Angiogenesis’ related genetic variants alter clinical features and prognosis of diffuse large B-cell lymphoma patients
Abstract
OBJECTIVES:
Single nucleotide variants (SNVs) in vascular endothelial growth factor A (VEGFA) and VEGFA receptor (KDR) genes confer different inherited abilities in angiogenesis (AG) pathway. We aimed in the present study to evaluate influence of six VEGFA and four KDR SNVs in clinical features and survival of diffuse large B-cell lymphoma (DLBCL) patients.
METHODS:
One hundred and sixty-eight DLBCL patients diagnosed between June 2009-September 2014 were enrolled in the study. Patients were homogeneously treated with R-CHOP. Genotypes were identified in genomic DNA by real-time polymerase chain reaction.
RESULTS:
Patients with VEGFA -634CC and +936CT or TT genotypes were at increased risk of showing grade III / IV toxicities and not achieving complete remission with treatment, and shorter event-free and overall survival were seen in patients with VEGFA -1154GA or AA genotype and VEGFA ATAGCC haplotype.
CONCLUSION:
Our data suggest that inherited abnormalities in AG’s gene modulate clinical features and prognosis of DLBCL patients homogeneously treated with R-CHOP.
1Introduction
Angiogenesis (AG), the process in which new vessels are formed from preexisting vasculature, is crucial for the growth and progression of diffuse large B cell lymphoma (DLBCL) [1]. The most important key regulator of AG is the vascular endothelial growth factor A (VEGFA), and its function is mediated by its binding to a specific receptor, the VEGFR2 or KDR [2]. Both proteins are encoded by polymorphic genes, having changes in their expressions or functions as consequence.
The ancestral C, G and C alleles of VEGFA -2578C/A (rs699947), -1154G/A (rs1570360) and +936C/T (rs3025039) single nucleotide variants (SNVs), respectively, were related to higher VEGFA production [3, 4], the variant T allele of VEGFA -2489C/T (rs1005230) was related to higher VEGFA transcription [5], while the roles of VEGFA -634G/C (rs2010963) and -460C/T (rs833061) distinct alleles were controversial [6]. Concerning KDR SNVs, the ancestral T and G alleles of -604T/C (rs2071559) and +1192G/A (rs2305948), respectively, were related to higher binding efficiency for VEGFA [7] and the variant A and T alleles of -271G/A (rs7667298) and +1719A/T (rs1870377), were related to higher binding efficiency [8], and higher KDR transcription [7], respectively.
Recently, VEGFA and KDR SNVs were associated with prognosis in DLBCL in two studies conducted in Asians [9, 10], but not in another study conducted in Caucasians [11]. Facing the paucity of information and possible relevance of the issue, the aim of the present study was to determine whether VEGFA -2578C/A, -2489C/T, -1154G/A, -634G/C, -460C/T and +936C/T, and KDR -604T/C, -271G/A, +1192G/A and +1719A/T SNVs affect clinicopathological features and outcome of DLBCL patients from an ethnically distinct population of southeastern region of Brazil.
2Patients and methods
2.1Study patients
This prospective study included 168 consecutive de novo DLBCL patients from the Haematology and Haemotherapy Centre of the University of Campinas and the A. C. Camargo Cancer Center. The patients were seen at diagnosis from June 2009 to September 2014.
All cases were diagnosed according to World Health Organization (WHO) classification criteria [12] and were staged using the Ann Arbor System [13] and the International Prognostic Index in rituximab era (R-IPI) criteria [14].
The immunohistochemistry panel used to define the cell of origin (COO) of the tumor was as follows: monoclonal antibodies against CD3 (PS1, 1/30, Novocastra, Newcastle upon Tyne, UK), CD10 (56C6, 1/20, Novocastra), CD 20 (L26, 1/300; Dako, Glostrup, Denmark) and MUM-1 (MUM1p, 1/40; Dako), and patients were classified as having germinal centre B-cell (GCB) or non-GCB subtype, as previously reported [15].
2.2Treatment protocol
Patients were treated with curative intent every 3 weeks for a total of six to eight cycles of R-CHOP, as decided by the clinician. Each cycle consisted of rituximab 375 mg/m2, cyclophosphamide 750 mg/m2, doxorubicin 50 mg/m2, and vincristine 1.4 mg/m2 (maximum dose: 2.0 mg) administered intravenously on day 1, and oral prednisone 100 mg from day 1 to day 5, and cycles were repeated every 21 days [16]. Dose reductions and treatment delays were based on the intensity of side effects, according to the Institutional protocol. Patients who failed initial therapy with R-CHOP received high-dose intravenous therapy (ICE, based on 3-weekly cycles of ifosfamide 5.000 mg/m2 on day 2 (24 h infusion), carboplatin area under the curve = 5 on day 2, etoposide 100 mg/m2 from day 1 to day 3 or DHAP, based on cisplatin 100 mg/m2 over 24 h on day 3, cytosine arabinoside 2 g/m2 every 12 h on day 4, and dexamethasone 40 mg from day 3 to day 6) followed by autologous stem cell transplantation [17]. Central nervous system (CNS) prophylaxis with intrathecal methotrexate 12 mg/m2 and dexamethasone 2 mg/ m2 was administered to patients with high (4 to 6) CNS-IPI score [18], and patients who presented CNS relapse received two cycles of R-DHAP alternating with high-dose methotrexate. Prophylactic granulocyte-colony stimulating factor (G-CSF) or antibiotics were not routinely administered.
2.3Toxicity and response criteria
Adverse events reported for each cycle of R-CHOP were obtained from patient information and clinic laboratorial exams, according to the Common Terminology Criteria for Advance Events of the National Cancer Institute (CTCAE v.4 2009). Response in patients who completed R-CHOP was classified as complete response (CR), partial response (PR), stable disease (SD) or progressive disease (PD) according to the International Working Group criteria [13]. Patients were followed up using contrast-enhanced computed tomography (CT) scan 3 months after the completion of therapy. The complete responders were reviewed every 3 months for the first 2 years and every 6 months from 3rd to 5th year. A full blood count and lactate dehydrogenase were checked at each follow-up, together with clinical history and examination for signs of recurrence. Re-imaging and further investigations were performed when there was a suspicion of recurrence.
2.4Genotyping
Genotypes were analyzed in DNA of peripheral blood samples by real-time polymerase chain reaction, using TaqMan SNP Genotyping Assays. All reactions were performed according to the manufacturer’s instructions, using positive and negative controls. 10%of genotype determinations were carried out twice in independent experiments with total concordance.
2.5Statistical analysis
The sample size of the study was based on the frequencies of VEGFA -2578C/A, -2489C/T, -1154G/A, 634G/C, -460C/T and +936C/T and KDR +1192G/A, +1719A/T, -604T/C and -271G/A SNVs genotypes found in healthy individuals from southeastern region of Brazil [19–21]. The procedure was realized according to a previous description [22], with the purpose of to ensure that the three genotypes (ancestral homozygote, heterozygote, and variant homozygote) were represented points. We identified the amount of 148 patients as the minimum desired sample size for the present study.
The pairwise linkage disequilibrium (LD) was performed to ensure that markers were appropriate for inclusion in haplotype estimates [23]. LD was measured by the disequilibrium coefficient (D′), and significance was considered at a D′≥80%. Only haplotypes with frequency higher than 10%were selected to analyses. Logistic regression model assessed associations between genotypes and clinicopathological features, and Bonferroni method was used to adjust values of multiple comparisons. Event-free survival (EFS) and overall survival (OS) encompassed time from diagnosis until relapse, progression, death due to tumor effects or last follow-up, and time from diagnosis until death by any cause or last follow-up, respectively. EFS and OS probabilities were estimated by the Kaplan-Meier method and compared by the log-rank test. Cox hazards model was used to identify variables predicting EFS and OS. Variables with P < 0.10 in univariate analyses were included in multivariate analyses. Significant results obtained in multivariate analyses were validated using a bootstrap resampling study to investigate the stability of risk estimates (1,000 replications). Differences were significant when P < 0.05. All procedures were carried out according to the Declaration of Helsinki, and the study was approved by the Ethics Committee of each institution.
3Results
The clinicopathological features of 168 DLBCL patients at diagnosis are presented in Table 1. The casuistic was composed by near two-thirds of cases with B symptoms and Ann Arbor III or IV stage, and one-third of cases with bulky disease, poor R-IPI and non-GCB subtype. 157 patients were submitted to R-CHOP; 11 patients were ineligible for treatment with R-CHOP due to advanced age and various comorbidities. Most patients who completed R-CHOP (88.4%) obtained CR or PR. Grade III or IV toxicity was seen in near one-third of cases; hematological toxicity was the most common one (anemia was seen in 19 cases, neutropenia in 39 cases, and thrombocytopenia in 13 cases), grade III cardiotoxicity was seen in one case and grade III nephrotoxicity was seen in another case.
Table 1
Clinicopathological features of 168 diffuse large B-cell lymphoma patients at diagnosis
| Variable | Median (range) or N (%) |
| Age (in years) | 57 (19–89) |
| Gender | |
| Female | 94 (55.9) |
| Male | 74 (44.1) |
| B symptoms | |
| Absent | 49 (29.2) |
| Present | 119 (70.8) |
| Bulky disease | |
| No | 111 (66.1) |
| Yes | 57 (33.9) |
| LDH (U/L) | 466 (31.0–3,797) |
| IHC-based subtype* | |
| GCB | 88 (71.5) |
| Non-GCB | 35 (28.5) |
| Ann Arbor | |
| I + II | 74 (44,1) |
| III + IV | 94 (55.9) |
| R-IPI | |
| Very good/good | 97 (57.7) |
| Poor | 71 (42.3) |
| Response to R-CHOP** | |
| Complete | 115 (83.3) |
| Partial | 7 (5.1) |
| Stable or progressive disease | 16 (11.6) |
| Toxicity grade III or IV*** | |
| Haematological | 49 (96.1) |
| Non-hematological | 2 (3.9) |
| Event-free survival (months) | 60.3 (1–135) |
| Overall survival (months) | 62.8 (1–135) |
| Final status | |
| Alive with disease | 9 (5.4) |
| Alive without disease | 96 (57.1) |
| Dead due to toxicity | 14 (8.3) |
| Dead by disease progression | 47 (28.0) |
| Dead by unrelated cause | 2 (1.2) |
R-IPI: International Prognostic Index in rituximab era; IHC: Immuno-histochemistry; GCB: germinal center B-cells; LDH: lactate dehydrogenase. *: IHC was performed only in patients with adequate lymph node tissue; **: Response to therapy was analyzed only in patients who completed the R-CHOP scheme; ***: toxicity was analyzed only in patients submitted at least for one cycle of R-CHOP. Cardiotoxicity and nephrotoxicity were computed as non-hematological toxicities
We found LD between VEGFA SNVs, and the ATAGCC haplotype (-2578A, -2489T, -1154A, -634G, -460C and +936C alleles; frequency: 18.8%) was included in analyses (Fig. 1). No associations of genotypes/haplotype and clinicopathological features (gender, B symptoms, histological subtype, Ann Arbor stage, and R-IPI) were observed in sample (Table 2). Grade III or IV toxicities, mainly myelosuppression, were more common in patients with VEGFA -634CC and +936CT or TT genotypes; these patients had 2.96 and 2.40-fold greater risk of presenting grade III or IV toxicity to R-CHOP than patients with the remaining genotypes, respectively (Table 3). An excess of VEGFA +936CT or TT genotype was seen in patients that did not achieve CR; these patients had 3.13-fold greater risk of not achieving CR with R-CHOP than patients with VEGFA +936CC genotype (Table 3).
Fig. 1
Linkage disequilibrium plot for diffuse large B cell lymphoma patients of single nucleotide variants (SNVs) in the vascular endothelial growth factor-A gene VEGFA -2578C/A (1, rs699947), -2489C/T (2, rs1005230), -460C/T (3, rs833061), -1154G/A (4, rs1570360), -634G/C (5, rs2010963) and +936C/T (6, rs3025039).
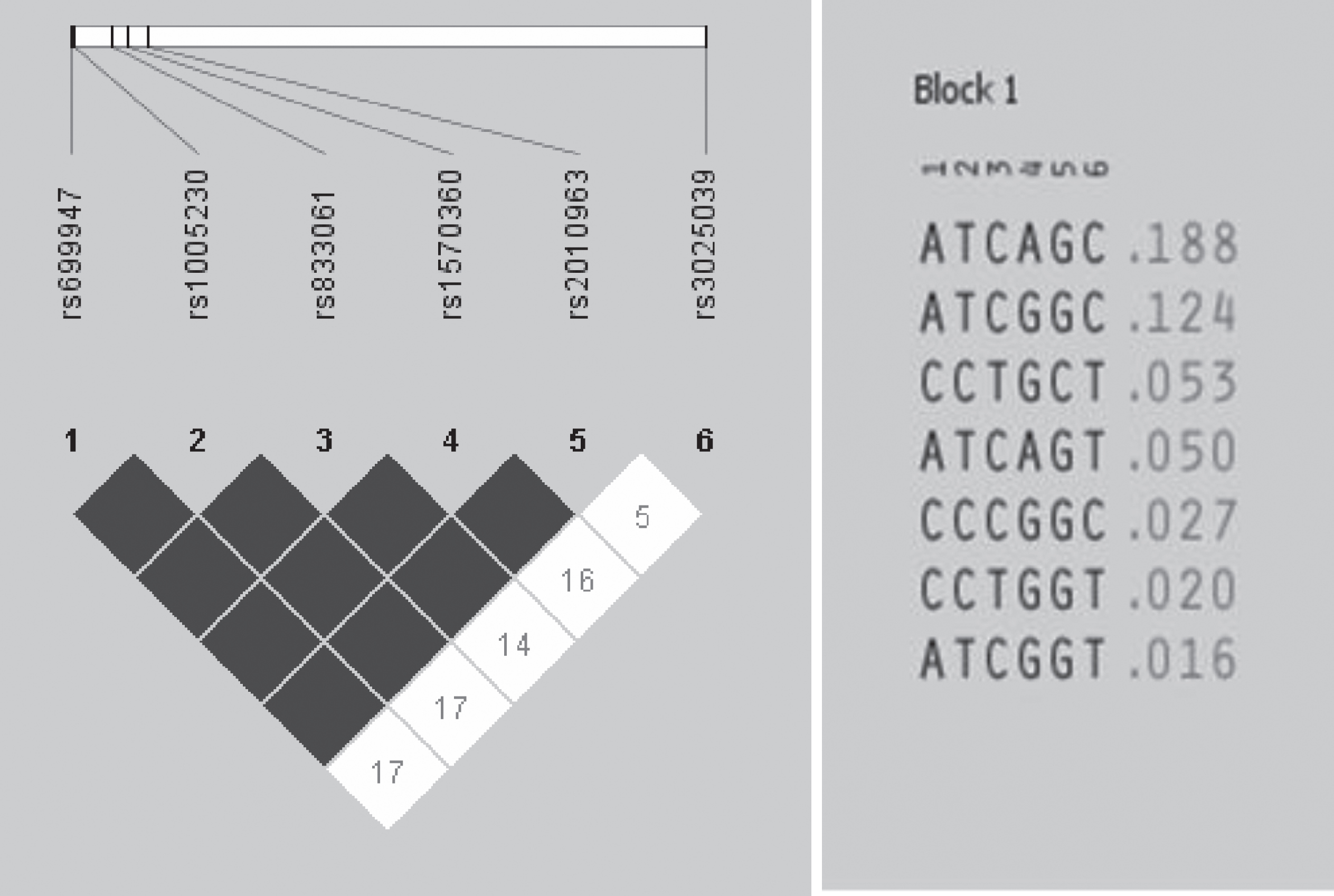
Table 2
Association of clinicopathological features and VEGFA -634G/C and +936C/T genotypes in diffuse large B-cell lymphoma patients
| Clinical feature | VEGFA 634 GG | VEGFA 634 GC | VEGFA 634 CC | P value | VEGFA 936 CC | VEGFA 936 CT | VEGFA 936 TT | P value |
| Number of patients (%) | 69 (41.1) | 76 (45.2) | 23 (13.7) | 122 (72.6) | 43 (25.6) | 3 (1.8) | ||
| Gender | ||||||||
| Male | 32 (43.2) | 32 (43.2) | 10 (13.5) | 0.87 | 51 (68.9) | 20 (27.0) | 3 (4.1) | 0.12 |
| Female | 37 (39.4) | 44 (46.8) | 13 (13.8) | 71 (75.5) | 23 (24.5) | 0 (0) | ||
| B symptoms | ||||||||
| Absent | 21 (42.9) | 23 (46.9) | 5 (10.2) | 0.70 | 36 (73.5) | 12 (24.5) | 1 (2.0) | 0.98 |
| Present | 48 (40.3) | 53 (44.5) | 18 (15.1) | 86 (72.3) | 31 (26.0) | 2 (1.7) | ||
| Subtype | ||||||||
| GCB | 37 (42.0) | 42 (47.7) | 9 (10.2) | 0.22 | 64 (72.7) | 22 (25.0) | 2 (2.3) | 0.83 |
| Non-GCB | 16 (45.7) | 12 (34.3) | 7 (20.0) | 23 (65.7) | 11 (31.4) | 1 (2.9) | ||
| Ann Arbor stage | ||||||||
| I/II | 23 (31.1) | 38 (51.3) | 13 (17.6) | 0.06 | 50 (67.5) | 21 (28.4) | 3 (4.1) | 0.10 |
| III/IV | 46 (48.9) | 38 (40.4) | 10 (10.7) | 72 (76.6) | 22 (23.4) | 0 (0) | ||
| R-IPI | ||||||||
| Low/ LI | 42 (43.3) | 41 (42.3) | 14 (14.4) | 0.66 | 71 (73.2) | 24 (24.7) | 2 (2.1) | 0.92 |
| Poor | 27 (38.0) | 35 (49.3) | 9 (12.7) | 51 (71.8) | 19 (26.8) | 1 (1.4) | ||
| Subtype | ||||||||
| GCB | 37 (42.0) | 42 (47.7) | 9 (10.2) | 0.22 | 64 (72.7) | 22 (25.0) | 2 (2.3) | 0.83 |
| Non-GCB | 16 (45.7) | 12 (34.3) | 7 (20.0) | 23 (65.7) | 11 (31.4) | 1 (2.9) |
GCB, germinal centre B-cell; R-IPI, International Prognostic Index in rituximab era; L, low; LI, low intermediate.
Table 3
Association of toxicity and response to therapy with VEGFA 634G/C and 936C/T genotypes in diffuse large B-cell lymphoma patients
| Variables | Patients | VEGFA -634G/C | VEGFA +936C/T | ||||
| N (%) | GG or GC N (%) | CC N (%) | P value | CC N (%) | CT or TT N (%) | P value | |
| Toxicity | |||||||
| Grade 0 to II | 106 (67.5) | 97 (70.8) | 9 (45.0) | 85 (73.9) | 21 (50.0) | ||
| Grade III or IV | 51 (32.5) | 40 (29.2) | 11 (55.0) | 0.03 | 30 (26.1) | 21 (50.0) | 0.007 |
| Complete response | |||||||
| Yes | 115 (83.3) | 102 (84.3) | 13 (76.5) | 89 (88.1) | 26 (70.3) | ||
| No | 23 (16.7) | 19 (15.7) | 4 (23.5) | 0.48 | 12 (11.9) | 11 (29.7) | 0.01 |
Toxicity was analyzed in 157 cases treated with R-CHOP with available exams; grade III or IV myelotoxicity was seen in 49 cases, cardiotoxicity in 2 cases, and nephrotoxicity in 1 case. Response was evaluated in 156 patients who completed R-CHOP; partial response and refractory disease were seen in 7 and 16 cases, respectively. Carriers of the VEGFA -634CC genotype were under a 2.96 (95%CI: 1.14–7.70)-fold increased risk of presenting grade III or IV toxicity during R-CHOP. Carriers of VEGFA +936CT or TT genotypes were under a 2.40 (95%CI: 1.14–5.04) more chance of presenting grade III or IV toxicity during treatment and a 3.13 (95%CI: 1.24–7.93) more chance of not achieving CR with R-CHOP. P significant values are presented in bold letters.
Median follow-up time was 74 months (range: 1–124). In last follow-up (December 2019), 105 patients were alive (9 with disease) and 63 patients died (14 due to toxicity, 47 of disease progression, and 2 by unrelated causes). The estimated 5-year EFS and OS were 61.6%and 65.1%, respectively. At 5-year of follow-up, EFS and OS were shorter in patients with B symptoms (Supplemental Figure 1A and 1B), non-GCB subtype (Supplemental Figure 1C and 1D), Ann Arbor stage III or IV (Supplemental Figure 1E and 1F), poor R-IPI (Supplemental Figure 1G and 1H), VEGFA -1154 GA or AA genotype (Fig. 2A and 2B), and VEGFA ATAGCC haplotype (Fig. 2C and 2D), and VEGFA +936CT or TT (47.3%vs 66.8%, P = 0.01) and KDR +1719 AA (20.0%vs 62.9%, P = 0.004) genotypes were related only to shorter EFS when compared to others (Kaplan-Meier estimates). Differences in EFS and OS of patients stratified by B symptoms, histological subtype, ECOG, and VEGFA -1154G/A genotypes and ATAGCC haplotype were seen in univariate and multivariate Cox analyses. Differences in EFS and OS of patients stratified by Ann Arbor staging and in EFS of patients stratified by VEGFA +936C/T and KDR +1719A/T genotypes were seen only in univariate Cox analysis.
Fig. 2
Kaplan-Meier estimates. A) Event-free survival (EFS) and B) overall survival (OS) of diffuse large B-cell lymphoma (DLBCL) patients stratified by genotypes of VEGFA 1154G/A single nucleotide variant (SNV); C) EFS and D) OS of DLBCL patients stratified by the presence or absence of VEGFA ATAGCC haplotype (-2578A, -2489T, -1154A, -634G, -460C and +936C alelles).
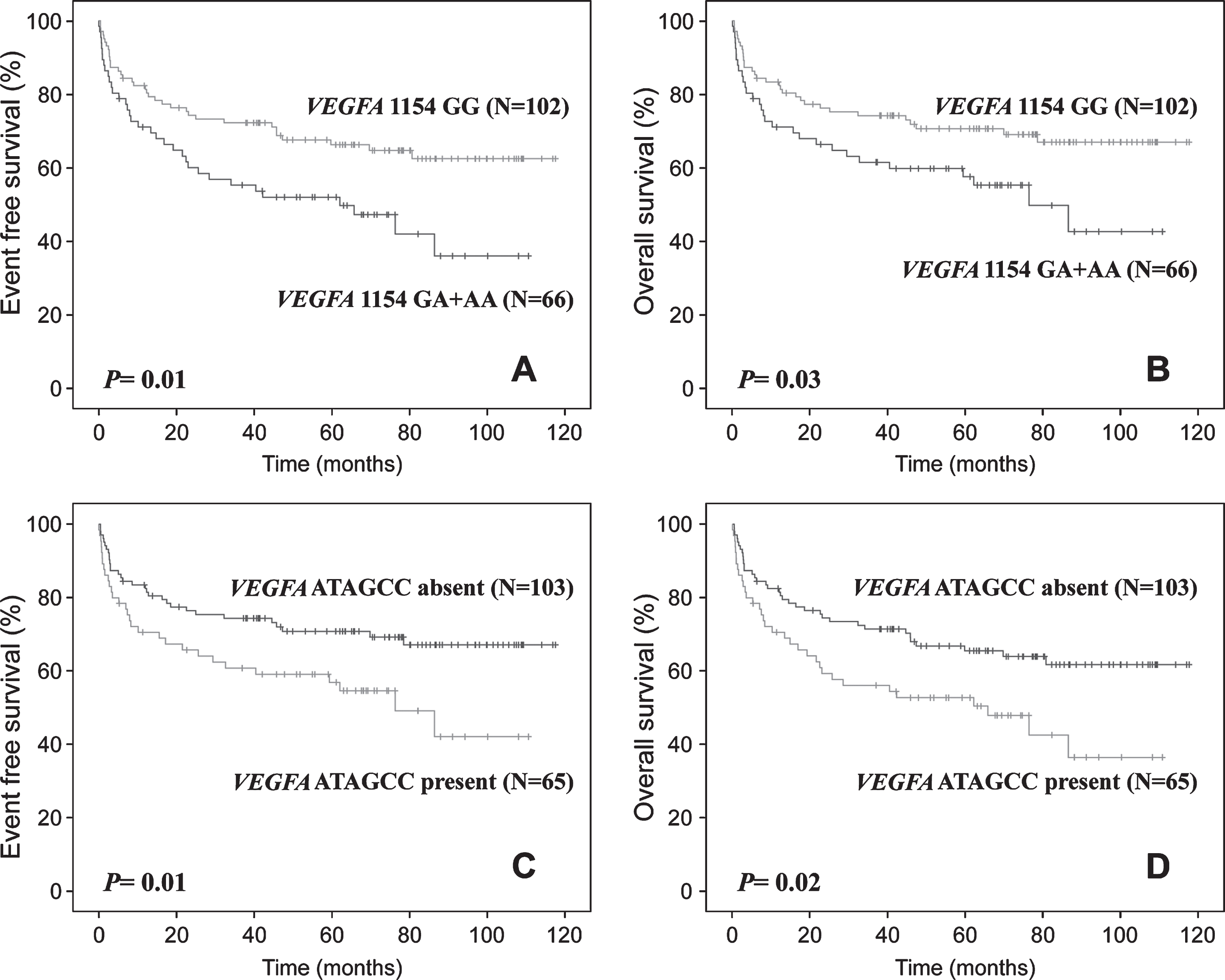
Patients with VEGFA +936CT or TT genotype and KDR +1719AA genotype had 1.78 and 3.52-fold greater risk of presenting disease progression in univariate Cox analysis, respectively. In multivariate analysis, patients with VEGFA -1154GA or AA genotype and ATAGCC haplotype had 2.64 and 2.52-fold greater risk of disease progression, and 2.97 and 2.33-fold greater risk of evolving to death than others, respectively. Associations found in multivariate analysis were confirmed by the bootstrapping method (Table 4).
Table 4
Association of clinicopathological features and VEGFA and KDR genotypes with survival in diffuse large B-cell lymphoma patients
| Characteristics | Event Free Survival | Overall Survival | ||||||||
| N event/ N total | Univariate HR (95%CI) | P value | Multivariate HR (95%CI) | P value | N event/ N total | Univariate HR (95%CI) | P value | Multivariate HR (95%CI) | P value | |
| B symptoms | ||||||||||
| Absent | 11/49 | Reference | Reference | 10/49 | Reference | Reference | ||||
| Present | 59/119 | 2.91 (1.52–5.55) | 0.001 | 2.35 (1.16–4.96) | 0.02 | 53/119 | 2.80 (1.42–5.52) | 0.003 | 2.12 (0.98–4.62) | 0.04 |
| Histological subtype* | ||||||||||
| GCB | 30/88 | Reference | Reference | 27/88 | Reference | Reference | ||||
| Non-GCB | 22/35 | 2.28 (1.31–3.97) | 0.003 | 2.00 (1.12–3.58) | 0.01 | 20/35 | 2.25 (1.26–4.02) | 0.006 | 1.96 (1.06–3.62) | 0.03 |
| Stage (Ann Arbor) | ||||||||||
| I or II | 25/74 | Reference | Reference | 21/74 | Reference | Reference | ||||
| III or IV | 45/94 | 1.62 (1.02–2.64) | 0.04 | 1.00 (0.53–1.91) | 0.98 | 42/94 | 1.78 (1.05–3.00) | 0.03 | 1.05 (0.53–2.07) | 0.87 |
| Age | ||||||||||
| ≤60 years | 36/96 | Reference | Reference | 31/96 | Reference | Reference | ||||
| > 60 years | 34/72 | 1.39 (0.87–2.23) | 0.16 | NA | 32/72 | 1.51 (0.92–2.48) | 0.10 | NA | ||
| Extranodal site | ||||||||||
| 0/1 | 22/61 | Reference | Reference | 21/61 | Reference | Reference | ||||
| > 1 | 48/107 | 1.20 (0.72–1.99) | 0.47 | NA | 42/107 | 1.09 (0.64–1.84) | 0.73 | NA | ||
| ECOG | ||||||||||
| 0/1 | 47/141 | Reference | Reference | 40/141 | Reference | Reference | ||||
| > 1 | 23/27 | 6.72 (4.02–11.21) | 0.001 | 4.20 (2.04–8.65) | 0.001 | 23/27 | 7.75 (4.57–13.14) | 0.001 | 4.84 (2.39–9.80) | 0.0001 |
| LDH | ||||||||||
| Normal | 13/42 | Reference | Reference | 11/42 | Reference | Reference | ||||
| Elevated | 57/126 | 1.70 (0.93–3.12) | 0.08 | 1.01 (0.50–2.02) | 0.97 | 52/126 | 1.82 (0.96–3.53) | 0.06 | 1.05 (0.49–2.24) | 0.88 |
| VEGFA -1154G/A | ||||||||||
| GG | 35/102 | Reference | Reference | 31/102 | Reference | Reference | ||||
| GA or AA | 35/66 | 1.75 (1.10–2.81) | 0.01a | 2.64 (1.29–5.39) | 0.007e | 32/66 | 1.77 (1.08–2.90) | 0.02g | 2.97 (1.35–6.32) | 0.005i |
| VEGFA +936C/T | ||||||||||
| CC | 45/124 | Reference | Reference | 41/124 | Reference | Reference | ||||
| CT or TT | 25/44 | 1.78 (1.09–2.91) | 0.02b | 1.27 (0.65–2.49) | 0.47 | 22/44 | 1.67 (0.99–2.80) | 0.05 | 1.25 (0.62–2.52) | 0.52 |
| KDR +1719A/T | ||||||||||
| AA | 5/5 | 3.52 (1.41–8.78) | 0.007c | 4.45 (1.00–14.1) | 0.05 | 4/5 | 2.65 (0.96–7.31) | 0.06 | 2.80 (0.78–10.06) | 0.11 |
| AT or TT | 65/163 | Reference | Reference | 59/163 | Reference | Reference | ||||
| VEGFA ATAGCC | ||||||||||
| Absent | 37/83 | Reference | Reference | 32/83 | Reference | Reference | ||||
| Present | 33/85 | 1.70 (1.06–2.72) | 0.02d | 2.52 (1.26–5.02) | 0.008f | 31/85 | 1.70 (1.04–2.79) | 0.03h | 2.33 (1.13–4.80) | 0.02j |
HR, hazard ratio; CI, confidence interval; NA, not applicable; GCB, germinal center B-cell; ECOG: Eastern Cooperative Oncology Group; LDH, lactate dehydrogenase. ATAGCC haplotype included VEGFA -2578A, -2489T, -1154A, -634G, -460C and +936C alelles. Ann Arbor stage was not included in multivariate analysis since it was included in R-IPI. P values < 0.05 were considered significant and are presented in bold letters. (a) Pbootstrap = 0.007; (b) Pbootstrap = 0.002; (c) Pbootstrap = 0.003; (d) Pbootstrap = 0.04; (e) Pbootstrap = 0.02; (f) Pbootstrap = 0.003; (g) Pbootstrap = 0.006; (h) Pbootstrap = 0.002; (i) Pbootstrap = 0.003; (j) Pbootstrap = 0.02.
4Discussion
We initially found that DLBCL patients enrolled in the present study presented similar clinicopathological aspects when compared to those of other countries [24, 25], and therefore the sample was adequate for evaluation of new prognostic factors in disease. Only B symptoms and histological GCB subtype were more common in our sample than in others and were described as characteristics of DLBCL in our country.
Secondly, we found no association of VEGFA and KDR genotypes and VEGFA haplotype with clinicopathological features of DLBCL, and VEGFA -2578C/A, -634G/C and +936C/T, and KDR +11926G/A SNVs were not associated with clinical features of DLBCL in study of Kim et al. [5]. Diao et al. [26] observed associations of VEGFA -2578C allele and +936TT genotype with tumor stage III or IV, and association of VEGFA +936T allele with IPI 4 was seen in study of Wróbel et al. [27], both conducted in non-Hodgkin lymphoma (LNH). Thus, ours and Kim’s data indicate that the analyzed SNVs do not alter clinicopathological features of DLBCL.
Thirdly, we observed that VEGFA -634CC and +936CT or TT genotypes were associated with higher toxicity to R-CHOP, mainly myelosuppression, and +936CT or TT genotype was also associated with poor response to chemoimmunotherapy. To the best of our knowledge, no studies focused on those issues previously. It is plausible that patients with VEGFA +936CT or TT genotype have worse response to therapy, since the T allele encodes less VEGFA than the C allele [3], having possible less tumor vasculature and less exposure of tumor cells to chemotherapy as consequence. However, association of the mentioned genotypes with higher toxicity it is not easily explained. One possibility is that the VEGFA +936T allele may modulate AG in different intensities in distinct organs, as proposed by Rutkowski et al. [28]. On the other hand, the roles of G and C alleles of VEGFA -634G/C SNV on VEGFA production are controversial [6]. Thus, the association of VEGFA -634CC genotype with high toxicity in our study may indicate that the C allele is enrolled with possible high VEGFA production and vessel formation, leading to high exposition of bone marrow cells to chemotherapy. However, associations of the VEGFA -634G/C and +936C/T SNVs with toxicity and response to R-CHOP need to be confirmed by further studies on the roles of ancestral and variant alleles in the VEGFA expression and micro vessel density in the tumor and bone marrow.
Finally, we observed that VEGFA -1154GA or AA and ATAGCC haplotype emerged as independent prognostic factors of short EFS and OS in our patients whereas VEGF +936CT or TT and KDR +1719AA were predictors of short EFS only. Kim et al. [9] found KDR 1719AT or TT genotype as independent prognostic factors related to worst EFS and OS in Korean DLBCL. Another recently published German work [11] did not find association between VEGFA -2578C/A, -1154G/A, -634G/C and +936C/T and KDR +1192G/A and +1719A/T SNVs with prognosis of DLBCL patients. Differences in results obtained in the present study and in Korean and German studies may be attributed to the ethnic heterogeneity of populations, since Brazilian population is highly heterogeneous (composed by indigenous and emigrants for Europe, Asia, and Africa) and mixed [29] while Korea and German populations are homogeneous. Imbalance of patients with unfavorable prognosis in studies may constitute another explanation for differences. According to our data, Yoon et al. [10] found VEGFA -1154 AA genotype as an independent prognostic factor with worst EFS and OS in Korean patients. Taken results together, the SNV alter survival in DLBCL, and this event does not depend on ethnic origin of patients. Since the VEGFA -1154A allele was related to low VEGFA production [4], we speculate that it may induce low number of vessels to supply the chemotherapy to tumor, resulting in disease progression and worse survival. Association of VEGFA ATAGCC haplotype and VEGFA +936CT or TT and VEGFR2 +1719AA genotypes with low VEGF production [7] also support our theory that low vessel formation impact negatively DLBCL prognosis. With these conflictual results, a meta-analysis composed by all studies is necessary to clarify the roles of VEGFA SNVs in survival of patients with DLBCL.
In the present study, VEGFA +936C/T SNV modulated CR but did not alter patients’ survival. Conversely, VEGFA -1154G/A SNV altered EFS and OS but did not influence CR rate. A possible explanation for these unexpected findings is based on the genetic variability of individuals who have the DLBCL. The VEGFA is highly polymorphic, with at least 30 functional SNVs in 5′-untranslated region (UTR), 3′-UTR, and promoter region [30]. Since it is already well known that a phenotype may not be determined by a single SNV, it is possible that the response pattern and survival of LDGCB patients of the present study had been determined by VEGFA +936C/T and VEGFA -1154G/A, respectively, but with the participation of other SNVs of the same gene or others [31].
It is also important to point that other AG-related parameters, such as other proangiogenic factors and their receptors, circulating endothelial cells, micro vessel density, and tumor microenvironment, have also been identified as prognostic indicators in different types of aggressive lymphomas [32], and may have contributed to the associations and lack of associations found in this study. Since bevacizumab, an angiogenic inhibitor tested in B-NHL have encountered disappointing outcomes [33, 34], the discovery of reliable angiogenic markers using non-invasive approaches is important for assessment of new AG-targeted therapies.
5Conclusion
Our data present preliminary evidence that inherited abnormalities in VEGFA and KDR genes are associated with poor outcome of patients with DLBCL. However, we are aware that these results should be validated in studies with larger numbers of patients from the various regions of the world, and by functional analyses of SNVs in tumor and bone marrow AG. If validated, the SNVs can be used to select patients with worse outcome that may receive distinct treatment.
Acknowledgments
We would like to thank “Fundação de Apoio à Pesquisa do Estado de São Paulo” (FAPESP) for the financial support (grant number 2015/15756-9).
Conflict of interest
The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplementary material
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/TUB-211510.
References
[1] | Ribatti D. , Nico B. , Ranieri G. , Specchia G. , Vacca A. , The role of angiogenesis in human non-Hodgkin lymphomas, Neoplasia 15: (3) ((2013) ), 231–238. |
[2] | Shibuya M. , Vascular endothelial growth factor (VEGF) and its receptor (VEGFR) signaling in angiogenesis: a crucial target for anti- and pro-angiogenic therapies, Genes Cancer 2: (12) ((2011) ), 1097–1105. |
[3] | Renner W. , Kotschan S. , Hoffmann C. , Obermayer-Pietsch B. and Pilger E. , A common 936 C/T mutation in gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels, J Vasc Res 37: (6) ((2000) ), 443–448. |
[4] | Shahbazi M. , Fryer A.A. , Pravica V. , Brogan I.J. , Ramsay H.M. , Hutchinson I.V. and Harden P.N. , Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection, J Am Soc Nephrol 13: (1) ((2002) ), 260–264. |
[5] | Kim Y.S. , Park S.W. , Kim M.H. , Jang E.J. , Park J.S. , Park S.J. , Baik H.W. , Chung G. and Hahm K.B. , Novel single nucleotide polymorphism of the VEGF gene as a risk predictor for gastroduodenal ulcers, J Gastroenterol Hepatol 23: (2) ((2008) ), 131–139. |
[6] | Stevens A. , Soden J. , Brenchley P.E. , Ralph S. and Ray D.W. , Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter, Cancer Res 63: (4) ((2003) ), 812–816. |
[7] | Wang Y. , Zheng Y. , Zhang W. , Yu H. , Lou K. , Zhang Y. , Qin Q. , Zhao B. , Yang Y. and Hui R. , Polymorphisms of KDR gene are associated with coronary heart disease, J Am Coll Cardiol 50: (8) ((2007) ), 760–767. |
[8] | An S.J. , Chen Z.H. , Lin Q.X. , Su J. , Chen H.J. , Lin J.Y. and Wu Y.L. , The -271> A > A polymorphism of kinase insert domain-containing receptor gene regulates its transcription level in patients with non-small cell lung cancer, BMC Cancer 9: ((2009) ), 144–150. |
[9] | Kim M.K. , Suh C. , Chi H.S. , Cho H.S. , Bae Y.K. , Lee K.H. , Lee G.W. , Kim I.S. , Eom H.S. , Kong S.Y. , Bae S.H. , Ryoo H.M. , Shin I.H. , Mun Y.C. , Chung H. and Hyun M.S. , VEGFA and VEGFR2 genetic polymorphisms and survival in patients with diffuse large B cell lymphoma, Cancer Sci 103: (3) ((2012) ), 497–503. |
[10] | Yoon K.A. , Kim M.K. , Eom H.S. , Lee H. , Park W.S. , Sohn J.Y. , Kim M.J. and Kong S.Y. , Adverse prognostic impact of vascular endothelial growth factor gene polymorphisms in patients with diffuse large B-cell lymphoma, Leuk Lymphoma 58: (11) ((2017) ), 2677–2682. |
[11] | Kaddu-Mulindwa D. , Rosolowski M. , Ziepert M. , Regitz E. , Assmann G. , Bewarder M. , Held G. , Pfreundschuh M. and Bittenbring J.T. , VEGFR2 and VEGFA2 polymorphisms are not associated with an inferior prognosis in Caucasian patients with aggressive B-cell lymphoma, Eur J Haematol 106: (1) ((2021) ), 100–104. |
[12] | Swerdlow S.H. , Campo E. , Harris N.L. , WHO classification of tumours of haematopoietic and lymphoid tissues. In: F.T. Bosman, E.S. Jaffe, S.R. Lakhani, H. Ohgaki, eds. World Health Organization Classification of Tumours. Lyon, France: IARC; (2008) . |
[13] | Cheson B.D. , Fisher R.I. , Barrington S.F. , Cavalli F. , Schwartz L.H. , Zucca E. and Lister T.A. , Alliance, Australasian Leukaemia and Lymphoma Group; Eastern Cooperative Oncology Group; European Mantle Cell Lymphoma Consortium; Italian Lymphoma Foundation; European Organisation for Research; Treatment of Cancer/Dutch Hemato-Oncology Group; Grupo Español de Médula Ósea; German High-Grade Lymphoma Study Group; German Hodgkin’s Study Group; Japanese Lymphoma Study Group; Lymphoma Study Association; NCIC Clinical Trials Group; Nordic Lymphoma Study Group; Southwest Oncology Group; United Kingdom National Cancer Research Institute. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification, J Clin Oncol 32: (27) ((2014) ), 3059–3068. |
[14] | Sehn L.H. , Berry B. , Chhanabhai M. , Fitzgerald C. , Gill K. , Hoskins P. , Klasa R. , Savage K.J. , Shenkier T. , Sutherland J. , Gascoyne R.D. and Connors J.M. , The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP, Blood 109: (5) ((2007) ), 1857–1861. |
[15] | Meyer P.N. , Fu K. , Greiner T.C. , Smith L.M. , Delabie J. , Gascoyne R.D. , Ott G. , Rosenwald A. , Braziel R.M. , Campo E. , Vose J.M. , Lenz G. , Staudt L.M. , Chan W.C. and Weisenburger D.D. , Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab, J Clin Oncol 29: (2) ((2011) ), 200–207. |
[16] | Pfreundschuh M. , Trümper L. , Osterborg A. , Pettengell R. , Trneny M. , Imrie K. , Ma D. , Gill D. , Walewski J. , Zinzani P.L. , Stahel R. , Kvaloy S. , Shpilberg O. , Jaeger U. , Hansen M. , Lehtinen T. , López-Guillermo A. , Corrado C. , Scheliga A. , Milpied N. , Mendila M. , Rashford M. , Kuhnt E. and Loeffler M. , MabThera International Trial Group. CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large-B-cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group, Lancet Oncol 7: (5) ((2006) ), 379–391. |
[17] | Gisselbrecht C. and Van E. , Den Neste, How I manage patients with relapsed/refractory diffuse large B cell lymphoma, Br J Haematol 182: (5) ((2018) ), 633–643. |
[18] | Schmitz N. , Zeynalova S. , Nickelsen M. , Kansara R. , Villa D. , Sehn L.H. , Glass B. , Scott D.W. , Gascoyne R.D. , Connors J.M. , Ziepert M. , Pfreundschuh M. , Loeffler M. and Savage K.J. , CNS International Prognostic Index: a risk model for CNS relapse in patients with diffuse large B-cell lymphoma treated with R-CHOP, J Clin Oncol 34: (26) ((2016) ), 3150–3156. |
[19] | Oliveira C. and Lourenço G.J. , Silva P.M.R. , Cardoso-Filho C. , Favarelli M.H.C. , Gonçales N.S.L. , Gurgel M.S.C. and Lima C.S.P. , Polymorphisms in the 5’ and 3’-untranslated regions of the VEGF gene are associated with breast cancer risk and clinicopathologic characteristics, Tumor Biol 32: (2) ((2011) ), 295–300. |
[20] | Brito A.B.C. , Lourenço G.J. , Oliveira G.B. , De Souza C.A. , Vassallo J. and Lima, C.S.P. , Associations of VEGF and VEGFR2 polymorphisms with increased risk and aggressiveness of multiple myeloma, Ann Hematol 93: (8) ((2014) ), 1363–1369. |
[21] | Vasconcelos V.C.A. , Lourenço G.J. , Brito A.B.C. and Vasconcelos V.L. , Maldaun M.V.C. , Tedeschi H. , Marie S.K.N. , Shinjo S.M.O. and Lima C.S.P. , Associations of VEGFA and KDR single nucleotide polymorphisms and increased risk and aggressiveness of high-grade gliomas, Tumor Biol 41: (9) ((2019) ), 1010428319872092. |
[22] | Beiguelman B. , Dinâmica dos Genes nas Famílias e Populações, Ribeir&o Preto, Sociedade Brasileira de Genética 2: ((1995) ), 1–472. |
[23] | Yang Y. , Li S.S. , Chien J.W. , Andriesen J. and Zhao L.P. , A systematic search for SNPs/haplotypes associated with disease phenotypes using a haplotype-based stepwise procedure, BMC Genet 9: ((2008) ), 90. |
[24] | Liu Y. and Barta S.K. , Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment, Am J Hematol 94: (5) ((2019) ), 604–616. |
[25] | Zhou Z. , Sehn L.H. , Rademaker A.W. , Gordon L.I. , Lacasce A.S. , Crosby-Thompson A. , Vanderplas A. , Zelenetz A.D. , Abel G.A. , Rodriguez M.A. , Nademanee A. , Kaminski M.S. , Czuczman M.S. , Millenson M. , Niland J. , Gascoyne R.D. , Connors J.M. , Friedberg J.W. and Winter J.N. , An enhanced International Prognostic Index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era, Blood 123: (6) ((2014) ), 837–842. |
[26] | Diao L.P. , Yu X.M. , Gao Y.H. , Li Y. , Liu H.S. , Liu L.H. , Zhou R.M. , Wang N. , Wu L.L. and Wang S.J. , Association of VEGF genetic polymorphisms with the clinical characteristics of non-Hodgkin’s lymphoma, J Cancer Res Clin Oncol 135: (11) ((2009) ), 1473–1481. |
[27] | Wróbel T. , Mazur G. , Dzietczenia J. , Gębura K. , Kuliczkowski K. and Bogunia-Kubik K. , VEGF and bFGF gene polymorphisms in patients with non-Hodgkin’s lymphoma, Biomed Res Int 2013: ((2013) ), 159813. |
[28] | Rutkowski P. , Bylina E. , Klimczak A. , Switaj T. , Falkowski S. , Kroc J. , Lugowska I. , Brzeskwiniewicz M. , Melerowicz W. , Osuch C. , Mierzejewska E. , Wasielewski K. , Woźniak A. , Grzesiakowska U. , Nowecki Z.I. , Siedlecki J.A. and Limon J. , The outcome and predictive factors of sunitinib therapy in advanced gastrointestinal stromal tumors (GIST) after imatinib failure: one institution study, BMC Cancer 12: ((2012) ), 107. |
[29] | Carvalho-Silva D.R. , Santos F.R. , Rocha J. and Pena S.D. , The phylogeography of Brazilian Y-chromosome lineages, Am J Hum Genet 68: (1) ((2001) ), 281–286. |
[30] | Vincenti V. , Cassano C. , Rocchi M. and Persico G. , Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3, Circulation 93: ((1996) ), 1493–1495. |
[31] | Erichsen H.C. and Chanock S.J. , SNPs in cancer research and treatment, Br J Cancer29; 90: (4) ((2004) ), 747–751. |
[32] | Jiang L. and Li N. , B-cell non-Hodgkin lymphoma: importance of angiogenesis and antiangiogenic therapy, Angiogenesis 23: (4) ((2020) ), 515–529. |
[33] | Ruan J. , Gregory S.A. , Christos P. , Martin P. , Furman R.R. , Coleman M. and Leonard J.P. , Long-term follow-up of R-CHOP with bevacizumab as initial therapy for mantle cell lymphoma: clinical and correlative results, Clin Lymphoma Myeloma Leuk 14: (2) ((2014) ), 107–113. |
[34] | Stopeck A.T. , Unger J.M. , Rimsza L.M. , LeBlanc M. , Farnsworth B. , Iannone M. , Glenn M.J. , Fisher R.I. and Miller T.P. , A phase 2 trial of standard-dose cyclophosphamide, doxorubicin, vincristine, prednisone (CHOP) and rituximab plus bevacizumab for patients with newly diagnosed diffuse large B-cell non-Hodgkin lymphoma: SWOG 0515, Blood 120: (6) ((2012) ), 1210–1217. |




