Plane-wave medical image reconstruction based on dynamic Criss-Cross attention and multi-scale convolution
Abstract
BACKGROUND:
Plane-wave imaging is widely employed in medical imaging due to its ultra-fast imaging speed. However, the image quality is compromised. Existing techniques to enhance image quality tend to sacrifice the imaging frame rate.
OBJECTIVE:
The study aims to reconstruct high-quality plane-wave images while maintaining the imaging frame rate.
METHODS:
The proposed method utilizes a U-Net-based generator incorporating a multi-scale convolution module in the encoder to extract information at different levels. Additionally, a Dynamic Criss-Cross Attention (DCCA) mechanism is proposed in the decoder of the U-Net-based generator to extract both local and global features of plane-wave images while avoiding interference caused by irrelevant regions.
RESULTS:
In the reconstruction of point targets, the experimental images achieved a reduction in Full Width at Half Maximum (FWHM) of 0.0499 mm, compared to the Coherent Plane-Wave Compounding (CPWC) method using 75-beam plane waves. For the reconstruction of cyst targets, the simulated image achieved a 3.78% improvement in Contrast Ratio (CR) compared to CPWC.
CONCLUSIONS:
The proposed model effectively addresses the issue of unclear lesion sites in plane-wave images.
1.Introduction
Traditional ultrasound imaging presents limitations regarding frame frequency. Consequently, the advent of plane-wave techniques, boasting superior imaging frame frequencies, has gained significant traction within the medical field. However, These techniques compromise image quality. Beam synthesis methods [1, 2, 3] offer a moderate enhancement in image quality. Nevertheless, the compounding process reduces the frame rate. With the emergence of deep learning in computer vision [4, 5], researchers have started exploring the adaptive learning capability of deep neural networks to obtain a nonlinear mapping relationship between low-resolution and high-resolution images, thereby enhancing the clarity of medical images.
Dong et al. first introduced CNNs for image super-resolution reconstruction, achieving superior results compared to interpolation-based methods. Attention mechanisms are widely used to mitigate the overemphasis on low-value feature information. Wei et al. proposed the CDC model, incorporating divide-and-conquer attention to achieve sub-regional image reconstruction [6]. Chen et al. [7] proposed a multi-attention augmented network, which stacks attention augmentation modules into a deep residual architecture to utilize the complementary information from multiple representation stages fully. Zhou et al. [8] proposed an Enhanced Generative Adversarial Network to enhance high-frequency information while obtaining multi-scale features. Wang et al. incorporated a higher-order degradation modeling process in image reconstruction, effectively enhancing the texture details of the images [9]. Compared to beam synthesis methods, reconstruction models based on neural networks produce visually more appealing images and are closer to practical application requirements. However, these methods often encounter inconsistent training results and slow convergence speeds.
This paper proposes an end-to-end plane-wave image reconstruction method. Specifically, the main contributions are as follows:
(1) We propose the MD-GAN model for plane-wave image reconstruction. The model employs a U-Net-based generator and a Patch discriminator. It incorporates a multi-scale convolution within the U-Net encoder to capture information from the input image at different frequencies. We propose Dynamic Criss-Cross Attention (DCCA) in the U-Net decoder to avoid interference caused by irrelevant regions in the single-beam plane wave image during reconstruction. Moreover, DCCA enhances the texture details and general information of the image.
(2) The MD-GAN incorporates the adversarial loss and integrates the L1-Structural Similarity index (L1-SSIM) loss and perceptual loss in the generator’s loss function.
(3) We conducted extensive experiments on the PICMUS 2016 dataset [10].
2.Methodology of this paper
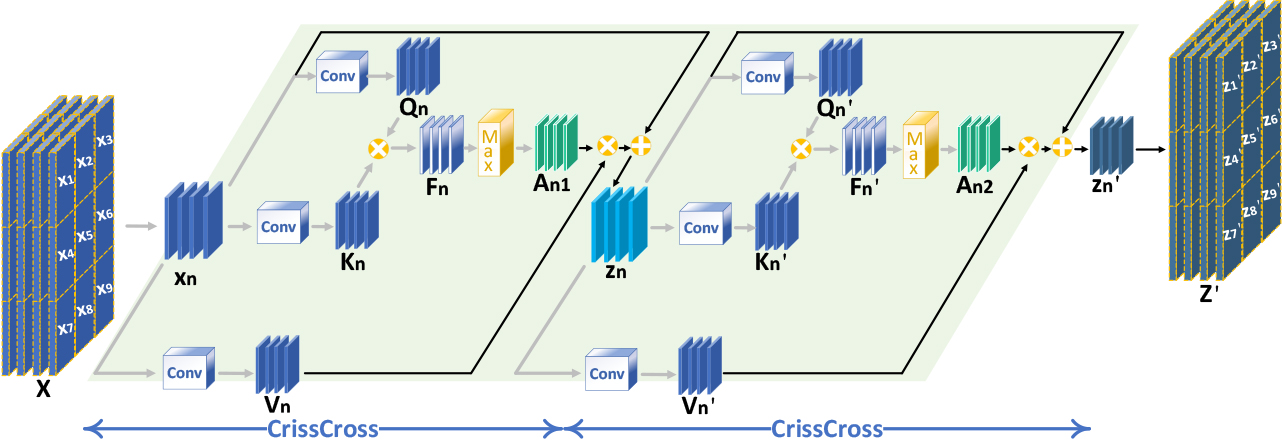
The single-beam plane-wave image exhibits significant scattering effects, blurred texture, and masked regions within the image. We propose the MD-GAN model to address the issues. The overall architecture of the model is depicted in Fig. 1. A U-Net-based generator and patch discriminator are used, with a single-beam plane-wave image as the input to the generator and 75 different beam plane-wave synthesized images as the actual labels. The generator generates the corresponding super-resolution images based on the input images, and the discriminator compares the generated images with the actual labels to discriminate the true from the false.
2.1Multi-scale convolution module
Inspired by the multidimensional feature extraction in the literature [11], this paper designs a multiscale module to capture the scattering features in each dimension through convolutional operations at different scales to alleviate scattering interference on image reconstruction. The structure of the multiscale convolution module is depicted in Fig. 2, and Table 1 presents the main parameters of each layer.
Table 1
Main parameters of multi-scale convolution module
| layer | Convolution parameters (kernel, step, padding) | The number of channels (in, out) |
| D0 | 1,1,0 | 1,2 |
| D1 | 2,2,0 | 2,16 |
| D2 | 4,2,1 | 18,16 |
| D3 | 6,2,2 | 18,16 |
| D4 | 8,2,3 | 18,16 |
Figure 1.
Schematic diagram of the overall structure of the modeU-Net Generator.
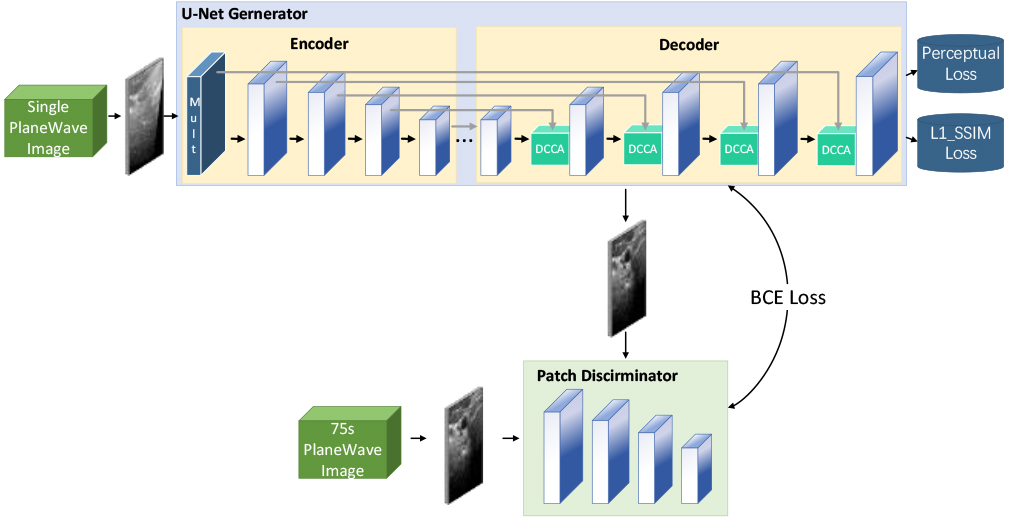
Figure 2.
Schematic diagram of multi-scale convolution module structure.
2.2Dynamic criss-cross attention
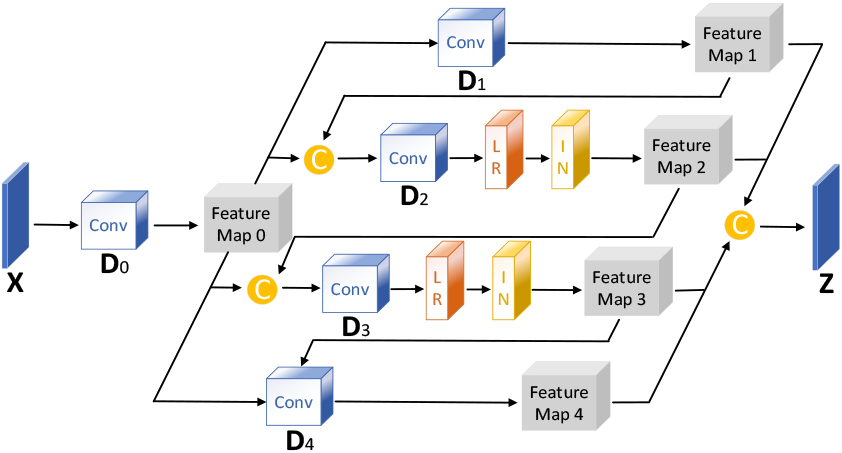
In image reconstruction, While the widely used self-attention mechanism comprehensively captures an image’s global structure, it fails to extract local image features. Referring to the enhancement of local features in reference [12], this study proposes the DCCA mechanism, which introduces a sliding window on top of Criss-Cross Self-Attention, and the introduction of the sliding window not only enhances the local features but also avoids the occlusion region in the single-beam plane-wave image from interfering with the reconstruction of the target region. DCCA establishes the connection between each pixel point in the sliding window by performing two cross operations inside the sliding window. By adjusting the sliding window’s size and step length, the proportion of global and local features extracted by DCCA can be fine-tuned. The larger the sliding window step size and the smaller the size, the higher the weight of local feature extraction, and vice versa, the smaller the step size and the larger the size, the higher the weight of global feature extraction. The implementation process of DCCA is depicted in Fig. 3.
Figure 3.
Schematic diagram of the Dynamic Criss-Cross Attention structure.
In the first step, a weight matrix is obtained. Three
In the second step, weights are applied to the input feature map. The weight vector
In the third step, the first and second steps are repeated. Ultimately, the feature region
The deep layer of the U-Net decoder utilizes DCCA with a relatively small sliding window size to focus on enhancing texture information. In contrast, the shallow layer employs DCCA with a relatively sizeable sliding window size to focus on the fusion of general information. The specific parameters of DCCA in the U-Net decoder are shown in Table 2.
Table 2
Main parameters of DCCA in decoder
| Layer | Sliding window parameters (length, width, step) |
| D1 | 9,9,9 |
| D2 | 7,7,7 |
| D3 | 5,5,5 |
| D4 | 3,3,3 |
2.3Loss function
This study introduces L1-SSIM loss and perceptual loss on top of the original adversarial loss. The combination of L1 loss and MS-SSIM loss (L1-SSIM loss) considers the generated image’s gray value and structural information. The perceptual loss function encourages the generated and actual images to contain similar high-level semantic information.
3.Experimental results and analysis
3.1Experimental details
For this study, we utilized the dataset from the 2016 Plane-Wave Imaging Challenge in Medical Ultrasound (PICMUS), comprising 360 image sets. The dataset included 120 sets of simulated images generated using Field II, 120 sets of experimental images obtained through the Verasonics Vantage 256 study scanner and L11 probe, and 120 sets of human carotid images. Image flipping operations were performed to augment the dataset and enhance its diversity, resulting in 1350 image sets. Subsequently, the dataset was divided into training and test sets in an 8:2 ratio.
Different evaluation measures were selected to assess the reconstruction performance for each type of image. The Full Width at Half Maximum (FWHM) and Contrast Ratio (CR) were utilized to evaluate the reconstruction of point and cyst target images, respectively. Furthermore, the PSNR and SSIM were employed to evaluate the reconstruction of actual images in vivo.
Full Width at Half Maximum: The FWHM indicates the beamwidth of the main lobe at -6 dB on the point spread function.
Contrast Ratio: The CR quantifies the level of differentiation between the target area and the background area.
Peak Signal-to-Noise Ratio: The PSNR measures the global pixel error between the generated and actual images to assess the degree of image reconstruction.
Structural Similarity Index: The SSIM is an index based on human visual perception, measuring the similarity between the generated and actual images.
3.2Comparative experiments
3.2.1Results of deep learning methods
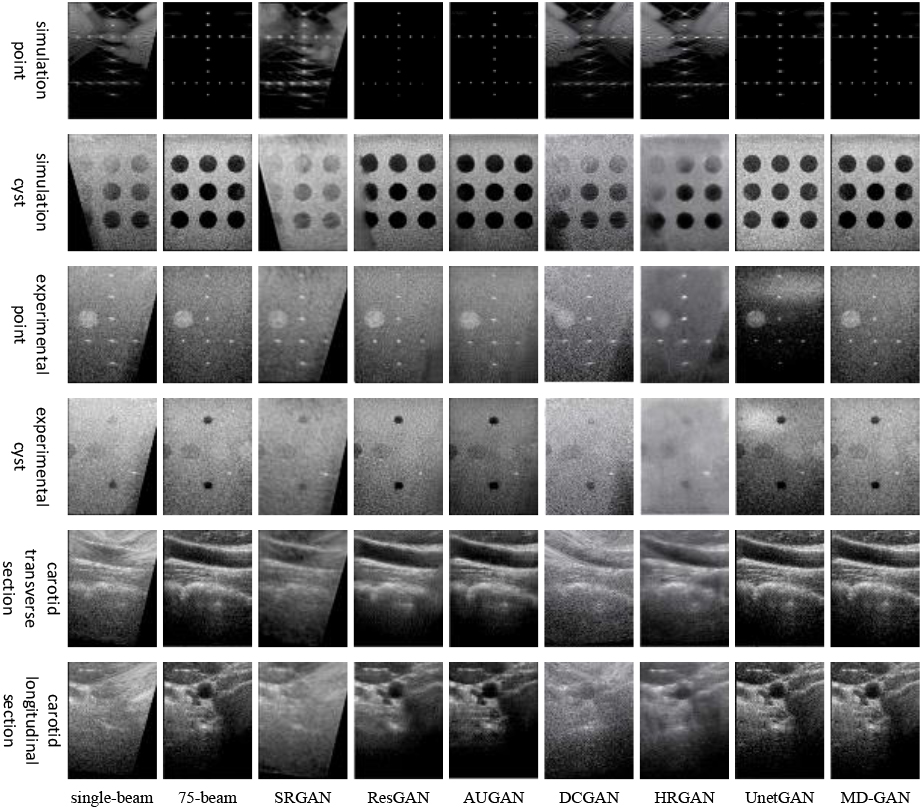
As depicted in Fig. 4, the MD-GAN model achieves the optimal visual outcome across various targets. This model effectively circumvents evident reconstruction traces in the masked areas of images, a predicament commonly associated with SRGAN and ResGAN models. Additionally, it successfully addresses the DCGAN model’s limitation in noise removal from single-beam plane wave images. It alleviates the issue of enhanced blurriness in texture details, as typically observed in images generated by the HRGAN model. Furthermore, it resolves the UnetGAN model’s challenge in accurately reconstructing the background region within images.
Table 3
A comparison of results with those of deep learning methods
| Method | FWHM (mm) | CR(dB) | In vivo target | |
|---|---|---|---|---|
| (sim/exp) | (sim/exp) | PSNR(dB) | SSIM | |
| HRGAN [13] | 0.3675/1.0356 | 16.9231/6.6463 | 22.1431 | 0.0929 |
| SRGAN [14] | 0.3749/0.7561 | 9.4526/8.0978 | 19.4147 | 0.0744 |
| DCGAN [15] | 0.3089/0.7207 | 11.5868/8.6473 | 19.0417 | 0.0721 |
| ResGAN [16] | 0.2916/0.4838 | 34.6607/25.2034 | 23.6817 | 0.1022 |
| AUGAN [17] | 0.2617/0.5223 | 36.8714/25.5976 | 15.5082 | 0.1286 |
| UNetGAN [18] | 0.3025/0.4922 | 29.8705/28.3958 | 30.2767 | 0.8339 |
| Our method | 0.2844/0.4822 | 36.8092/25.9520 | 33.5419 | 0.9577 |
Table 3 illustrates that compared to the ResGAN model, MD-GAN achieves reductions of 0.0072 mm and 0.0016 mm in FWHM for simulated and experimental point targets, respectively. Furthermore, the contrast of simulated and experimental cyst targets generated by MD-GAN reaches 36.8092 dB and 25.9520 dB, surpassing most prevalent deep-learning reconstruction methods. In the context of in vivo carotid artery images, the reconstructed images produced by MD-GAN exhibit substantial improvements, with a 10.78% increase in PSNR and a 14.85% enhancement in SSIM compared to the suboptimal UnetGAN.
Table 4
A comparison of results with those of traditional methods
| Method | FWHM(mm) (sim/exp) | CR(dB) (sim/exp) | In vivo target PSNR(dB) |
|---|---|---|---|
| CPWC | 0.2943/0.5321 | 35.4682/25.0031 | |
| MV | 0.2941/0.5333 | 35.5730/25.5150 | 25.9980 |
| CF | 0.2920/0.4301 | 34.7891/24.9383 | 34.5332 |
| Our method | 0.2844/0.4822 | 36.8092/25.9520 | 33.5419 |
3.2.2Results of non-deep learning methods
As shown in Table 4, the FWHM of the simulated point target was reduced by 0.0076 mm compared to the CF method. The simulated and experimental cyst target’s CR was improved by 3.48% and 1.71% compared to the MV method. The proposed model maintained a higher PSNR value for the generated images in the reconstruction of in vivo actual images, which was significantly better than the MV method.
3.2.3Model scale comparison
As shown in Table 5, MD-GAN has a certain advantage in terms of the model scale, and compared with the SRGAN model, it is less computationally intensive and more time efficient despite the more significant number of parameters.
Table 5
A comparison of model scale
| Method | Parameters | MACs(G) |
|---|---|---|
| HRGAN [13] | 873.4534K | 172.1326 |
| SRGAN [14] | 839.6215K | 165.1523 |
| DCGAN [15] | 35.1916G | 72.3446 |
| ResGAN [16] | 31.4203G | 142.7519 |
| AUGAN [17] | 43.8314G | 53.8906 |
| UNetGAN [18] | 42.6963M | 53.2867 |
| Our method | 43.6180M | 63.1105 |
Figure 4.
Comparison plots of experimental results of deep learning methods.
3.3Ablation experiments
3.3.1Ablation experiment of the generator part
The effectiveness of the U-Net network improvement in this model was evaluated through ablation experiments in the generator section, as presented in Fig. 5. The scatter in the generated images is significantly reduced by incorporating the multi-scale convolution module. Moreover, the addition of DCCA resulted in significantly enhanced simulated point targets. The color of the obtained cyst target was also deepened by adding DCCA to the reconstruction of the cyst target. Furthermore, in reconstructing the actual image in vivo, the addition of DCCA was able to reconstruct the carotid artery image with a more complete overall structure.
Table 6
Results of generator ablation experiments
| Generator | FWHM (mm) | CR(dB) | In vivo target | |
| (sim/exp) | (sim/exp) | PSNR(dB) | SSIM | |
| DCCA | 0.2883/0.4871 | 35.0633/25.2113 | 30.1439 | 0.9113 |
| Mult | 0.2936/0.4896 | 29.4425/23.4765 | 32.1721 | 0.9362 |
| DCCA | 0.2844/0.4822 | 36.8092/25.9520 | 33.5419 | 0.9577 |
Figure 5.
Plot of the ablation experiment’s results for the generator part.
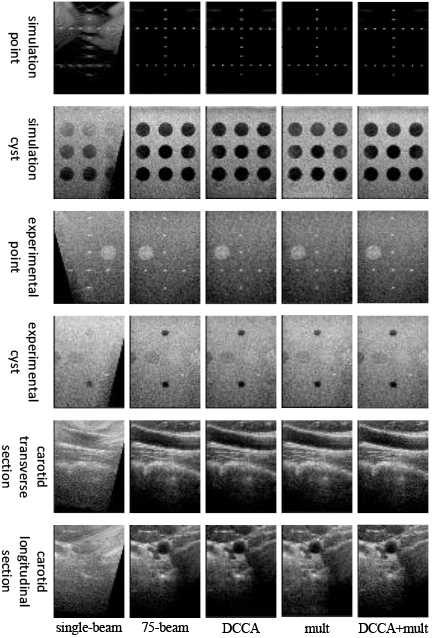
As presented in Table 6, In the case of point targets, incorporating DCCA resulted in a reduction of 0.0092 mm in the FWHM of simulated point targets. Cyst targets, including DCCA, yielded a significant enhancement in CR for both simulated and experimental cyst targets, with improvements of 25.02% and 10.54%, respectively. In the reconstruction of real in vivo images, utilizing the multi-scale convolution module led to an increase in PSNR of 3.3980 dB and an improvement in SSIM of 0.0464 for the carotid reconstruction images.
3.3.2Loss function ablation experiment
Figure 6 illustrates the experimental results. Only the original adversarial loss function was retained, resulting in a generated image that failed to restore the overall structure resembling the actual image. On this basis, introducing perceptual loss still did not lead to the reconstruction of the overall structure of the image. However, combining the adversarial loss with L1-SSIM loss proved effective in reconstructing the image’s global information and texture details. Moreover, the combination of perceptual, adversarial, and L1-SSIM loss yielded plane-wave ultrasound images with a more apparent texture and higher contrast.
Table 7
Results of loss function ablation experiments
| Loss | FWHM (mm) | CR(dB) | In vivo target | |
|---|---|---|---|---|
| (sim/exp) | (sim/exp) | PSNR(dB) | SSIM | |
| BCE | 0.7282/1.1354 | 5.5708/5.9618 | 18.1301 | 0.0338 |
| BCE | 0.7801/1.3501 | 10.0051/3.2537 | 16.1572 | 0.0366 |
| BCE | 0.2897/0.4888 | 34.7758/25.8869 | 31.2932 | 0.9169 |
| BCE | 0.2844/0.4822 | 36.8092/25.9520 | 33.5419 | 0.9577 |
Table 7 demonstrates that solely employing the traditional adversarial loss leads to inferior metrics for point target images, cyst target images, and in vivo actual images. By introducing perceptual loss on top of this, most of the experimental metrics will be reduced. Integrating L1-SSIM with the adversarial loss yields improved experimental metrics across all image types. In this model, retaining L1-SSIM, perceptual loss, and adversarial loss further enhances the experimental indexes.
3.3.3Ablation experiments of sliding windows in DCCA
In this study, ablation experiments were conducted to investigate the impact of sliding windows of DCCA in the U-Net decoder. Specifically, as depicted in Fig. 7, the reconstructed simulated point targets appeared noticeably weaker when the 7
Table 8
Results of the DCCA’s sliding window ablation experiments
| Sliding window | FWHM (mm) | CR(dB) | In vivo target | |
| (sim/exp) | (sim/exp) | PSNR(dB) | SSIM | |
| 3*3+5*5+7*7 | 0.2847/0.4836 | 34.6919/25.7749 | 31.5622 | 0.9561 |
| 3*3+5*5+9*9 | 0.2864/0.5061 | 34.4011/25.7141 | 32.2631 | 0.9447 |
| 3*3+7*7+9*9 | 0.2891/0.4885 | 36.1980/25.9410 | 29.5940 | 0.9170 |
| 5*5+7*7+9*9 | 0.2937/0.4993 | 34.9729/25.8108 | 32.4483 | 0.9444 |
| 3*3+5*5+7*7 +9*9 | 0.2844/0.4822 | 36.8092/25.9520 | 33.5419 | 0.9577 |
Figure 6.
Plot of the ablation experiment’s results for the loss function part.
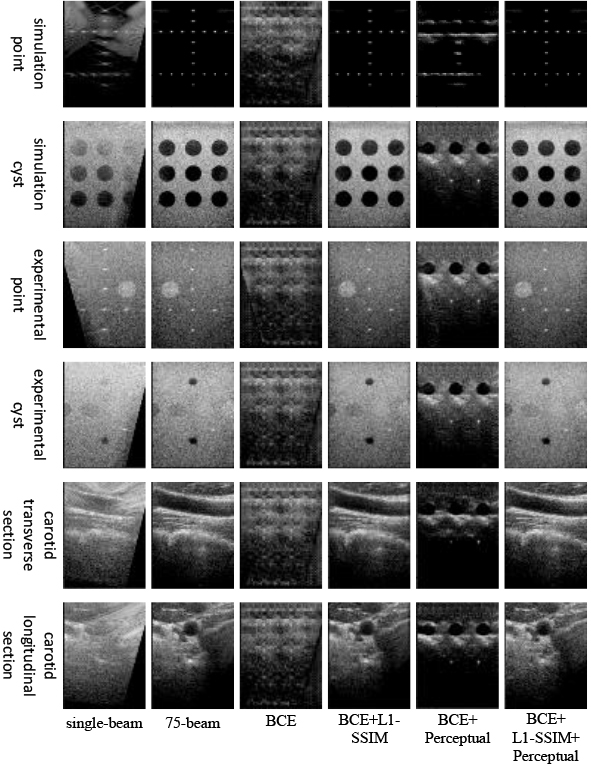
Figure 7.
Plots of experimental results for ablation of sliding windows in DCCA.
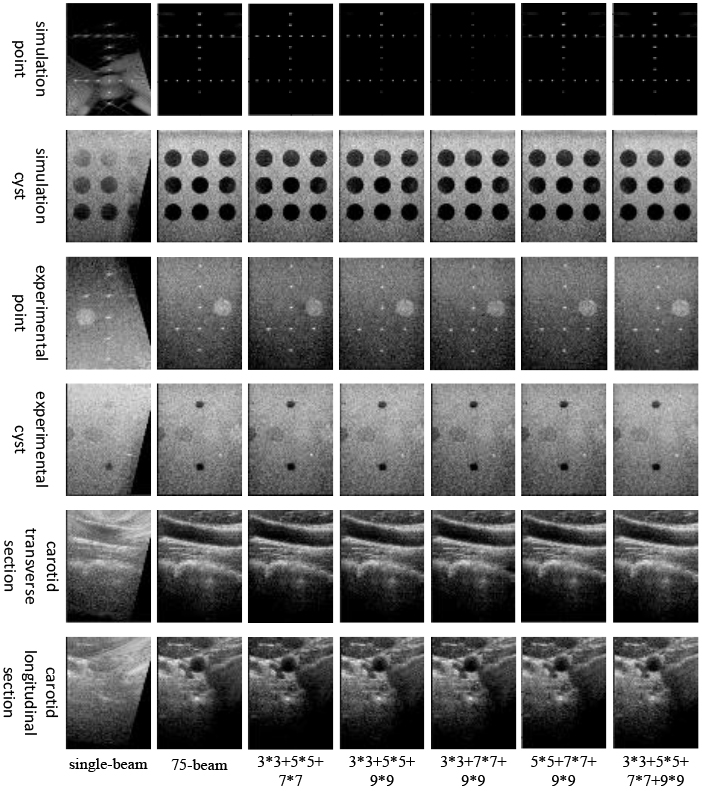
Table 8 presents the effects of sliding windows in DCCA on various target reconstructions. For point target reconstruction, including a 3
4.Discussion
This study proposes the MD-GAN image reconstruction method, which utilizes the U-Net generator and Patch discriminator. The U-Net generator adds the multi-scale convolution module and DCCA to separate the image’s high and low-frequency information while enhancing the image’s local and global features and suppressing the interference of irrelevant regions to the reconstructed target. Moreover, the loss function of this model is enriched with perceptual loss and L1-SSIM loss to enhance further the quality of the images generated.
4.1Contribution of modules
In medical image reconstruction, the use of self-attention, although better at establishing the overall structure of the image, can be influenced by the occluded areas of the single-beam plane-wave image in the process of extracting global features, resulting in the overall darkness of the reconstructed experimental and in vivo images. The DCCA proposed in this paper effectively solves the above problems. The introduction of the sliding window in DCCA can avoid the interference of irrelevant regions to the target region in the reconstruction process, increase the local feature extraction ability while retaining the traditional self-attention global feature extraction ability, enhance the cyst target and simulation point target while ensuring the reconstruction effect of the background region, improve the CR of the cyst target image, and reduce the FWHM of the simulation point target image.
The addition of a multi-scale convolution module to the encoder of the U-Net network aids in separating the high and low-frequency information from the input image through the use of convolutional kernels of varying sizes. This separation facilitates the determination of the boundary of the simulated point target and eliminates scatter around the simulated point target. The multi-scale convolution module, which combines DCCA in the decoder, can more effectively utilize the separated high and low-frequency information, reducing the FWHM of the point target image and an improvement in the PSNR of the carotid artery image.
In this study, the loss function of the MD-GAN model introduces L1-SSIM loss and perceptual loss on top of the original adversarial loss function. The L1-SSIM loss can guarantee the grayscale value of the generated image while building a clear image structure, ensuring that the generated image maintains high structural similarity with the actual image. Additionally, the perceptual loss encourages the generated image to contain similar high-level semantic information as the actual image. The original adversarial loss alone is insufficient to establish the actual structure of the image. However, the combination of adversarial loss, L1-SSIM loss, and perceptual loss leads the model to converge in the optimal direction, resulting in the generation of plane-wave ultrasound images with complete structure and precise details.
4.2Model quality
The MD-GAN model exhibits excellent performance in point target images, cyst target images, and in vivo carotid artery images, and it surpasses several other evaluation models in terms of visual effects and various evaluation metrics. However, it is essential to note that deep learning-based reconstruction models achieve image-to-image reconstruction by learning the nonlinear relationship between low-resolution and high-resolution images without fully utilizing beam information. Consequently, the FWHM of the generated experimental point target images and the PSNR of carotid artery images still do not outperform the performance of beam-based reconstruction methods.
Deep learning-based methods demonstrate substantial fluctuations in overall reconstruction results in point target images. Specifically, SRGAN yields noticeably poor reconstruction results. DCGAN and HRGAN models generate the point target image with significant scatter and unsatisfactory performance across all experimental indexes. While ResGAN manages to reduce the FWHM of both simulated and experimental point target images, evident reconstruction traces are present in the occluded areas of the images. The UNetGAN model exhibits a limited ability to extract low-frequency information and fails to accurately reconstruct the background area of the experimental point target image. Introducing an attention mechanism in AUGAN enhances the model’s ability to extract high-frequency features and performs better in reconstructing simulated point target images. However, the model has limited capability for extracting global features, resulting in poor reconstruction quality in the masked region of the experimental point target image. By integrating DCCA with a relatively sizeable sliding window size into the shallow layer of the decoder, MD-GAN effectively enhances the low-frequency information extracted from the multi-scale convolution module, thereby achieving high-quality reconstruction of occluded and background regions. The utilization of the multi-scale convolution module separates the high and low-frequency information, resulting in accurate reconstruction of point target boundaries and effectively eliminating scattered spots surrounding the point target. Furthermore, incorporating DCCA with a relatively small sliding window size in the deep layer of the decoder enhances local features, resulting in a more prominent point target. Consequently, the model successfully reduces the FWHM of the generated image while producing reconstructed images with clear targets and complete structures.
Regarding reconstructing cyst target images, the SRGAN model yields terrible visual effects and image metrics. DCGAN and HRGAN generate images with low CR, and there are severe noise and image blurring problems, respectively. While ResGAN manages to improve the CR of cyst images, it fails to recover obscured parts of the images. The introduction of attention in the AUGAN model significantly deepens the cyst target in the reconstructed image and improves the CR of the simulated cyst target. However, the model exhibits a poor ability to acquire low-frequency information, resulting in overall darkness in the generated cyst images and slight reconstruction traces. The UNetGAN model struggles to accurately extract global features of the image, leading to too bright and too dark areas in the background of the generated image. Although high CR is achieved in reconstructing the experimental cyst target, the model needs to improve the quality of the generated images. The MD-GAN model adds DCCA with increasing sliding window size sequentially from deep to shallow layers of the U-Net decoder. This approach increases the weight of local feature extraction before increasing the weight of global information extraction, ensuring that the generated image possesses a more precise overall structure while highlighting the cyst target and improving the CR of the cyst target image.
The MD-GAN model outperforms the other evaluated models in reconstructing in vivo carotid artery images, primarily due to the rest of other evaluation models’ inability to effectively extract low-frequency features and accurately reconstruct the global structure. MD-GAN combines a multi-scale convolution module with DCCA to capture low-frequency information precisely and introduces the L1-SSIM loss function to positively guide the structural similarity between the generated and actual images. Nevertheless, compared to traditional beam synthesis methods like CF and GCF, the reconstruction effect of MD-GAN in vivo carotid artery images still has room for improvement.
5.Conclusion
In this paper, we have proposed a novel end-to-end plane-wave image reconstruction network called MD-GAN. This network utilizes a U-Net-based generator and Patch discriminator, incorporating a multi-scale convolution module in the encoder of the U-Net generator to extract information of different frequencies. Additionally, DCCA is introduced in the decoder of the U-Net generator. It enhances local features, integrates global information, and suppresses the interference of unrelated regions with the reconstructed target.
Experimental results demonstrate the effectiveness of our proposed method. However, deep learning methods face challenges in effectively utilizing acoustic emission and echo information during image reconstruction. Addressing this limitation will be a crucial focus of our future research.
Conflict of interest
None to report.
References
[1] | Zhang H, Zhu Y, Zhou B, et al. Multibeam minimum variance beamforming for ring array ultrasound imaging. Physics in Medicine and Biology. (2023) . |
[2] | Nair AA, Tran TD, Bell MAL. Robust short-lag spatial coherence imaging. IEEE transactions on ultrasonics. (2017) ; 65: (3): 366-377. |
[3] | Hashemseresht M, Afrakhteh S, Behnam H. High-resolution and high-contrast ultrafast ultrasound imaging using coherent plane wave adaptive compounding. Biomedical Signal Processing and Control. (2022) ; 73: : 103446. |
[4] | Jiang K, Wang Z, Yi P, et al. Hierarchical dense recursive network for image super-resolution. Pattern Recognition. (2020) ; 107: : 107475. |
[5] | Jiang M, Zhi M, Wei L, et al. FA-GAN: Fused attentive generative adversarial networks for MRI image super-resolution. Computerized Medical Imaging and Graphics. (2021) ; 92: : 101969. |
[6] | Wei P, Xie Z, Lu H, et al. Component divide-and-conquer for real-world image super-resolution. Computer Vision–ECCV. (2020) ; 101-117. |
[7] | Chen R, Zhang H, Liu J. Multi-attention augmented network for single image super-resolution. Pattern Recognition. (2022) ; 122: : 108349. |
[8] | Wang Q, Zhou H, Li G, et al. Single Image Super-Resolution Method Based on an Improved Adversarial Generation Network. Applied Sciences. (2022) ; 12: (12): 6067. |
[9] | Wang X, Xie L, Dong C, et al. Real-esrgan: Training real-world blind super-resolution with pure synthetic data. Proceedings of the IEEE/CVF international conference on computer vision. (2021) ; 1905-1914. |
[10] | Liebgott H, Rodriguez-Molares A, Cervenansky F, et al. Plane-wave imaging challenge in medical ultrasound. IEEE International Ultrasonics symposium. (2016) ; 1-4. |
[11] | Lyu Y, Xu Y, Jiang X, et al. AMS-PAN: Breast ultrasound image segmentation model combining attention mechanism and multi-scale features. Biomedical Signal Processing and Control. (2023) ; 81: : 104425. |
[12] | Gao Z, Wang L, Wu G. Lip: Local importance-based pooling. Proceedings of the IEEE/CVF International Conference on Computer Vision. (2019) ; 3355-3364. |
[13] | Zhang X, Li J, He Q, et al. High-quality reconstruction of plane-wave imaging using generative adversarial network. (2018) ; IEEE International Ultrasonics Symposium (IUS). 2018: : 1-4. |
[14] | Ledig C, Theis L, Huszár F, et al. Photo-realistic single image super-resolution using a generative adversarial network. Proceedings of the IEEE conference on computer vision and pattern recognition. (2017) ; 4681-4690. |
[15] | Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv preprint arXiv1511.06434, (2015) . |
[16] | He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition. (2016) 770-778. |
[17] | Tang J, Zou B, Li C, et al. Plane-wave image reconstruction via generative adversarial network and attention mechanism. IEEE Transactions on Instrumentation and Measurement. (2021) ; 70: : 1-15. |
[18] | Hao X, Su X, Wang Z, et al. UNetGAN: A robust speech enhancement approach in time domain for extremely low signal-to-noise ratio condition. arXiv preprint arXiv2010.15521, (2020) . |




