Biomedical named entity recognition based on fusion multi-features embedding
Abstract
BACKGROUND:
With the exponential increase in the volume of biomedical literature, text mining tasks are becoming increasingly important in the medical domain. Named entities are the primary identification tasks in text mining, prerequisites and critical parts for building medical domain knowledge graphs, medical question and answer systems, medical text classification.
OBJECTIVE:
The study goal is to recognize biomedical entities effectively by fusing multi-feature embedding. Multiple features provide more comprehensive information so that better predictions can be obtained.
METHODS:
Firstly, three different kinds of features are generated, including deep contextual word-level features, local char-level features, and part-of-speech features at the word representation layer. The word representation vectors are inputs into BiLSTM as features to obtain the dependency information. Finally, the CRF algorithm is used to learn the features of the state sequences to obtain the global optimal tagging sequences.
RESULTS:
The experimental results showed that the model outperformed other state-of-the-art methods for all-around performance in six datasets among eight of four biomedical entity types.
CONCLUSION:
The proposed method has a positive effect on the prediction results. It comprehensively considers the relevant factors of named entity recognition because the semantic information is enhanced by fusing multi-features embedding.
1.Introduction
With the rapid development of biomedical technology, biomedical literature is also growing at an exponential rate. For example, since the global COVID-19 outbreak in March 2020, PMC has published more than 280,000 papers related to COVID-19. So, it is very important to quickly and efficiently extract disease-related information. The Biomedical Named Entity Recognition (BNER) is the first step and the most important step in biomedical semantic information extraction [1, 2, 3]. BNER is a prerequisite and critical part of building medical knowledge graphs, medical question and answer systems, medical text classification in biomedical field [4, 5]. In addition, highly accurate entity extraction largely guarantees the high reliability and applicability of the constructed knowledge graphs and medical Q&A systems [6, 7, 8]. It is the basic step of many downstream text mining applications and lays a foundation for further mining the rich information in the biomedical literature.
BNER has received attention from researchers and it is a interesting research topic in Natural Language Processing (NLP). BNE mainly includes gene, protein, DNA, RNA, disease, drug, and chemical substance [9]. Until now, various text mining methods are applied to identify BNEs, such as lexicon and rule-based methods [10, 11, 12], statistical machine learning methods [13, 14, 15, 16, 17, 18, 19], deep learning methods [20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31].
The deep learning approach uses end-to-end model training and automatic feature extraction to avoid tedious manual extraction by acquiring features and distributed data representations with good generalization capabilities. Compared to the lexicon and rule-based methods or statistical machine learning methods, deep learning neural network-based methods have the advantage of no longer relying on manual features and domain knowledge, reducing the cost of manual feature extraction, having more robust generalization, and effectively improving system efficiency [2, 3, 4].
In the last few years, the performance of Long Short-Term Memory Networks [18] (LSTM) and Conditional Random Fields [19] (CRF) for NER has improved considerably. Yao et al. [20] first built a multi-layer neural network on unlabeled biomedical texts to implement word embedding vectors for biomedical words on a large-scale corpus. Huang et al. [21] proposed a BiLSTM-CRF model for predicting sequence labels. Lyu et al. [22] used the BiLSTM-RNN model to combine biomedical word embedding and character embedding for entity recognition. Although their work has achieved some success, the word vector methods used are too simple to study the deep meaning of biomedical texts.
Gridach et al. [26] first used character-level embedding to represent features in the biomedical domain and constructed the hybrid model BiLSTM-CRF for named entity recognition. Liu et al. [27] extracted lexical and morphological features of words, and a multi-channel convolutional neural network was post-connected to the BiLSTM-CRF model for entity extraction. Patel et al. [28] adopted a method based on Flair and GloVe embeddings and a bidirectional LSTM-CRF-based sequence tagger to train a BNER model. Yoon et al. [29] build a model using multiple BiLSTM-CRFs that are constructed on top of multiple individual task NER models (STMs). They can exchange information to each other for better prediction result. Wang et al. [30] constructed a multi-task learning framework that shares character-level information and word-level information of related BNEs to achieve significant performance gains. Sachan et al. [31] proposed BiLM-NER model, a bi-directional language model (BiLM) which is trained on unlabeled data, a better method is adopted to initialize the parameters of NER model. In addition, recent work on building tools in the biomedical field, such as HunFlair [32] and BERN [33], has achieved good results.
However, direct biomedical application methods has the following limitations: (i) Word representation models like Word2Vec [34], ELMo [35], BERT [36], and ALBERT [37] are mainly trained on datasets with general non-domain specific texts (e.g., Wikipedia). The general word and sentence patterns are different from the biomedical text patterns. It is difficult to catch the biomedical semantic information from the text. (ii) The representation layer in previous work [26, 27, 28, 29, 30, 31] is simple and does not effectively capture local and global information of words. A single word vector representation inevitably avoids the problem of multiple word meanings.
To solve these problems, we propose an end-to-end approach based on biomedical fuse multi-feature embedding to handle the shortcomings mentioned above for BNER. Our goal is to extract a variety of features representing biomedical named entities from different dimensions, so as to improve the effect of entity recognition. Our contributions are as follows:
A multi-feature embedding based BNER method to capture features from biomedical texts is proposed. Combining deep contextual word-lever features, local char-level features, and part-of-speech features of biomedical texts enhances the semantic information representation of words to identify entities effectively.
We have validated the effectiveness of our method on eight datasets of four entity types in the biomedical field: disease, drug/chemical, gene/protein, and species, and all results show that our model is better than other published methods.
2.Method
Our proposed model consists of two main components: (i) Multi-Feature Representation layer for enhancing the semantic information, (ii) Downstream named entity recognition task layer for capturing long dependent information, and getting globally optimal tagging sequence. The whole workflow is shown in Fig. 1.
Figure 1.
Multi-Feature Embedding BiLSTM-CRF architecture.
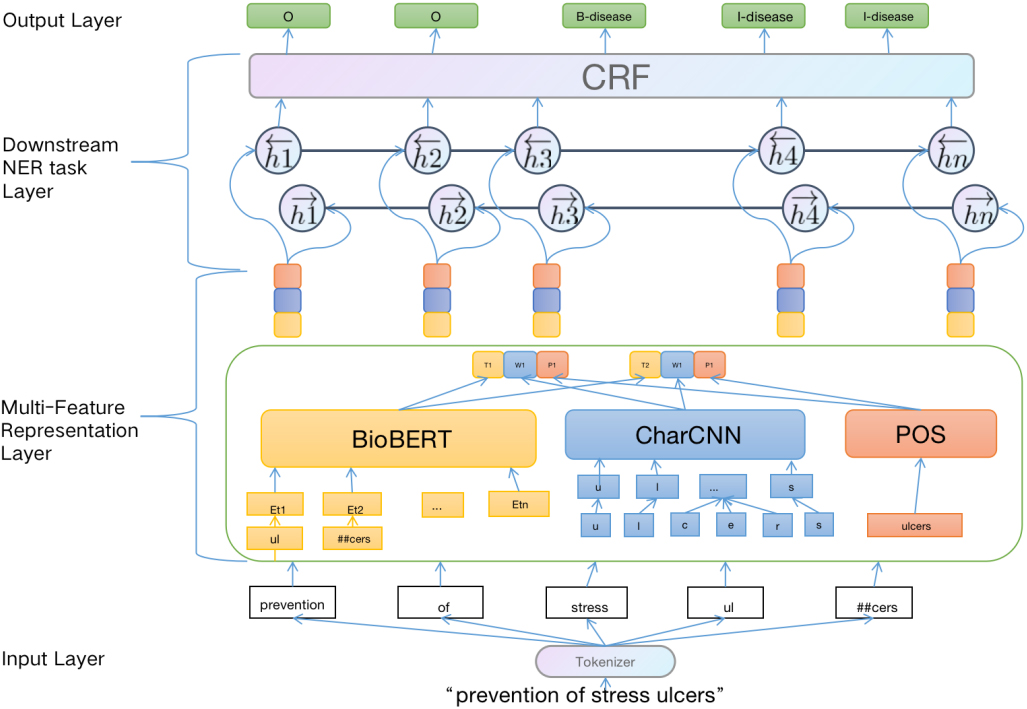
2.1Multi-feature representation layer
The NER task is to label the entities in a given sentence. More formally, given the input sentence
To extract the coverage information of the input sentence as much as possible, we apply the following three feature extraction methods from different perspectives to represent the input sequence: deep context word-level features, local char-level features, and part-of-speech features.
2.1.1Deep context word-level features
At the text representation layer, we adopt the BERT architecture [36] as our model to obtain deep contextual word-level features of biomedical texts. Since what needs to be identified in this study are entities in the biomedical domain, we employ BioBERT [38], which is a model pre-trained on a large-scale biomedical corpus based on the BERT architecture. A language model trained with masks using bidirectional transformers does a good job of contextualizing words. Below we will briefly introduce the core architecture in the BERT model: the Transformer Encoder.
In order to better represent biomedical text, twelve Transformer Encoders are used in the pre-trained language model BioBERT. In the Encode layer, use a multi-headed self-attention mechanism instead of the traditional LSTM model. The formula for the self-attention mechanism is shown in Eq. (1):
(1)
where,
The multi-headed self-attention mechanism learns multiple
(2)
(3)
2.1.2Local char-level features
To fully consider the internal information of biomedical texts, Character-level Convolutional Neural Network [40] (CharCNN) is used to extract the local char-level features of each word in the biomedical text. In this paper, the character-level convolutional neural network contains three components: a character vector layer, a one-dimensional convolutional layer, and a maximum pooling layer.
The character vector layer transforms the characters in each word of the input sequence into its corresponding character vector, which constitutes the character vector matrix of the word that will be continuously updated and learned during the model training. The alphabet used in our models consists of 70 characters [30] as shown in Table 1.
Table 1
The alphabet used in all of our CharCNN models
| Character type | Chararcters |
|---|---|
| English letters | abcdefghijklmnopqrstuvwxyz |
| Digits | 0123456789 |
| Others | -,;.!?:”’/ |
In the convolution layer, multiple layers of convolution kernels are used on the character vector matrix to extract the local features of the words. Finally, the local char-level feature vectors of biomedical text words are obtained by using maximum pooling.
2.1.3Part-of-speech features
To represent the syntactic pattern characteristics of the words in the input sentence, such as the dependencies between the suffixes and the output labels, we use the NLTK to get the part-of-speech tag sequence of the text sentence.
Lexicality, as the fundamental grammatical property of words, is a crucial feature of words and utterances. The part-of-speech tag sequence contains the syntactic information of the words in the sentence.
2.2Design downstream named entity recognition task layer
2.2.1BiLSTM for long dependency information
To process the long dependency and contextual information in the input text sequence, we apply Bidirectional Long Short-Term Memory Networks (BiLSTM) to access more semantic dependencies.
LSTM introduces a gating mechanism in the RNN structure to selectively change what needs to be retained and capture long-range associative information, which effectively overcomes the problems of traditional RNN models. The specific calculation process of LSTM is given in Eq. (2.2.1):
(4)
where,
The BiLSTM model which uses a forward and backward LSTM module can consider the contextual information to extract bi-directional sentence features to obtain better results, which is more suitable for the characteristics of BNER.
2.2.2CRF for globally optimal tagging sequence
To solve the problem of possible inter-dependencies and mutual constraints in the tag sequences, such as B-gene tags should not be followed by I-chem tags, we adopt the Conditional Random Field (CRF) in this research.
Although the BiLSTM model can recognize entity boundaries, it does not fully learn the label dependencies between entity sequences. CRF is a probabilistic graphical model proposed based on the EM [41] model and the HMM [42] model commonly used for tagging sequences such as named entity recognition, target recognition, and linguistic annotation. It can get a globally optimal tagging sequence by considering the relationship of adjacent tags and using a state transfer matrix to decode jointly.
Assume that the output sequence of the BiLSTM model is X and one of the predicted sequences is Y. then the evaluation score P (X, Y) can be obtained by Eq. (5).
(5)
where,
3.Experiment
3.1Experiment datasets
To verify the universality and effectiveness of the proposed model, our method is evaluated in eight datasets containing four entity types: gene/protein, drug/chemical, disease and species. Datasets including gene and protein include BC2GM and JNLPBA corpus. Datasets including drug and chemical include BC4CHEMD and BC5CDR-chem corpus. Datasets including disease include NCBI and BC5CDR-disease corpus. Datasets including species include LINNAEUS and Species-800 corpus. A brief explanation of the dataset used in this paper is as follows:
• BC5CDR: This dataset is provided by BioCreative V Chemical Disease Relation Extraction (BC5CDR) Task [43]. The dataset has two subtasks, the first is to identify chemical entities, and the second is to identify disease entities.
• BC4CHEMD: This dataset is provided by BioCreative Community Challenge IV Task [44]. It is a data manually marked by expert chemistry from 10,000 PubMed abstracted references for the development and evaluation of chemical NER tools, and eventually contains about 80,000 chemical entities.
• NCBI Disease: This dataset is provided by Doğan et al. [45], which includes 6892 disease mentions from 793 abstracts. It is worth mentioning that the dataset is labeled at the Mention and conceptual levels, with the characteristics of large scale and high quality, and it aligns the disease name with the corresponding disease concept ID.
• JNLPBA: This dataset was provided by Kim et al. [46] and annotated on 2400 abstracts in MEDLINE database, resulting in a total of 22,402 sentences containing five entity types: DNA, RNA,cell-type, cell-line and protein.
• BC2GM: This dataset is provided by Smith et al. [47] at 2008 the BioCreative II Gene Mention Task. The entire dataset contains 20,000 sentences, and participants were asked to identify the genes mentioned in the sentences by giving the beginning and end characters of the sentences.
• LINNEAUS: This dataset is proposed by Gerner et al. [48], which is an open-source species name recognition and normalization software system. To verify the system, the authors created a LINNEAUS dataset of 100 full-text documents randomly selected from the PMC and manually annotated, resulting in 4259 species references.
• Species-800: This dataset is proposed by Pafilis et al. [49]. and the species entities were annotated manually on 800 PubMed abstracts.
Biomedical NLP researchers widely use these datasets for testing BNER models. The above datasets used in our paper are publicly available and can be downloaded from the following url:https://github.com/cambridgeltl/MTL-Bioinformatics-2016. In order to identify named entities, we use the IOB marking scheme. The detailed data of all datasets are listed in Table 2.
Table 2
Statistics of datasets
| Entity type | Dataset | The number of annotations |
|---|---|---|
| Disease | NCBI-Disease | 6,881 |
| BC5CDR-Disease | 12,694 | |
| Chemical/Drug | BC4CHEMD | 79,842 |
| BC5CDR-Chem | 15,411 | |
| Gene/Peotein | BC2GM | 20,703 |
| JNLPBA | 35,460 | |
| Species | LINNAEUS | 4,077 |
| Species-800 | 3,708 |
3.2Experiment evaluation measure
To evaluate the performance of our method, we adopt three evaluation measures: precision (P): indicates the proportion of items that should be retrieved among all retrieved items, recall (R): indicates the proportion of all retrieved items to all items that should be retrieved, F1-score (F1): indicates the weighted harmonic average of Precision and Recall. In order to more accurately identify entities in biomedical literature, we believe that the entity predicted by the model is correct only when the entity type and boundary are completely matched.
3.3Analysis of experiment result
3.3.1Comparison with different representation
In order to test the multi-feature embedding representation, our proposed method compare with two main representation methods, Word2Vec and BioBERT. The benchmark methods of downstream NER tasks all adopt method BiLSTM-CRF.
As shown in Fig. 2, the performance of the models based on the pre-training BioBERT is better than Word2Vec. The main reason is the latter uses the word vectors obtained by traditional statics embedding method and cannot handle the problems of ambiguity and polysemy.
In addition, our model outperforms in terms of F1 values due to the character-level local features and lexical features incorporated. That is because CharCNN module can extract local character-level features, which can not only represent morphological information to a certain extent, but also model biological entities with mixed case, special characters and fuzzy boundaries. The addition of lexical features adds more linguistic information to the text. Thus, the combined feature embedding method proposed in this paper improves the accuracy of the model for entities.
3.3.2Comparison with state-of-the-art studies
To evaluate the performance of the proposed method, we compare with the state-of-the-art methods including CollaboNet [29], MTM [30], BiLM [31], MTL-LS [52], BioBERT [38], DTranNER [50], BioBERT-MRC [51], Bio-XLNet-CRF [53], MT-BioNER [54].
Table 3
Comparison of BNER methods for “Gene/Protein”
| Models | Datasets | |||||
|---|---|---|---|---|---|---|
| BC2GM | JNLPBA | |||||
| P | R | F1 | P | R | F1 | |
| CollaboNet | 0.8049 | 0.7899 | 0.7973 | 0.7443 | 0.8322 | 0.7858 |
| MTM | 0.8210 | 0.8370 | 0.8407 | 0.7091 | 0.7634 | 0.7352 |
| BiLM | 0.8181 | 0.8157 | 0.8169 | 0.7139 | 0.7906 | 0.7503 |
| MTL-LS | – | – | 0.8294 | – | – | 0.7766 |
| BioBERT | 0.8432 | 0.8512 | 0.8472 | 0.7224 | 0.8356 | 0.7749 |
| DTranNER | 0.8421 | 0.8484 | 0.8456 | – | – | – |
| BioBERT-MRC | – | – | 0.8511 | – | – | 0.7845 |
| Bio-XLNet-CRF | – | – | 0.8252 | – | – | 0.7586 |
| MT-BioNER | 0.8442 | 0.8514 | 0.8478 | – | – | – |
| Ours | 0.9188 | 0.9147 | 0.9167 | 0.8662 | 0.9239 | 0.8930 |
Figure 2.
Comparison of experimental results of Word2Vec, BioBERT and our method.
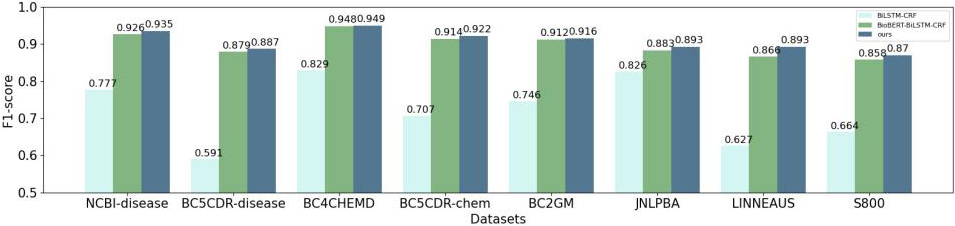
Table 4
Comparison of BNER methods for “Disease”
| Models | Datasets | |||||
|---|---|---|---|---|---|---|
| NCBI-Disease | BC5CDR-Disease | |||||
| P | R | F1 | P | R | F1 | |
| CollaboNet | 0.8548 | 0.8727 | 0.8636 | 0.8561 | 0.8261 | 0.8408 |
| MTM | 0.8586 | 0.8642 | 0.8614 | 0.8373 | 0.8293 | 0.8333 |
| BiLM | 0.8641 | 0.8831 | 0.8734 | 0.8810 | 0.9049 | 0.8928 |
| MTL-LS | – | – | 0.8897 | – | – | 0.8734 |
| BioBERT | 0.8822 | 0.9125 | 0.8971 | 0.8647 | 0.8784 | 0.8715 |
| DTranNER | 0.8821 | 0.8904 | 0.8862 | 0.8675 | 0.8740 | 0.8722 |
| BioBERT-MRC | – | – | 0.8939 | – | – | 0.8756 |
| Bio-XLNet-CRF | – | – | 0.8861 | – | – | 0.8656 |
| MT-BioNER | 0.8890 | 0.9094 | 0.8991 | – | – | – |
| Ours | 0.9273 | 0.9444 | 0.9357 | 0.9011 | 0.8752 | 0.8878 |
Results are summarized in Tables 3–6. The table reports three indicators on each dataset: precision (P), recall(R) and F1 score (F1). We use bold to show the best score, and use the underline to show the second best score. As a result, our proposed method acquire the best performance on six datasets among eight..
The reasons why our model achieves better performance are (i) It enhances the semantic representation of the entity by fusing features of different perspectives. (ii) We use multiple layers of convolution kernels in CharCNN, which can catch the non-surface information of the word from multiple angles. In addition, character level has the advantage that it can be easily generalized to all languages.
Table 5
Comparison of BNER methods for “Species”
| Models | Datasets | |||||
|---|---|---|---|---|---|---|
| LINNAEUS | Species-800 | |||||
| P | R | F1 | P | R | F1 | |
| BioBERT | 0.9077 | 08583 | 0.8824 | 0.7280 | 0.7536 | 0.7406 |
| MTL-LS | – | – | 0.8506 | – | – | – |
| Bio-XLNet-CRF | – | – | 0.8856 | – | – | – |
| Ours | 0.9489 | 0.8509 | 0.8937 | 0.8549 | 0.8864 | 0.8702 |
Table 6
Comparison of BNER methods for “Chemical/drug”
| Models | Datasets | |||||
|---|---|---|---|---|---|---|
| BC4CHEMD | BC5CDR-chem | |||||
| P | R | F1 | P | R | F1 | |
| CollaboNet | 0.9078 | 0.8701 | 0.8885 | 0.9426 | 0.9238 | 0.9331 |
| MTM | 0.9130 | 0.8753 | 0.8937 | 0.9309 | 0.8956 | 0.9129 |
| BiLM | – | – | – | 0.8810 | 0.9049 | 0.8928 |
| MTL-LS- | – | – | 0.9247 | – | – | 0.9314 |
| BioBERT | 0.9280 | 0.9192 | 0.9236 | 0.9368 | 0.9326 | 0.9347 |
| DTranNER | 0.9194 | 0.9204 | 0.9109 | 0.9428 | 0.9404 | 0.9416 |
| BioBERT-MRC | – | – | 0.9270 | – | – | 0.8939 |
| Bio-XLNet-CRF | – | – | 0.9273 | – | – | 0.9341 |
| MT-BioNER | – | – | 0.8890 | 0.9094 | 0.899 | |
| Ours | 0.9482 | 0.9512 | 0.9497 | 0.9117 | 0.9347 | 0.9227 |
As shown in Tables 3–6, it can be seen that the F1 score of our model is the highest in the medical datasets of (i) Gene/protein-related datasets, our model achieves 91.67% (6.56% improvement) in the BC2GM and 89.30% (10.72% improvement) in the JNLPBA. (ii) Disease-related datasets achieves 93.57% (5.66% improvement) in the NCBI. (iii) Chemical/drug-related datasets achieves 94.97% (2.27% improvement) in the NCBI. (iv) Species-related datasets achieves 89.37% (1.13% improvement) in the LINNAEUS and 87.02% (12.96% improvement) in the Species-800. Compared with other recent methods, the results are competitive and definite improvements can be seen on most datasets
On BC5CDR dataset, the result is not the best one because our method differentiates between subsets in BC5CDR dataset. BC5CDR dataset contains two subsets, BC5CDR-disease and BC5CDR-Chem. In the experiment, we split it into the recognition tasks of two entity categories, resulting in the loss of the relationship between the entity types in sentences.
The above excellent results are attributed to the fact that the model proposed in this paper can extract deep context word-level, local char-level, and part-of-speech features, so that the training of the word vector can better represent the grammatical and semantic information, thus improving the performance of entity recognition.
4.Conclusion
In this paper we proposed a method that fuses multi-feature embedding representations to represent words for named entity recognition in the biomedical domain. Enhanced semantic information representation of words is achieved by fusing deep contextual features, character-level local features, and lexical features of biomedical texts. Our presentation layer can help solve some problems in linguistics such as polysemy, semantics, out of vocabulary, and special entity within biomedical texts. Experiments are conducted to validate the model’s effectiveness proposed in this paper on eight datasets of four entity types in the biomedical domain. As a result, our model accomplished good performance on six BNER datasets.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Nos 61911540482 and 61702324) and the Shanghai Sailing Program (No. 21YF1416700).
Conflict of interest
None to report.
References
[1] | Kocaman V, Talby D. Biomedical named entity recognition at scale//International Conference on Pattern Recognition. Springer, Cham, (2021) ; 635-646. |
[2] | Song B, Li F, Liu Y, et al. Deep learning methods for biomedical named entity recognition: a survey and qualitative comparison. Briefings in Bioinformatics. (2021) ; 22: (6): bbab282. |
[3] | Wang Y, Tong H, Zhu Z, et al. Nested Named Entity Recognition: A Survey. ACM Transactions on Knowledge Discovery from Data (TKDD), (2022) . |
[4] | Muralikrishnan RK, Gopalakrishna P, Sugumaran V. Biomedical Named Entity Recognition (NER) for Chemical-Protein Interactions. (2021) . |
[5] | Bonner S, Barrett IP, Ye C, et al. A review of biomedical datasets relating to drug discovery: A knowledge graph perspective. arXiv preprint arXiv2102.10062, (2021) . |
[6] | Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Briefings in Bioinformatics. (2005) ; 6: (1): 57-71. |
[7] | Alshahrani M, Thafar MA, Essack M. Application and evaluation of knowledge graph embeddings in biomedical data. PeerJ Computer Science. (2021) ; 7: : e341. |
[8] | Fukuda K, Tsunoda T, Tamura A, et al. Toward information extraction: identifying protein names from biological papers//Pac symp biocomput. (1998) ; 707: (18): 707-718. |
[9] | Song M, Yu H, Han WS. Developing a hybrid dictionary-based bio-entity recognition technique. BMC Medical Informatics and Decision Making. (2015) ; 15: (1): 1-8. |
[10] | Gorinski PJ, Wu H, Grover C, et al. Named entity recognition for electronic health records: a comparison of rule-based and machine learning approaches. arXiv preprint arXiv1903.03985, (2019) . |
[11] | Erickson GM. An oligopoly model of dynamic advertising competition. European Journal of Operational Research. (2009) ; 197: (1): 374-388. |
[12] | Friedman C, Alderson PO, Austin JHM, et al. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association. (1994) ; 1: (2): 161-174. |
[13] | Li Y, Lin H, Yang Z. Incorporating rich background knowledge for gene named entity classification and recognition. BMC Bioinformatics. (2009) ; 10: (1): 1-15. |
[14] | Zhou GD, Su J. Exploring deep knowledge resources in biomedical name recognition//Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications (NLPBA/BioNLP). (2004) ; 99-102. |
[15] | Liao Z, Wu H. Biomedical named entity recognition based on skip-chain Crfs//2012 international conference on industrial control and electronics engineering. IEEE, (2012) ; 1495-1498. |
[16] | Finkel JR, Dingare S, Nguyen H, et al. Exploiting context for biomedical entity recognition: From syntax to the web//Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications (NLPBA/BioNLP). (2004) ; 91-94. |
[17] | Settles B. Biomedical named entity recognition using conditional random fields and rich feature sets//Proceedings of the international joint workshop on natural language processing in biomedicine and its applications (NLPBA/BioNLP). (2004) ; 107-110. |
[18] | Shi X, Chen Z, Wang H, et al. Convolutional LSTM network: A machine learning approach for precipitation nowcasting. Advances in neural information processing systems, (2015) ; 28. |
[19] | Lafferty J, McCallum A, Pereira FCN. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. (2001) . |
[20] | Yao L, Liu H, Liu Y, et al. Biomedical named entity recognition based on deep neutral network. Int J Hybrid Inf Technol. (2015) ; 8: (8): 279-288. |
[21] | Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging. arXiv preprint arXiv1508.01991, (2015) . |
[22] | Lyu C, Chen B, Ren Y, et al. Long short-term memory RNN for biomedical named entity recognition. BMC Bioinformatics. (2017) ; 18: (1): 1-11. |
[23] | Sun C, Yang Z, Wang L, et al. Biomedical named entity recognition using BERT in the machine reading comprehension framework. Journal of Biomedical Informatics. (2021) ; 118: : 103799. |
[24] | Gao S, Kotevska O, Sorokine A, et al. A pre-training and self-training approach for biomedical named entity recognition. PloS One. (2021) ; 16: (2): e0246310. |
[25] | Bin HU, Tianyu G, Geng D, et al. Faster biomedical named entity recognition based on knowledge distillation. Journal of Tsinghua University (Science and Technology). (2021) ; 61: (9): 936-942. |
[26] | Gridach M. Character-level neural network for biomedical named entity recognition. Journal of Biomedical Informatics. (2017) ; 70: : 85-91. |
[27] | Liu J, Chen S, He Z, et al. Learning BLSTM-CRF with Multi-channel Attribute Embedding for Medical Information Extraction//CCF International Conference on Natural Language Processing and Chinese Computing. Springer, Cham. (2018) ; 196-208. |
[28] | Patel H. Bionerflair: biomedical named entity recognition using flair embedding and sequence tagger. arXiv preprint arXiv2011.01504, (2020) . |
[29] | Yoon W, So CH, Lee J, et al. Collabonet: collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinformatics. (2019) ; 20: (10): 55-65. |
[30] | Wang X, Zhang Y, Ren X, et al. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics. (2019) ; 35: (10): 1745-1752. |
[31] | Sachan DS, Xie P, Sachan M, et al. Effective use of bidirectional language modeling for transfer learning in biomedical named entity recognition//Machine learning for healthcare conference. PMLR. (2018) ; 383-402. |
[32] | Weber L, Sänger M, Münchmeyer J, et al. HunFlair: an easy-to-use tool for state-of-the-art biomedical named entity recognition. Bioinformatics. (2021) ; 37: (17): 2792-2794. |
[33] | Sung M, Jeong M, Choi Y, et al. BERN2: an advanced neural biomedical named entity recognition and normalization tool. arXiv preprint arXiv2201.02080, (2022) . |
[34] | Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. arXiv preprint arXiv1301.3781, (2013) . |
[35] | Matthew E. Peters, Mark Neumann, Mohit Iyyer, Matt Gardner, Christopher Clark, Kenton Lee, and Luke Zettlemoyer. Deep contextualized word representations. CoRR, abs/1802.05365, (2018) . |
[36] | Devlin J, Chang MW, Lee K, et al. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv1810.04805, (2018) . |
[37] | Lan Z, Chen M, Goodman S, et al. Albert: A lite bert for self-supervised learning of language representations. arXiv preprint arXiv1909.11942, (2019) . |
[38] | Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. (2020) ; 36: (4): 1234-1240. |
[39] | Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Advances in neural information processing systems, (2017) , 30. |
[40] | Zhang X, Zhao J, LeCun Y. Character-level convolutional networks for text classification. Advances in neural information processing systems, (2015) , 28. |
[41] | Dempster AP, Nan ML, Donald BR. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society: Series B (Methodological). (1977) ; 39: (1): 1-22. |
[42] | Gales M, Young S. The application of hidden Markov models in speech recognition[M]. Now Publishers Inc, (2008) . |
[43] | Li J, Sun Y, Johnson RJ, et al. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database, (2016) , 2016. |
[44] | Krallinger M, Rabal O, Akhondi SA, et al. Overview of the BioCreative VI chemical-protein interaction Track//Proceedings of the sixth BioCreative challenge evaluation workshop. (2017) ; 1: : 141-146. |
[45] | Doğan RI, Leaman R, Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. Journal of Biomedical Informatics. (2014) ; 47: : 1-10. |
[46] | Kim JD, Ohta T, Tsuruoka Y, et al. Introduction to the bio-entity recognition task at JNLPBA//Proceedings of the international joint workshop on natural language processing in biomedicine and its applications. (2004) ; 70-75. |
[47] | Smith L, Tanabe LK, Kuo CJ, et al. Overview of BioCreative II gene mention recognition. Genome Biology. (2008) ; 9: (2): 1-19. |
[48] | Gerner M, Nenadic G, Bergman CM. LINNAEUS: a species name identification system for biomedical literature. BMC Bioinformatics. (2010) ; 11: (1): 1-17. |
[49] | Pafilis E, Frankild SP, Fanini L, et al. The SPECIES and ORGANISMS resources for fast and accurate identification of taxonomic names in text. PloS One. (2013) ; 8: (6): e65390. |
[50] | Hong SK, Lee JG. DTranNER: biomedical named entity recognition with deep learning-based label-label transition model. BMC Bioinformatics. (2020) ; 21: (1): 1-11. |
[51] | Sun C, Yang Z, Wang L, et al. Biomedical named entity recognition using BERT in the machine reading comprehension framework. Journal of Biomedical Informatics. (2021) ; 118: : 103799. |
[52] | Chai Z, Jin H, Shi S, et al. Hierarchical shared transfer learning for biomedical named entity recognition. BMC Bioinformatics. (2022) ; 23: (1): 1-14. |
[53] | Chai Z, Jin H, Shi S, et al. Noise Reduction Learning based on XLNet-CRF for Biomedical Named Entity Recognition. IEEE/ACM Transactions on Computational Biology and Bioinformatics, (2022) . |
[54] | Tong Y, Chen Y, Shi X. A multi-task approach for improving biomedical named entity recognition by incorporating multi-granularity information//Findings of the Association for Computational Linguistics: ACL-IJCNLP. (2021) ; 2021: : 4804-4813. |




