Suppression of Krüppel-like factor 5 basal expression by CREB1 binding to far distal element
Abstract
BACKGROUND:
Krüppel-like factor 5 (KLF5) is a transcription factor regulating the proliferation and differentiation of epithelial cells, and its uncontrolled expression is closely associated with carcinoma progression. Sp3 binding to the minimal essential region (MER) of KLF5 gene is critical for KLF5 basal expression, but the expression control mechanism is unknown.
OBJECTIVE:
This study aimed to identify a regulatory region for KLF5 basal expression and the binding protein in carcinoma cells by analyzing the promoter upstream region.
METHODS:
Reporter assays determined the silencer region. The protein binding to the region was identified by database analysis and ChIP assay. The protein mediating the interaction between the region and the MER was confirmed through chromosome conformation capture (3 C) on ChIP assay. The effects of the protein on KLF5 expression were analyzed using qRT-PCR and western blot.
RESULTS:
Reporter assay localized the 425-region from upstream KLF5 gene as the silencer. Database analysis and ChIP assay found CREB1 binding to the 425-region. CREB1 siRNA or mutation of CREB1-binding site in the 425-region increased luciferase activities and decreased the binding to 425-region. 3 C on ChIP assay showed that CREB1 mediated interaction of the 425-region and the MER. CREB1 overexpression decreased endogenous KLF5 expression and luciferase activity.
CONCLUSIONS:
The 425-region is the silencer of KLF5 basal expression, and CREB1 binding suppresses the expression.
1Introduction
Development and maintenance of epithelial tissues require various regulatory factors involved in the proliferation and differentiation of cells and the presence of epithelial stem cells. In stratified squamous epithelium, keratinocytes express transcription factors according to the basal cell layer and suprabasal cell layer to organize tissue integrity. Their uncontrolled expression is frequently observed in squamous cell carcinomas and is associated with the progression [1–4]. Previous studies documented that an aggressive subset of carcinomas occasionally overexpress transcriptional factors that are usually expressed in basal cells [5–9]. It emphasizes the importance of transcriptional regulation of target genes in the pathophysiology of epithelial tissue.
Krüppel-like factor (KLF) consists of three C2H2-zinc finger domains and transcriptionally regulates various developmental and cellular phenomena [10]. Among 17 members of the family, KLF5 is rich in intestinal epithelial cells and controls proliferation and differentiation balance of the cells [2, 10]. It is also expressed in the basal cell layer of squamous cell epithelium and rapidly disappears in suprabasal differentiating cells [11]. KLF5 overexpression in mouse epidermis results in basal cell hyperplasia [12], and the expression is frequently up-regulated in patients with poorly differentiated and aggressive carcinomas [8, 13–18]. Thus, the mechanism of KLF5 gene transcription is important for understanding normal and carcinoma cell states.
We have shown that the minimal essential region (MER) required for KLF5 basal expression is located downstream of the transcription start site (+145 to +330) and that Sp3 binding to the GC box in the MER is critical for basal expression [19]. However, nothing is known about mechanisms to up- or down-regulate the expression. This study shows that the 425-region from -2,001 to -1,577 acts as a silencer through binding with CREB1.
2Materials and methods
2.1Cells and constructs
Human oral carcinoma cell lines (HSC2, HO-1-u-1, HSC3, KOSC2 and Ca9-22), MCF7 (breast carcinoma), DLD1 (colon carcinoma), T24 (urinary bladder carcinoma), HT1080 (fibrosarcoma), and HEK293T (embryonic kidney) were obtained from Cell Resource Center for Biomedical Research, Cell Bank (Tohoku University, Sendai, Japan), Health Science Research Resources Bank (Osaka, Japan) or RIKEN Cell Bank (Tsukuba, Japan).
CREB1 cDNA with Myc-tag at the 3’ end (Origene Technologies, Rockville, MD, USA) was cloned into pCI-neo plasmid (Promega, Maddison, WI, USA). KLF5 gene promoter (-2,894 to +424, transcription start site designated to as +1, GenBank Accession Number NM_001730) was amplified by PCR using a human bacterial artificial chromosome clone RP11-138K4 and KOD-FX DNA polymerase (TOYOBO, Osaka, Japan). Fragments with different upstream sites and commonly stopping at +424 (clones 1–6, Fig. 1B) were generated using PCR primers with KpnI and XhoI-sites at forward and reverse primers, respectively, and were ligated to upstream of luciferase-reporter gene in pGL4.10 plasmid (Promega). Clone Δ5 lacking a region from -1,576 to +144 (Fig. 2) and constructs mutating putative CREB1-binding sites (Mut #1, #2, #3 and #(2 + 3); Fig. 4B) were generated by PCR. Primer sequences are listed in Table 1, and all constructs were used after confirmation of DNA sequences. Short interfering RNAs (siRNAs, Invitrogen, Carlsbad, CA, USA) for CREB1 (s3489, s3490), CEBPB (s2891, s2892), or p63 (s16411, s229400) and a negative control siRNA (siCtrl, Silencer Select Negative Control siRNA #2) were used.
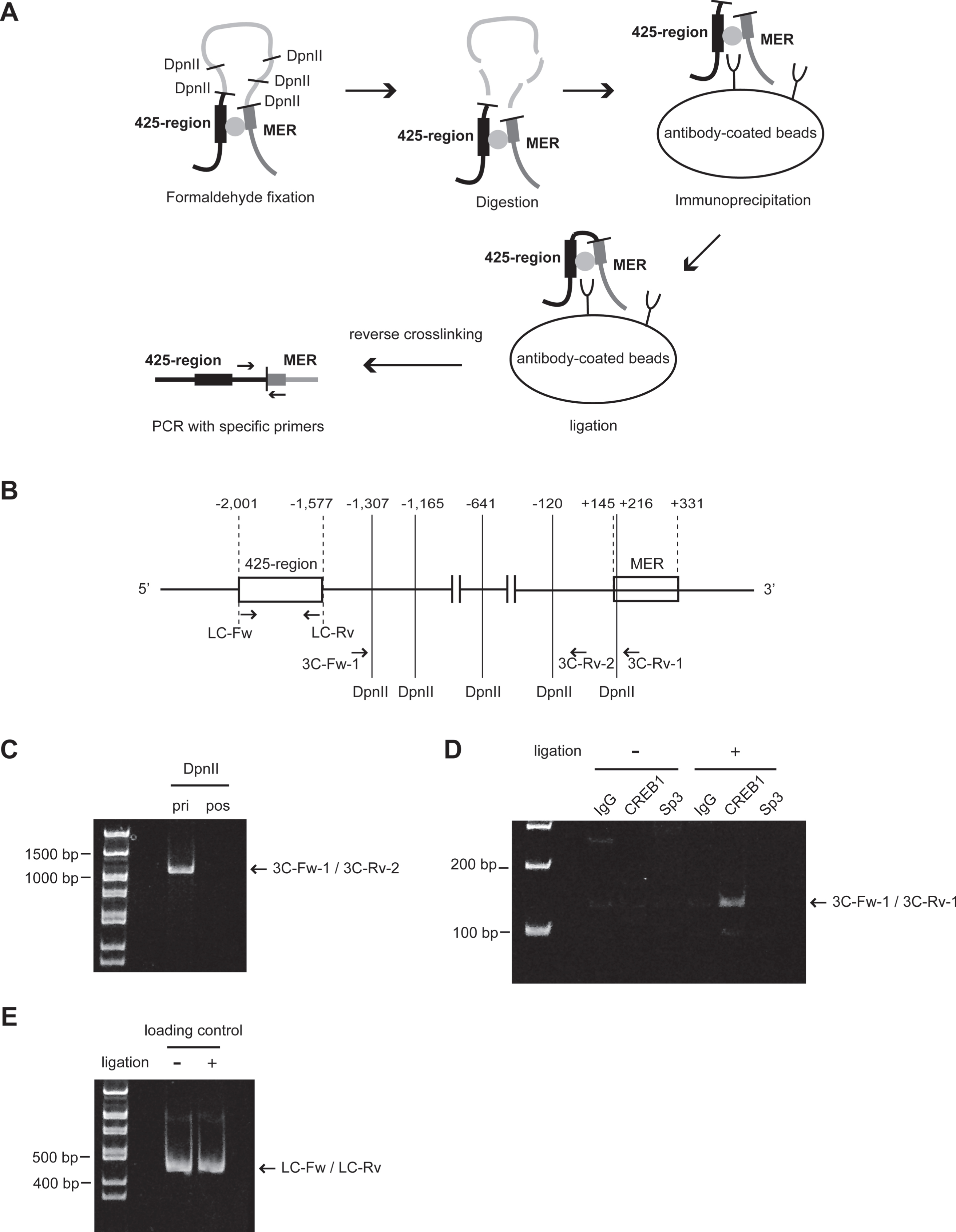
Fig. 1
Identification of KLF5 gene silencer region. (A) Conservation of a 3 kb genomic region from -2,894 to +424 in mammals visualized using ECR Browser (https://ecrbrowser.dcode.org/). The X-axis indicates genomic coordinates. The Y-axis indicates the degree of conservation between the human and mammalian genome. Horizontal blue lines above the graph, with strand/transcriptional orientation represent by arrows. Coding exons are depicted in blue, untranslated regions in yellow, intergenic regions in red, transposable elements and simple repeats in green and introgenic regions in salmon pink. Exon1 represents KLF5 (GenBank Accession Number NM_001730). The 3 kb region contained introgenic regions of KLF5 (NM_001286818). (B) Relative luciferase activities (RLA) in HSC2 cells. Luciferase-reporter plasmids conjugated with various length of KLF5 upstream region were constructed (left panel), and RLA standardized by renilla control plasmid were measured (right panel). (C) RLA of KLF5 upstream region deletion constructs in other oral carcinoma cell lines. (D) RLA of KLF5 upstream region deletion constructs in other types of malignant tumor cell lines and HEK293T cells. The data are presented as means±SD of quadruplicate experiments (n = 4). Statistical significances were compared with the former clones in each cell line (*, P < 0.05, **, P < 0.01 and ***, P < 0.001).
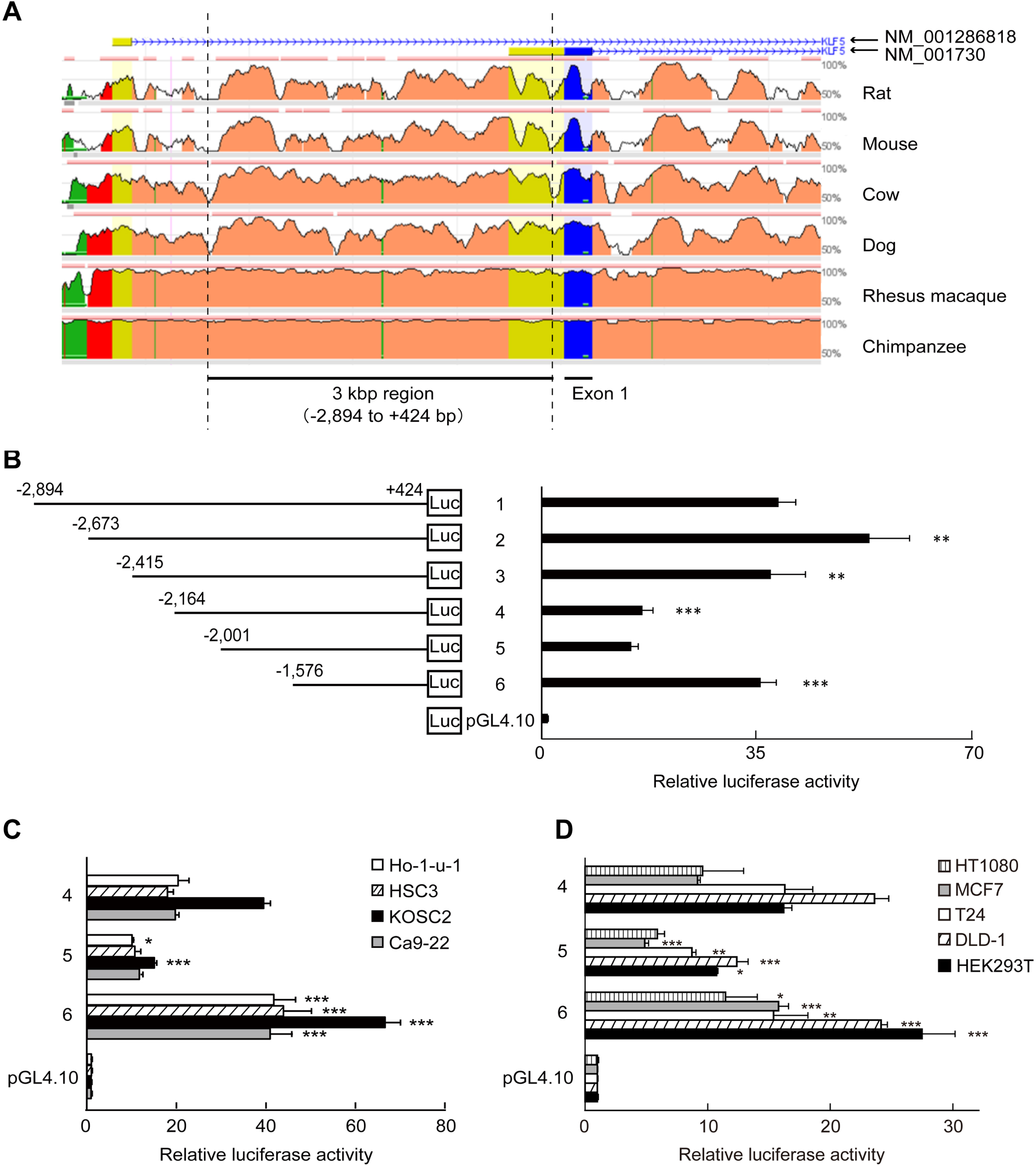
Fig. 2
The inhibition of MER activity of KLF5 gene by the 425-region. Clones containing MER of KLF5 gene or other fragments were transfected into HSC2 cells. RLA were normalized by renilla control plasmid. Clone Δ5 was a construct that ligated the 425-region (-2,001 to -1,577) and the MER (+145 to+331). Clone MER and clone ΔMER were constructs that contained the MER and deleted the MER from clone MER, respectively. The data are presented as means±SD of quadruplicate experiments (n = 4). *, P < 0.05 compared to clone MER.
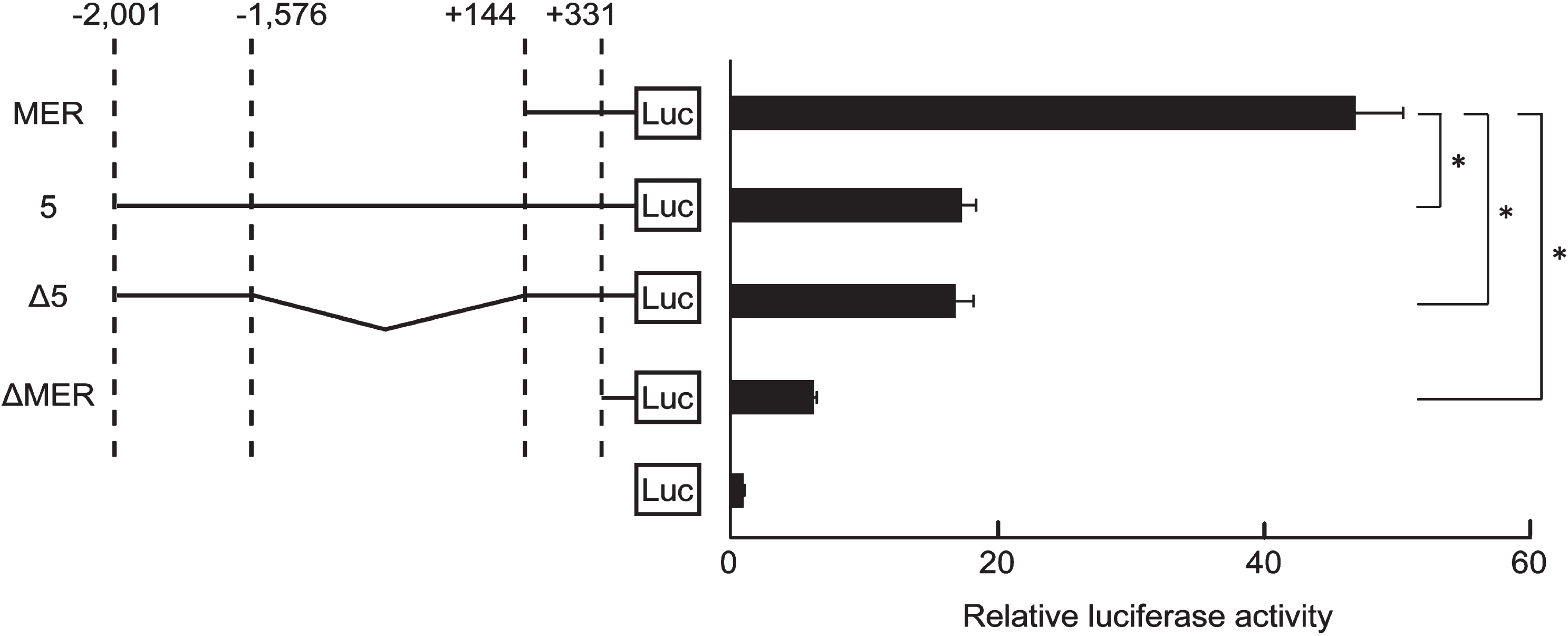
Fig. 4
Insertion of mutation at CREB1-binding sites in the 425-region. (A) Binding of CREB1 to the 425-region of the endogenous genome in HSC2 cells was analyzed by ChIP assay using anti-CREB1 antibody at each concentration. Input DNA (Input) and precipitates from non-immune IgG (IgG) were used as positive and negative controls in ChIP assay, respectively. 1.5μg, 0.75μg and 0.375μg were amounts of anti-CREB1 antibody used in ChIP assay. NC and KLF5 BAC were used for negative and positive controls in end-point PCR, respectively. Water or KLF5 BAC clone were used as the PCR templates. (B) RLA of clone Δ5 and mutants at putative CREB1-binding sites (Mut #1, Mut #2, Mut #3 and Mut # (2 + 3)). These clones were transfected into HSC2. Luc indicates Luciferase (Luc)-reporter gene in pGL4.10 plasmid. Squares are the CREB1-binding sites, and filled squares are mutation CREB1 binding sites. The data were represented as means±SD (n = 4). *, P < 0.05 compared to Δ5. (C) The primer settings for quantitative ChIP were designed to include the plasmid DNA sequence. The total length of PCR product was 477 bp containing the 425-region. Squares filled in gray (#1, #2 and #3) are the CREB1-binding sites. Fw and Rv indicate forward and reverse primers, respectively. (D) Relative folds of immunoprecipitation of CREB1 to clone Δ5 with (Mut #1, Mut #2, Mut #3 and Mut # (2 + 3)) or without mutation were standardized by IgG (n = 4). pGL4.10 was mock. *, P < 0.05 compared to Δ5.
Table 1
PCR primer sequences
| Subject | Primers | Sequencea | |
| Reporter assayb | clone 1 | F | 5′-GAGGTACCTCTATATTAATTTCAAGC-3′ |
| clone 2 | F | 5′-GAGGTACCCCCCCGCATATTCATC-3′ | |
| clone 3 | F | 5′-GAGGTACCTATCTTCCCTAATG-3′ | |
| clone 4 | F | 5′-GAGGTACCCACATGGCAAATG-3′ | |
| clone 5 | F | 5′-GAGGTACCTTTAAAACCTGCTACTG-3′ | |
| clone 6 | F | 5′-GAGGTACCAGGTTTGATGTTATCAG-3′ | |
| clone 1–6 | R | 5′-GATCTCGAGCGGGTGGACTCCTCA-3′ | |
| Mutagenesisc | clone Δ5 | F | 5′-TGCCAATCAGGCGAT-3′ |
| clone Mut #1 | F | 5′-ACCCCAACGCTAGGCTGAGATCTATATTATGAA-3′ | |
| clone Mut #2 | F | 5′-CGGGCCACTGAAGCTCAGCTAAAAAGACAAAAGGA-3′ | |
| clone Mut #3 | F | 5′-ATTAAGAATTATCTAGCTGAGATTTTGCCAATCAG-3′ | |
| clone Δ5 | R | 5′-AAATGTGATGTAGATAATTC-3′ | |
| clone Mut #1 | R | 5′-TTCATAATATAGATCTCAGCCTAGCGTTGGGGT-3′ | |
| clone Mut #2 | R | 5′-TCCTTTTGTCTTTTTAGCTGAGCTTCAGTGGCCCG-3′ | |
| clone Mut #3 | R | 5′-CTGATTGGCAAAATCTCAGCTAGATAATTCTTAAT-3′ | |
| CREB1 cDNAd | CREB1-Myc | F | 5′-GAGCTAGCATGACCATGGAATCTGGAGC-3′ |
| CREB1-Myc | R | 5′-CAGAATTCTTACAGATCCTCTTCTGAGATG-3′ | |
| ChIPe | Endogenous the 425-region | F | 5′-TTTAAAACCTGCTACTGTATTG-3′ |
| clone Δ5 | F | 5′-CAGTGCAAGTGCAGGTGCCAGAAC-3′ | |
| Endogenous the 425-region | R | 5′-AAATGTGATGTAGATAATTC-3′ | |
| clone Δ5 | R | 5′-AAATGTGATGTAGATAATTC-3′ | |
| 3C on ChIPf | 3C-Fw-1 | F | 5′-GCAAAAGCACTTATTAAATAATAC-3′ |
| LC-Fw | F | 5′-TTTAAAACCTGCTACTGTATTG-3′ | |
| 3C-Rv-1 | R | 5′-ACTACTGACACTTGACGCCC-3′ | |
| 3C-Rv-2 | R | 5′-ACAACTTCTCTGACAGATTG-3′ | |
| LC-Rv | R | 5′-AAATGTGATGTAGATAATTC-3′ |
aForward primer (F), reverse primer (R). bKpnI-susceptible site XhoI-susceptible sequence in forward and reverse primers were underlined, respectively. cSubstitution of nucleotide in CREB1-binding site mutants (Mut #1 to #3) were shown in bold. dNheI-susceptible site EcoRI-susceptible sequence in forward and reverse primers were underlined, respectively. ePrimers for amplification of endogenous the 425-region and clone Δ5 CREB1-binding region were used for conventional and quantitative ChIP assay, respectively. f3 C on ChIP assay primers for confirmation of the interaction of the 425-region with MER of KLF5 gene, and of DpnII digestion. LC-Fw and LC-Rv primers were used for amplifying loading control products.
The study was approved by The Nippon Dental University School of Life Dentistry at Tokyo Safety Committee on Genetic Recombination Experiments (approval no. #56) and carried out according to the guidelines of the committee.
2.2Transfection for siRNAs or cDNA
Cells were plated on 1.5 x 105 cells per well on 6-well plates and were transfected 24 hours after plating using siRNAs or 2.5μg cDNA by Lipofectamine 3000 (Invitrogen). The following concentrations of siRNA were used: CREB1, 25 nM; CEBPB, 25 nM; p63, 10 nM. After 48 hours of the transfection, the cells were harvested and subsequently analyzed.
2.3Reporter assay
Cells were seeded into 24-well plates at a density of 40,000 cells per well. After 24 hours, the cells were transfected with 0.5μg luciferase-reporter clones (Fig. 1A), 0.1μg renilla control plasmids (pGL4.74, Promega) and 0.25μg cDNA or siRNA by Lipofectamine 3000 (Invitrogen). After 48 hours of the transfection, normalized relative luciferase activities (RLA) were measured using Dual-Luciferase Reporter Assay system (Promega).
2.4Western blot
Total cell proteins were size fractionated by SDS-polyacrylamide gel electrophoresis and electrotransferred to PVDF membranes. The membranes were probed with antibodies to CREB1 (ab32515), CEBPB (ab32358), p63 (ab53039), KLF5 (ab137676; abcam, Cambridge, UK), Myc tag (#2276; Cell Signaling Technology, Danvers, MA, USA) or β-actin (C-15; Sigma-Aldrich, St. Louis, MO, USA), followed by horseradish peroxidase-labeled secondary antibodies. Binding was detected by ECL Select (GE Healthcare Life Science, Buckinghamshire, UK) and visualized using Ez-Capture MG (ATTO, Tokyo, Japan).
2.5Chromatin immunoprecipitation (ChIP) assays
ChIP assay was conducted as previously described [20]. HSC2 cells were transfected with constructs of mutation of putative CREB1-binding site (Mut #1, #2, #3 and #(2 + 3)) or pGL4.10 plasmid alone. After 48 hours of the transfection, cells were collected and subjected to ChIP assays. For quantitation of immunoprecipitation, KOD SYBR qPCR Mix (TOYOBO) and StepOne Real-Time PCR Systems (Applied Biosystems, Bedford, MA, USA) were used. Primer sequences are shown in Table 1.
2.6Chromosome conformation capture (3 C) on ChIP assay
To analyze the interaction between the 425-region and the MER mediated by CREB1 and/or Sp3, chromosome conformation capture (3 C) on ChIP assay was performed [21]. Cross-linked genomic DNA of HSC2 cells was digested by DpnII (New England Biolabs, Ipswich, MA, USA) overnight at 37°C. Dynabeads M-280 (Invitrogen) preincubated with anti-CREB1, anti-Sp3 or normal rabbit IgG were reacted with the genomic DNA overnight at 4°C. Samples added to 10% SDS were incubated for 30 min at 65°C to elute DNA from Dynabeads, and then treated with T4 DNA ligase for 4 hours at 16°C. Subsequently, the samples were treated with Proteinase K (Wako, Osaka, Japan; overnight at 65°C) and RNase A (Nippon Gene, Tokyo, Japan; 1 hour at 37°C). DNA was purified by phenol/chloroform treatment. Each sample was subjected to conventional end point PCR with KOD-FX DNA polymerase (TOYOBO). Products were analyzed by polyacrylamide gel electrophoresis. Primer sequences are shown in Table 1.
2.7Quantitative reverse transcription PCR (qRT-PCR)
Total RNA reverse-transcribed into cDNA was subjected to qPCR using KOD SYBR qPCR Mix (TOYOBO) and StepOne Real-time PCR system (Applied Biosystems). The primer sequences are listed in Table 1, and a TaqMan probe for KLF5 (Hs00156145_m1, Applied Biosystems) was used. Expression levels normalized with GAPDH (TaqMan Endogenous Control Human GAPDH; Applied Biosystems) were determined by the standard curve method (2-ΔΔCt).
2.8Database and statistical analyses
ECR browser (https://ecrbrowser.dcode.org/) was used to identify evolutionary conserved regions in KLF5 gene. Putative transcription factor binding sites were searched with TFBIND (https://tfbind.hgc.jp/), Promoter Scan (https://www-bimas.cit.nhi.gov/molbio/proscan/), and JASPAR (https://jaspardev.genereg.net). Data were analyzed using BellCurve for Excel version 3.20 (Social Survey Research Information, Tokyo, Japan). Student’s t-test was used for data sets containing two experimental groups, and Tukey-Kramer’s HSD test or Dunnett’s test for multiple group comparison. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
3Results
3.1Identification of the silencer region for KLF5 expression
Database analysis around human KLF5 gene showed that a 3 kbp region from -2,894 to +424 upstream exon 1 of KLF5 (NM_001730) was highly conserved in mammals (Fig. 1A). It suggests that this region is involved in the regulation of gene expression. Then, luciferase-reporter constructs with different lengths of the region were prepared and subjected to reporter assay using HSC2 oral carcinoma cells (Fig. 1B). To define the KLF5 gene regulatory region, we considered the clones showing a significant difference of relative luciferase activity (RLA) than the prior clones. As shown in Fig. 1B, RLA was gradually lowered by shorting the region but rapidly elevated in clone 6 compared to clone 5 (P < 0.001), and it was reproduced in other oral carcinoma cell lines, malignant tumor cell lines, and HEK293T cells (Fig. 1C and D). These results indicate that the 425-region between -2,001 and -1,577 suppresses KLF5 gene expression.
3.2CREB1 involvement in KLF5 down-regulation
To examine whether the 425-region affected KLF5 basal expression, we generated clone Δ5 that had a deletion in a region spanning -1,576 and +144: the clone ligating the 425-region and MER of KLF5 gene (Fig. 2). RLA of clone Δ5 and full-length clone 5 were dramatically decreased compared to clone MER, and RLA of clone ΔMER deleting the MER was further decreased (Fig. 2). Additionally, there was no difference between RLA of clone Δ5 and full-length clone 5 (Fig. 2). These results indicate that a region spanning from -1,576 to+144 does not inhibit MER activity, and factors binding to the 425-region are important for KLF5 down-regulation. Database analysis showed possible binding sites for CREB1, CEBPB, and p63 in the 425-region (Fig. 3A). After confirmation of down-regulation of their proteins by siRNAs (Fig. 3B), RLA of clone Δ5 were measured. siRNA for CREB1 but not CEBPB and p63 significantly increased the RLA, and p63 for siRNA, s229400, decreased the RLA (Fig. 3C).
Fig. 3
Putative transcription factor binding sites in the 425-region. (A) DNA sequence of the 425-region. The 425-region contained putative CREB1-, CEBPB-, and p63-binding sites. The 425-region contained three putative CREB1-binding sites. (B) Expression of endogenous proteins after siRNA transfection was analyzed by western blot. CREB1 and CEBPB isoforms are indicated by arrows. Non-specific bands are indicated by arrowheads. β-actin was used as an internal control. (C) RLA of clone Δ5 at depletion of CREB1, CEBPB, or p63. HSC2 cells transfected with CREB1 siRNA (s3489 and s3490), CEBPB siRNA (s2891 and s2892), p63 siRNA (s16411 and s229400), or negative control siRNA (siCtrl) were used. A reporter plasmid, pGL4.10, was used as control for reporter assays. The data are represented as means±SD (n = 4). *, P < 0.05 compared to siCtrl.
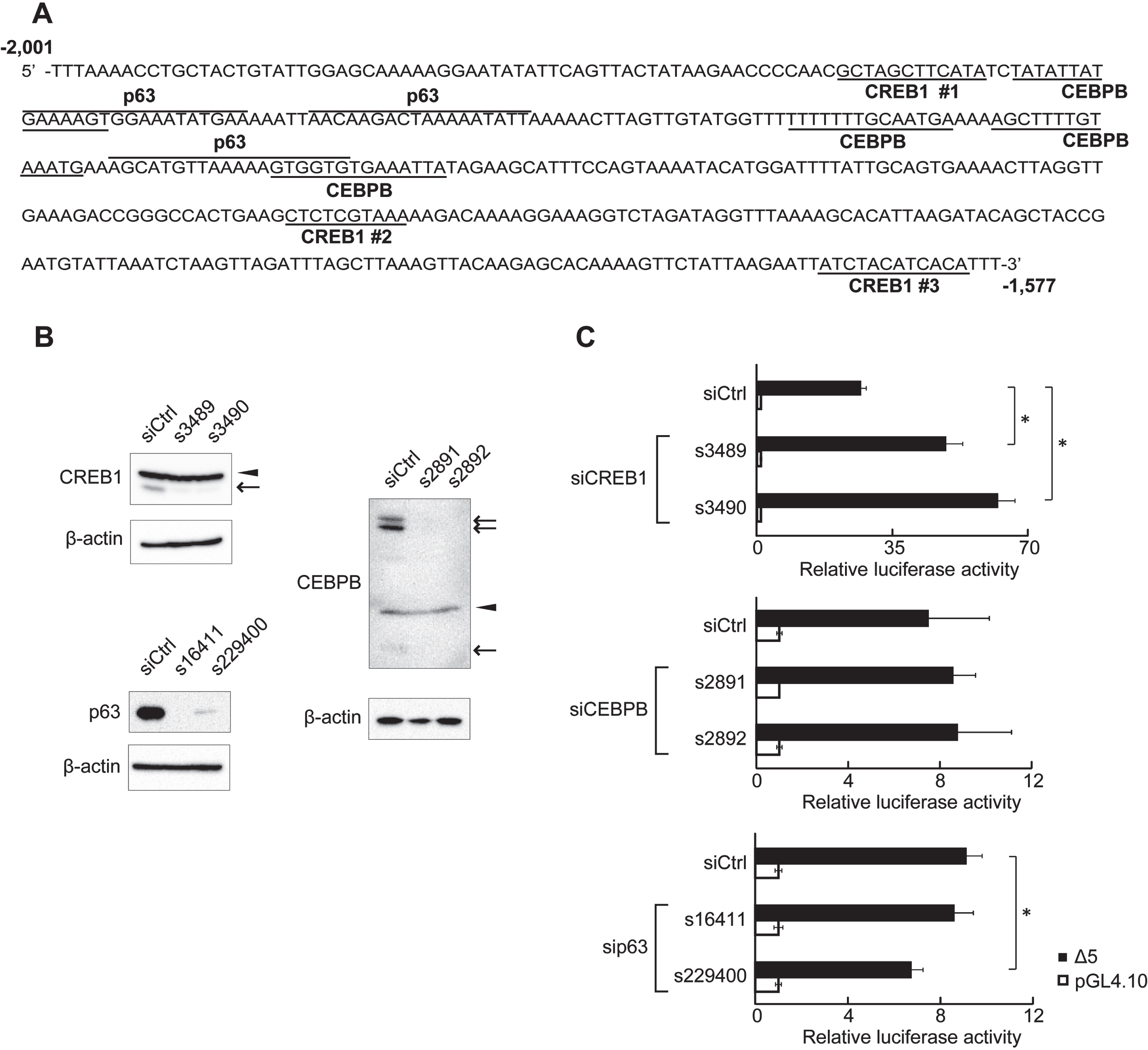
CREB1 binding to the 425-region of the endogenous genome was detected by ChIP assay. The 425-region was dose-dependently precipitated by anti-CREB1 antibody (Fig. 4A). The 425-region included three putative CREB1-binding sites (Fig. 3A). We mutated each of the sites (Mut #1, #2 and #3) in clone Δ5, and then carried out reporter assays to examine the effects of the binding sites on the MER activity (Fig. 4B). In comparison to clone Δ5, RLA was increased in clone Mut #2 or clone Mut #3 but not in clone Mut #1, and most significantly clone Mut # (2 + 3). CREB1 binding to each clone was quantified by ChIP assay and the binding to all mutant clones was decreased at almost the same level (Fig. 4C and D). These results indicate that CREB1 binds to all putative CREB1-binding sites and that suppression of KLF5 gene expression largely depends on its binding to #2 and #3 sites.
3.3Interaction of the 425-region with MER of KLF5 gene through binding of CREB1
We examined whether the interaction of the 425-region with the MER was mediated by CREB1 and/or Sp3 by 3 C on ChIP assay. A brief overview of the assay and primer settings are depicted in Fig. 5A and B. Genomic DNA spanned by 3C-Fw-1 to 3C-Rv-2 primers and including DpnII sites was amplified before DpnII digestion but not after the digestion (Fig. 5 C). A specific 3 C on ChIP product using 3C-Fw-1 and 3C-Rv-1 primers was detected under conditions with ligation and anti-CREB1 antibody but not with anti-Sp3 antibody (Fig. 5D). A product amplified by LC-Fw to LC-Rv was used as loading control (Fig. 5E).
Fig. 5
Chromosome conformation capture (3 C) on ChIP assay to determine the interaction between the 425-region and MER of KLF5 gene. (A) Procedure of 3 C on ChIP assay. (B) The schematic of KLF5 gene promoter with the position and orientation of the primer sets indicated. 3C-Fw-1 and 3C-Rv-1 primers were designed to amplify a novel ligation product formed between the restriction fragments from the 425-region and the MER. 3C-Fw-1 and 3C-Rv-2 primers were used for the confirmation of DpnII digestion. LC-Fw and LC-Rv primers were designed for loading control of chromatin with ligation or without ligation after the digestion. (C) The confirmation of chromatin digested with DpnII before immunoprecipitation. A primer set (3C-Fw-1 and 3C-Rv-2) was used. pri, prior to DpnII digestion; pos, posterior to DpnII digestion. (D) The result of 3 C on ChIP assay using anti-CREB1 antibody or anti-Sp3 antibody. IgG was used as a negative control in the immunoprecipitation step. A primer set (3C-Fw-1 and 3C-Rv-1) was used. (E) The confirmation of the amount of chromatin for use with ligation or without ligation after the digestion. A primer set (LC-Fw and LC-Rv) was used.
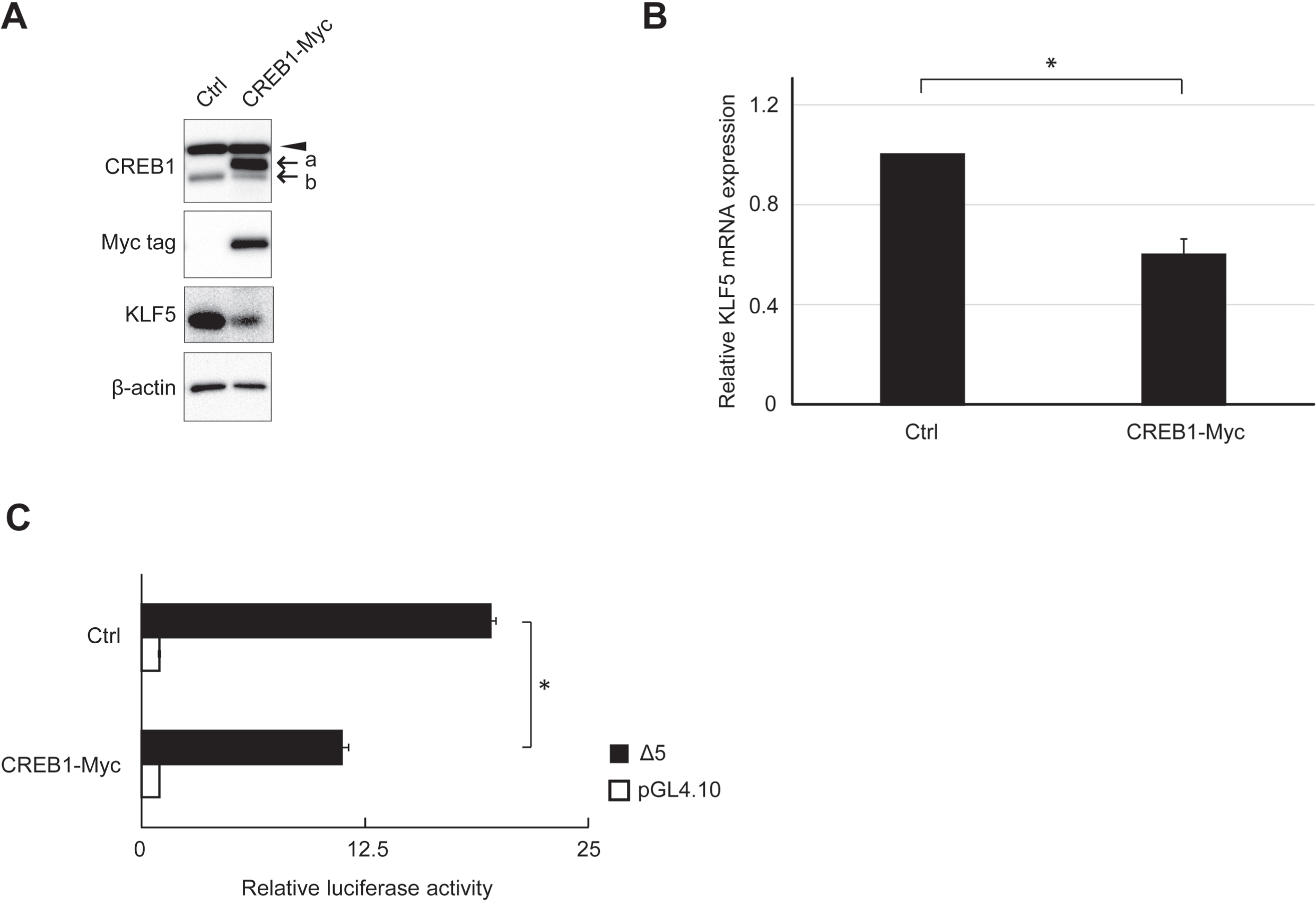
3.4The effect of CREB1 on endogenous KLF5 expression
The effects of CREB1 cDNA transfection on endogenous KLF5 protein and mRNA expression levels were analyzed. KLF5 protein and mRNA expression levels were apparently decreased by CREB1 cDNA transfection (Fig. 6A and B). The reporter assay using clone Δ5 showed a significant decrease in RLA by CREB1 cDNA transfection (Fig. 6C).
Fig. 6
Effect of CREB1 expression on endogenous KLF5 expression. (A) Expression of endogenous proteins after CREB1 cDNA-tagged Myc tag (CREB1-Myc) transfection was analyzed by western blot. β-actin was used as an internal control. The exogenous and endogenous CREB1 are indicated by “a” and “b”, respectively. Exogenous CREB1 was also probed by anti-Myc tag antibody. Non-specific bands are indicated by arrowheads. (B) Quantitative analysis of KLF5 mRNA expression in HSC2 cells after CREB1 cDNA (CREB1-Myc) or pCI-neo vector (Ctrl) transfection. The result shows that the relative fold of the expression to cells transfected pCI-neo vector. *, P < 0.05 compared to Ctrl. (C) RLA of clone Δ5 at overexpression of CREB1. HSC2 cells transfected with CREB1 cDNA (CREB1-Myc) or pCI-neo vector (Ctrl) were used. The reporter plasmid pGL4.10 was used as the control of reporter assay. The data are represented as means±SD (n = 4). *, P < 0.05 compared to Ctrl.
4Discussion
KLF5 that controls the proliferation and differentiation of epithelial cells is preferentially expressed in basal cells of stratified squamous epithelium and rapidly disappears in suprabasal cells [2, 22, 23]. KLF5 expression is higher in proliferating cells than in differentiated cells and increases the growth rate in epithelia, such as the esophagus and intestine [24–28]. KLF5 is frequently overexpressed in a subset of patients with aggressive carcinoma and is thought to play an essential role in cancer progression [8, 13, 15, 16, 29]. KLF5 basal expression requires Sp3 binding to the MER of KLF5 gene [19], but other elements for fine-tuning the expression are almost unknown. The present study shows that the 425-region from -2,001 to -1,577 acts as a silencer and CREB1 binding to the 425-region represses KLF5 gene expression through interacting with the MER.
Gene expression is controlled by physical interactions of transcriptional regulators binding to gene promoters and far distal elements [30–33]. In this study, reporter assays using clones with different length of KLF5 upstream sequence showed that the 425-region strongly and commonly repressed reporter gene activities in oral carcinoma cells and other cell lines. The 425-region decreased the basal expression of KLF5 reporter gene in a clone directly ligated with the MER (clone Δ5) and comparably in a clone spanning -1,576 and+144 (clone 5). It is apparent that the 425-region represses KLF5 basal expression.
Database analysis suggested potential binding sites for CREB1, p63 and CEBPB in the 425-region. Because siRNAs for p63 and CEBPB did not up-regulate RLA of clone Δ5, they may be not directly involved in the suppression of KLF5 basal expression. In contrast, CREB1 siRNA increased RLA. The 425-region encoded three different CREB1-binding sites, and quantitative ChIP assay showed that mutation of each site decreased CREB1 binding to the 425-region. In the reporter assay, mutation at CREB1 #1 site did not affect RLA in contrast to be increased RLA by #2 and #3 mutations. These results strongly suggest that CREB1 can bind to all the binding sites and the binding to #2 and #3 sites plays a dominant role in suppressing KLF5 basal expression.
CREB1-mediated interaction of the 425-region with the MER was analyzed by 3 C on ChIP assay. CREB1 is a member of CREB/ATF family and is known to dimerize with this member [34, 35]. Its interaction with other transcription factors frequently regulates target gene expression [36–39]. Sp3 up- or down-regulates various genes by incorporating a transcription regulatory complex [40–42]. Because Sp3 binding to the MER is essential for KLF5 basal expression, we hypothesized that the interaction of CREB1 on the 425-region and Sp3 on the MER forms the transcriptional repressor complex. In fact, CREB1 was detected in the complex, but Sp3 was undetectable. In human keratinocytes, promoter regulation of loricrin (LOR) as a keratinocyte differentiation marker requires interactions of CREB/ATF family and Sp family on the promoter [36]. In poorly differentiated keratinocytes, the interactions of CREB/ATF family and Sp family by binding to their respective elements or the dimerization of CREB/ATF family and other positive regulators prevents the binding of factors required for LOR gene expression to the promoter. In terminally differentiated keratinocytes, the levels of CREB proteins are decreased, and relative levels of positive regulators required for promoter activation are increased, resulting in the positive regulators binding to the promoter. CREB1 overexpression suppressed KLF5 basal expression and endogenous KLF5 expression in this study. These suggest possibilities that the CREB1 complex mediating the 425-region and the MER is formed with other MER-binding protein(s) and binds to the Sp3-binding site in the MER, or that the complex incorporates other positive regulators of KLF5 gene expression. These possibilities can lead to the inhibition of Sp3 binding to the MER.
KLF5 preferentially localizes to the basal cell layer of stratified squamous epithelium and CREB1 to basal and spinous layers [19, 43]. In oral carcinoma, KLF5 has high expression at the periphery of carcinoma cell nests containing poorly differentiated cells, whereas has low expression near the center of the nest consisting of well differentiated cells [8]. Calcium-induced mouse keratinocyte differentiation is associated with increased expression of CREB1 [43]. In this study, exogenous CREB1 induced the repression of KLF5 protein and mRNA expression in oral carcinoma cells. Moreover, interaction partners of CREB1 are frequently different in undifferentiated and differentiated cells [44, 45]. These findings suggest that differentiation-induced alteration in CREB1 expression may contribute to the regulation of KLF5 gene expression. Previous studies have reported that KLF5 expression is associated with poor prognosis in patients with carcinoma. The high expression of KLF5 correlated with poor patient survival in pancreatic, colorectal, breast and gastric carcinomas [13, 15, 16, 46]. Knockdown of KLF5 reduced tumor sphere formation in breast cancer cells [16]. These facts indicate that KLF5 is a tumor activator. However, KLF5 is also reported as a tumor suppressor [47]. Thus, the role of KLF5 seems to be controversial.
In this study, we found that the 425-region from -2,001 to -1,577 acts as a silencer and CREB1 binding to the 425-region represses KLF5 basal expression through interacting with the MER. It will help to understand the two-face KLF5 function and elucidate epithelial and carcinoma cell physiology.
Acknowledgments
This work was supported by JSPS KAKENHI Grant Numbers JP17H07147 and JP21K16957 (N.M.).
Author contributions
CONCEPTION: Nozomi Mihara and Kazushi Imai.
INTERPRETATION OR ANALYSIS OF DATA: Nozomi Mihara.
PREPARATION OF THE MANUSCRIPT: Nozomi Mihara and Kazushi Imai.
REVISION FOR IMPORTANT INTELLECTUAL CONTENT: Nozomi Mihara.
SUPERVISION: Nozomi Mihara.
Conflict of interest
The authors have no conflicts of interest to report.
References
[1] | Dang DT , Pevsner J , Yang VW . The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. (2000) ;32: (11-12):1103–21. doi: 10.1016/S1357-2725(00)00059-5 |
[2] | McConnell BB , Yang VW . Mammalian Krüppel-Like factors in health and diseases. Physiol Rev. (2010) ;90: (4):1337–81. doi: 10.1152/physrev.00058.2009 |
[3] | Ratushny V , Gober MD , Hick R , Ridky TW , Seykora JT . From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J Clin Invest. (2012) ;122: (2):464–72. doi: 10.1172/JCI57415.464 |
[4] | Hedberg ML , Berry CT , Moshiri AS , Xiang Y , Yeh CJ , Attilasoy C , et al. Molecular Mechanisms of Cutaneous Squamous Cell Carcinoma. Int J Mol Sci. (2022) ;23: (7):3478. doi: 10.3390/ijms23073478 |
[5] | Grandis JR , Melhem MF , Barnes EL , Tweardy DJ . Quantitative immunohistochemical analysis of transforming growth factor- α and epidermal growth factor receptor in patients with squamous cell carcinoma of the head and neck. Cancer. (1996) ;78: (6):1284–92. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1284::AID-CNCR17>3.0.CO;2-X |
[6] | Chen I-H , Chang JT , Liao C-T , Wang H-M , Hsieh L-L , Cheng A-J . Prognostic significance of EGFR and Her-2 in oral cavity cancer in betel quid prevalent area. Br J Cancer. (2003) ;89: (4):681–6. doi: 10.1038/sj.bjc.6601171 |
[7] | Huang S-F , Chuang W-Y , Chen I-H , Liao C-T , Wang H-M , Hsieh L-L . EGFR protein overexpression and mutation in areca quid-associated oral cavity squamous cell carcinoma in Taiwan. Head Neck. (2009) ;31: (8):1068–77. doi: 10.1002/HED |
[8] | Shibata M , Chiba T , Matsuoka T , Mihara N , Kawashiri S , Imai K . Krüppel-like factors 4 and 5 expression and their involvement in differentiation of oral carcinomas. Int J Clin Exp Pathol. (2015) ;8: (4):3701–9. |
[9] | Calderon-Aparicio A , Yamamoto H , De Vitto H , Zhang T , Wang Q , Bode AM , et al. RCC2 promotes esophageal cancer growth by regulating activity and expression of the Sox2 transcription factor. Molecular Cancer Research. (2020) ;18: (11):1660–74. doi: 10.1158/1541-7786.MCR-19-1152 |
[10] | Palioura D , Lazou A , Drosatos K . Krüppel-like factor (KLF) An emerging foe of cardiovascular health. J Mol Cell Cardiol. (2022) ;163: :56–66. doi: 10.1016/j.yjmcc.2021.10.002 |
[11] | McConnell BB , Ghaleb AM , Nandan MO , Yang VW . The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays. (2007) ;29: (6):549–57. doi: 10.1002/bies.20581 |
[12] | Sur I , Rozell B , Jaks V , Bergström A , Toftgard R . Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J Cell Sci. (2006) ;119: (17):3593–601. doi: 10.1242/jcs.03070 |
[13] | Soon M-S , Hsu L-S , Chen C-J , Chu P-Y , Liou J-H , Lin S-H , et al. Expression of Kruppel-like factor 5 in gastric cancer and its clinical correlation in Taiwan. Virchows Archiv. (2011) ;459: (2):161–6. doi: 10.1007/s00428-011-1111-0 |
[14] | Jia L , Zhou Z , Liang H , Wu J , Shi P , Li F , et al. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene. (2016) ;35: (16):2040–51. doi: 10.1038/onc.2015.263 |
[15] | Li Y , Kong R , Chen H , Zhao Z , Li L , Li J , et al. Overexpression of KLF5 is associated with poor survival and G1/S progression in pancreatic cancer. Aging. (2019) ;11: (14):5035–57. doi: 10.18632/aging.102096 |
[16] | Takagi Y , Sakai N , Yoshitomi H , Furukawa K , Takayashiki T , Kuboki S , et al. High expression of Krüppel-like factor 5 is associated with poor prognosis in patients with colorectal cancer. Cancer Sci. (2020) ;111: (6):2078–92. doi: 10.1111/cas.14411 |
[17] | Siraj AK , Pratheeshkumar P , Divya SP , Parvathareddy SK , Alobaisi KA , Thangavel S , et al. Krupple-Like Factor 5 is a Potential Therapeutic Target and Prognostic Marker in Epithelial Ovarian Cancer. Front Pharmacol. (2020) ;11: :598880. doi: 10.3389/fphar.2020.598880 |
[18] | Chen P , Qian X-K , Zhang Y-F , Sun X-G , Shi X-J , Gao Y-S . KLF5 promotes proliferation in gastric cancer via regulating p21 and CDK4. Eur Rev Med Pharmacol Sci. (2020) ;24: (8):4224–31. doi: 10.26355/eurrev_202004_21002 |
[19] | Mihara N , Chiba T , Yamaguchi K , Sudo H , Yagishita H , Imai K . Minimal essential region for krüppel-like factor 5 expression and the regulation by specificity protein 3-GC box binding. Gene. (2017) ;601: :36–43. doi: 10.1016/j.gene.2016.12.002 |
[20] | Kimura H , Hayashi-Takanaka Y , Goto Y , Takizawa N , Nozaki N . The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct Funct. (2008) ;33: (1):61–73. doi: 10.1247/csf.07035 |
[21] | Sexton T , Kurukuti S , Mitchell JA , Umlauf D , Nagano T , Fraser P . Sensitive detection of chromatin coassociations using enhanced chromosome conformation capture on chip. Nat Protoc. (2012) ;7: (7):1335–50. doi: 10.1038/nprot.2012.071 |
[22] | Choi S , Myers JN . Molecular pathogenesis of oral squamous cell carcinoma: Implications for therapy. J Dent Res. (2008) ;87: (1):14–32. doi: 10.1177/154405910808700104 |
[23] | Diakiw SM , D’Andrea RJ , Brown AL . The double life of KLF Opposing roles in regulation of gene-expression, cellular function, and transformation. IUBMB Life. (2013) ;65: (12):999–1011. doi: 10.1002/iub.1233 |
[24] | Goldstein BG , Chao H-H , Yang Y , Yermolina YA , Tobias JW , Katz JP . Overexpression of Krüppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. (2007) ;292: (6):1784–92. doi: 10.1152/ajpgi.00541.2006 |
[25] | Chanchevalap S , Nandan MO , Merlin D , Yang VW . All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Krüppel-like factor 5. FEBS Lett. (2004) ;578: (1-2):99–105. doi: 10.1016/j.febslet.2004.10.079 |
[26] | Sun R , Chen X , Yang VW . Intestinal-enriched Krüppel-like Factor (Krüppel-like Factor 5) is a Positive Regulator of Cellular Proliferation. J Biol Chem. (2001) ;276: (10):6897–900. doi: 10.1074/jbc.C000870200 |
[27] | Conkright MD , Wani MA , Anderson KP , Lingrel JB . A gene encoding an intestinal-enriched member of the Krüppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. (1999) ;27: (5):1263–70. doi: 10.1093/nar/27.5.1263 |
[28] | Ohnishi S , Laub F , Matsumoto N , Asaka M , Ramirez F , Yoshida T , et al. Developmental expression of the mouse gene coding for the Kruppel-like transcription factor KLF5. Dev Dyn. (2000) ;217: :421–9. doi: 10.1002/(SICI)1097-0177(200004)217:4<421::AID-DVDY9>3.0.CO;2-1 |
[29] | Chaib H , Cockrell EK , Rubin MA , Macoska JA . Profiling and verification of gene expression patterns in normal and malignant human prostate tissues by cDNA microarray analysis. Neoplasia. (2001) ;3: (1):43–52. doi: 10.1038/sj.neo.7900126 |
[30] | West AG , Fraser P . Remote control of gene transcription. Hum Mol Genet. (2005) ;14: :R101–11. doi: 10.1093/hmg/ddi104 |
[31] | Delgado MD , León J . Gene expression regulation and cancer. Clinical and Translational Oncology. (2006) ;8: (11):780–7. doi: 10.1007/978-3-319-24951-3_4 |
[32] | Dean A . On a chromosome far, far away: LCRs and gene expression. Trends in Genetics. (2006) ;22: (1):38–45. doi: 10.1016/j.tig.2005.11.001 |
[33] | Thomas MC , Chiang C-M . The General Transcription Machinery and General Cofactors. Crit Rev Biochem Mol Biol. (2006) ;41: (3):105–78. doi: 10.1080/10409230600648736 |
[34] | Hai T , Curran T . Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. (1991) ;88: (9):3720–4. doi: 10.1073/pnas.88.9.3720 |
[35] | van Dam H , Castellazzi M . Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene. (2001) ;20: (19):2453–64. 10.1038/sj.onc.1204239 |
[36] | Jang S-I , Steinert PM . Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1, and AP2 families. J Biol Chem. (2022) ;277: (44):42268–79. doi: 10.1074/jbc.M205593200 |
[37] | Rozenberg JM , Bhattacharya P , Chatterjee R , Glass K , Vinson C . Combinatorial recruitment of CREB, C/EBPβ and c-Jun determines activation of promoters upon keratinocyte differentiation. PLoS One. (2013) ;8: (11):e78179. doi: 10.1371/journal.pone.0078179 |
[38] | Hummler E , Cole TJ , Blendy JA , Ganss R , Aguzzi A , Schmid W , et al. Targeted mutation of the CREB gene: Compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. (1994) ;91: (12):5647–51. doi: 10.1073/pnas.91.12.5647 |
[39] | Steven A , Friedrich M , Jank P , Heimer N , Budczies J , Denkert C , et al. What turns CREB on? And off? And why does it matter? Cell Mol Life Sci. (2020) ;77: :4049–67. doi: 10.1007/s00018-020-03525-8 |
[40] | Majello B , De Luca P , Hagen G , Suske G , Lania L . Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. (1994) ;22: (23):4914–21. doi: 10.1093/nar/22.23.4914 |
[41] | Hagen G , Dennig J , Preiß A , Beato M , Suske G . Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. (2498) ;270: (42):9–94. doi: 10.1074/jbc.270.42.24989 |
[42] | Valin A , Gill G . Regulation of the dual-function transcription factor Sp3 by SUMO. Biochem Soc Trans. (2007) ;35: (6):1393–6. doi: 10.1042/BST0351393 |
[43] | Rutberg SE , Adams TL , Olive M , Alexander N , Vinson C , Yuspa SH . CRE DNA binding proteins bind to the AP-1 target sequence and suppress AP-1 transcriptional activity in mouse keratinocytes. Oncogene. (1999) ;18: (8):1569–79. doi: 10.1038/sj.onc.1202463 |
[44] | Hurst HC , Totty NF , Jones NC . Identification and functional characterisation of the cellular activating transcription factor 43 (ATF-43) protein. Nucleic Acids Res. (1991) ;19: (17):4601–9. doi: 10.1093/nar/19.17.4601 |
[45] | Ellis MJC , Hurst HC , Goodbourn S . A novel cyclic AMP response element-binding protein-1 (CREB-1) splice product may down-regulate CREB-1 activity. J Mol Endocrinol. (1995) ;14: (2):191–8. doi: 10.1677/jme.0.0140191 |
[46] | Tong D , Czerwenka K , Heinze G , Ryffel M , Schuster E , Witt A , et al. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. (2006) ;12: (8):2442–8. doi: 10.1158/1078-0432.CCR-05-0964 |
[47] | Chen C , Bhalala HV , Qiao H , Dong J-T . A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. (2002) ;21: (47):6567–72. doi: 10.1038/sj.onc.1205817 |




