POLR3A-mutated Wiedemann-Rautenstrauch fibroblasts display differential profile of intron retention and expression of TP53 isoforms
Abstract
BACKGROUND:
Wiedemann-Rautenstrauch Syndrome (WRS) is a neonatal progeroid syndrome for which biallelic pathogenic variants in RNA polymerase III subunit A (POLR3A) have recently been described. POLR3 is a 17 subunits protein complex responsible for the transcription of short RNAs including all the transfer RNAs (tRNAs), the 5 S subunit of ribosomal RNA, the short nuclear RNA U6, among other regulatory RNAs.
OBJECTIVE:
We aim to evaluate the impact of POLR3A pathogenic variants on the relative expression of the short nuclear RNA U6, p53 isoforms and on the differential profile of intron retention in fibroblasts derived from patients with WRS and control fibroblasts.
METHODS:
RNA was extracted by the TRIzol method; intron retention analysis was performed by using IRFinder from an mRNA sequencing (RNA-Seq) platform; P53 isoforms, short nuclear RNA U6 and additional genes related to cell senescence were measured by RT-PCR.
RESULTS:
No significant differences were found in the percentage of intron retention (control: 7.8%, WRS1 : 6.3%and WRS2 : 8.14%). Genes showing higher intron retention profile in both groups were mainly related to RNA binding pathways, cell cycle regulation, positive regulation of transcription, positive regulation of inflammatory pathways, negative regulation of apoptosis, RNA transcription, mitochondria, and regulation of translation initiation. However, in WRS fibroblasts the genes with more intron retention were those related to the immune response and mitochondrial function; while in control those related to the response to oxidative stress had the most introns retained. WRS1 showed higher expression of short nuclear RNA U6 compared to control and WRS2; while both WRS cells showed higher expression of p53β and lower percentage of Δ133p63α, consistent with a higher expression of the cellular senescence markers p16 and p21.
CONCLUSIONS:
These results demonstrated the important role of POLR3A in the maintenance of cellular homeostasis and highlight its potential role in cell senescence in WRS.
1Introduction
Wiedemann-Rautenstrauch Syndrome (WRS) is a neonatal progeroid syndrome first described in 1977 by Thomas Rautenstrauch and Friedemann Snigula, and is clinically characterized by premature aging signs since birth, lipodystrophy, prominent veins, triangular face, apparent macrocephaly, wide fontanelle, low-set ears, neonatal teeth, micrognathia, thin skin and short stature [2, 25].
WRS is caused by biallelic pathogenic variants in the gen POLR3A, which codes for the RNA Polymerase III subunit A [26]. The POLR3A is a 17 subunits protein that synthesize short RNAs including all transfer RNAs (tRNAs), the 5 S subunit of ribosomal RNA, the short nuclear RNA U6, among other regulatory RNAs [26]. Recently, it has been described that the activity of POLR3 is required for proper repair of double-strand breaks mediated by homologous recombination, by protecting the remaining single strand of DNA at the 3’ end, preventing it from being degraded by endonucleases [21]. This suggests a potential pathway by which decreased function of POLR3A leads to genomic instability, cell senescence and the progeroid phenotype in WRS [3].
The spliceosome is a large ribonucleoprotein complex that removes introns from the pre-mRNA. It is composed by 5 short nuclear RNAs (U1, U2 U4, U5 and U6) and other regulatory and structural proteins [7]. The short nuclear RNA U6, one of the POLR3 regulated transcripts, is a key component of the spliceosome, which in conjunction with U2 catalyze the phosphodiester bond between the exonic ends. Changes in expression of U4 and U6 are related to intron retention, while changes in expression of U1 and U2 are related with exon skipping events [9].
Aging is characterized by splicing dysfunction caused by changes in the expression of the splicing factors and the spliceosome components, alteration in the subcellular location of the splicing factors, spliceosome complex dysfunction, loss of the RNA control mechanisms and translation of aberrant mRNAs [4]. Consistently, intron retention has been described as a post-transcriptional signature associated with progressive aging, in different types of cells and species [1, 12, 13], affecting the cellular homeostasis and giving rise to dysfunctional proteins or causing protein deficiency because of nonsense-mediated decay [1].
TP53 gene codifies for 13 different P53 isoforms depending on the use of different promotors or the alternative splicing of intron 2 and intron 9 [33]. P53α, the classic P53 isoform, is a transcriptional factor that upregulate the expression of genes related to cell cycle, apoptosis, DNA repair, cellular senescence, and metabolism [11]. P53β is a transcript produced by the alternative splicing of intron 9 that causes a premature stop codon in exon 9 and the loss of a long fraction of the exon 9 and exons 10 and 11 [33]. P53β increase the transcriptional activity of p21, raising the activity of the P53 pathway and leading to cellular senescence [16]. Δ133p53α is a transcript produced using the promotor P2, an internal promotor located in intron 4, initiating the translation in the codon 133 in exon 5. This protein functions as an inhibitor of cellular senescence by a competitive mechanism that inhibits p53α function [34].
A study in human fibroblasts found that increased expression of p53α and decreased expression of Δ133p53α induce cellular senescence; while higher expression of Δ133p53α inhibited the appearance of the senescent phenotype [10]. These same results were replicated in T lymphocytes [30], astrocytes [30], pluripotent stem cells [14], and even in fibroblasts from patients with Hutchinson-Gilford progeria [34]. The degradation of Δ133p53α occurs via autophagy. A decrease in STUB1 expression, an autophagy regulatory protein, leads to an increase in the recruitment of Δ133p53α to the autophagosome and increased its degradation [10]. This regulatory mechanism of Δ133p53α seems to be relevant during replicative senescence. The expression of P53β is regulated by splicing regulatory factors. When SRSF7 is recruited during P53 splicing, the P53β isoform is generated; while if SRSF3 or SRSF1 are recruited, the P53α isoform is formed. A positive relationship between radiation-induced DNA damage and SRSF7 expression has been found [6], which could explain why when genomic instability increases there is also an increase in P53β expression and cellular senescence [34].
In the present paper, we analyzed the impact of the pathogenic variants in POLR3A over the expression of RNU6, intron retention profile and differential expression of P53 isoforms in primary fibroblasts from patients with WRS and a healthy control.
2Material and methods
2.1Cell culture
Primary human fibroblasts were obtained from forearm skin biopsies of a 21 years old female patient (WRS1) with confirmed diagnosis of WRS (POLR3A c. 3772_2773delCT; p. Leu1258Glyfs*12), a 6 years old female patient (WRS2) with confirmed diagnosis of WRS (POLR3A c.3G > T; p. Met1Leu*) and a healthy 21 years old female (control). Even though, only one pathogenic variant had been found in each patient, patients present a clinical phenotype consistent with classic WRS. This study was approved by the Institutional Review Board of the National University of Colombia, according to the ethical standard set by the Declaration of Helsinki and later amendment and all patients or the legally authorized representatives of patients unable to give consent for themselves, signed informed consent before the sample collection. The fibroblasts were cultured under standard conditions in Dulbecco’s modified Eagle’s medium, supplemented with 10%fetal bovine serum and 1%penicillin/streptomycin at 37°C in a humidified 5%CO2 incubator. The medium was changed every time the cells reached 70%confluence.
2.2RT-qPCR
RNA was extracted using TRIzol reagent (ThermoFisher Scientific, Rockford IL, USA) according to manufacturer’s protocol and was quantified using a NanoDrop Spectrophotometer (ThermoFisher Scientific, USA). 500 ng of total RNA was used for reverse transcription assays with Luna Universal One-Step RT-qPCR kit (New England Biolabs) according to the manufacturer’s protocol. RT-qPCR was performed in a 10μL reaction in a CFX96 Touch Real-Time PCR Detection Systems (Bio-Rad) with 50 cycles. Primers used were as follows: RNU6 forward was 5’-CAGCACATATACTAAAATTGGAACG-3’, RNU6 reverse was 5’-ACGAATTTGCGTGTCATCC-3’, RNU48 forward was 5′-TGATGATGACCCCAGGTAACTC-3′, RNU48 reverse was 5′-GAGCGCTGCGGTGATG-3, GSTM1 forward was 5’- TACTCTGAGCCCTGCTCGGT-3, GSTM1 reverse was 5’- TGTATTCCAGGAGCAGGCGG-3’, p16 forward was 5’-CCAACGCACCGAATAGTTACG-3’, p16 reverse was 5’-GCGCTGCCCATCATCATG-3’, p21 forward was 5’-TGGAGACTCTCAGGGTCGAAA-3’, p21 reverse was 5’-AGATGGTGATGGGCTTCCC-3’, U2AF1 forward was 5’- CCAGTCTGCTGACGGTTTG-3, U2AF1 reverse was 5’- AGGTGGTCTCCCAGGTTGT-3, SRSF3 forward was 5’-AGCTGATGCAGTCCGAGAG-3’, SRSF3 reverse was 5’-GGTGGGCCACGATTTCTAC-3’, SRSF7 forward was 5’-CGGTACGGAGGAGAAACCAAG-3’, SRSF7 reverse was 5’-AGCCACAAATCACCTTTCCATC-3’, STUB1 forward was 5’-TCAAGGAGCAGGGCAATCGT-3’, STUB1 reverse was 5’-CAGCGGGTTCCGGGTGAT-3’, GAPDH forward was 5’- TGGTATCGTGGAAGGACTCA-3' and GAPDH reverse was 5’-CCAGTAGAGGCAGGGATGAT-3'. Data analysis was performed in CFX Manager Software (BioRad) and IBM SPSS Statistics 28.0.1. The DDCt method was used for relative expression quantification. Expression of each gene under each of the conditions was assayed in triplicate.
2.3RNA-Seq and Intron retention analysis
PolyA mRNA was purified from the RNA extracted and sequenced by next generation sequencing using Illumina NovaSeq 6000 (Macrogen, Seoul, South Korea). RNA-Seq data were delivered in FASTA, sequences were aligned using STAR aligner v27.0a, using as reference genome the human genome GRCh38 [8]. BAM unsorted data were upload to IRFinder for the intron retention analysis [23]. The differential intron retention expression was analyzed in DESeq2 in R, according to instructions in IRFinder protocol. KEGG metabolic pathways and Gene Ontology enriched terms were analyzed using DAVID [28].
2.4Intron retention events verification
RNA extracted was submitted to reverse transcription to generate cDNA using Superscript III kit (Invitrogen). cDNA was quantified using a NanoDrop Spectrophotometer (ThermoFisher Scientific, USA). For amplification, 40 cycles of PCR were realized in a CFX96 Touch Real-Time PCR Detection Systems (Bio-Rad) using the following primers: PILRB forward was 5-‘ATCCCCTGTTAGAGACGGGC-3’, PILRB reverse was 5’- GAACTCAGGCATAGGGGAGC-3’, K7EP35 forward was 5’-CCTGTCTGCTGTGATTCAGG-3’, K7EP35 reverse was 5’GGAGGTTGCAGTGAGCTGAG-3’, GSTM1 forward was 5’-TTAGGCCTGTCTGCGGAATC-3’, GSTM1 reverse was 5’-GAGATGGAGGGTCCCTGACT-3’, OAS1 was 5’-CCTCCAGCTGCAACCATG-3’ and OAS1 reverse was 5’-GTCTGCTTCCTAGCCATCTG-3’. Later a 2%agarose gel electrophoresis was performed using the PCR products. Experiments were performed by triplicate.
2.5P53 isoforms analysis
RNA extracted was used to generate cDNA using Superscript III kit (Invitrogen). cDNA was quantified using a NanoDrop Spectrophotometer (ThermoFisher Scientific, USA). To measure the expression of each p53 isoform, a nested PCR, a 2%agarose electrophoresis, and quantification of each isoform were performed following the instructions in Khoury’s Protocol [15]. Expression of each isoform under each condition was assayed in triplicate.
2.6Statistics
Statistical significance was determined using a Student’s T tests for normal distributions and Mann-Whitney U tests for non-normal distributions using IBM SPSS Statistics 28.0.1.
3Results
3.1Cellular senescence biomarkers are increased in WRS cells
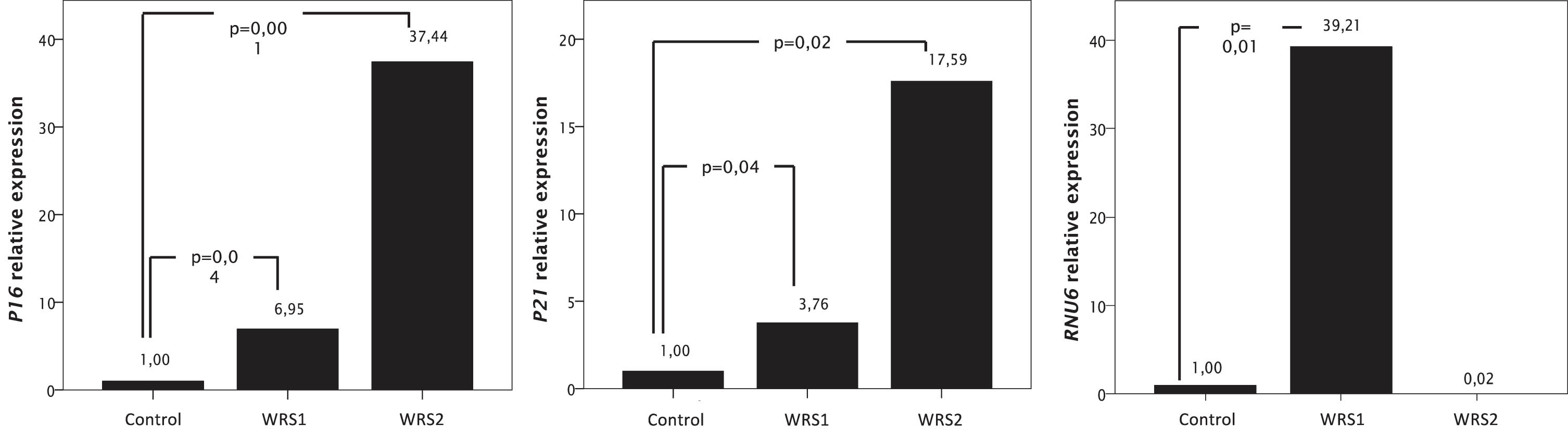
To analyze if WRS cells present early characteristics of cellular senescence, we measured the relative expression of P16 and P21 by RT-qPCR (Fig. 1). WRS fibroblasts with pathogenic variants in POLR3A showed a significant increase in the relative expression of mRNA for P16 and P21 which are associated with the activation of the P53 pathway. In WRS1 cells there is an increased expression of P16 of almost 7 times; while in WRS2 cells, expression of P16 increases more than 37 times, relative to control cells (Fig. 1).
Fig. 1
mRNA relative expression of genes related with cellular senescence (P16 and P21), and RNU6, one of the transcripts of the RNA Polymerase III, measured by RT-PCR in WRS and control fibroblasts. Data were normalized to GAPDH expression for the P16 and P21 expression and to RNU48 for the RNU6 expression. Increased expression of RNU6 in WRS1 cells compared to control cells diminished the expression of genes related to cellular senescence.
3.2Intron retention profile and relative expression of RNU6 and U2AF1 in WRS cells
Using RNA-Seq data we examined the global intron retention rate in WRS1, WRS2 and control cells. In control cells 873 of the 11,186 expressed genes displayed intron retention (7.8%); in WRS1 cells 857 genes of the 13,655 expressed genes presented intron retention (6.3%); and in WRS2 cells 941 genes of the 11,564 expressed genes (9.14%) presented intron retention. Genes showing higher intron retention profile in both groups were mainly related to RNA binding pathways, cell cycle regulation, positive regulation of transcription, positive regulation of inflammatory pathways, negative regulation of apoptosis, RNA transcription, mitochondria, and regulation of translation initiation. The metabolic pathways particularly affected by differential intron retention in WRS1 and WRS2 cells are related to the immune response, antigen presentation and mitochondrial function (Table 1).
Table 1
| Enriched terms in transcripts with increased intron retention in WRS cells | ||
| Term | p value | adjusted p value |
| Protein binding | 2.40E-16 | 6.40E-15 |
| Nucleoplasm | 1.70E-13 | 1.10E-10 |
| Nucleus | 1.90E-11 | 6.20E-09 |
| Membrane | 2.00E-10 | 4.30E-08 |
| RNA binding | 2.40E-08 | 1.10E-05 |
| Cytoplasm | 8.40E-08 | 1.40E-05 |
| Cytosol | 2.20E-06 | 2.90E-04 |
| Nuclear speck | 2.90E-05 | 3.10E-03 |
| Mitochondrion | 7.40E-05 | 5.90E-03 |
| Endoplasmic reticulum | 7.40E-05 | 5.90E-03 |
| Macromolecular complex | 8.70E-05 | 6.20E-03 |
| Regulation of cell cycle | 6.00E-06 | 1.70E-02 |
| Protein ubiquitination | 1.00E-05 | 1.70E-02 |
| Protein processing in endoplasmic reticulum | 8.30E-05 | 2.30E-02 |
| Apoptosis | 1.50E-04 | 2.30E-02 |
| Positive regulation of transcription | 2.10E-05 | 2.30E-02 |
| Regulation of transcription | 3.40E-05 | 2.60E-02 |
| mRNA transcription from RNA polymerase II promoter | 4.70E-05 | 2.60E-02 |
| Positive regulation of NF-kappaB signaling | 5.80E-05 | 2.70E-02 |
| Ribonucleoprotein complex | 5.50E-04 | 3.60E-02 |
| Ubiquitin-protein transferase activity | 1.50E-04 | 4.60E-02 |
| Enriched terms in transcripts with increased intron retention in control cells | ||
| Term | p value | adjusted p value |
| Protein binding | 2.20E-18 | 2.00E-15 |
| Nucleoplasm | 2.70E-14 | 1.70E-11 |
| Nucleus | 6.30E-12 | 1.90E-09 |
| Membrane | 2.90E-10 | 6.00E-08 |
| RNA binding | 7.70E-07 | 6.30E-05 |
| Cytosol | 1.20E-06 | 1.90E-04 |
| Regulation of cell cycle | 4.20E-06 | 7.50E-03 |
| Protein ubiquitination | 4.70E-06 | 7.50E-03 |
| Regulation of transcription | 7.10E-06 | 7.60E-03 |
| Positive regulation of transcription | 1.40E-05 | 1.10E-02 |
| Macromolecular complex | 1.50E-05 | 1.60E-03 |
| Positive regulation of NF-kappaB signaling | 2.10E-05 | 1.40E-02 |
| mRNA transcription from RNA polymerase II promoter | 2.70E-05 | 1.40E-02 |
| Negative regulation of apoptotic process | 4.00E-05 | 1.80E-02 |
| Ubiquitin-protein transferase activity | 5.20E-05 | 1.50E-02 |
| Apoptosis | 7.30E-05 | 2.10E-02 |
| Nuclear speck | 9.20E-05 | 8.10E-03 |
| Regulation of translational initiation | 9.50E-05 | 3.80E-02 |
| Endoplasmic reticulum | 1.50E-04 | 1.10E-02 |
The intron retention differences between groups were confirmed by PCR analysis in four genes: K7EP35 (Fig. 2A), OAS1 (Fig. 2B), PILRB (Fig. 2C) and GSTM1 (Fig. 2D) (Fig. 2). We observed that WRS cells present intron retention in K7EP35 while control cells did not. K7EP35 codes for NADH dehydrogenase 1 alpha subcomplex subunit 11. The NADH dehydrogenase complex or complex I is the first enzyme in the electron transport chain, and it catalyzes electron transfer from NADH to ubiquinone. This suggests that there may be a deficiency in energy generation in WRS cells [17].
Fig. 2
Intron retention analysis by PCR and agarose electrophoresis and mRNA relative expression of U2AF1 and GSTM1 in control and WRS cells. A) Migration pattern of the PCR products of the K7EP35 gene. Molecular weight marker in the first lane, one single band with a molecular weight of 320 bp in the second lane that corresponds to the expression of the transcript without intron retention in the control cells and the PCR products of the WRS1 and WRS2 cells in the third and fourth lane, respectively, with two bands observed, the smaller one corresponding to the amplicon without the retained intron and with a molecular weight of 320 bp, and the larger one corresponding to the amplicon with the retained intron and with a molecular weight of 499 bp. B) Migration pattern in agarose gel electrophoresis of the PCR products of the OAS1 gene. The molecular weight marker in the first lane, the PCR products of the control cells with no bands observed in the second lane, and one band of 498 pb observed in the third (WRS1) and fourth (WRS2) lane that corresponds to an OAS1 intron. C) Migration pattern on agarose gel electrophoresis of the PCR products of the PILRB gene. The molecular weight marker in the first lane. No band was observed in the second and third lane (control cells and WRS1 cells), which refers to the fact that there was no retention of any intron, and a band of 432 bp in the fourth lane, which corresponds to the amplicon with the retained intron of PILRB in WRS2 cells. D) Migration pattern in agarose gel electrophoresis of the PCR products of the GSTM1 gene. The molecular weight marker in the first lane. One single band of 303 bp observed in the second lane, that corresponds to GSTM1 with intron retention in control cells. And a single band of 96 bp in the third (WRS1) and the fourth (WRS2) lane, which refers to the GSTM1 amplicon without the retained intron. E) Relative expression of U2AF1 and E) GSTM1 normalized to GAPDH expression, showing and increased expression in WRS cells compared to control cells, with a greater difference in WRS2 cells.
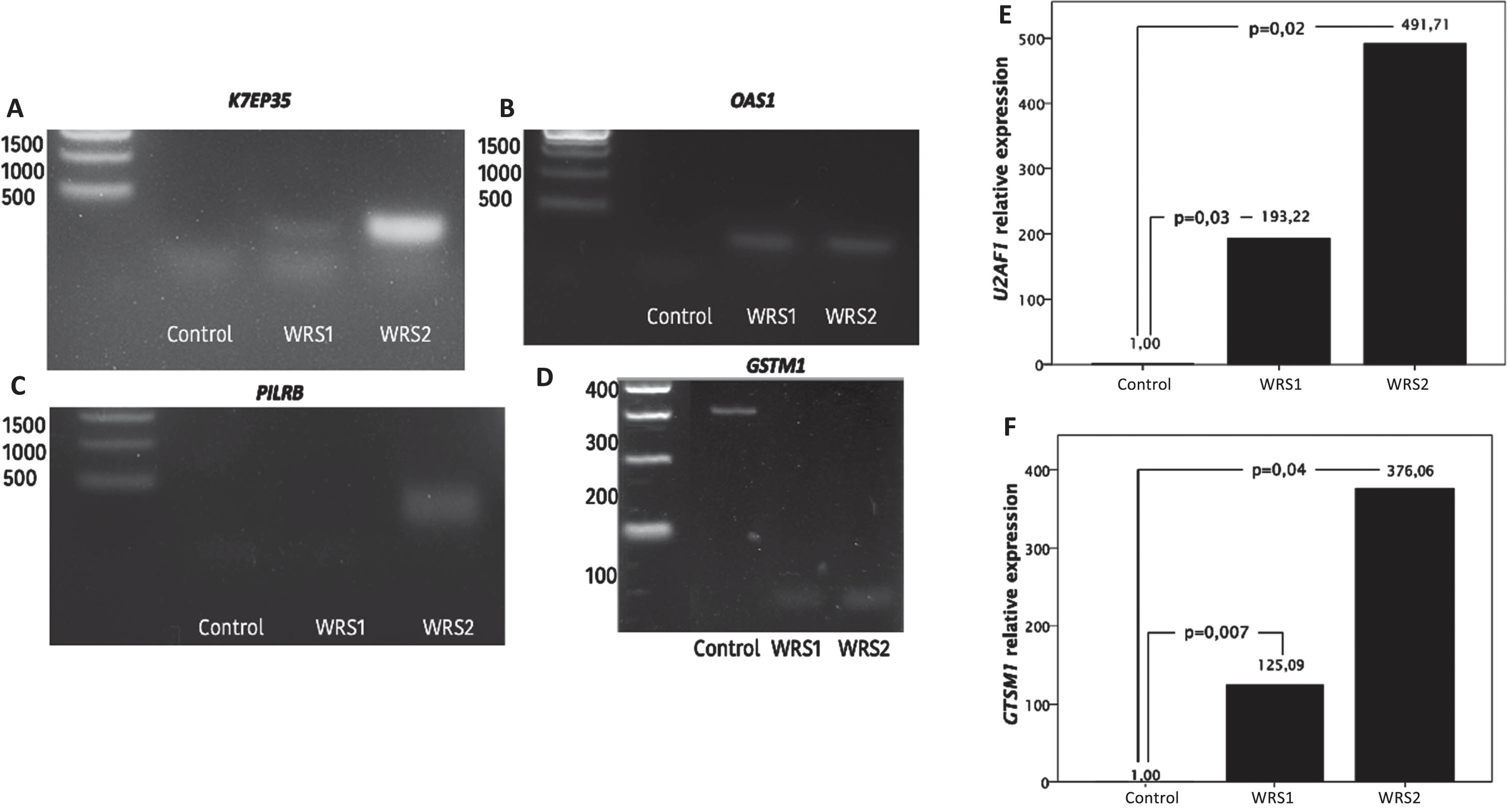
As well, there was a higher intron retention in OAS1 and PILRB in WRS cells. OAS1 codes for 2’-5’-oligoadenylate synthetase 1, one of the interferon-induced genes that has a significant role in antiviral function [20]. Interestingly, POLR3 has been described to be important in antiviral response, acting as a sensor of foreign DNA and enhancing the innate immune response against viral DNA [18]. PILRB codes for paired immunoglobin like type 2 receptor beta, which is involved in the activation of many pathways of the immune system.
In addition, GSTM1 had a higher intron retention in control cells than in WRS cells. GSTM1 codes for glutathione-S-transferase μ-1, an enzyme with protective effects against oxidative damage [19]. We found that GSTM1 expression was markedly rise in both WRS cells, it was increased 125 times in WRS1, while it was 376 times in WRS2 cells, relative to control cells (Fig. 2F), suggesting that WRS cells are exposed to a high oxidative stress.
We also searched for differential expression of splicing factors in WRS cells using RNA-Seq data and found that U2AF1 mRNA expression was upregulated in WRS cells. We verified changes in relative expression of U2AF1 between cells by RT-qPCR (Fig. 2E). We found that both WRS cells showed an increase in the U2AF1 mRNA expression, the difference in WRS2 cells being greater (while in WRS1 cells the U2AF1 expression was 193 times increased, in WRS2 cells the U2AF1 expression was 491 times increased compared to control cells).
3.3WRS cells have increased expression of P53α and a decreased expression of Δ133p53α
Considering the importance of the p53 pathway on cellular senescence and the evidence that the expression of specific isoforms of p53 influence the development of cellular senescence, we measured the expression of 6 different p53 isoforms. In WRS2 P53β expression was significantly increased, while Δ133p53α was significantly reduced (P < 0,05); WRS1 cells showed a similar expression level of P53β and Δ133p53α as WRS2, but these changes were not statistically significant compared to control cells (Fig. 3A). Interestingly, in WRS cells the expression of P53α was significantly reduced compared to control, while the expression of P53β, P53γ and Δ133p53β was higher (Fig. 3A).
Fig. 3
Relative expression of TP53 isoforms measured by agarose electrophoresis of nested PCR products and mRNA relative expression of SRSF3, SRSF7 and STUB1; involved in p53b and D133p53a regulation. A) Relative expression of TP53 isoforms (one asterisk (*), p < 0,05) showing and increased expression of p53b and a decreased expression of D133p53a in WRS2 cells. B) mRNA relative expression of SRSF3, SRSF7 and STUB1 normalized to GAPDH expression. WRS cells showed an increased expression of SRSF7 related to a rise in p53b expression, although the expression of STUB1 was markedly augmented in WRS cells, it was not enough to increase the expression of D133p53a.
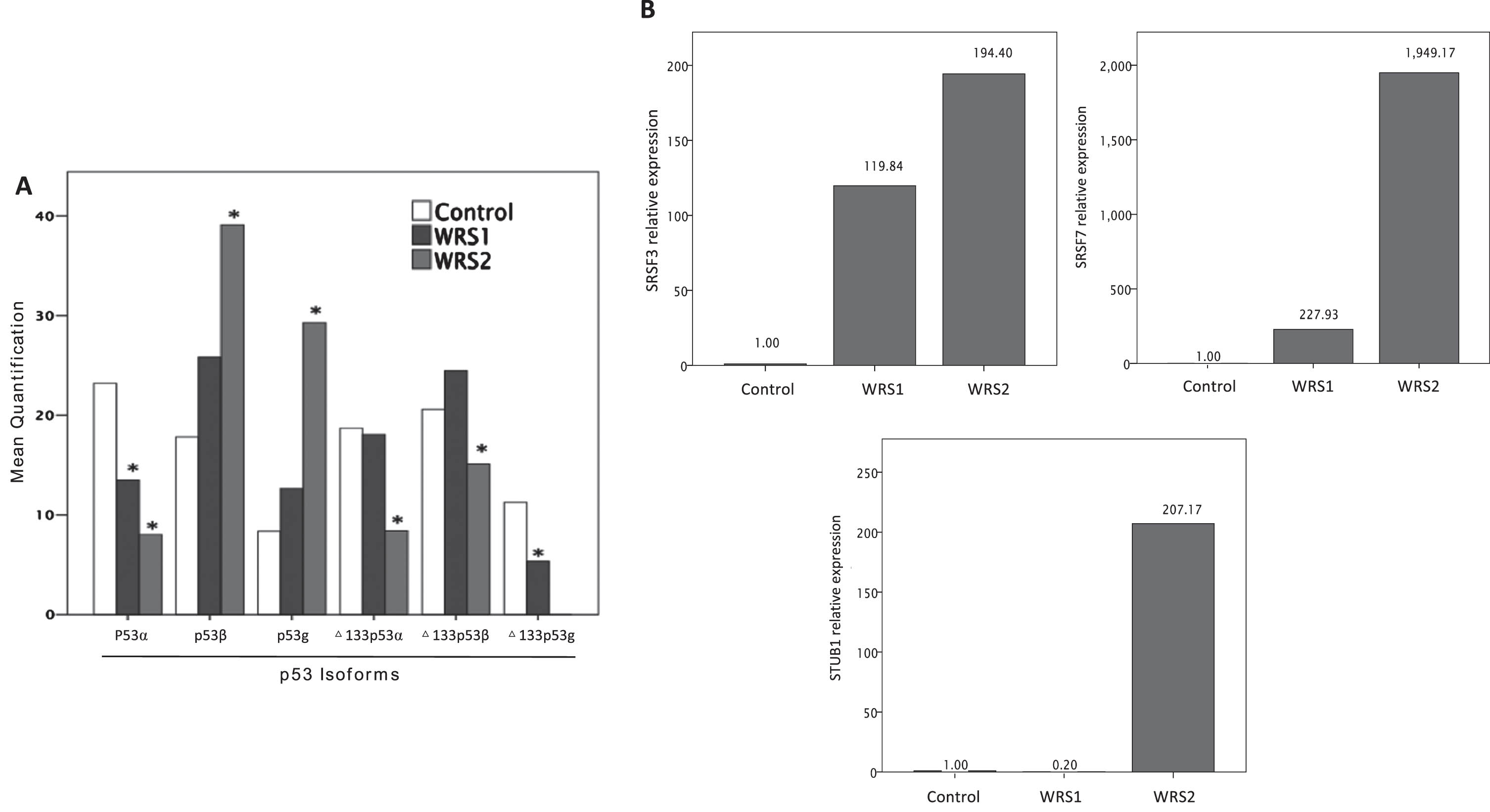
Further, we analyzed the expression of genes that have been described to regulate the expression of different P53 isoforms, in particular the regulators of the P53β and Δ133p53a such as SRSF3, SRSF7 and STUB1 by using RT-qPCR. WRS cells present a marked increase in the expression level of SRSF3 and SRSF7, while the expression of STUB1 increase only in WRS2 cells (Fig. 3B).
4Discussion
WRS is defined as a condition with an autosomal recessive inheritance, with biallelic pathogenic variants in POLR3A gene, which could be homozygous, but more commonly are in compound heterozygous disposition. It is interesting that in the WRS patients, only one pathogenic variant has been identified, however the patients show a complete phenotype of the disease.
In WRS1 the identified pathogenic variant is POLR3A: c. 3772_3773delCT (p. Leu1258Glyfs*12), which could exert a negative dominance effect over the wild type allele. It is expected that an altered function in POLR3A should affect the transcriptional function of POLR3 on its promoters, affecting the mRNA expression of RNU6 causing a dysfunction in splicing homeostasis. However, what we found was that WRS1 patient expressed less intron retained genes than the control (6.3%vs. 7.8%), and surprisingly, showed an increased expression in RNU6 (39.21 times the expression in the control). We suspect this could be a compensatory effect based on the expression of the healthy allele, that allow the cell to maintain the splicing homeostasis. Nevertheless, this compensatory effect is not enough to abate the cellular senescence, proven in the increased expression in P16 and P21 genes. This could be explained because of a genomic instability caused by a dysfunctional homologous recombination repair of double strand breaks, that could increase the expression in SRSF7 causing a rise in expression of p53β, empowering the p53 pathway related to cellular senescence. Additionally, WRS1 cells showed less expression of Δ133p53α and of STUB1 compared to control. Other important finding was that although the percentage of intron retention events was not substantially different from the control cells, the genes expressed with retained introns were different between both groups, made evident by the rise in intron retention expression in genes related to mitochondrial function and cellular replication, associated with fewer intron retention events in genes related to immune function and oxidative stress response. The altered mitochondrial function caused by these changes may be associated with the increased in expression of genes related to oxidative stress response, like GSTM1 (125 more times than in control cells), and with the activation of the cellular senescence pathway.
In WRS2 cells, the identified pathogenic variant is POLR3A: c. 3G > T (p. Met1Leu*), this change affects the start codon, inhibiting the expression of this allele. We suspect that the mechanism of the disease in this patient is haploinsufficiency, which can cause a decreased function in POLR3A and POLR3. In fact, WRS2 cells showed a rise in intron retention events (9.6%vs. 7.8%), along with less expression of RNU6 compared to control (0.02 times). The intron retention events in WRS2 cells were more present in genes related with mitochondrial function and cellular replication, while the genes with less intron retention were related to the oxidative stress response and the immune function. Therefore, WRS2 cells showed 376 times more expression of GSTM1 compared to control. Even though, RNU6 expression was decreased in WRS2 cells, these cells showed an increase in the expression of U2AF1 (491 times more than in control cells), as a compensatory effect. These cells also showed a marked increase in p16 and p21 genes, related to the cellular senescence (37 more times than in control, and 17 times more than in control, respectively). In terms of the p53 pathway, these cells showed a marked increase in genes related with genomic instability and regulation of the p53 isoform expression; SRSF3 (194 more times than in control) and SRSF7 (1949 times more than in control), being related to a significant increase in p53β expression. Even though WRS2 cells showed an increased expression of STUB1, this was not enough to increase Δ133p53α expression, which was significantly decreased compared to control. All results together make the cellular phenotype more severe in WRS2 cells compared to WRS1 cells, being consistently associated with the severity of the patient’s phenotype, WRS1 being one of the oldest patients known with WRS.
Different studies have described the association between intron retention events with aging and cellular senescence, in cell culture and in both animal models and in humans [1, 24, 34], and is most probably associated to an altered cellular response to stress [24]. We have previously described that WRS cells showed an early entrance to a senescence phenotype as has been described similarly for cells derived from other progeroid syndromes [5]. Although WRS cells did not show a significant overall increase in intron retention rate, it is striking that in control cells, genes related to oxidative stress response have higher intron retention, while genes related to mitochondrial function have higher intron retention in WRS cells. This could be related to an altered mitochondrial function, as in both WRS cells, genes with more retained introns were related to mitochondrial function and cellular replication.
Intron-retained transcripts can have multiple fates. A subset of these transcripts is retained within the nucleus while another subset is exported to the cytoplasm. In the cytoplasm Intron-retained transcripts could act in different ways: 1) as signals that direct the subcellular localization of transcripts and proteins; 2) could be converted to new protein isoforms; 3) function as a post-transcriptional regulatory mechanism of other RNAs; and 4) may be degraded through the nonsense mediated decay system [31]. In the nucleus, transcripts with retained introns are most probably waiting for a signal to continue the splicing process, once a particular stress situation has ended [24]. A study in mice described that during the response to heat-shock stress there is a large content of RNA with retained introns within the nucleus and that the genes with the highest percentage of intron retention score were related to tRNAs synthesis, to nuclear pores and to the spliceosome machinery; whereas genes related to protein folding and antioxidant response were spliced as usual [24]. For future experiments, it could be interesting to look at the cellular location of the genes expressed with retained introns.
Genomic instability and oxidative stress are two of the main inducers of cellular senescence [5]. Increased oxidative stress is associated with increases in nuclear and mitochondrial DNA damage, which leads to activation of signaling pathways related to DNA damage response. Damaged mitochondrial DNA impairs the normal function of oxidative phosphorylation leading to further increase in oxidative stress, decreased mitochondrial membrane potential and decrease in the production of ATP [32]. During cellular senescence there is an increase in mitochondrial biogenesis and mitochondrial mass without an improvement in the overall mitochondrial function. These changes could be the reason of the higher intron retention rates in genes related to mitochondrial function observed in WRS cells, while there was a lower intron retention and a higher mRNA expression in GSTM1 which is related to response to an increased oxidative stress.
POLR3 function is important for proper nucleolus function and ribosome biogenesis [29]. A previous study has shown that WRS fibroblasts have an increased number of nucleoli, as well as an increased nucleolar area [3], suggesting that alterations in the nucleolus could be central in the initiation of senescence in WRS cells. Alterations in nucleolar structure and function affects the glutathione transferase activity by decreasing its function and increasing oxidative stress [35]. An increase in nucleolar exposure to oxidative stress has been shown to change the localization of nucleophosmin and invert the redox state of the nucleolus, altering the stability of P53, which in turns promote its activation and induction of cellular senescence [35].
Intron retention also play an important role during embryogenesis [22], neurogenesis, and neuronal homeostasis [27]. Although the mechanisms by which intron retention occurs globally have not been clearly described, there appears to be an association between intron retention and changes in the expression of spliceosome components and in the expression of RNA-binding proteins [9]. A recent study in fibroblasts showed that cells with knock-down of U2AF1 protein had greater intron retention events and a premature onset of cell senescence [36]. We showed that although WRS cells expressed senescence markers, we did not find significant differences in the rate of intron retention. This observation could be related to the higher expression of U2AF1 and U6 nuclear short RNA, that in conjunction could act as a compensatory mechanism leading to decrease in intron retention. However, it is difficult to understand how WRS1 cells can increase the expression of the short nuclear RNA U6 considering the decreased function of POLR3 and that the promoter of RNU6 is exclusive of POLR3 and not POLR2 as demonstrated by chromatin immunoprecipitation experiments [7]. One possibility is that there seems to be a compensatory increase in the expression of POLR3 in cells from WRS1 patient [3].
Previously it has been described that POLR3 plays an important role during repair of DNA double-strand breaks mediated by homologous recombination, and alterations in this function is related to an increase in genomic instability [21]. We have demonstrated that WRS cells have higher immuno-staining for phosphorylated histone 2 (pH2AX), a marker for double-strand DNA breaks [3]. Genomic instability is a potent inducer of P53α and P53β expression associated to increased expression of the ribosomal protein L26 (RPL26), which is a positive regulator of P53α by negatively regulating the expression of the MDM2 ubiquitin ligase. Binding of RPL26 to the P53 transcript favors the recruitment of SRSF7 which in turn favors alternative splicing and increase generation of the P53β isoform [6]. P53β leads to increase in the expression of P21 further promoting cellular senescence [16].
5Conclusions
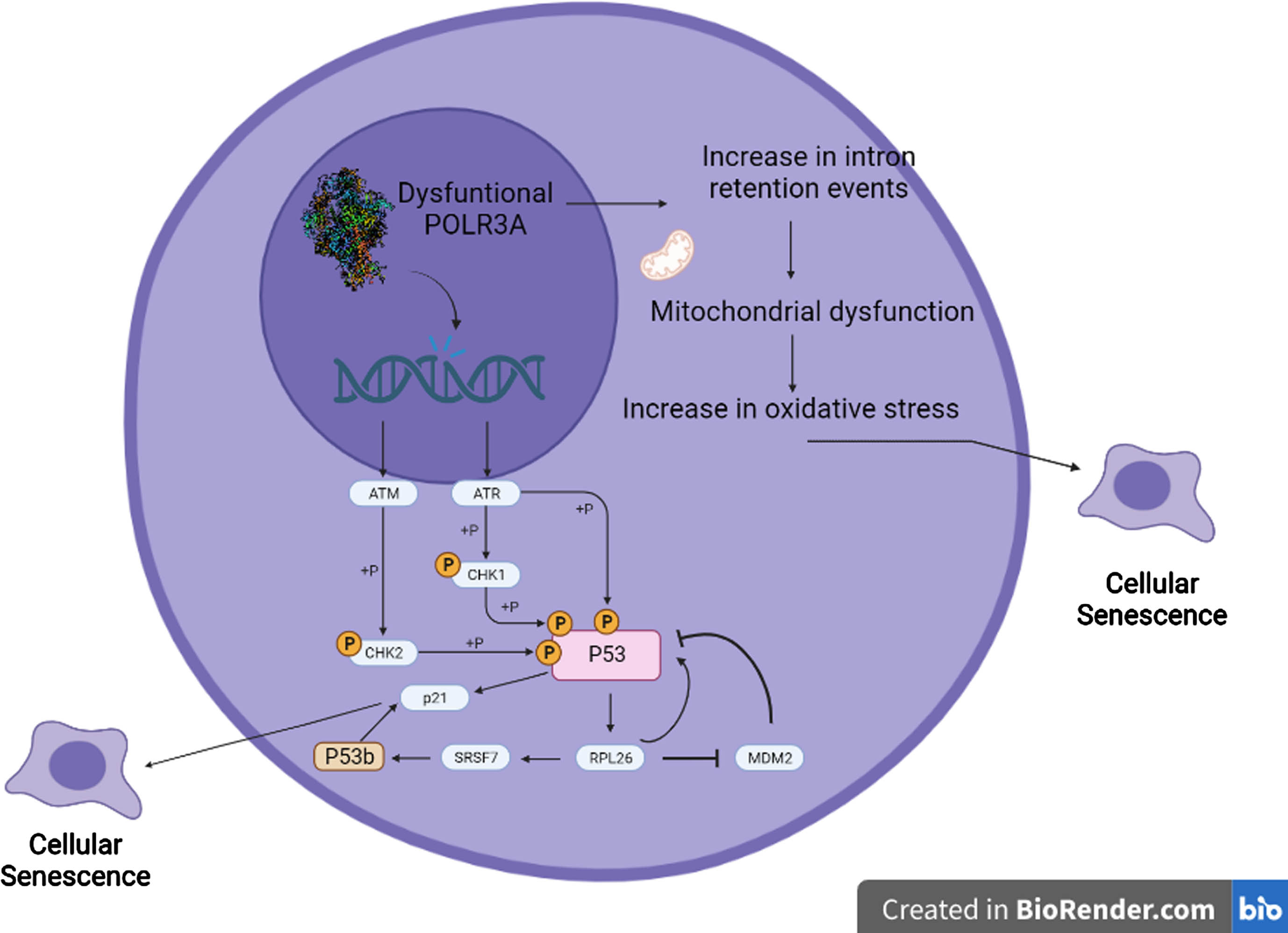
The present study furthers our knowledge about the pathophysiological pathways that can lead to cellular senescence in fibroblasts with altered POLR3A function. Decreased function of POLR3A is related to an increase in genomic instability that affects the homeostasis of the splicing machinery and the expression of P53 isoforms, contributing to the phenotype of WRS cells (Fig. 4). Although both WRS patients present a phenotype consistent with WRS the molecular mechanisms that trigger the clinical picture are different, as there are different mechanisms by which POLR3A dysfunction can lead to cellular senescence, depending on the specific pathogenic variant and its mechanism of disease (negative dominance effect vs. haploinsufficiency). Understanding the processes that occur in the cells of patients with progeroid syndromes helps to understand the pathophysiological mechanisms of normal aging.
Fig. 4
Proposed model of the interaction between POLR3A, p53b, intron retention and the process of cellular senescence. RNA Polymerase III dysfunction due to pathogenic variants in POLR3A cause alterations in homologous recombination DNA repair process, resulting in increased genomic instability. This genomic instability leads to activation of pathways related to DNA damage, including ATM and ATR, which ultimately lead to p53 phosphorylation and activation. Activation of p53 increases the expression of the ribosomal protein RPL26. RPL26 is a negative regulator of the expression of the ubiquitin ligase MDM2, which induces the degradation of both p53 and RPL26 through the proteasome. When RPL26 is bound to the p53 transcript, it recruits SRSF7, which promotes alternative splicing to form the p53β isoform. p53β increases the transcriptional activity of p21 via the p53 pathway, further promoting cellular senescence. Because of changes in RNU6 expression and in splicing homeostasis caused by POLR3A dysfunction, there is a rise in intron retention events specifically in genes related to mitochondrial function, leading to mitochondrial dysfunction, and increased oxidative stress, which can ultimately lead to cellular senescence.
Acknowledgments
Funded by Minciencias (110184466884) and DIEB-Universidad Nacional de Colombia.
Author contributions
All authors fulfil the criteria for authorship. P.V.G. and D.F.S-G performed most of the experiments and statistical analyses, and participated in manuscript writing and critical revision, G.A and H.A. conceived and designed the research and made critical revisions of the manuscript for key intellectual content. G.A supervised the experiments and the research execution. All authors read and approved the final version of the manuscript and have agreed to the authorship and order of authorship for this manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
The data supporting the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
[1] | Adusumalli S. , et al.., Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease, Aging Cell 18: (3) ((2019) ), e12928. |
[2] | Arboleda G. , et al.., Neonatal progeroid syndrome (Wiedemann-Rautenstrauch syndrome): Report of three affected sibs, American Journal of Medical Genetics Part A 155A: (7) ((2011) ), 1712–1715. |
[3] | Báez-Becerra C.T. , et al.., Nucleolar disruption, activation of P53 and premature senescence in POLR3A-mutated Wiedemann-Rautenstrauch syndrome fibroblasts, , Mechanisms of Ageing and Development 192: ((2020) ), 111360. |
[4] | Bhadra M. , et al.., Alternative splicing in aging and longevity, Human Genetics 139: (3) ((2020) ), 357–369. |
[5] | Carrero D. , Soria-Valles C. , Lopez-Otin C. Hallmarks of progeroid syndromes: Lessons from mice and reprogrammed cells, Disease Models& Mechanisms 9: (7) ((2016) ), 719–735. |
[6] | Chen J. , et al.., DNA-Damage-Induced Alternative Splicing of p53, Cancers 13: (2) ((2021) ), 251. |
[7] | Didychuk A.L. , Butcher S.E. , Brow D.A. The life of U6 small nuclear RNA, from cradle to grave, RNA 24: (4) ((2018) ), 437–460. |
[8] | Dobin A. , et al.., STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29: (1) ((2013) ), 15–21. |
[9] | Dvinge H. , et al.., RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing, Genome Research 29: (10) ((2019) ), 1591–1604. |
[10] | Fujita K. , et al.., P53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence, Nature Cell Biology 11: (9) ((2009) ), 1135–1142. |
[11] | Gudikote J.P. , et al.., Inhibition of nonsense-mediated decay rescues p53β/γ isoform expression and activates the p53 pathway in MDM2-overexpressing and select p53-mutant Cancers, The Journal of Biological Chemistry 297: (5) ((2021) ), 101163. |
[12] | Harries L.W. , et al.., Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing, Aging Cell 10: (5) ((2011) ), 868–878. |
[13] | Holly A.C. , et al.., Changes in splicing factor expression are associated with advancing age in man, Mechanisms of Ageing and Development 134: (9) ((2013) ), 356–366. |
[14] | Horikawa I. , et al.., Δ133p53 represses p53-inducible senescence genes and enhances the generation of human induced pluripotent stem cells, Cell Death & Differentiation 24: (6) ((2017) ), 1017–1028. |
[15] | Khoury M.P. , et al.., Detecting and quantifying p53 isoforms at mRNA level in cell lines and tissues, Methods in Molecular Biology 962: ((2013) ), 1–14. |
[16] | Kim S. , An S.S.A. Role of p53 isoforms and aggregations in cancer, Medicine 95: (26) ((2016) ), e3993. |
[17] | Kuang Y. , et al.., NADH dehydrogenase subunit 1/4/5 promotes survival of acute myeloid leukaemia by mediating specific oxidative phosphorylation, Molecular Medicine Reports 25: (6) ((2022) ), 195. |
[18] | Lata E. , et al.., RNA Polymerase III Subunit Mutations in Genetic Diseases, Frontiers in Molecular Biosciences 8: ((2021) ), 696438. |
[19] | Le T.H. GSTM1 Gene, Diet, and kidney disease: Implication for Precision Medicine? Recent Advances in Hypertension, Hypertension 78: (4) ((2021) ), 936–945. |
[20] | Leisching G. , Wiid I. , Baker B. OAS1, 2, and 3: Significance During Active Tuberculosis? , The Journal of Infectious Diseases 217: (10) ((2018) ), 1517–1521. |
[21] | Liu S. , et al.., RNA polymerase III is required for the repair of DNA double-strand breaks by homologous recombination, Cell 184: (5) ((2021) ), 1314–1329.e10. |
[22] | Liu S. , et al.Deterioration of the human transcriptome with age due to increasing intron retention and spurious splicing, bioRxiv (2022). |
[23] | Middleton R. , et al.., IRFinder: assessing the impact of intron retention on mammalian gene expression, Genome Biology 18: ((2017) ), 1–11. |
[24] | Okada N. , et al.., Intron retention as a new pre-symptomatic marker of aging and its recovery to the normal state by a traditional Japanese multi-herbal medicine, Gene 794: ((2021) ), 145752. |
[25] | Paolacci S. , et al.., Wiedemann-Rautenstrauch syndrome: A phenotype analysis, American Journal of Medical Genetics Part A 173: (7) ((2017) ), 1763–1772. |
[26] | Paolacci S. , et al.., Specific combinations of biallelic POLR3A variants cause Wiedemann-Rautenstrauch syndrome, Journal of Medical Genetics 55: (12) ((2018) ), 837–846. |
[27] | Petrić Howe M. ,et al.,. Physiological intron retaining transcripts in the cytoplasm abound during human motor neurogenesis, Genome Research 32: (10) ((2022) ), 1808–1825. |
[28] | Sherman B.T. , et al.., DAVID: a web server for functional enrichment analysis and functional annotation of gene lists update (2021 update), Nucleic Acids Research 50: (1) ((2022) ), 216–221. |
[29] | Tiku V. , Antebi A. Nucleolar Function in Lifespan Regulation, Trends in Cell Biology 28: (8) ((2018) ), 662–672. |
[30] | Turnquist C. , et al.., P53 isoforms regulate astrocyte-mediated neuroprotection and neurodegeneration, Cell Death and Differentiation 23: (9) ((2016) ), 1515–1528. |
[31] | Vanichkina D.P. , et al.., Challenges in defining the role of intron retention in normal biology and disease, Seminars in Cell & Developmental Biology 75: ((2018) ), 40–49. |
[32] | Vasileiou P.V.S. , et al.., Mitochondrial Homeostasis and Cellular Senescence, Cells 8: (7) ((2019) ), 686. |
[33] | Vieler M. , Sanyal S. P53 Isoforms and Their Implications in Cancer, Cancers 10: (9) ((2018) ), 288. |
[34] | von N. et al.,Muhlinen P53 isoforms regulate premature aging in human cells, Oncogene 37: (18) ((2018) ), 2379–2393. |
[35] | Yang K. , et al.., A redox mechanism underlying nucleolar stress sensing by nucleophosmin, Nature Communications 7: ((2016) ), 13599. |
[36] | Yao J. , et al.., Prevalent intron retention fine-tunes gene expression and contributes to cellular senescence, Aging Cell 19: (12) ((2020) ), e13276. |




