problems and has outperformed state-of-the-art (SOTA) approaches to classify lung diseases with nominal parameters. In addition to the methods listed above, there is still unresolved research on classifying covid-19 patients. To define covid-19 patients as well as other classes such as pneumonia, tuberculosis, and normal, a novel neural network framework is proposed in this research. This paper’s key impact is as follows:
• Development of a novel neural network with a sharpened cosine similarity layer to extract features named SCS-Net with a minimal number of trainable parameters for reliable and accurate classification.
• An effective sharpened cosine similarity layer is proposed instead of the convolution layer to make the results bounded and decrease the model’s variance to be generalized.
• The chest X-ray dataset is used in extensive research to evaluate the accuracy of the proposed model and existing methods for binary class and multiclass classification labeling (covid-19, pneumonia, tuberculosis, and normal).
• A fast decision network is being deployed for the pre-screening and early detection of covid-19 and other lung disorders.
The article is organized as follows: Section 2 presents the DL approach for covid-19 diagnosis along with the research’s motivation and related analysis. The tools and techniques used for the proposed work are described in Section 3. The experimental findings and a discussion are presented in Section 4. Conclusions and recommendations for further study and development are presented in Section 5.
2Background
The section initially presents the motivation behind the proposed covid-19 classification model. After that, the recent advancements in covid-19 classification models and shortcomings of the existing approaches are presented.
2.1Motivation of the research
According to earlier findings, most of the covid-19 infected patients are identified as having pneumonia and tuberculosis disease. As a result, radiographic testing plays a more significant role in early childhood infection diagnosis of covid-19 infection. It is challenging to categorize a patient based solely on CXR scans. The deep learning architectures currently used for classification have high model complexity and numerous training parameters. A novel neural network with nominal training parameters and greater accuracy is suggested to solve this issue using the SCS layer. The neural network model with SCS layer is considered for covid-19 classification to deploy on computing- or power-limited devices to make rapid decisions.
2.2Related work
The influence of artificial intelligence on the clinical care of covid-19 is briefly discussed in this section, along with studies that used CXR/CT scans to diagnose pneumonia, tuberculosis, and covid-19. In the recent decade, AI has shown considerable promise in medical image processing [15]. DL has been used widely in medical domains [16], including disease prediction [17], identification of pulmonary nodules [18], and classification of benign/malignant cancers [19]. A technique to create artificial CXR images called auxiliary classifier GAN based on covidGAN was proposed by Waheed et al. (2020) [20]. The CNN model is trained using synthetic images. The approach classified Covid-CXR and Normal-CXR with a 95% accuracy rate. A covid-DeepNet model was suggested by Panthakkan et al. to swiftly detect covid-19 and non-covid pneumonia infections from lung X-rays of critically sick patients (2021) [21]. The model can quickly test for covid-19 and has a classification accuracy of 99.67%. The major drawback are large number of layers and nodes which leads to large number of parameters. Waisy et al. (2021) suggested a COVID-DeepNet model with contrast constrained adaptive histogram equalization and Butterworth filter to improve contrast and remove noises in order to properly predict covid-19 infection, and obtains the accuracy of 99.93% [22]. The biggest downside of the Covid-DeepNet system is that it was trained to divide the input CX-R picture into two different groups (e.g., healthy and Covid-19 infected).
To distinguish between covid and non-covid, Turkoglu et al. (2021) suggested a multiple kernel extreme learning deep neural network model and achieved an accuracy of 98.36% [23]. This method’s drawback is that it can only be used for binary categorization. To improve model performance, Murugan et al. (2021) combine the backpropagation approach and the Whale Optimization Algorithm (WOA) [24]. The method experimented with different pre-trained models like Alex net, GoogLeNet, squeeze net, resnet50, and resnet50 with WOA. The resnet50 with WOA achieved an accuracy of 98.78%, but the number of trainable parameters is vast. Based on the U-net concept, Tianbo et al. (2021) created an architecture that included down sampling, skip links, and ultimately connected layers. [25]. Using a publicly available dataset that contained binary and multiclass classification, the model detects COVID-19 with an accuracy of 99.53% and 95.35% in binary and multiclass tasks, respectively. Deep transfer learning combined with super pixel-based segmentation was offered by Prakash et al. (2021) for detecting and localization of COVID-19 [26]. The method’s overall accuracy for binary classification was 99.53%, while for multiclass, it was 99.79%.
Agarwal S et al. in 2022 have proposed model for four classes each: normal, covid-19, bacterial pneumonia, and viral pneumonia [27]. Set 1 of the datasets comprises normal, covid-19, and pneumonia. They looked at various pre-trained CNN models, including mobilenetv2, inceptionv3, Xception, VGG16, InceptionResNetV2, NASNet mobile, resnet50, and DenseNet121, which are improved by swapping out the architecture’s last layers. Based on the results, densenet121 had an accuracy of 97% for the set 1 dataset, while mobilenetv2 had an accuracy of 81% on the set2 dataset. The disadvantage of this work is that the accuracy level for four class classifications is poor due to the model’s inability to discriminate features.
A covidDetNet architecture with nine convolutional layers and one fully connected layer was proposed by Naeem et al. (2022). Two activations are utilized in this architecture. Leaky ReLU and ReLU, along with batch and channel normalization [28]. The architecture has classified the covid, pneumonia, and normal, which obtained an accuracy of 98.40%. The model complexity is more regarding the number of parameters to be trained. Abhijit et al. (2022) suggested combining image segmentation and classification architecture using deep learning algorithms to identify lung abnormalities [29]. The approach was carried out in three phases. The segmentation technique was first carried out using CXR images created using conditional GAN architecture. Then segmented image features are extracted using VGG 19 model, which is merged with the BRISK key point extraction and classified with a random forest algorithm in the final layer. The testing accuracy of image classification is 96.6% when utilizing the architecture of the Visual Geometry Group-19 model in conjunction with the binary robust invariant scalable key points approach. Syed et al. (2022) proposed a novel image classification strategy for determining whether or not a patient is infected with covid19 [30]. To boost accuracy, the last layer of SoftMax is substituted with a KNN classifier in this technique. The suggested evolutionary approach is utilized to automatically discover the ideal values for CNN’s hyperparameters, resulting in a considerable improvement in classification accuracy. Larger and more diversified databases are still needed for validation before claiming that deep learning may help clinicians recognize COVID-19 in X-ray images since cross-dataset analysis shows that even cutting-edge models lack generalization power.
S.K. Ghosh et al. (2022) introduced the enhanced residual network (ENResNet) for the visual categorization of COVID-19 from chest X-ray images. The ENResNet is used to build the residual images and is normalized using patches [31]. The ENResNet achieves a classification accuracy of 99.7% in binary and 98.4% in multiclass detection. Tripti Goelr et al. (2022) proposed a two-step DL design for COVID-19 diagnosis leveraging CXR images [32]. The two steps of the recommended DL architecture are feature extraction and classification. The “Multi-Objective Grasshopper Optimization Algorithm (MOGOA)” is used to optimize the DL network layers; as a result, these networks are known as “Multi-COVID-Net.” A covid fuzzy ensemble network (COFE-Net) was presented by Banerjee et al. (2022) in ensemble deep learning architectures [33]. The classification accuracy for the COFE-Net model is 99.49% for the binary class and 96.39% for the multiclass. The proposed network was compared to an existing pre-trained network to establish the network’s efficiency. SARS Net was introduced by Kumar et al. (2022) to classify COVID-19 using the images CXR [34]. In this paper, SARS-Net combines a 2L-GCN model and a CNN model. The model’s accuracy and sensitivity on the validation set were 97.60% and 92.90%, respectively. Gurkan Kavuran et al. (2022) introduced the MTU-COVNet hybrid architecture to abstract visual features from volumetric thoracic CT images to identify COVID-19 [35]. Then features are optimized from the concentrated layers and obtain overall accuracy of 97.7%.
2.3Limitations of the existing studies
Several researchers have tried to use contemporary DL frameworks to automate the analysis of pneumonia and COVID-19. However, the following drawbacks and difficulties affect several of the notable algorithms:
• There is an inconsistency between the models’ speed, resilience, and accuracy.
• The lack of generalization techniques for accurate model predictions.
• Lack of regularization techniques in models to avoid overfitting
• Models’ trainable model parameters are high in numbers, leading to high training time and difficulty in model deployment.
3Tools and techniques
3.1Description of the datasets
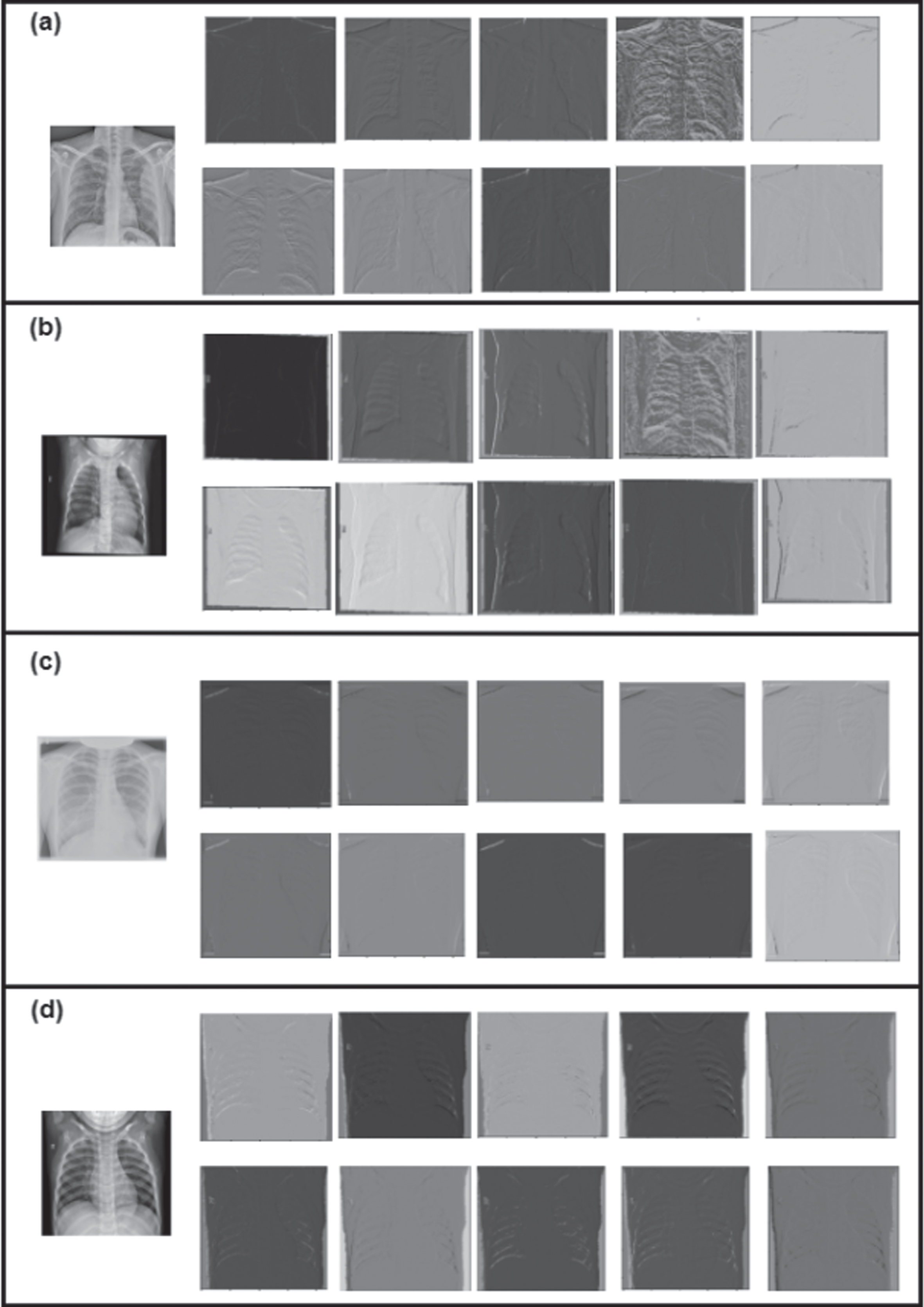
The CXR datasets are collected via Kaggle and GitHub and include diseases such as Covid-19, Normal, Pneumonia, and Tuberculosis in binary and multiclass form [36–39]. The images have different dimensions and are resized to 224 by 224 pixels. In cases of viral pneumonia, the white opacity surrounding the lungs on an X-ray image reduces and is almost non-existent in Covid-19 images. Model X-ray images from the database are shown in Fig. 1. The distribution of categories across the obtained dataset is depicted in Fig. 2.
Fig. 1
Samples from dataset a) Covid-19 (b) Normal (c) Pneumonia (d) Tuberculosis.
Fig. 2
Dataset distribution of multiclass experiment.
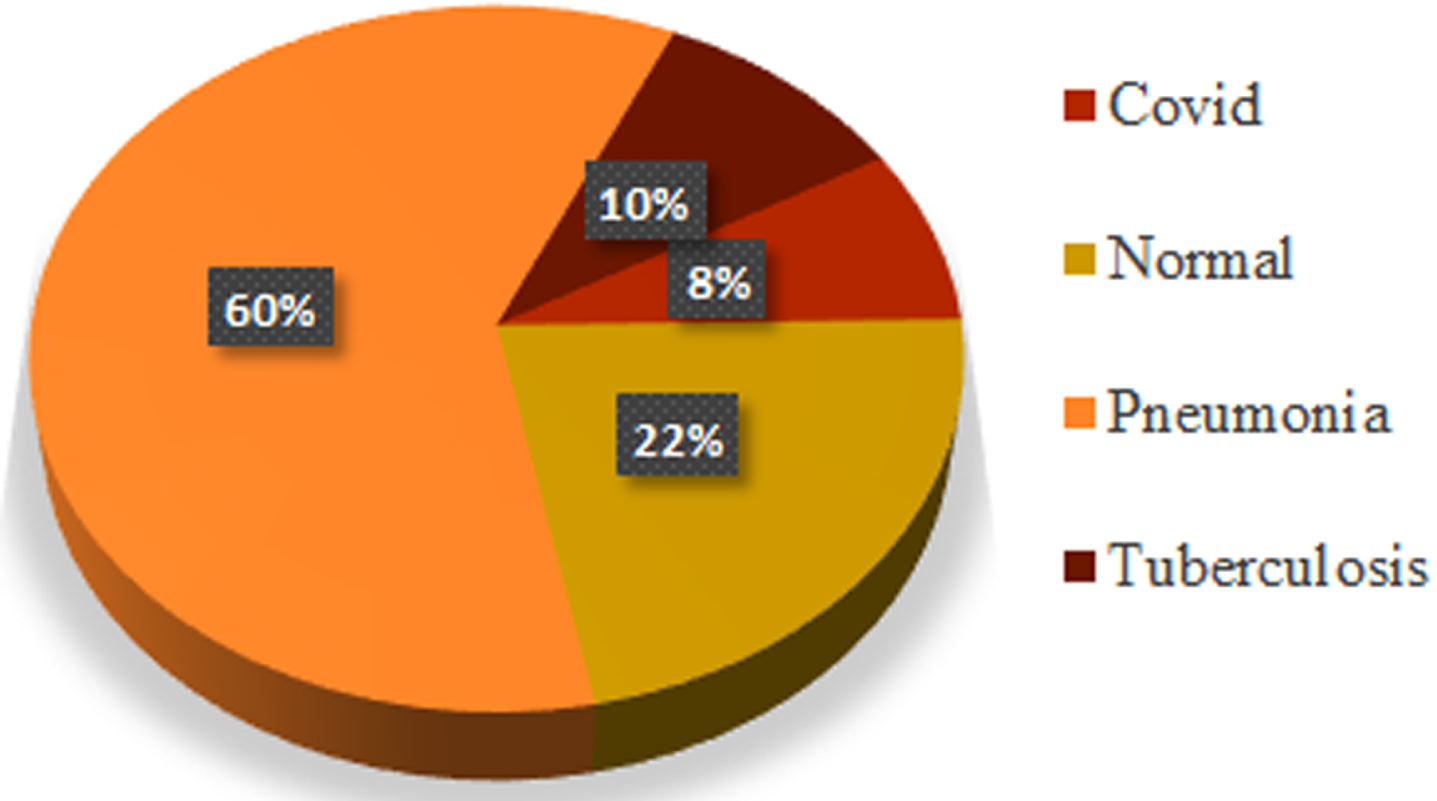
In total, there are 7135 images in the datasets shown in Table 1, of which 576 are COVID-19 images, 1583 are normal images, 4273 are pneumonia images, and 703 are tuberculosis images. The dataset was split into 710 images for validation and 6425 for training. The validation data includes 61 COVID-19 images, 174 normal images, 411 images of pneumonia, and 64 images of tuberculosis—a total of 1564 normal and 573 COVID-19 images used for binary classification. For data validation, 161 normal images and 54 COVID-19 images were employed. Different image augmentation techniques, including rotation, random crop, width shift range, and height shift range, to prevent the model from overfitting are used to enhance model performance.
Table 1
Dataset dissemination
| Normal | COVID-19 | Pneumonia | Tuberculosis | |
| Before Augmentation | 576 | 1583 | 4273 | 703 |
| After Augmentation | 2880 | 3915 | 21365 | 3515 |
3.2Problem with CNN
Convolutional layers, which include a collection of filters whose parameters are learned throughout the training, are the essential building component of a convolutional neural network. While each filter convolves with the image, the feature map is produced. A feature map is created when a filter is applied to the input, revealing the locations and intensities of a detected feature in the input. The kernel and an image are confined to two values, a high and a low. Positive values imply “prefer the signal value to be as high as possible,” which is ideal for understanding a convolutional kernel. Negative values indicate “prefer as low a frequency value as possible,” and zeroes suggest that “it doesn’t matter what the value.” The convolution layer output size is computed using equation 1. Convolution results from a sliding dot product between the kernel and an image portion. But dot product alone is a poor metric and results in a poor feature detector [40]. The fundamental shortcoming of the convolution layer is that it does not encode information when the objects’ location and orientation change. Furthermore, the intrinsic data of the image is entirely lost, and the information is sent to the same neurons, rendering them incapable of dealing with this type of information.
(1)
Where, W = Size of the input, F = Size of the filter, P = Padding, S = Stride
Equation 2 represents a convolution operation, a strided dot product between an input and a kernel. CNN has a very high level of accuracy when it comes to image recognition tasks and automatically selects critical information without human intervention. The convolution layer’s primary issue is spatially invariant to input data. Furthermore, a lot of training data is required because the convolution layer does not encode the location and orientation of the object. Again, the Max pooling layer operation makes a CNN much slower. CNNs have millions of parameters, and a tiny dataset would lead them to overfit since they require vast data to make them more generalized. To solve this issue, we replace the convolution layer with a sharpened cosine similarity layer.
(2)
Where, X = Input, Y = Kernel
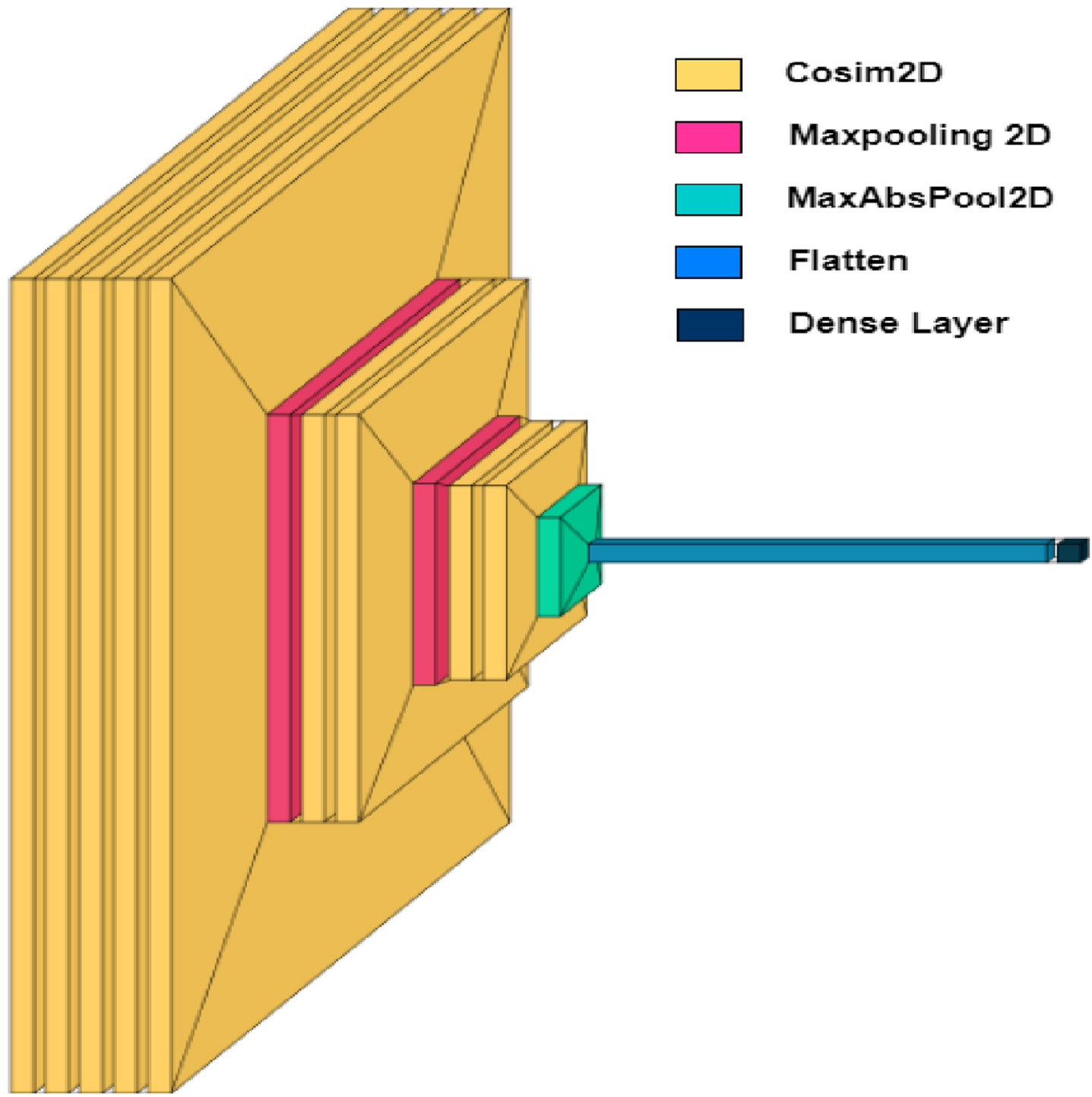
3.3Proposed SCS-Net
The flowchart of the proposed work is shown in Fig. 3, and the pipeline starts X-ray machine used to capture the X-ray of the patients and stored in the database. The produced images from the X-ray are used to train the model for classification of binary class and multiclass classification labeling (covid-19, pneumonia, tuberculosis, and normal. The images are preprocessed to fit the model for training. Train data and validation data are split from the dataset. The SCS-Net architecture is developed with different layers consisting of a cosine similarity layer, max-pooling layer, and flattened and dense layer. Figure 4 depicts the SCS-Net architecture with layers combined. The training set is employed to create the SCS-Net model. After the model has been trained, validation data are used to evaluate it, and performance metrics, including accuracy, precision, recall, F1 measure, and Cohen’s kappa score, are generated. Moreover, the SCS-Net model’s accuracy is evaluated in various samples of state-of-the-art architecture. Instead of applying a convolution layer, we employed the SCS layer to build the feature map. SCS is a feature-building method for neural networks that is an alternative approach to convolution. Similar to convolution, SCS is a stride operation that takes features out of an image. But before calculating the dot product, the image patch and kernel are adjusted to have a magnitude of 1, producing a cosine similarity, also known as cosine normalization [41]. The cosine of the angle formed by the two-dimensional signal and kernel vectors is given in equation 3.
(3)
(4)
(5)
Fig. 3
Flowchart for SCS-Net model.
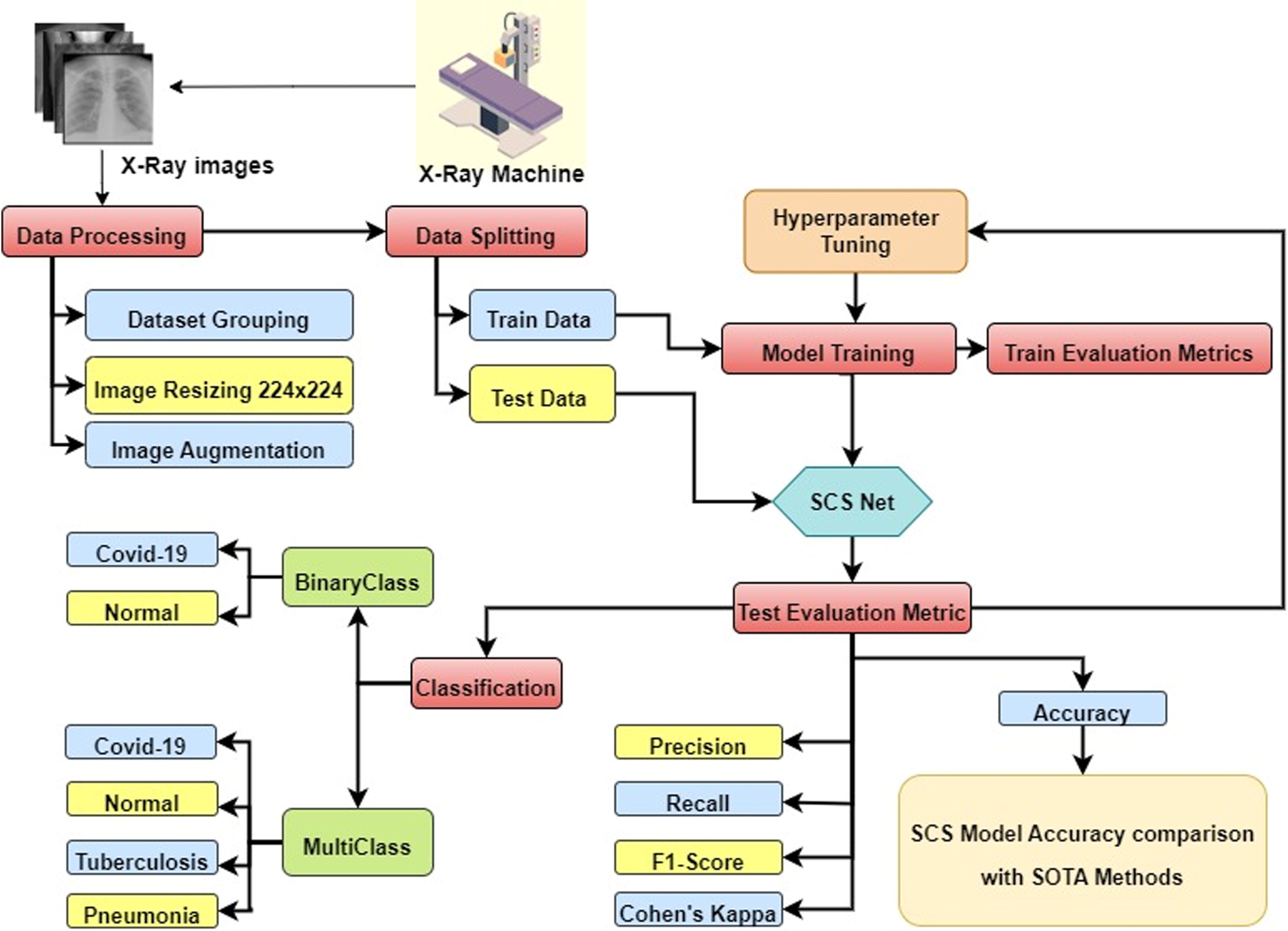
Fig. 4
Overall architecture of SCS-Net.
Where, X = Input, Y = Kernel, Q = Scale invariance, P = Sharpening exponents.
Convolution-like stride operations, such as SCS, extract features from images. The Sharpened Cosine Distance (SCD) is described in equation (4), and the Sharpened Cosine Similarity (SCS) is in equation (5). The sharpening exponents must be learned for each unit. This peak can be sharpened by raising it to a power of an exponent [42]. It performs better than a convolution neural network with 10x-100x fewer parameters and doesn’t require normalization or activation functions after SCS layers. Since the cosine has a wide range, two very different vectors might have a high cosine similarity. It can be sharpened by keeping the sign while increasing the size of the result to a power P. If the signal or kernel magnitude ever approaches 0, this measure may become numerically unstable, increasing the input strength by a modest amount, Q. The kernel magnitude doesn’t grow too narrow.
The benefits of sharpened cosine similarity are as follows:
• Unscaled inputs can be handled by using sharpened cosine similarity.
• The architecture’s simplicity and parameter efficiency seem to be the main advantages of SCS.
• The SCS Net provides better accuracy with an optimal number of model parameters.
• SCS layers are not needed an activation layer, normalization, or dropout layer.
• Maximum absolute pooling is used instead of Maxpooling to choose the element with the most significant magnitude, even if the values are negative.
One of the essential constraints of the SCS is training the model on GPUs and TPUs by raising operations to the power of P often don’t parallelize efficiently. On GPUs, reducing the parameters greatly accelerates performance, but SCS loses its “sharpened” quality.
3.3.1Absolute max-pooling, flatten and dense layer
Pooling is the process of down sampling the dimension of the data. This specific filter is updated in backpropagation until and unless we have an appropriate value for extracting the features in the particular images. Even if the element has a negative, it chooses the highest magnitude. The max-pooling equation is given in equation 6.
(6)
Where, N = Image size, P = size of padding, F = size of filter, S = Stride.
Flatten layer turns data into a one-dimensional column vector to pass the input to a dense layer. The Dense layer contains a fully connected neuron with SoftMax activation for multiclass and sigmoid for binary classification.
3.4Model parameter tuning
The SCS-Net model is trained using 40 epochs, 64 batch sizes, and a 0.0001 learning rate. The momentum and RMS prop optimizer combined model are trained using the Adam optimizer. The Keras tuner library is used to fine-tune the hyperparameters to make the proposed SCS-Net robust. The key reason for utilizing the Adam optimizer is that it uses very little memory and is computationally efficient. The learning rate affects the pace of learning of the DL model, which determines the number of movements required to minimize the loss function value. The momentum enhances both model training speed and accuracy. For binary classification, the sigmoid function is utilized at the output layer, while SoftMax activation is used for multiclass classification.
4Experimental design
An Intel i9 processor, 64GB of RAM, and a 24 GB RTX6000 GPU are employed in the experiment. Model development and training were carried out using the Keras and TensorFlow frameworks. Table 2 shows the model shapes and parameters for binary and multiclass classification. The SCS-Net model for 4 class classification contains 30,620 trainable parameters and for binary classification contains 16,038 trainable parameters.
Table 2
SCS-Net architecture parameters
| Model: “Sequential” | Model: “Sequential” | ||||
| Number of Classes: 4 | Number of Classes: 2 | ||||
| Layer (type) | Output Shape | Param # | Layer (type) | Output Shape | Param# |
| Cos_sim2d(CosSim2D) | (None,222,222,10) | 290 | Cos_sim2d(CosSim2D) | (None,222,222,10) | 290 |
| Cos_sim2d_1(CosSim2D) | (None,222,222,10) | 120 | Cos_sim2d_1(CosSim2D) | (None,222,222,10) | 120 |
| Cos_sim2d_2(CosSim2D) | (None,222,222,10) | 120 | Cos_sim2d_2(CosSim2D) | (None,222,222,10) | 120 |
| Cos_sim2d_3(CosSim2D) | (None,222,222,12) | 144 | Cos_sim2d_3(CosSim2D) | (None,222,222,12) | 144 |
| Cos_sim2d_4(CosSim2D) | (None,222,222,12) | 168 | Cos_sim2d_4(CosSim2D) | (None,222,222,12) | 168 |
| Max_pooling2d | (None,111,111,12) | 0 | Max_pooling2d | (None,111,111,12) | 0 |
| (MaxPooling2D) | (MaxPooling2D) | ||||
| Cos_sim2d_5(CosSim2D) | (None,111,111,24) | 22 | Cos_sim2d_5(CosSim2D) | (None,111,111,24) | 22 |
| Cos_sim2d_6(CosSim2D) | (None,111,111,8) | 208 | Cos_sim2d_6(CosSim2D) | (None,111,111,8) | 208 |
| Max_pooling2d_1 | (None,55,55,8) | 0 | Max_pooling2d_1 | (None,55,55,8) | 0 |
| (MaxPooling2D) | (MaxPooling2D) | ||||
| Cos_sim2d_7(CosSim2D) | (None,53,53,32) | 44 | Cos_sim2d_7(CosSim2D) | (None,53,53,32) | 44 |
| Cos_sim2d_8(CosSim2D) | (None,53,53,10) | 340 | Cos_sim2d_8(CosSim2D) | (None,53,53,10) | 340 |
| Max-abs_pool2d | (None,27,27,10) | 0 | Max-abs_pool2d | (None,27,27,10) | 0 |
| (MaxAbsPool2D) | (MaxAbsPool2D) | ||||
| Flatten | (None,7290) | 0 | Flatten | (None,7290) | 0 |
| Dense | (None,4) | 29164 | Dense | (None,2) | 14,582 |
| Total params: 30,620 | Total params: 16,038 | ||||
| Trainable params: 30,620 | Trainable params: 16,038 | ||||
| Non-trainable params: 0 | Non-trainable params: 0 | ||||
4.1Results and discussion
The experiments are conducted for binary classification and multiclass classification. We have used the 10% dataset for validation and the 90% dataset for training. DL methods use this split ratio frequently for model training and validation [43–45]. The model is trained using the same parameter with a 40-epoch, a 0.0001 learning rate, and a 64-batch size. For training and validation, the 224x224 CXR images are utilized. The model keeps track of the validation accuracy and modifies the initial learning for better feature extraction by using checkpoint callbacks. An Adam optimizer with a momentum of 0.9, a binary cross-entropy loss function for binary classes, and a categorical cross-entropy loss function for multiclass classification are utilized for training the SCS-Net model.
Figure 5(a) and 5(b) show the training and validation accuracy of binary and multiclass classification. Figure 6(a) and 6(b) show the loss of binary and multiclass classification training and validation. Accuracy, precision, recall, F1-score, and kappa score are the metrics used to evaluate the model performance. The formulae for the performance metrics are given in Table 3. Figure 7 shows the confusion matrix of the SCS-Net for multiclass and binary classification, which is used to determine performance metrics for SCS-Net. The SCS-Net model is developed to achieve a minimal false-positive class for binary classification; we achieved only four images that fall into the false positive category for covid-19 disease, and in multiclass, seven images fall into false positive classes for covid-19 disease. In Table 4, the proposed work is compared to different SOTA models reported in XiangYu [46], Stephan [47], Gaobo Liang [48], Gayathri J [49], Soni M [50], Mehedi M [51], Lokeswari V [52], Shikhar J [53], Emtiaz Hussai [54], Tanvir M [55] and Ahmed S Elkorany [56] by accuracy for both binary and multi-classification. The proposed SCS-Net model based on cosine similarity operation that can identify distinct features between covid-19 and other diseases is analyzed. To demonstrate the SCS-Net performance, it is compared with a few published experiments that mainly use CNN architecture. Our SCS-Net obtained an accuracy of 97.67 % (binary class) and 94.05 % (4 class), which is better than SOTA models for covid-19 classification. The following factors contribute to the SCS-Net model’s higher performance. SCS-net models use cosine similarity functions to learn the data projection. The SCS-Net model is more beneficial in model-trainable parameters and also learns non-linear transformations.
Fig. 5
Training and validation accuracy of SCS-Net model (a) Binary Class (b) Multiclass.
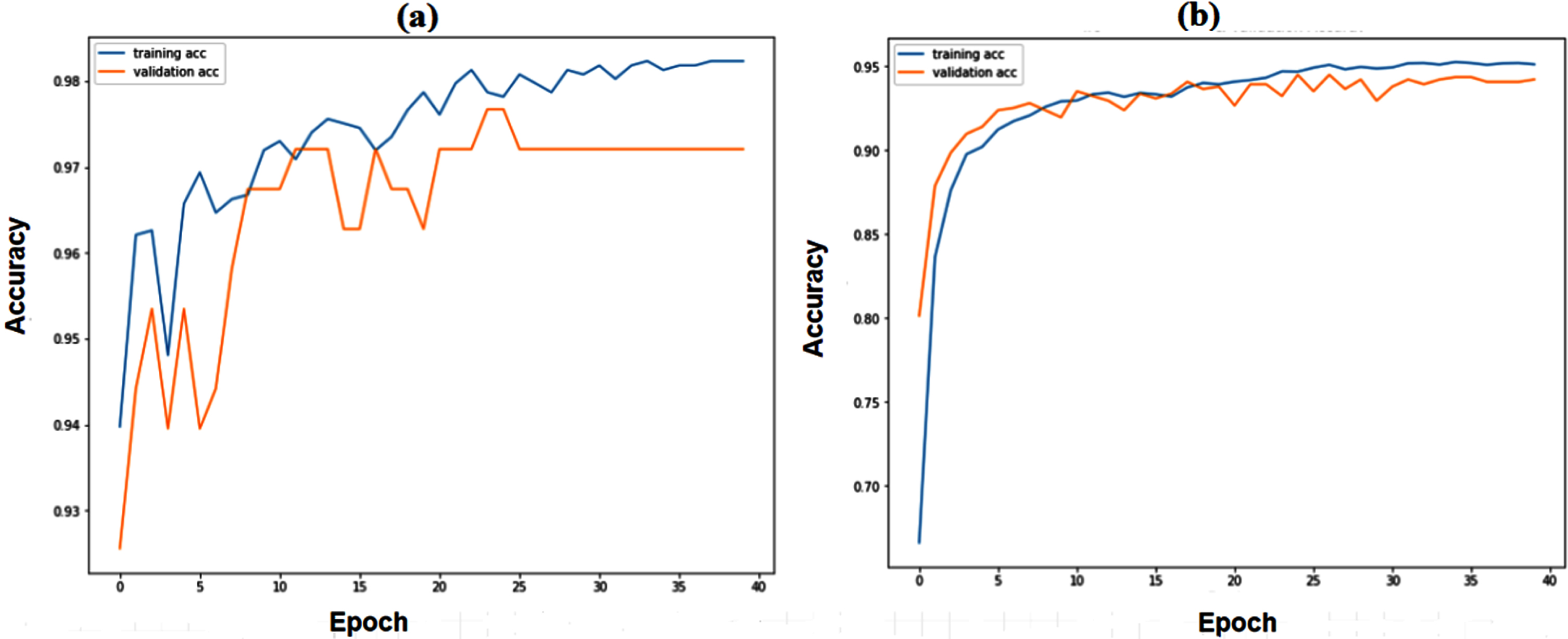
Fig. 6
Training and validation Loss of SCS-Net model (a) Binary Class (b) Multiclass.
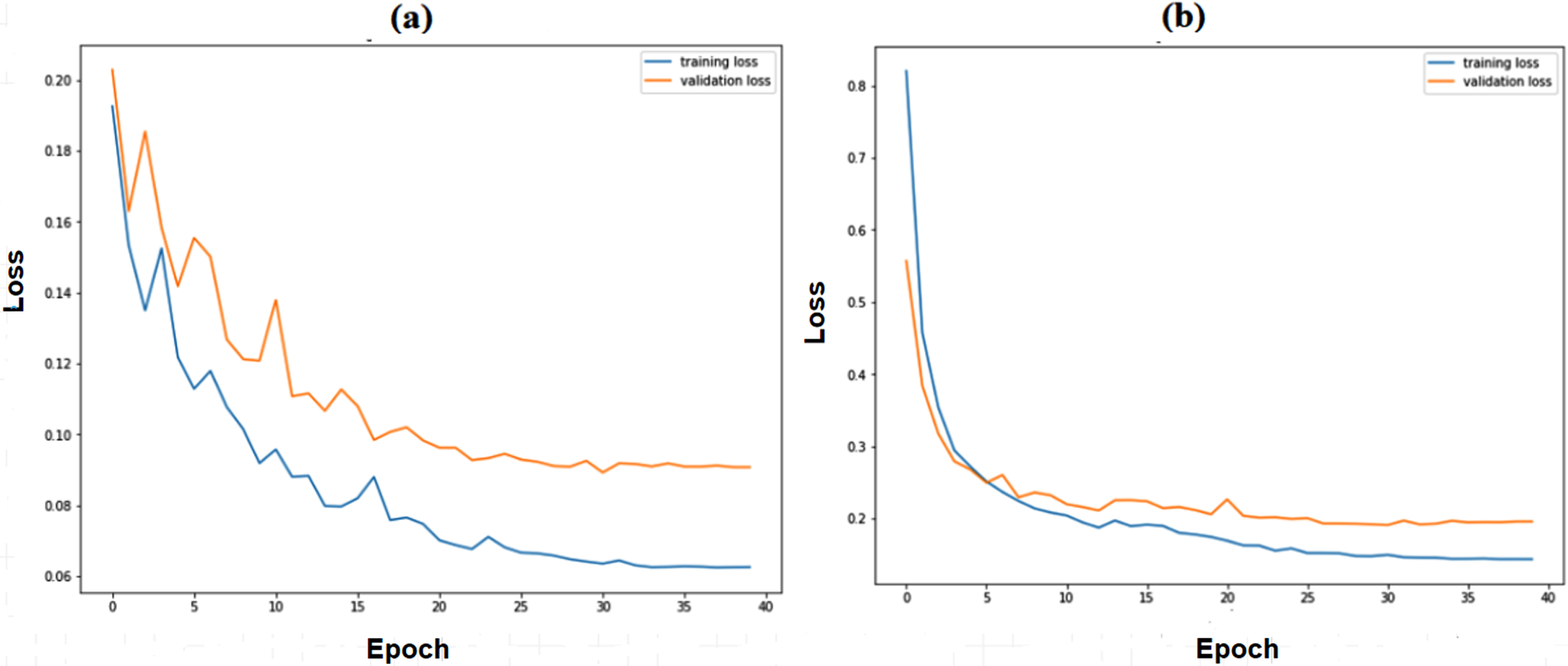
Table 3
Performance Metrics
| Metrics | Formulae |
| tp = true positive, tn = true negative, | |
| fp = false positive, fn = false negative | |
| Accuracy |
|
| Precision |
|
| Recall |
|
| F1-Score |
|
Table 4
Performance Analysis between SCS-Net model and other models for binary and multiclass
| Ref | Classification | Accuracy (%) |
| Binary class | ||
| SCS-Net Model | 2 Class | 97.67 |
| XiangYu [46] | 2 Class | 96.62 |
| Stephan [47] | 2 Class | 94.81 |
| Gaobo Liang [48] | 2 Class | 90.54 |
| Gayathri J [49] | 2 class | 95.78 |
| Soni M [50] | 2 class | 93.00 |
| Mehedi M [51] | 2 class | 92.70 |
| Multi class | ||
| SCS-Net Model | 4 Class | 94.50 |
| Lokeswari V [52] | 4 class | 88.00 |
| Shikhar J [53] | 4 Class | 87.13 |
| Emtiaz Hussai [54] | 4 Class | 91.20 |
| Tanvir M [55] | 4 Class | 90.20 |
| Ahmed S Elkorany [56] | 4 Class | 94.40 |
Fig. 7
(a) Multi-class Classification (b) Binary Class Classification.
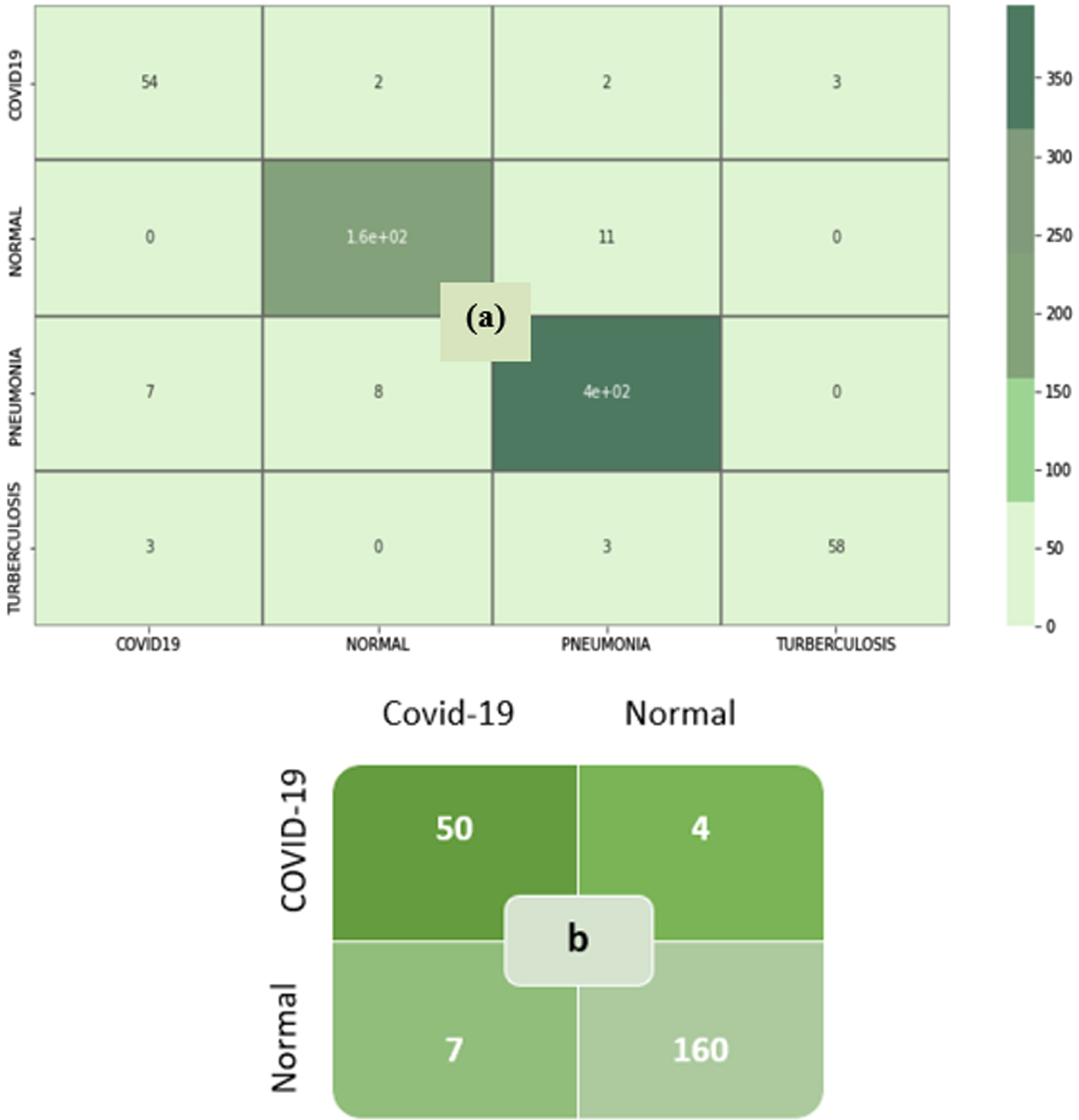
Moreover, compared to conventional CNN model, the SCS-Net model achieved good performance with substantially 10X-100X parameters. The SCS-Net architecture is also trained on an imbalanced dataset, but it proves to be a better model when counting the number of false positives among the classes. As seen in Table 5, model training parameters are significantly less for both binary and multiclass classification. It shows each label’s performance metrics on recall, precision, f1-score, overall accuracy, kappa score, and trainable parameter. The SCS layer proved worthy in extracting the distinct features between different classes and proved better in model training parameters.
Table 5
Performance Analysis of binary and multiclass classification of SCS-Net
| Experiment | Classes | Precision (%) | Recall (%) | F1-score (%) | Kappa score (%) | Accuracy (%) | Parameter |
| Binary Class | Covid-19 | 98.0 | 93.0 | 95.0 | 93.70 | 97.67 | 16,038 |
| Normal | 98.0 | 99.0 | 98.0 | ||||
| Multiclass | Covid-19 | 84.0 | 89.0 | 86.0 | 90.70 | 94.05 | 30,620 |
| Normal | 94.0 | 94.0 | 94.0 | ||||
| Pneumonia | 96.0 | 96.0 | 96.0 | ||||
| Tuberculosis | 95.0 | 91.0 | 93.0 |
The Covid-19, tuberculosis, pneumonia, and normal classes processed using the SCS layer’s filter are captured in the feature space. Visualize a feature map for a specific image to explain the observed features. In contrast to feature maps towards the model output, which capture more distinctive features, feature maps nearer the input are thought to identify fine-grained information. While the deeper layer networks learn more sophisticated features like pathological lesions, the first layer networks always learn basic features like edges, lines, patterns, and color. Later layers incorporate features from earlier layers to create new features. To analyze the feature maps of the proposed SCS Net model are shown in Fig. 8. Figure 8(a) shows the feature map of covid-19 class, Fig. 8(b) shows the feature maps of the pneumonia class, 8(c) shows the feature maps of the tuberculosis class, and finally, 8(d) shows the feature maps of normal class at the SCS layer. The SCS-Net feature map visualization demonstrates that the model can recognize features that can distinguish between the classes of covid-19, pneumonia, tuberculosis, and normal class.
Fig. 8
Feature map visualization of SCS layer.
Finally, our technique is cost-effective and time-efficient, and healthcare practitioners can quickly adopt it, enabling faster covid-19 screening processes and earlier isolation of patients. If a CXR image is available, real-time screening of covid-19 patients using SCS-Net techniques may be achievable with minimal human intervention. Furthermore, AI-based screens can be tailored to the level of understanding of the end user, and technicians may not be required to be trained in complex computational procedures. The following are the study’s drawbacks, which present the immediate potential for additional research.
• The model is accomplished on a minimal dataset and may be tested with a diverse dataset in the future.
• Different image modalities of covid-19 disease will be trained and tested in SCS-Net.
5Conclusion and future scope
Even without user intercession, covid-19 detection using computational methods from CXR images can accomplish with an accuracy of 97.67% and a kappa score of 93.70% in binary and multiclass classification models of 94.05% and a kappa score of 90.70%, respectively. These systems are sophisticated to operate with smaller datasets, and such methods are critical in situations where test kits are scarce. This work proposed SCS-Net using sharpened cosine similarity to perform binary class (covid-19 and normal and multiclass datasets (covid-19, Normal, Tuberculosis, and Pneumonia). It proves that the SCS-Net model provides better accuracy, recall, precision, F1-Score, and Kappa score with few training parameters. The SCS-Net better discriminates the covid-19 cases in both binary and multiclass experiments. It can be achieved by the best combinations of SCS layers and model parameters for training the SCS-Net architecture. As a result, for the initial evaluation of covid-19, the SCS-Net can be useful for radiologists and other healthcare professionals. The SCS-Net has much practical application potential and can help front-line medical staff diagnose covid-19 accurately and quickly.
In the future, the proposed work can be extended to use different image modalities like CT scans, CXR images, and ultrasound to predict covid-19. The GAN will be incorporated in SCS-Net model to tackle dataset balance issue. The vision transformers model may also be explored in the future to classify covid-19 diseases. Furthermore, we will strive to test our model with diverse datasets.
Declaration of competing interest
The authors declare that they have no established conflicting financial interests or personal relationships that may have influenced the research presented in this paper.
Data availability
The corresponding author will provide the datasets used for the current study upon reasonable request.
References
[1] | Gayathri J.L. , Abraham B. , Sujarani M.S. and Nair M.S. , A computer-aided diagnosis system for the classification of COVID-19 and non-COVID-19 pneumonia on chest X-ray images by integrating CNN with sparse autoencoder and feed forward neural network, Comput. Biol. Med 141: (November 2021) ((2022) ), 105134. doi: 10.1016/j.compbiomed.2021.105134. |
[2] | Abdi A. , Jalilian M. , Sarbarzeh P.A. and Vlaisavljevic Z. , Diabetes and COVID-19: A systematic review on the current evidences, Diabetes Res. Clin. Pract. 166: ((2020) ), 108347. doi: 10.1016/j.diabres.2020.108347. |
[3] | Mamalakis M. , et al., DenResCov-19: A deep transfer learning network for robust automatic classification of COVID-19, pneumonia, and tuberculosis from X-rays, Comput. Med. Imaging Graph. 94: (April) ((2021) ), 102008, doi: 10.1016/j.compmedimag.2021.102008. |
[4] | Maghdid H.S. , Ghafoor K.Z. , Sadiq A.S. , Curran K. and Rabie K. , A Novel AI-enabled Framework to Diagnose Coronavirus COVID-19 using Smartphone Embedded Sensors: Design Study, pp. 1–5. |
[5] | Malhotra A. , et al., Multi-task driven explainable diagnosis of COVID-19 using chest X-ray images, Pattern Recognit. 122: ((2022) ), 108243, doi: 10.1016/j.patcog.2021.108243. |
[6] | Zheng C. , Deng X. , Fu Q. and Zhou Q. , Deep Learning-based Detection for COVID-19 from Chest CT usingWeak Label (2020), 1–13. |
[7] | Kassania S.H. , Kassasni P.H. , Wesolowski M.J. , Schneider K.A. and Deters R. , Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: A machine learning based approach, arXiv 2019: ((2020) ), 1–18. |
[8] | Saygılı A. , A new approach for computer-aided detection ofcoronavirus (COVID-19) from CT and X-ray images using machinelearning methods, Appl. Soft Comput. 105: ((2021) ), 107323, doi: 10.1016/j.asoc.2021.107323. |
[9] | Chandran V. , et al., Diagnosis of Cervical Cancer based on Ensemble Deep Learning Network using Colposcopy Images , 2021: ((2021) ). |
[10] | Pathan S. , Siddalingaswamy P.C. and Ali T. , Automated Detection of Covid-19 from Chest X-ray scans using an optimized CNN architecture, Appl. Soft Comput. 104: ((2021) ), 107238, doi: 10.1016/j.asoc.2021.107238. |
[11] | Sabeena Beevi K. , Nair M.S. and Bindu G.R. , Automatic mitosis detection in breast histopathology images using Convolutional Neural Network based deep transfer learning, Biocybern. Biomed. Eng. 39: (1) ((2019) ), 214–223, doi: 10.1016/j.bbe.2018.10.007. |
[12] | Afshar P. , Heidarian S. , Naderkhani F. , Oikonomou A. , Plataniotis K.N. and Mohammadi A. , COVID-CAPS: Acapsule network-based framework for identification of COVID-19cases from X-ray images, Pattern Recognit. Lett. 138: ((2020) ), 638–643, doi: 10.1016/j.patrec.2020.09.010. |
[13] | Li J. , et al., Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19, Pattern Recognit 114: ((2021) ), 107848, doi: 10.1016/j.patcog.2021.107848. |
[14] | Wang L. , et al., Trends in the application of deep learning networks in medical image analysis: Evolution between 2012 and 2020, Eur. J. Radiol. 146: ((2022) ), 110069, doi: 10.1016/j.ejrad.2021.110069. |
[15] | Shi F. , et al., Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19, arXiv (2020), 1–11. |
[16] | Nabavi S. , et al., Medical imaging and computational image analysis in COVID-19 diagnosis: A review, Comput. Biol. Med. 135: (November 2020) ((2021) ), 104605, doi: 10.1016/j.compbiomed.2021.104605. |
[17] | Bharati S. , Podder P. and Mondal M.R.H. , Hybrid deep learning for detecting lung diseases from X-ray images, Informatics Med. Unlocked 20: ((2020) ), 100391, doi: 10.1016/j.imu.2020.100391. |
[18] | Hertel R. and Benlamri R. , COV-SNET: A deep learning model for X-ray-based COVID-19 classification, Informatics Med. Unlocked 24: (May) ((2021) ), 100620, doi: 10.1016/j.imu.2021.100620. |
[19] | Alirezazadeh P. , Hejrati B. , Monsef-Esfahani A. and Fathi A. , Representation learning-based unsupervised domain adaptation for classification of breast cancer histopathology images, Biocybern. Biomed. Eng. 38: (3) ((2018) ), 671–683, doi: 10.1016/j.bbe.2018.04.008. |
[20] | Waheed A. , Goyal M. , Gupta D. , Khanna A. , Al-Turjman F. and Pinheiro P.R. , CovidGAN: Data Augmentation Using Auxiliary Classifier GAN for Improved Covid-19 Detection, IEEE Access 8: ((2020) ), 91916–91923, doi: 10.1109/ACCESS.2020.2994762. |
[21] | Panthakkan A. , Anzar S.M. , Al Mansoori S. and Al Ahmad H. , A novel DeepNet model for the efficient detection of COVID-19 for symptomatic patients, Biomed. Signal Process. Control 68: (May) ((2021) ), 102812, doi: 10.1016/j.bspc.2021.102812. |
[22] | Al-Waisy A.S. , et al., COVID-DeepNet: Hybrid Multimodal Deep Learning System for Improving COVID-19 Pneumonia Detection in Chest X-ray Images, Comput. Mater. Contin. 67: (2) ((2021) ), 2409–2429, doi: 10.32604/cmc.2021.012955. |
[23] | Turkoglu M. , COVID-19 Detection System Using Chest CT Images and Multiple Kernels-Extreme Learning Machine Based on Deep Neural Network, Irbm 42: (4) ((2021) ), 207–214, doi: 10.1016/j.irbm.2021.01.004. |
[24] | Murugan R. , Goel T. , Mirjalili S. and Chakrabartty D.K. , WOANet: Whale optimized deep neural network for the classification of COVID-19 from radiography images, Biocybern. Biomed. Eng. 41: (4) ((2021) ), 1702–1718, doi: 10.1016/j.bbe.2021.10.004. |
[25] | Wu T. , Tang C. , Xu M. , Hong N. and Lei Z. , ULNet for the detection of coronavirus (COVID-19) from chest X-ray images, Comput. Biol. Med. 137: (September) ((2021) ), 104834. doi: 10.1016/j.compbiomed.2021.104834. |
[26] | Prakash N.B. , Murugappan M. , Hemalakshmi G.R. , Jayalakshmi M. and Mahmud M. , Deep transfer learning for COVID-19 detection and infection localization with superpixel based segmentation, Sustain. Cities Soc. 75: (March) ((2021) ). doi: 10.1016/j.scs.2021.103252. |
[27] | Aggarwal S. , Gupta S. , Alhudhaif A. , Koundal D. , Gupta R. and Polat K. , Automated COVID-19 detection inchest X-ray images using fine-tuned deep learningarchitectures, Expert Syst. 39: (3) ((2022) ), 1–17, doi: 10.1111/exsy.12749. |
[28] | Ullah N. , et al., A Novel CovidDetNet Deep Learning Model for Effective COVID-19 Infection Detection Using Chest Radiograph Images, Appl. Sci. 12: (12) ((2022) ), doi: 10.3390/app12126269. |
[29] | Bhattacharyya A. , Bhaik D. , Kumar S. , Thakur P. , Sharma R. and Pachori R.B. , A deep learning based approach for automatic detection of COVID-19 cases using chest X-ray images, Biomed. Signal Process. Control 71: (PB) ((2022) ), 103182, doi: 10.1016/j.bspc.2021.103182. |
[30] | Jalali S.M.J. , Ahmadian M. , Ahmadian S. , Hedjam R. , Khosravi A. and Nahavandi S. , X-ray image based COVID-19 detection using evolutionary deep learning approach, Expert Syst. Appl 201: (March) ((2022) ), 116942, doi: 10.1016/j.eswa.2022.116942. |
[31] | Ghosh S.K. and Ghosh A. , ENResNet: A novel residual neural network for chest X-ray enhancement based COVID-19 detection, Biomed. Signal Process. Control 72: (PA) ((2022) ), 103286, doi: 10.1016/j.bspc.2021.103286. |
[32] | Goel T. , Murugan R. , Mirjalili S. and Chakrabartty D.K. , Multi-COVID-Net: Multi-objective optimized network for COVID-19 diagnosis from chest X-ray images, Appl. Soft Comput. 115: ((2022) ), 108250, doi: 10.1016/j.asoc.2021.108250. |
[33] | Banerjee A. , Bhattacharya R. , Bhateja V. , Singh P.K. , Lay-Ekuakille A. and Sarkar R. , COFE-Net: An ensemble strategy for Computer-Aided Detection for COVID-19, Meas. J. Int. Meas. Confed. 187: (September 2021) ((1102) ), 110289, doi: 10.1016/j.measurement.2021.110289. |
[34] | Kumar A. , Tripathi A.R. , Satapathy S.C. and Zhang Y.D. , SARS-Net: COVID-19 detection from chest x-rays by combining graph convolutional network and convolutional neural network, Pattern Recognit. 122: ((2022) ), 108255, doi: 10.1016/j.patcog.2021.108255. |
[35] | Kavuran G. , İn E. , Geçkil A.A. , Şahin M. and Berber N.K. , MTU-COVNet: A hybrid methodology for diagnosing the COVID-19 pneumonia with optimized features from multi-net, Clin. Imaging 81: (September 2021) ((2022) ), 1–8, doi: 10.1016/j.clinimag.2021.09.007. |
[36] | Mooney P. , Chest X-Ray Images (Pneumonia). https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia. |
[37] | Tawsifur Rahman A.K. and Dr. Muhammad Chowdhury , Tuberculosis (TB) Chest X-ray Database. https://www.kaggle.com/datasets/tawsifurrahman/tuberculosis-tb-chest-xray-dataset. |
[38] | Patel P. , Chest X-ray (Covid-19 & Pneumonia). https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia. |
[39] | Covid-chestxray-dataset. https://github.com/ieee/covid-chestxray-dataset. |
[40] | Pisoni R. , Sharpened Cosine Distance as an Alternative for Convolutions (2022). https://www.rpisoni.dev/posts/cossim-convolution/. |
[41] | Luo C. , Zhan J. , Xue X. , Wang L. , Ren R. and Yang Q. , Cosine normalization: Using cosine similarity instead of dot product in neural networks, (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), Lect. Notes Comput. Sci. 11139: (LNCS) ((2018) ), 382–391, doi: 10.1007/978-3-030-01418-6_38. |
[42] | Pisoni R. , Sharpened Cosine Similarity: Part 2. https://www.rpisoni.dev/posts/cossim-convolution-part2/. |
[43] | Ahsan M.M. , Alam T.E. , Trafalis T. and Huebner P. , Deep MLP-CNN model using mixed-data to distinguish between COVID-19 and Non-COVID-19 patients, Symmetry (Basel) 12: (9) ((2020) ), doi: 10.3390/sym12091526. |
[44] | Venkatesan C. , Sumithra M.G. and Murugappan M. , NFU-Net: An Automated Framework for the Detection of Neurotrophic Foot Ulcer Using Deep Convolutional Neural Network, Neural Process. Lett. (2022), doi: 10.1007/s11063-022-10782-0. |
[45] | Murugan S. , et al., DEMNET: A Deep Learning Model for Early Diagnosis of Alzheimer Diseases and Dementia from MR Images, IEEE Access 9: ((2021) ), 1–1, doi: 10.1109/access.2021.3090474. |
[46] | Yu X. , Lu S. , Guo L. , Wang S.H. and Zhang Y.D. , ResGNet-C: A graph convolutional neural network for detection of COVID-19, Neurocomputing 452: ((2021) ), 592–605, doi: 10.1016/j.neucom.2020.07.144. |
[47] | Stephen O. , Sain M. , Maduh U.J. and Jeong D.U. , An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare, , J. Healthc. Eng. 2019: ((2019) ), doi: 10.1155/2019/4180949. |
[48] | Liang G. and Zheng L. , A transfer learning method with deep residual network for pediatric pneumonia diagnosis, Comput. Methods Programs Biomed 187: ((2020) ), doi: 10.1016/j.cmpb.2019.06.023. |
[49] | Gayathri J.L. , Abraham B. , Sujarani M.S. and Nair M.S. , A computer-aided diagnosis system for the classification of COVID-19 and non-COVID-19 pneumonia on chest X-ray images by integrating CNN with sparse autoencoder and feed forward neural network, Comput. Biol. Med. 141: (December 2021) ((2022) ), 105134, doi: 10.1016/j.compbiomed.2021.105134. |
[50] | Soni M. , Singh A.K. , Babu K.S. , Kumar S. , Kumar A. and Singh S. , Convolutional neural network based CT scan classification method for COVID-19 test validation, Smart Heal. 25: (June) ((2022) ), 100296, doi: 10.1016/j.smhl.2022.100296. |
[51] | Masud M. , A light-weight convolutional Neural Network Architecture for classification of COVID-19 chest X-Ray images, Multimed. Syst. (0123456789) ((2022) ), 1–10, doi: 10.1007/s00530-021-00857-8. |
[52] | Venkataramana L. , et al., Classification of COVID-19 from tuberculosis and pneumonia using deep learning techniques, 1: ((2022) ), 3, doi: 10.1007/s11517-022-02632-x. |
[53] | Johri S. , Goyal M. , Jain S. , Baranwal M. , Kumar V. and Upadhyay R. , A novel machine learning-based analytical framework for automatic detection of COVID-19 using chest X-ray images, Int. J. Imaging Syst. Technol. 31: (3) ((2021) ), 1105–1119, doi: 10.1002/ima.22613. |
[54] | Hussain E. , Hasan M. , Rahman M.A. , Lee I. , Tamanna T. and Parvez M.Z. , CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images, Chaos, Solitons and Fractals 142: ((2021) ), 110495, doi: 10.1016/j.chaos.2020.110495. |
[55] | Mahmud T. , Rahman M.A. and Fattah S.A. , CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization, Comput. Biol. Med. 122: (May) ((2020) ), 103869, doi: 10.1016/j.compbiomed.2020.103869. |
[56] | Elkorany A.S. and Elsharkawy Z.F. , COVIDetection-Net: A tailored COVID-19 detection from chest radiography images using deep learning, Optik (Stuttg). 231: (January) ((1664) ), 166405, doi: 10.1016/j.ijleo.2021.166405. |




