Unfavorable prognosis and clinical consequences of APOBEC3B expression in breast and other cancers: A systematic review and meta-analysis
Abstract
INTRODUCTION:
Controversy exists regarding the association of apolipoprotein B mRNA editing enzyme catalytic subunit 3B APOBEC3B, (A3B) overexpression and poor prognosis, metastasis, and chemotherapy drug resistance in cancers. Here we conducted a systematic review and meta-analysis to determine its prognostic value and clinicopathological features in breast cancer and some other malignancies.
MATERIALS AND METHODS:
PubMed, Scopus, Cochrane Library, Web of Science, and EMBASE were searched up to Feb 2022 for the association of A3B with breast, ovarian, gastrointestinal and lung cancers. The pooled hazard ratios with 95% confidence interval (CI) were evaluated to assess disease-free survival (DFS), overall survival (OS), and recurrence-free survival (RFS) in cancers under study.
RESULTS:
Over 3700 patients were included in this meta-survey. Elevated levels of A3B were significantly related to low OS (pooled HR = 1.30; 95% CI:1.09–1.55, P < 0.01), poor DFS (pooled HR = 1.66; 95% CI:1.17–2.35, P < 0.01) and poor RFS (HR = 1.51, 95% CI:1.11–2.04, P = 0.01). Subgroup analysis revealed that high A3B expression was associated with poor OS in lung (HR = 1.85, 95% CI: 1.40–2.45), and breast cancers (HR = 1.38, 95% CI: 1.00–1.89). High expression of A3B did not display any significant association with clinicopathologic features.
CONCLUSION:
APOBEC3B overexpression is related to poor OS, DFS and RFS only in some cancer types and no generalized role could be predicted for all cancers.
1Introduction
Cancer is regarded as major cause of mortality and morbidity all over the world [1]. In the year 2020, 19.3 million new cancer patients and 10.0 million cancer-associated deaths were reported worldwide. According to GLOBOCAN, 28.4 million new cancer cases are predicted for the year 2040 worldwide [2]. Despite many advances in cancer diagnosis and treatment, the cancer mortality rate is still high and cancer remains a major health problem all over the world [3–5]. It is generally believed that the accumulation of diverse mutations are important driving force for the development of neoplasia [6]. Mutations may originate from exogenous sources which are categorized as environmentally induced or rooted intracellularly [7]. DNA repair systems and their enzyme components are speculated as endogenous mutation sources. Based on the analyses of data from The Cancer Genome Atlas (TCGA), it is reported that APOBEC cytidine deaminase mutagenesis patterns are evident in cancers and could be implicated in the induction of somatic mutations leading to genome instability and carcinogenesis [8, 9].
APOBEC3 or A3 proteins are DNA cytidine deaminases that have a remarkable role in mutations in many different types of cancers [10, 11]. This family is specific to mammals, and in humans comprises seven members (A, B, C, D, F, G, and H) which are highly homologous. Their genes are located in a tandem cluster on chromosome 22q13; the proteins are involved in innate immunity and anti-viral immune response [12–14]. Current evidence, mainly resulting from studies on yeasts, suggest that APOBEC3A and APOBEC3B are responsible for somatic mutations in human cancers [15, 16] but due to the conflicting outcomes from studies performed on tumors, the mutagenic nature and role of APOBEC subtypes in various human cancers are not fully elucidated yet [17, 18].
Out of the seven members of the APOBEC3 enzyme family, APOBEC mutagenesis in tumors is attributed to A3A and A3B enzymes. Positive connection between mRNA level of A3A and A3B enzymes and APOBEC-signature mutations in tumors as well as in vitro overexpression studies strongly confirmed that high activity of these enzymes is mutagenic and genotoxic [7, 19]. Conclusions regarding the role of A3A and A3B expression in mutagenetic processes were mainly drawn through RNA-seq studies despite the fact that these highly homologous APOBECs are poorly resolved and their mRNAs cannot be confidently quantified by short sequencing reads. The dominant mutagenic potentials of A3A or A3B expression in different cancers are highly debated. For example, Banday et al. [20] based on their observations reported that A3B and not A3A is expressed in bladder cancer cell lines, which show a high load of APOBEC-signature mutations, demonstrating that in some cancer cell lines APOBEC-mediated mutagenesis is dominantly caused by the A3B activity. Tsubo M et al. [21] quantified A3B mRNA expression by RT-qPCR in fresh frozen tumors and paired adjacent normal breast tissue samples and reported that A3B mRNA expression was significantly higher in tumors comparing to the adjacent normal tissues.
Recently the clinical significance of A3B is extensively investigated because of its relevance to survival endpoints including DFS, OS, RFS in cancer patients [22, 23]. A3B is a unique member of the family with permanent nuclear localization [24]. This gene has cytidine deaminase activity and contains one catalytically active C-terminal domain (CTD) that bound to ssDNA and N-terminal pseudo-catalytic domain (NTD) [16, 25].
A3B is considered to induce mutation in breast cancer and overexpression of APOBEC3B is reported in variety of cell lines [26]. Also, A3B is introduced as the main factor contributing to mutations in various human cancers [16]. The expression of A3B is linked to the accumulation of somatic mutations during tumorigenesis and progression [26–28]. Recent studies have reported that A3B is significantly up-regulated in many cancers like ovarian, cervix, breast, lung, head & neck, and bladder [29, 30]. Abnormal overexpression of A3B promotes mutations in the PIK3CA gene in several human cancers [31, 32]. Through the knockdown experiments, it is revealed that endogenous A3B correlates with higher levels of genomic uracil, elevated frequencies of pathogenic mutations, and C-to-T transitions. Also, experimentally A3B overexpression results in cell cycle deviation and cell death, DNA fragmentation and aggregation of γH2AX. Phosphorylation of H2AX in the Ser-139 residue causes the formation of γH2AX which is biomarker of DNA damage and genotoxicity [26, 33, 34].
There are different perspectives on the link between A3B and the cell cycle. Lackey et al. reported that the expression pattern of A3B is similar during the mitosis phase and S phase [35]. But, Hirabayashi et al. found that A3B mRNA is the only member of the APOBEC3 family that is differentially expressed at the G2/M-phase in myeloma and bone marrow mononuclear cells [36]. Ma et al. hypothesized that A3B expression induces human hepatocellular carcinoma (HCC) tumorigenesis through deaminase-independent activity and increases cycle progression in HCC cells [37]. McCann et al. stated that A3B can bind to CDK4 and inhibit CDK4-dependent nuclear translocation of cyclin D1, thus delaying cell cycle progression. Overexpression of A3B leads to G1/S phase stalling and increases A3B access to its substrate (ssDNA). Thus, it may enhance the likelihood of mutation and genomic DNA deamination [38]. In addition, pan-cancer analysis showed that the expression of A3B is related to cell cycle-regulated genes [39]. Although these evidences proposed that cell cycle-dependent activities are linked to the regulation of expression of A3B, little is known about the mechanism of cell cycle-dependent regulation of A3B expression and many questions remain unresolved.
Recently, Du et al. argued that high expression of APOBEC3B was linked to worse OS and DSF in patients affected with ovarian cancer [40]. Besides, A3B upregulation can induce the expression of some immunotherapy response genes like; T-cell infiltration and PD-L1 in non-small cell lung cancer (NSCLC) patients, so it can be used as a predictive biomarker [41].
Although deregulation of APOBEC3B has been reported in many cancer types, the conclusions about the prognostic value of A3B for patients remain controversial. A large number of studies reported that high A3B expression is linked with adverse outcomes or poor prognosis in cancer patients [40, 42–44]. However, several studies reported that A3B expression improved OS in patients with cancer or have no correlations with disease-free or overall survival [32, 45, 46]. To date, no meta-analysis has been performed to comprehensively assess the prognostic significance of APOBEC3B expression in cancer patients. Therefore, we performed a systematic review and meta-analysis based on previously published data to assess the link between APOBEC3B expression and the clinicopathological factors and survival in patients with diverse types of cancer.
2Materials and methods
2.1Study strategy
We conducted the present study based on the standard guidelines (PRISMA Checklist) [47]. Two researchers (SJ and RS) separately searched the databases of the Cochrane Library, PubMed, Scopus, Web of Science, and EMBASE to collect all related studies published up to Feb 2022. The following Medical Subject Heading terms were used in this study: (“Cancer” OR “Tumor” OR “Carcinoma” OR “Neoplasm” OR “malignancy”) AND (“APOBEC3B” OR “A3B” OR “Apolipoprotein B mRNA Editing Enzyme”). First the abstract and the title of the publications were retrieved and the full text of relevant articles were reviewed to make sure that the data of interest were included. Any conflicts were solved through discussion with a third researcher.
2.2Inclusion and exclusion criteria
Studies included in this analysis were selected using the following criteria: 1) Studies were published as full paper in English 2) Any type of human malignancy or cancer was pathologically proven in patients 3) Expression of APOBEC3B was evaluated in human tissues using any technique 4) Patients were divided into two groups (high /low or positive /negative) according to the expression levels of A3B and the correlations between A3B expression and clinicopathological data and/or survival outcome was investigated 4) Studies calculated hazard ratio (HR) for the prognostic outcome and odds ratios (ORs) for clinicopathological characteristics or provided sufficient data to extract them. Studies with insufficient data, non-human samples, reviews, reports, letters, and meeting abstracts were excluded.
2.3Data extraction
Two researchers (SJ and RS) independently collected the following data from the studies: the first author’s name, year of publication, tumor type, sample size, follow-up time, country/geographic region of the population, detection method, cut-off values, source of HRs, outcome and other related data. Overall survival, progression-free survival (PFS), disease-free survival and relapse-free survival (RFS) were considered as outcome. Hazard ratio (HR) and 95% confidence interval (CI) were extracted from text or calculated from the survival curves by using Tierney’s method. Any disagreements were solved through discussion.
2.4Quality assessment
Two authors (SJ and RS) evaluated the quality of articles independently using the Newcastle-Ottawa scale (NOS) rating system (0–9 stars). Studies with at least six scores were identified as high-quality studies. Any disagreements were solved by discussion.
2.5Statistical analysis
To evaluate the connection between A3B and the clinicopathological factors in patients with cancer, ORs with corresponding 95% confidence intervals [48] were calculated. Our analysis was performed by pooling the HRs and 95% CIs of survival outcomes (OS, DFS, RFS) directly from the included studies. Some studies depicted the prognosis outcomes as Kaplan-Meier curves and in these cases, by the Get Data Graph Digitizer (version 2.24) the curves were read to contract HR estimates based on the Tierney et al. method [49]. For checking statistical heterogeneity between studies, the Higgins I2 statistic and Chi-Square test were applied. The random-effect model and subgroup analysis were applied to find the heterogeneity. To find any possible publication, bias Funnel plots combined with Begg’s and Egger’s linear regression tests were used. To evaluate the influence of each study on the combined effect values, sensitivity analysis was employed. The statistical analyses were carried out using the Stata13.0 software (Stata Corporation, College Station, TX) and metapackage under R 3.6.3 [50].
3Results
3.1Characteristics of studies
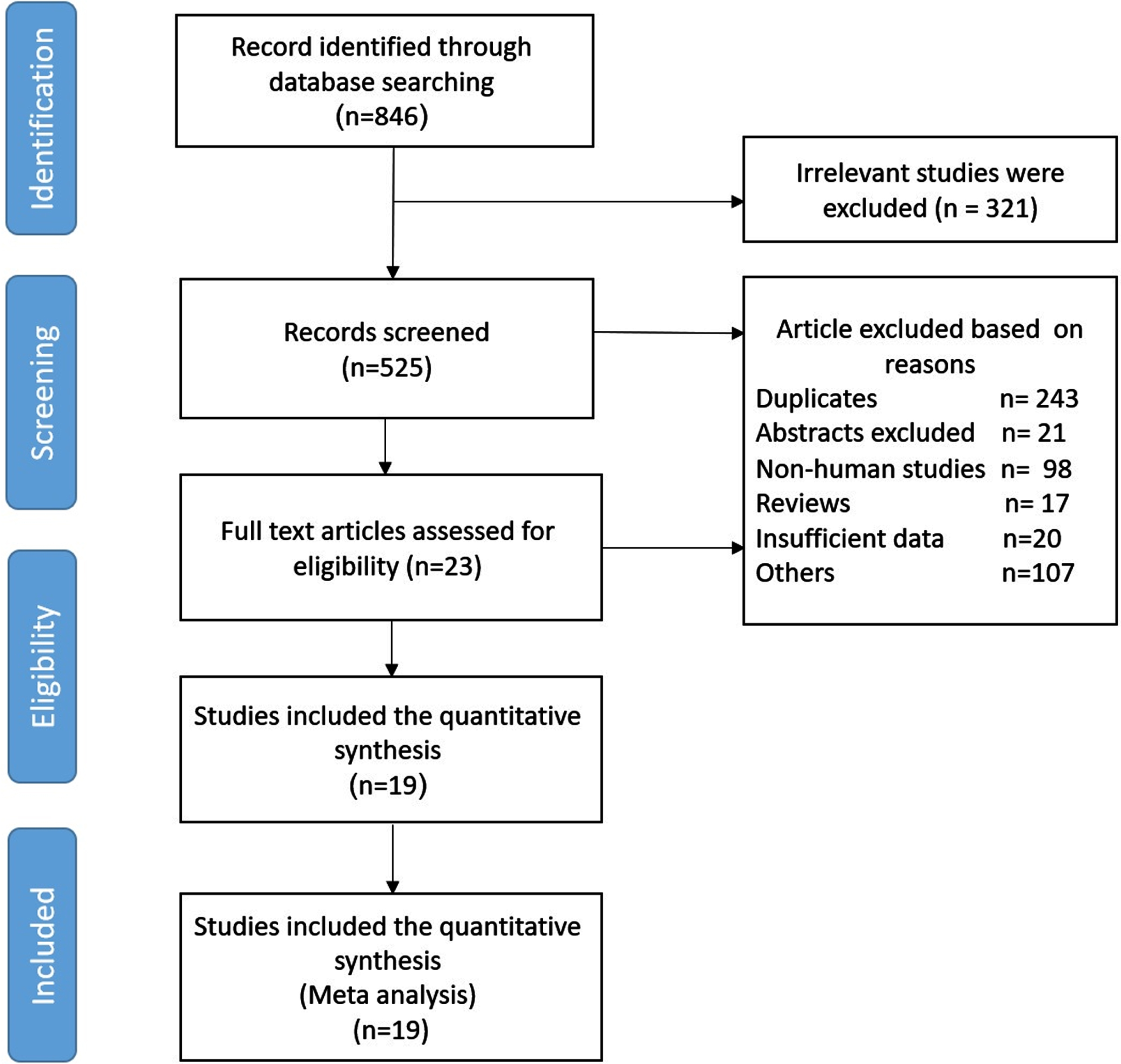
A total of 846 articles were obtained using the primary search according to PRISMA guidelines. Based on our defined exclusion criteria, 827 papers were excluded. Detailed excluding criteria and the number of papers failed to attain eligibility criteria are as follow: irrelevant studies (n = 321), duplicates (n = 243), reviews (n = 17), congress and scientific meeting abstracts (n = 21), letters (n = 4), noncancerous studies (n = 99), animal studies (n = 98), cell line studies (n = 4), and studies lacking sufficient data (n = 20). Finally, 19 studies were included for this meta-analysis. The flow diagram of article selection is shown in Fig. 1. Among the 19 studies, 4 were from Japan, nine from China, three from the USA, the other three from Germany, the Netherlands and South Korea. The included studies contained seven types of cancers, namely breast cancer (n = 5), ovarian cancer (n = 4), non-small cell lung cancer (n = 2), nasopharyngeal carcinoma (n = 1), renal cell carcinoma (n = 1), gastric cancer (n = 2), squamous cell carcinomas (n = 2), neuroendocrine tumors (n = 1), bladder cancer (n = 1). In 11 studies the A3B expression level was detected by immunohistochemistry (IHC) and eight studies performed real time quantitative PCR (qRT-PCR) to detect A3B expression. The cut-off values for A3B expression were different among studies due to various definitions.
Fig. 1
Flow diagram of the study selection process in meta-analysis.
3.2Study quality
NOS scores of all included studies (n = 19) ranges from 6 to 8. More information about the papers included in this study is given in Table 1.
Table 1
Characteristic of the included studies
| Study | Year | Country | Cancer Type | Sample size | Follow-up (month) | Detection method | Cut-off value | Evaluation of expression (H or L /+or –) | Expression associated with poor prognosis | Source of HR | Outcome |
| Kim H, et al. [69] | 2021 | Korea | Metastatic Urothelial Carcinoma | 94 | NA | IHC | NA | High/low | Low | 1 | OS, PFS |
| Xia S, et al. [70] | 2021 | China | Gastric cancer | 482 | 42 | IHC | Mean | High/low | High | 1 | OS, DFS |
| Feng C, et al. [71] | 2021 | China | NPC | 103 | 60 | IHC | Median | High/low | High | NA | NA |
| Serebrenik A, et al. [45] | 2020 | USA | CCOC | 48 | 24 | IHC | Median | High/low | Low | 1 | OS PFS |
| Mao Y, et al. [44] | 2020 | China | Breast Cancer | 116 | 97 | IHC | Median | High/low | High | 1,2 | OS, RFS, DSF, DMFS |
| Gara SK, et al. [42] | 2020 | USA | ACC | 38 | NA | RT-qPCR | Median | High/low | High | 1 | OS |
| Feng C, et al. [72] | 2019 | China | GEP-NENs | 158 | NA | IHC | Median | High/low | High | NA | NA |
| Yan Du, et al. [29] | 2018 | China | Ovarian cancer | 88 | 74.77 | IHC | Positive≥25% | Positive/Negative | High | 1 | OS, DFS |
| Fujiki Y, et al. [46] | 2018 | Japan | Breast Cancer | 173 | 63 | RT-qPCR | NA | High/low | High | 2 | RFS |
| Rüder U, et al. [73] | 2018 | Germany | Ovarian cancer | 469 | NA | IHC, mRNA | Median | Positive/Negative | High | 1 | OS PFS |
| Jiang Ch, et al. [74] | 2018 | China | NSCLC | 203 | 60 | IHC | Median | High/low | High | 1 | OS |
| Tokunaga E, et al. [75] | 2016 | Japan | Breast Cancer | 305 | 59.16 | RT-qPCR | Median | High/low | High | 1 | RFS |
| Leonard B, et al. [76] | 2016 | USA | HGSOC | 354 | NA | RT-qPCR | Median | High/low | Low | 1 | OS PFS |
| Yan Sh, et al. [43] | 2016 | China | NSCLC | 221 | 60 | IHC | Median | High/low | High | 1 | OS, DFS |
| Kosumi K, et al. [32] | 2016 | Japan | ESCC | 147 | 45.6 | IHC | Median | High/low | High | 2 | OS, DFS |
| Xu L, et al. [77] | 2015 | China | RCC | 299 | 67 | IHC | Median | High/low | High | 1 | RFS |
| Zang J, et al. [78] | 2015 | China | Gastric cancer | 236 | NA | IHC, RT-qPCR | Median | High/low | High | NA | NA |
| Tsuboi M, et al. [21] | 2015 | Japan | Breast Cancer | 93 | NA | RT-qPCR | Mean | High/low | High | 2 | OS, DFS |
| Sieuwerts AM, et al. [79] | 2014 | Netherlands | Breast Cancer | 149 | NA | RT-qPCR | Median | High/low | High | 1 | OS, DFS, MFS |
NA: Not available, NPC: Nasopharyngeal carcinoma, NSCLC Non-small-cell lung carcinoma, CCOC: Clear cell ovarian carcinoma, RCC: Renal cell carcinoma, HGSOC: High-grade serous ovarian carcinoma, ACC: Adrenocortical carcinoma, GEP-NENs: Neuroendocrine neoplasms, HGSOC: Ovarian carcinoma, ESCC: Esophageal squamous cell carcinoma.
3.3Relationship of A3B overexpression and prognosis outcome
A total of 13 studies with 2502 patients had reported the relationship between OS and A3B expression level. The random-effects model was used due to heterogeneity (I2 = 93%, P < 0.01).
Poor prognosis was significantly associated with higher A3B expression (pooled HR = 1.30; 95% CI:1.09–1.55) (Fig. 2). Subgroup analysis showed that A3B expression was associated with OS of patients with lung cancer (HR = 1.85, 95% CI:1.40–2.45), and breast cancer (HR = 1.38, 95% CI:1.00–1.89). For the rest of cancers, no statistically significant results were obtained (Table 2).
Fig. 2
Association between A3B expression and survival outcomes.
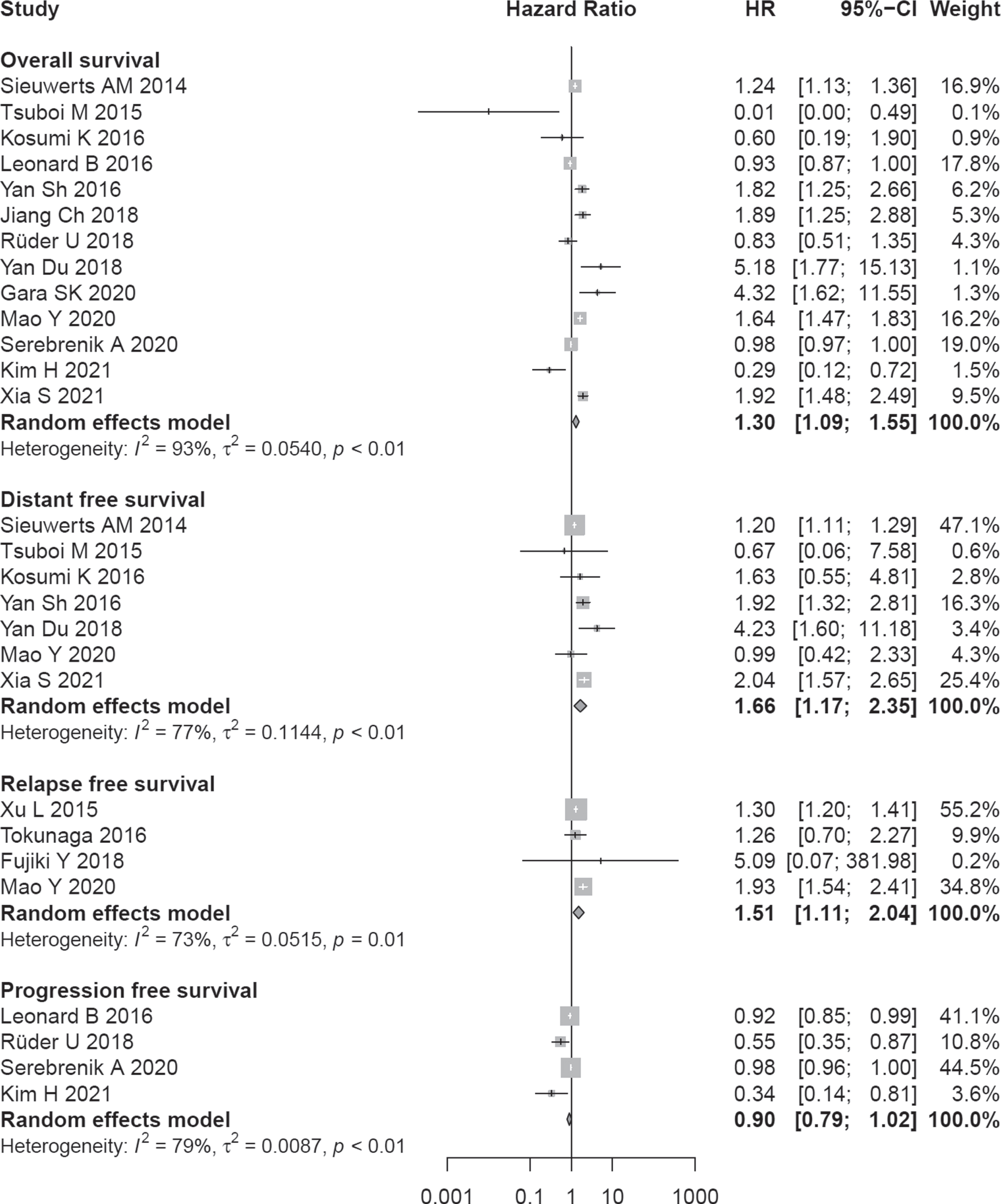
Table 2
Results of subgroup analysis of pooled HRs with regard to overall survival
| n. study | HR | 95% CI | I2 % | p Heterogeneity | ||
| Overall | 13 | 1.30 | (1.10–1.55) | 93.4 | 0.004 | |
| Region | East Asia | 8 | 1.50 | (1.08–2.08) | 76.6 | < 0.001 |
| USA | 3 | 0.98 | (0.87–1.09) | 81.9 | 0.004 | |
| Europe | 2 | 1.09 | (0.76–1.57) | 60.9 | 0.110 | |
| Detection Method | ||||||
| IHC | 8 | 1.41 | (1.02–1.97) | 95.1 | < 0.001 | |
| RT-qPCR | 5 | 1.10 | (0.83–1.47) | 89.5 | < 0.001 | |
| Cancer Type | ||||||
| Ovarian | 4 | 0.97 | (0.87–1.08) | 75.0 | 0.007 | |
| Breast | 3 | 1.38 | (1.00–1.89) | 90.4 | < 0.001 | |
| Gastrointestinal | 2 | 1.24 | (0.41–3.74) | 73.1 | 0.054 | |
| Lung | 2 | 1.85 | (1.40–2.45) | 0.0 | 0.896 | |
| Others | 2 | 1.12 | (0.08–15.66) | 93.6 | < 0.001 |
CI = confidence interval, HR = hazard ratio, calculated by random effects meta-analysis.
To differentiate between RT-qPCR and IHC techniques used to determine A3B expression, subgroup analysis was performed. Only high expression of A3B detected by IHC method (8 studies) was associated with overall survival (HR = 1.41, 95% CI:1.02–1.97). Subgroup analysis based on geographic regions showed correlation in patients from the East Asia region with 8 studies (HR = 1.50, 95% CI:11.08–2.08) (Table 2). Sensitivity analysis demonstrated that our result was stable based on estimates of the HRs (Fig. S1a). There was no evidence of publication bias by the Egger test (P = 0.123) and the Begg test (P = 0.329) (Fig. S1b).
A total of 7 studies with 1296 patients had reported the relationship between poor DFS and A3B expression level. The random-effects model was used due to heterogeneity (I2 = 77%, P < 0.01). Poorer outcome in terms of DFS was significantly associated with higher A3B expression (pooled HR = 1.66; 95% CI:1.17–2.35) (Fig. 2). Sensitivity analysis displayed that the result was stable based on estimates of the HRs (Fig. S2a). There was no evidence of publication bias by the Egger test (P = 0.201) and the Begg test (P = 0.881) (Fig. S2b).
A poor prognosis in terms of RFS also was associated with higher expression of A3B (HR = 1.51, 95% CI:1.11–2.04, no. of studies = 4). A total of 4 studies reported the relationship between poor PFS and A3B expression level. The random-effects model was used due to heterogeneity (I2 = 79%, P < 0.01). Poorer outcome in terms of PFS was not associated with higher A3B expression (pooled HR = 0.90; 95% CI:0.79–1.02) (Fig. 2).
3.4Association of A3B expression with age and gender groups
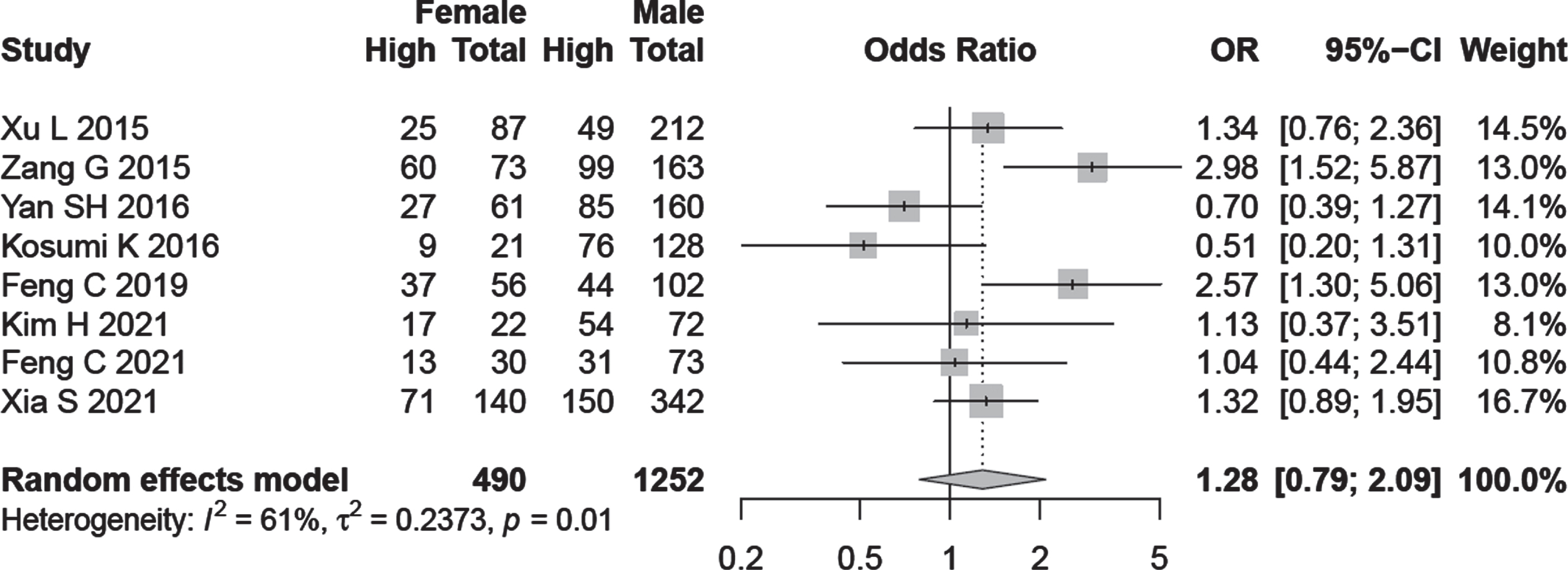
Subgroup analysis on the basis of age and gender was performed in relation to A3B expression but the results were not statistically significant (OR = 0.87, 95% CI:0.59–1.26, OR = 1.28, 95% CI:0.79–2.09) respectively (Fig. 3).
Fig. 3
Forest plot regarding the association of A3B expression with age and gender.
3.5Association of A3B expression and clinical stage
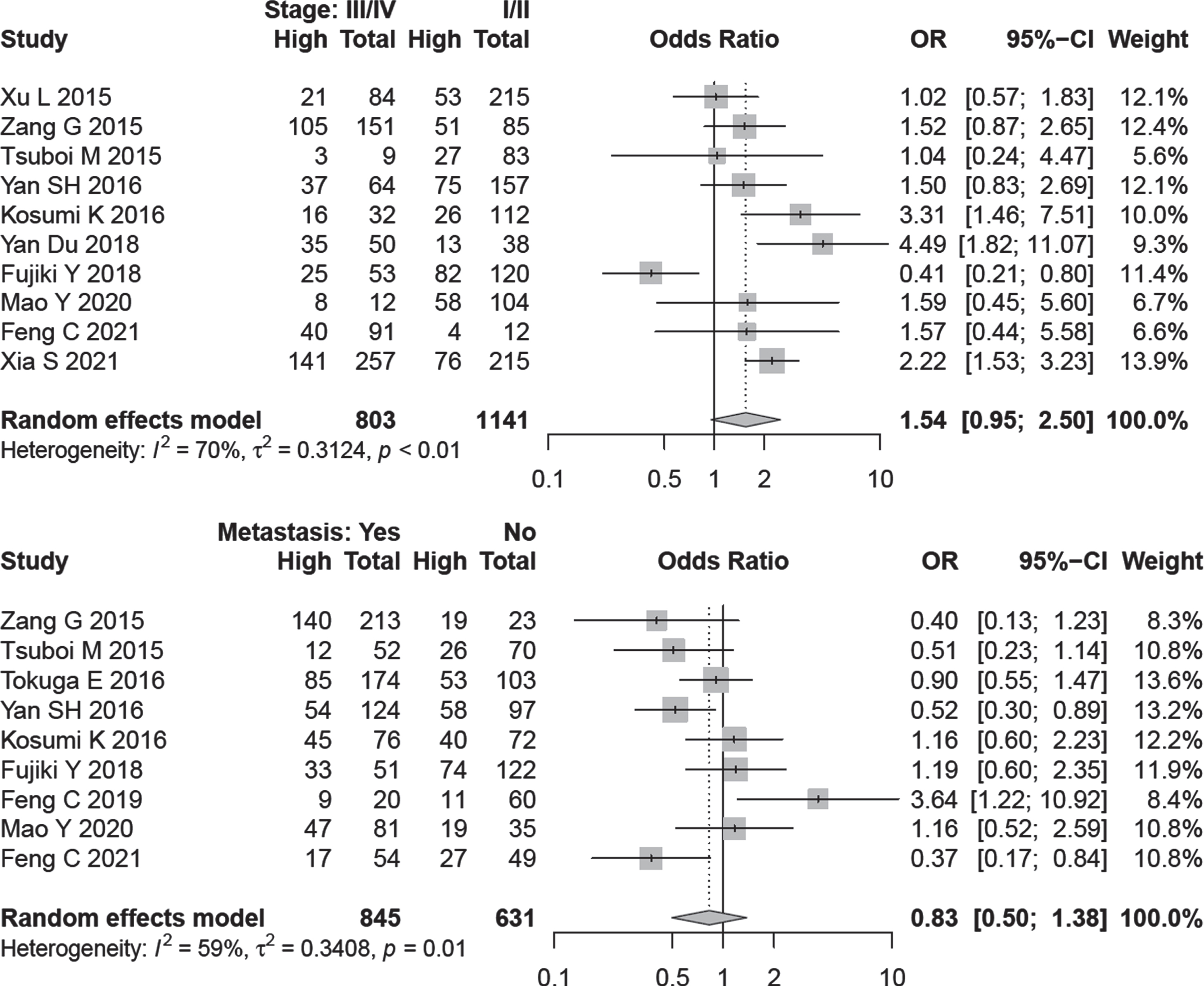
OR values and corresponding CIs were pooled to evaluate the relationship between A3B expression and clinicopathological data. From the data of 10 studies, A3B expression was not associated with cancer stage (OR = 1.54, 95% CI: 0.95–2.50, I2 = 70%) (Fig. 4). In a subgroup analysis, higher expression of A3B detected by IHC method was associated with advanced tumor stage in all cancer types (OR = 1.89, 95% CI: 1.17–3.07, I2 = 50.6%, no. of studies = 6). There was no evidence of publication bias by Begg (P = 0.788) and Egger’s test (P = 0.830) (Fig. S3)
Fig. 4
Forest plot for the association between A3B expression and lymph node metastasis /cancer stage.
3.6Association of A3B expression and lymph node metastasis
Nine studies comprising 1476 patients were evaluated to assess A3B expression and lymph node dissemination. A random-effect model was applied because of inter-study heterogeneity (I2 = 59.0%, P = 0.01). The outcome is indicated that A3B expression has no relation with lymph node metastasis (OR = 0.83, 95% CI: 0.50–1.38) (Fig. 4).
The determined OR values remained the same by excluding any single article (Fig. S4). Begg funnel plot was symmetric (P = 1.0) and Egger test did not show a significant publication bias (P = 0.756) (Fig. S4).
3.7Association of A3B expression with grade and tumor size
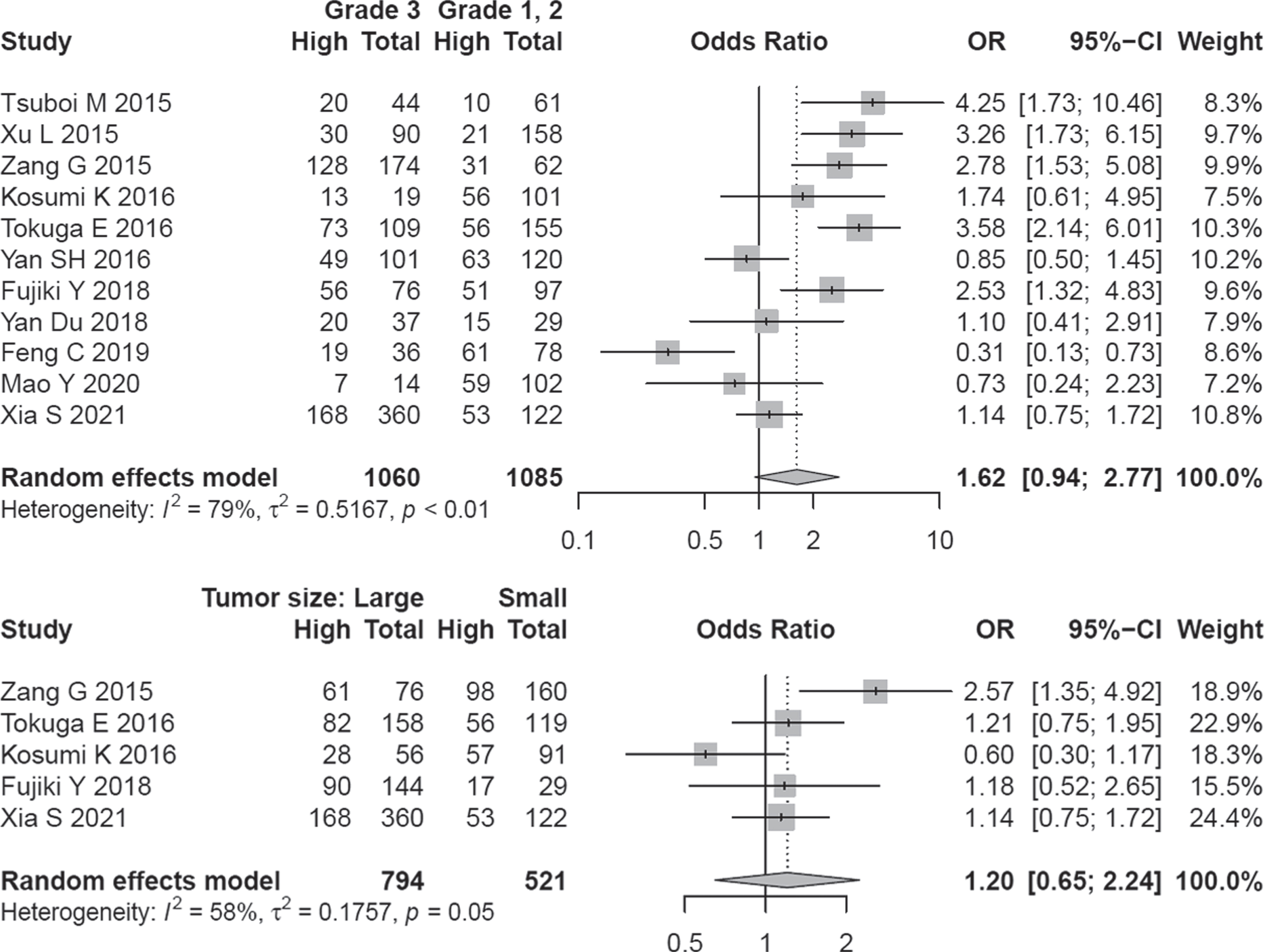
Eleven studies covering 2145 patients were analyzed for assessing the relationship of A3B expression and the clinical grade of tumors. A random-effect model was applied because of the inter-study heterogeneity (I2 = 79.1%, P < 0. 01). Although half of the studies implied a direct association of clinical grade with higher A3B expression, overall effect size did not show a significant association (OR = 1.62, 95% CI: 0.94–2.77) (Fig. 5).
The determined OR values remained the same by excluding any single article (Fig. S5). Begg funnel plot was symmetric (P = 0.484) and Egger test did not show a significant publication bias (P = 0.822) (Fig. S5). A3B expression was not significantly correlated with tumor size (OR = 1.20, 95% CI = 0.45–2.24) (Fig. 5).
Fig. 5
Forest plot regarding the association of A3B expression and grade / tumor size.
4Discussion
Cancer is the most important challenge for life expectancy in both developing and developed countries of the world [1, 51]. Despite advances in clinical and molecular research to find effective biomarkers with prognostic significance in cancers, but steady rise in cancer incidence is indicative of the urgent need for reliable and effective cancer screening/prognosis biomarkers [52]. APOBEC3B is one of the main sources of mutagenesis in several human cancers including lung, bladder, head and neck, cervix, breast, etc [30, 53, 54]. Recently, many studies reported an abnormal expression of A3B in multiple cancer types which may contribute to the poor prognosis and therapeutic resistance [41, 55, 56]. Although the significance of A3B as a mutation source is becoming well-acknowledged, the prognostic value of A3B expression in cancers remains controversial. In this meta-analysis, we investigated the prognostic value and clinicopathological features of A3B expression in cancer patients. We evaluated survival data of 3776 patients in 19 studies. Our meta-analysis revealed that increased A3B expression was closely associated with poor OS, poor DFS and poor RFS in some of cancers.
To reduce the heterogeneity of studies, the sub-group and meta-regression analysis were conducted by factors including study region, cancer type, detection method and expression measurement method either by protein or mRNA quantification. Results from stratified analysis by study region showed that the associations between high A3B expression and poor OS were significant in the East Asia region but not in non-Asian populations, suggesting that the relation between A3B expression and poor OS could be ethnic-dependent. In the subgroup analysis based on cancer type, APOBEC3B expression was associated with poor OS in lung and breast cancer but this was not true in the case of ovarian cancer. This may be indicative of that A3B might have different biological behaviors and clinical characteristics in various cancer types.
Results from stratified analysis by method showed that the associations between high A3B expression and poor OS were significant in the studies that used the IHC method but not RT-qPCR. There is a possible explanation for these results. APOBEC3 family have considerable nucleotide identity at the mRNA level so these high identities make some complication to the quantification of the mRNA expression [57–59]. Some of the included studies used RT-qPCR and their result may be affected by these high identities.
In addition, the role of A3B in cancer metastasis and progression should also be considered. For example, A3B overexpression can cause genomic instability in human cells by inducing C-to-T transitions which might lead to tumorigenesis [6]. Additionally, Yamazaki et al. found that highly expressed A3B in myeloma cells causes DNA substitutions and deletions. This shows that A3B could induce mutations in the tumor genome in a way that bypasses the DNA repair mechanism, potentially leading to genomic instability in myeloma [60]. Saito et al. also observed that irradiation can induce APOBEC3B expression and may lead to radiation-induced mutation in HepG2, HeLa and SAS cells [61].
The role of A3B expression in different malignancies suggested that common signaling pathways in various cancer types are likely to be involved in dysregulation of A3B expression. Maruyama et al. found that high expression of A3B has been induced by the PKC/IKK/NFkB signaling pathway in various cancer cells, which might lead to the accumulation of somatic mutations and genetic diversity in many common cancers [62]. Also, P53 is a serious inhibitor for A3B expression in cells and limits its mutagenicity. In cancer cells due to the loss of p53 through mutation, A3B overexpression can elevate the power of mutagenesis and tumor evolution [63].
Chemotherapy is the first-line approach to destroy cancer cells for many cancers. Recent, studies showed that A3B expression is correlated with resistance to chemotherapy. Periyasamy et al. found that chemotherapy can promote A3B expression via DNA-PKcs and ATM-mediated activation of the NF-κB pathways and therefore decrease the efficacy of chemotherapeutic treatments [64]. Serebrenik et al. suggested that in ovarian cancer, APOBEC3B is a predictive biomarker of response to platinum-based chemotherapy [45]. Furthermore, in ER-positive breast cancer, A3B expression can induce tamoxifen resistance. Such observations suggest that A3B has a role in the progression of tumors and inhibiting APOBEC3B expression might improve the efficacy of cancer treatments [65].
Mutation in A3B gene was reported to play role in malignancy development. For example, Radmanesh et al. demonstrated that truncating variant at the A3B locus (c.783delG) is related to the risk of breast cancer and early onset of the disease [66]. The results of one meta-analysis revealed that 30 kb APOBEC3B deletion can decrease A3B expression and does not link to the risk of breast and ovarian cancers in the European population [67]. However, Cescon et al. proved that in breast cancer, A3B overexpression reflects mitosis and cell cycle-related gene expression, whereas a deletion polymorphism is linked to immunological activation. The authors proposed that the A3Bdel polymorphism could be utilized to predict cancer immunotherapy [68].
In spite of the inspiring results, there are several limitations in the current meta-analysis that need to be taken into consideration. First, the cut-off value and detection method for APOBEC3B expression varied among the included studies, which may cause heterogeneity and bias. Second, all the studies included in our meta-analysis were in English, which might bring selection bias. Third, only a few included studies evaluated PFS and DFS, which might result in bias. Fourth, most of the eligible studies were conducted in Asia, this might also lead to bias and our results might be more relevant to Asian patients. Fifth, immune infiltration into the tumor microenvironment may contribute to an increase of APOBEC3B expression in qPCR measurements and may lead to bias. Finally, some HRs and 95% CIs were not mentioned directly in the included studies. Then, we extracted them from Kaplan–Meier curves which could not be precise enough.
In conclusion, to the best of our knowledge, the present study is the first comprehensive meta-analysis exploring the prognostic value of APOBEC3B overexpression in various cancers. The results indicated that high APOBEC3B expression is significantly related to poor clinical outcomes in cancer patients especially breast cancer. Therefore, APOBEC3B overexpression might be used as an adverse prognostic biomarker for cancer patients. However, further high quality studies are needed to strengthen these findings.
Acknowledgments
We are thankful to Professor Roya Kelishadi, Director of Research Institute for Primordial Prevention of Non-Communicable Disease, Isfahan University of Medical Sciences, for her support throughout the course of this study.
Financial support of Vice-Chancellor for research, Isfahan University of Medical Sciences, through grant number 397811 is gratefully acknowledged.
Author contributions
CONCEPTION: SJ and RS
DATA CURATION: SJ and MY
ANALYSIS OF DATA: SJ, MY, RN and SGH
PREPARATION OF THE MANUSCRIPT: SJ and RS
REVISION FOR IMPORTANT INTELLECTUAL CONTENT: SJ, RS, RN and SGH
Conflict of interest
The authors have declared no conflict of interest.
Supplementary material
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/TUB-211577.
References
[1] | Bray F , Ferlay J , Soerjomataram I , Siegel RL , Torre LA , Jemal A . Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) ;68: (6):394–424. doi:10.3322/caac.21492 |
[2] | Sung H , Ferlay J , Siegel RL , Laversanne M , Soerjomataram I , Jemal A , et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) . doi:10.3322/caac.21660 |
[3] | Hulvat MC . Cancer Incidence and Trends. Surg Clin North Am. (2020) ;100: (3):469–81. doi:10.1016/j.suc.2020.01.002 |
[4] | Lichtenstein AV . Strategies of the War on Cancer: To Kill or to Neutralize? Front Oncol. (2019) ;8: (667). doi:10.3389/fonc.2018.00667 |
[5] | Siegel RL , Miller KD , Jemal A . Cancer statistics, 2018. CA Cancer J Clin. (2018) ;68: (1):7–30. doi:10.3322/caac.21442 |
[6] | Shinohara M , Io K , Shindo K , Matsui M , Sakamoto T , Tada K , et al. APOBEC3B can impair genomic stability by inducing base substitutions in genomic DNA in human cells. Scientific Reports. (2012) ;2: (1):806. doi:10.1038/srep00806 |
[7] | Kuong KJ , Loeb LA . APOBEC3B mutagenesis in cancer. Nat Genet. (2013) ;45: (9):964–5. doi:10.1038/ng.2736 |
[8] | Stransky N , Egloff AM , Tward AD , Kostic AD , Cibulskis K , Sivachenko A , et al. The mutational landscape of head and neck squamous cell carcinoma. Science. (2011) ;333: (6046):1157–60. doi:10.1126/science.1208130 |
[9] | Barbieri CE , Baca SC , Lawrence MS , Demichelis F , Blattner M , Theurillat JP , et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. (2012) ;44: (6):685–9. doi:10.1038/ng.2279 |
[10] | Koning FA , Newman EN , Kim EY , Kunstman KJ , Wolinsky SM , Malim MH . Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. (2009) ;83: (18):9474–85. doi:10.1128/JVI.01089-09 |
[11] | Stenglein MD , Burns MB , Li M , Lengyel J , Harris RS . APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nature Structural & Molecular Biology. (2010) ;17: (2):222–9. doi:10.1038/nsmb.1744 |
[12] | Olson ME , Harris RS , Harki DA . APOBEC Enzymes as Targets for Virus and Cancer Therapy. Cell Chemical Biology. (2018) ;25: (1):36–49. doi:10.1016/j.chembiol.2017.10.007 |
[13] | Harris RS , Dudley JP . APOBECs and virus restriction. Virology. (2015) ;479-480: :131–45. doi:10.1016/j.virol.2015.03.012 |
[14] | Hultquist JF , Lengyel JA , Refsland EW , LaRue RS , Lackey L , Brown WL , et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. (2011) ;85: (21):11220–34. doi:10.1128/JVI.05238-11 |
[15] | Law EK , Levin-Klein R , Jarvis MC , Kim H , Argyris PP , Carpenter MA , et al. APOBEC3A catalyzes mutation and drives carcinogenesis in vivo . J Exp Med. (2020) ;217: (12). doi:10.1084/jem.20200261 |
[16] | Zou J , Wang C , Ma X , Wang E , Peng G . APOBEC3B, a molecular driver of mutagenesis in human cancers. Cell Biosci. (2017) ;7: :29. doi:10.1186/s13578-017-0156-4 |
[17] | Chan K , Roberts SA , Klimczak LJ , Sterling JF , Saini N , Malc EP , et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. (2015) ;47: (9):1067–72. doi:10.1038/ng.3378 |
[18] | Cortez LM , Brown AL , Dennis MA , Collins CD , Brown AJ , Mitchell D , et al. APOBEC3A is a prominent cytidine deaminase in breast cancer. PLoS Genet. (2019) ;15: (12):e1008545. doi:10.1371/journal.pgen.1008545 |
[19] | Langenbucher A , Bowen D , Sakhtemani R , Bournique E , Wise JF , Zou L , et al. An extended APOBEC3A mutation signature in cancer. Nat Commun. (2021) ;12: (1):1602. doi:10.1038/s41467-021-21891-0 |
[20] | Rouf Banday A , Onabajo OO , Lin SH-Y , Obajemu A , Vargas JM , Delviks-Frankenberry KA , et al. Targeting natural splicing plasticity of APOBEC3B restricts its expression and mutagenic activity. Communications Biology. (2021) ;4: (1):386. doi:10.1038/s42003-021-01844-5 |
[21] | Tsuboi M , Yamane A , Horiguchi J , Yokobori T , Kawabata-Iwakawa R , Yoshiyama S , et al. APOBEC3B high expression status is associated with aggressive phenotype in Japanese breast cancers. Breast Cancer. (2016) ;23: (5):780–8. doi:10.18632/oncotarget.25495 |
[22] | Venkatesan S , Angelova M , Puttick C , Zhai H , Caswell DR , Lu WT , et al. Induction of APOBEC3 Exacerbates DNA Replication Stress and Chromosomal Instability in Early Breast and Lung Cancer Evolution. Cancer Discov. (2021) ;11: (10):2456–73. doi:10.1158/2159-8290.CD-20-0725 |
[23] | Kim YS , Sun S , Yoon JS , Ko YH , Won HS , Kim JS . Clinical implications of APOBEC3A and 3B expression in patients with breast cancer. PLoS One. (2020) ;15: (3):e0230261. doi:10.1371/journal.pone.0230261 |
[24] | Lackey L , Demorest ZL , Land AM , Hultquist JF , Brown WL , Harris RS . APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol. (2012) ;419: (5):301–14. doi:10.1016/j.jmb.2012.03.011 |
[25] | Hou S , Silvas TV , Leidner F , Nalivaika EA , Matsuo H , Kurt Yilmaz N , et al. Structural Analysis of the Active Site and DNA Binding of Human Cytidine Deaminase APOBEC3B. J Chem Theory Comput. (2019) ;15: (1):637–47. doi:10.1021/acs.jctc.8b00545 |
[26] | Burns MB , Lackey L , Carpenter MA , Rathore A , Land AM , Leonard B , et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. (2013) ;494: (7437):366–70. doi:10.1038/nature11881 |
[27] | Cescon DW , Haibe-Kains B , Mak TW . APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A. (2015) ;112: (9):2841–6. doi:10.1073/pnas.1424869112 |
[28] | Harris RS . Cancer mutation signatures, DNA damage mechanisms, and potential clinical implications. Genome Med. (2013) ;5: (9):87. doi:10.1186/gm490 |
[29] | Du Y , Tao X , Wu J , Yu H , Yu Y , Zhao H . APOBEC3B up-regulation independently predicts ovarian cancer prognosis: a cohort study. Cancer Cell International. (2018) ;18: (1):78. doi:10.1186/s12935-018-0572-5 |
[30] | Burns MB , Temiz NA , Harris RS . Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. (2013) ;45: (9):977–83. doi:10.1038/ng.2701 |
[31] | Henderson S , Chakravarthy A , Su X , Boshoff C , Fenton TR . APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. (2014) ;7: (6):1833–41. doi:10.1016/j.celre2014.05.012 |
[32] | Kosumi K , Baba Y , Ishimoto T , Harada K , Nakamura K , Ohuchi M , et al. APOBEC3B is an enzymatic source of molecular alterations in esophageal squamous cell carcinoma. Med Oncol. (2016) ;33: (3):26. doi:10.1007/s12032-016-0739-7 |
[33] | Mah LJ , El-Osta A , Karagiannis TC . γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. (2010) ;24: (4):679–86. doi:10.1038/leu.2010.6 |
[34] | Rahmanian N , Shokrzadeh M , Eskandani M . Recent advances in γH2AX biomarker-based genotoxicity assays: A marker of DNA damage and repair. DNA Repair. (2021) ;108: :103243. doi:10.1016/j.dnare2021.103243 |
[35] | Lackey L , Law EK , Brown WL , Harris RS . Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. (2013) ;12: (5):762–72. doi:10.4161/cc.23713 |
[36] | Hirabayashi S , Shirakawa K , Horisawa Y , Matsumoto T , Matsui H , Yamazaki H , et al. APOBEC3B is preferentially expressed at the G2/M phase of cell cycle. Biochem Biophys Res Commun. (2021) ;546: :178–84. doi:10.1016/j.bbrc.2021.02.008 |
[37] | Ma W , Ho DW , Sze KM , Tsui YM , Chan LK , Lee JM , et al. APOBEC3B promotes hepatocarcinogenesis and metastasis through novel deaminase-independent activity. Mol Carcinog. (2019) ;58: (5):643–53. doi:10.1002/mc.22956 |
[38] | McCann JL , Klein MM , Leland EM , Law EK , Brown WL , Salamango DJ , et al. The DNA deaminase APOBEC3B interacts with the cell-cycle protein CDK4 and disrupts CDK4-mediated nuclear import of Cyclin D1. J Biol Chem. (2019) ;294: (32):12099–111. doi: 10.1074/jbc.RA119.008443 |
[39] | Ng JCF , Quist J , Grigoriadis A , Malim MH , Fraternali F . Pan-cancer transcriptomic analysis dissects immune and proliferative functions of APOBEC3 cytidine deaminases. Nucleic Acids Res. (2019) ;47: (3):1178–94. doi:10.1093/nar/gky1316 |
[40] | Du Y , Tao X , Wu J , Yu H , Yu Y , Zhao H . APOBEC3B up-regulation independently predicts ovarian cancer prognosis: a cohort study. Cancer Cell Int. (2018) ;18: :78. doi:10.1186/s12935-018-0572-5 |
[41] | Wang S , Jia M , He Z , Liu XS . APOBEC3B and APOBEC mutational signature as potential predictive markers for immunotherapy response in non-small cell lung cancer. Oncogene. (2018) ;37: (29):3924–36. doi:10.1038/s41388-018-0245-9 |
[42] | Gara SK , Tyagi MV , Patel DT , Gaskins K , Lack J , Liu Y , et al. GATA3 and APOBEC3B are prognostic markers in adrenocortical carcinoma and APOBEC3B is directly transcriptionally regulated by GATA3. Oncotarget. (2020) ;11: (36):3354–70. doi:10.18632/oncotarget.27703 |
[43] | Yan S , He F , Gao B , Wu H , Li M , Huang L , et al. Increased APOBEC3B Predicts Worse Outcomes in Lung Cancer: A Comprehensive Retrospective Study. J Cancer. (2016) ;7: (6):618–25. doi:10.7150/jca.14030 |
[44] | Mao Y , Lv M , Zhang Y , Nie G , Cui J , Wang Y , et al. APOBEC3B expression and its prognostic potential in breast cancer. Oncol Lett. (2020) ;19: (4):3205–14. doi:10.3892/ol.2020.11433 |
[45] | Serebrenik AA , Argyris PP , Jarvis MC , Brown WL , Bazzaro M , Vogel RI , et al. The DNA Cytosine Deaminase APOBEC3B is a Molecular Determinant of Platinum Responsiveness in Clear Cell Ovarian Cancer. Clin Cancer Res. (2020) ;26: (13):3397–407. doi:10.1158/1078-0432.CCR-19-2786 |
[46] | Fujiki Y , Yamamoto Y , Sueta A , Yamamoto-Ibusuki M , Goto-Yamaguchi L , Tomiguchi M , et al. APOBEC3B gene expression as a novel predictive factor for pathological complete response to neoadjuvant chemotherapy in breast cancer. Oncotarget. (2018) ;9: (55):30513–26. doi:10.18632/oncotarget.25495 |
[47] | Moher D , Liberati A , Tetzlaff J , Altman DG . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. (2009) ;151: (4):264–9, w64. doi:10.7326/0003-4819-151-4-200908180-00135 |
[48] | Cisler JM , Begle AM , Amstadter AB , Resnick HS , Danielson CK , Saunders BE , et al. Exposure to interpersonal violence and risk for PTSD, depression, delinquency, and binge drinking among adolescents: data from the NSA-R. J Trauma Stress. (2012) ;25. doi:10.1002/jts.21672 |
[49] | Tierney JF , Stewart LA , Ghersi D , Burdett S , Sydes MR . Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. (2007) ;8: (1):1–16. doi:10.1186/1745-6215-8-16 |
[50] | Balduzzi S , Rücker G , Schwarzer G . How to perform a meta-analysis with R: a practical tutorial. Evidence-based mental health. (2019) ;22: (4):153–60. doi:10.1136/ebmental-2019-300117 |
[51] | Han L , Wang B , Wang R , Wang Z , Gong S , Chen G , et al. Prognostic and Clinicopathological Significance of Long Non-coding RNA PANDAR Expression in Cancer Patients: A Meta-Analysis. Front Oncol. (2019) ;9: :1337. doi:10.3389/fonc.2019.01337 |
[52] | Poojary M , Jishnu PV , Kabekkodu SP . Prognostic Value of Melanoma-Associated Antigen-A (MAGE-A) Gene Expression in Various Human Cancers: A Systematic Review and Meta-analysis of 7428 Patients and 44 Studies. Mol Diagn Ther. (2020) ;24: (5):537–55. doi:10.1007/s40291-020-00476-5 |
[53] | Zou J , Wang C , Ma X , Wang E , Peng G . APOBEC3B, a molecular driver of mutagenesis in human cancers. Cell & Bioscience. (2017) ;7: (1):29. doi:10.1186/s13578-017-0156-4 |
[54] | Roberts SA , Lawrence MS , Klimczak LJ , Grimm SA , Fargo D , Stojanov P , et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature Genetics. (2013) ;45: (9):970–6. doi:10.1038/ng.2702 |
[55] | Swanton C , McGranahan N , Starrett GJ , Harris RS . APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer Discov. (2015) ;5: (7):704–12. doi:10.1158/2159-8290.CD-15-0344 |
[56] | Driscoll CB , Schuelke MR , Kottke T , Thompson JM , Wongthida P , Tonne JM , et al. APOBEC3B-mediated corruption of the tumor cell immunopeptidome induces heteroclitic neoepitopes for cancer immunotherapy. Nat Commun. (2020) ;11: (1):790. doi:10.1038/s41467-020-14568-7 |
[57] | Burns MB , Leonard B , Harris RS . APOBEC3B: pathological consequences of an innate immune DNA mutator. Biomed J. (2015) ;38: (2):102–10. doi:10.4103/2319-4170.148904 |
[58] | Henderson S , Fenton T . APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol Med. (2015) ;21: (5):274–84. doi:10.1016/j.molmed.2015.02.007 |
[59] | Refsland EW , Stenglein MD , Shindo K , Albin JS , Brown WL , Harris RS . Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. (2010) ;38: (13):4274–84. doi:10.1093/nar/gkq174 |
[60] | Yamazaki H , Shirakawa K , Matsumoto T , Hirabayashi S , Murakawa Y , Kobayashi M , et al. Endogenous APOBEC3B Overexpression Constitutively Generates DNA Substitutions and Deletions in Myeloma Cells. Scientific Reports. (2019) ;9: (1):7122. doi:10.1038/s41598-019-43575-y |
[61] | Saito Y , Miura H , Takahashi N , Kuwahara Y , Yamamoto Y , Fukumoto M , et al. Involvement of APOBEC3B in mutation induction by irradiation. Journal of Radiation Research. (2020) ;61: (6):819–27. doi:10.1093/jrr/rraa069 |
[62] | Maruyama W , Shirakawa K , Matsui H , Matsumoto T , Yamazaki H , Sarca AD , et al. Classical NF-κB pathway is responsible for APOBEC3B expression in cancer cells. Biochem Biophys Res Commun. (2016) ;478: (3):1466–71. doi:10.1016/j.bbrc.2016.08.148 |
[63] | Periyasamy M , Singh AK , Gemma C , Kranjec C , Farzan R , Leach DA , et al. p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res. (2017) ;45: (19):11056–69. doi:10.1093/nar/gkx721 |
[64] | Periyasamy M , Singh AK , Gemma C , Farzan R , Allsopp RC , Shaw JA , et al. Induction of APOBEC3B expression by chemotherapy drugs is mediated by DNA-PK-directed activation of NF-κB. Oncogene. (2021) ;40: (6):1077–90. doi:10.1038/s41388-020-01583-7 |
[65] | Law EK , Sieuwerts AM , LaPara K , Leonard B , Starrett GJ , Molan AM , et al. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci Adv. (2016) ;2: (10):e1601737. doi:10.1126/sciadv.1601737 |
[66] | Radmanesh H , Spethmann T , Enßen J , Schürmann P , Bhuju S , Geffers R , et al. Assessment of an APOBEC3B truncating mutation, c.783delG, in patients with breast cancer. Breast Cancer Res Treat. (2017) ;162: (1):31–7. doi:10.1007/s10549-016-4100-9 |
[67] | Klonowska K , Kluzniak W , Rusak B , Jakubowska A , Ratajska M , Krawczynska N , et al. The 30 kb deletion in the APOBEC3 cluster decreases APOBEC3A and APOBEC3B expression and creates a transcriptionally active hybrid gene but does not associate with breast cancer in the European population. Oncotarget. (2017) ;8: (44):76357–74. doi:10.18632/oncotarget.19400 |
[68] | Cescon DW , Haibe-Kains B , Mak TW . APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proceedings of the National Academy of Sciences. (2015) ;112: (9):2841–6. doi:10.1073/pnas.1424869112 |
[69] | Kim H , Kim O , Lee MA , Lee JY , Hong SH , Ha US , et al. Prognostic Impact of APOBEC3B Expression in Metastatic Urothelial Carcinoma and Its Association with Tumor-Infiltrating Cytotoxic T Cells. Curr Oncol. (2021) ;28: (3):1652–62. doi:10.3390/curroncol28030154 |
[70] | Xia S , Gu Y , Zhang H , Fei Y , Cao Y , Fang H , et al. Immune inactivation by APOBEC3B enrichment predicts response to chemotherapy and survival in gastric cancer. Oncoimmunology.. (2021) ;10: (1):1975386. doi:10.1080/2162402X.2021.1975386 |
[71] | Feng C , Zhang Y , Huang J , Zheng Q , Yang Y , Xu B . The Prognostic Significance of APOBEC3B and PD-L1/PD-1 in Nasopharyngeal Carcinoma. Appl Immunohistochem Mol Morphol. (2021) ;29: (3):239–44. doi:10.1097/PAI.0000000000000852 |
[72] | Feng C , Zheng Q , Yang Y , Xu M , Lian Y , Huang J , et al. APOBEC3B High Expression in Gastroenteropancreatic Neuroendocrine Neoplasms and Association With Lymph Metastasis. Appl Immunohistochem Mol Morphol. (2019) ;27: (8):599–605. doi:10.1097/PAI.0000000000000695 |
[73] | Rüder U , Denkert C , Kunze CA , Jank P , Lindner J , Jöhrens K , et al. APOBEC3B protein expression and mRNA analyses in patients with high-grade serous ovarian carcinoma. Histol Histopathol. (2019) ;34: (4):405–17. doi:10.14670/HH-18-050 |
[74] | Jiang C , Zhang Y , Li Y , Lu J , Huang Q , Xu R , et al. High CEP55 expression is associated with poor prognosis in non-small-cell lung cancer. Onco Targets Ther. (2018) ;11: :4979–90. doi:10.2147/OTT.S165750 |
[75] | Tokunaga E , Yamashita N , Tanaka K , Inoue Y , Akiyoshi S , Saeki H , et al. Expression of APOBEC3B mRNA in Primary Breast Cancer of Japanese Women. PLoS One. (2016) ;11: (12):e0168090. doi:10.1371/journal.pone.0168090 |
[76] | Leonard B , Starrett GJ , Maurer MJ , Oberg AL , Van Bockstal M , Van Dorpe J , et al. APOBEC3G Expression Correlates with T-Cell Infiltration and Improved Clinical Outcomes in High-grade Serous Ovarian Carcinoma. Clin Cancer Res. (2016) ;22: (18):4746–55. doi:10.1158/1078-0432.CCR-15-2910 |
[77] | Xu L , Chang Y , An H , Zhu Y , Yang Y , Xu J . High APOBEC3B expression is a predictor of recurrence in patients with low-risk clear cell renal cell carcinoma. Urol Oncol. (2015) ;33: (8):340.e1–8. doi:10.1016/j.urolonc.2015.05.009 |
[78] | Zhang J , Wei W , Jin HC , Ying RC , Zhu AK , Zhang FJ . The roles of APOBEC3B in gastric cancer. Int J Clin Exp Pathol. (2015) ;8: (5):5089–96. doi:10.1080/2162402X.2021.1975386 |
[79] | Sieuwerts AM , Willis S , Burns MB , Look MP , Meijer-Van Gelder ME , Schlicker A , et al. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer. (2014) ;5: (6):405–13. doi:10.1007/s12672-014-0196-8 |



