MR brain tissue classification based on the spatial information enhanced Gaussian mixture model
Abstract
BACKGROUND:
Classifying T1-weighted Magnetic Resonance brain scans into cerebrospinal fluid, gray matter and white matter is one of the most critical tasks in neurodegenerative disease analysis. Since manual delineation is a labor-intensive and time-consuming process, automated methods have been widely adopted for this purpose. One group of commonly used method by biomedical researchers are based on Gaussian mixture model. The main drawbacks of this model include complex computational cost and parameter selection with the presence of imaging defects such as intensity inhomogeneity and noise.
OBJECTIVE:
To alleviate these aspects, an improved Gaussian mixture model-based method is proposed in this work.
METHODS:
Standard mixture model was used to formulate individual voxel intensity. A set of spatial weightings were created to represent local tissue characteristics. The emphasis of this method is its “lite” and robust implementation mode highlighted by a dedicated entropy term. The Expectation-Maximization algorithm was then iteratively executed to estimate model parameters. The Maximum a Posteriori criterion was employed to determine for each voxel if it belongs to a certain tissue.
RESULTS:
The proposed method was validated on both simulated and real MR scans. The averaged Dice coefficient of segmented brain tissues on each dataset ranged between [66.41, 87.42] for cerebrospinal fluid, [80.57, 85.35] for gray matter, and [83.17, 85.63] for white matter.
CONCLUSIONS:
Experiments illustrated the effectiveness and reliability in tissue classification against imaging defects compared with manually constructed reference standard.
1.Introduction
Neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease, are prevalent chronic neuro-system disorders among middle-aged and elderly people. The disorders are complicated and progressive, constituting the leading cause of death world-wide. The societal burden of such diseases is enormous [1].
Magnetic Resonance Imaging, in particular the T1-weighted MRI, is of vital importance in medical practice to track the mechanisms underlying neuro-degenerative diseases. It is generally believed that the cerebral atrophy could characterize such disorders as a loss of neurons and connection between structures. Quantification of tissue atrophic changes, i.e., cerebrospinal fluid (CSF), gray matter (GM) and white matter (WM), plays a prominent role to investigate brain disorder state, progression, and treatment strategy [2].
Manual tissue delineation by radiologists is immensely time-consuming and laborious. Therefore, much attention has been attracted to the research of automated and accurate MR segmentation methods [1, 2].
There are already many techniques to obtain segmentation results, which could be roughly categorized into region-based, shape-based, machine learning-based and classification-based ones. Although published for years, they still suffer from a variety of issues. Region-based methods run growing model through voxels under tissue connection and intensity homogeneous criterion [3]. Shape-based methods build specific anatomical structures by employing a prior information concerning contour, location, and orientation [4]. Both mentioned above are challenged due to the sensitivity to imaging noise and intensity inhomogeneity, or the so-called bias field, which leads to less accurate tissue boundaries. Machine learning-based methods use classifiers or deep models to separate tissues by dimensions of image features; however, their implementation requires extremely large computation resources [5].
In classification-based segmentation, unsupervised clustering has been widely adopted such as Fuzzy C-Means and finite mixture models, among which the Gaussian mixture model, GMM, is the most recognized [6]. GMM methods formulate tissue intensity as independent Gaussian distribution, and mix several distributions as a uniformity. Thus, they provide the capability to characterize individual voxel into multiple tissues before final decision. For further performance enhancement or time-cost reduction, tons of efforts have been made over the past two decades to research the extension of GMM. Image spatial information has been designed for representing tissue prior distributions inferred from neighborhood voxels instead of constant values. Some recent publications further improve previous works. For instance, Guerrout et al. [7] described the hidden Markov random field theory and Broyden-Fletcher-Goldfarb-Shanno for tissue labeling. Azimbagirad et al. [8] proposed a method based on intensity histogram dedicated to Alzheimer’s diseased brains. Ji et al. [9] used rough conception to incorporate with spatial relationship for overcoming noise.
Unfortunately, GMM methods are still challenged by mentioned imaging defects. Various initialization parameter choices result in quite different segmentations, and complex pipeline settings also increase computation consumption. This work proposes an improved GMM-based tissue classification method to alleviate the disadvantages of existing similar methods. First, standard GMM was reviewed for demonstrating the basic theory. Second, a novel spatial weighting was introduced to determine the tissue prior distribution in GMM; this spatial term was inspired by entropy measuring volume label complexity, which played an important role in ensuring result quality and acceleration. Finally, an Expectation-Maximization (EM) algorithm estimated the model parameters iteratively under the Maximum a Posteriori (MAP) criterion.
The contributions of this model can be summarized as these statements: (1) high accuracy benefits from iterative correction of mis-labeled voxels from biased boundaries and isolated “noisy” tissues; (2) a “lite” implementation allows low computational consumption; (3) all model parameters were self-adaptively obtained for better robustness to imaging defects without complex tuning steps.
The remainder of this paper is organized as follows. Section 2 details the methods. Section 3 describes the data, experiments and provides quantitative results and discussion. Finally, Section 4 draws conclusions.
2.Method
2.1Standard Gaussian mixture model
Let
(1)
with
The overall prior probability of tissue
(2)
Hence, the joint probability of
(3)
From the Bayesian rule [10], the posterior probability of
(4)
Finally,
(5)
2.2Entropy-enhanced spatial information
In most cases, T1-weighted MR imaging suffers from bias field and noise during scanning. Such kind of defects will significantly impact the classification accuracy. The Markov Random Field has revealed that a voxel label could only be determined by its neighborhoods [7]. Thus, most of the attention in this work has been drawn to enhance the GMM via optimal choices of the prior distribution in Eq. (2) by maximizing the spatial image information.
As mis-classification takes place, wrong labels intersperse in real labels, resulting in intricate tissue boundaries. Therefore, this subsection elaborates on an entropy weighting
(6)
while
To completely take use of the information from the non-isotropic scan itself as well as the classification procedure, both prior and posterior probability distribution terms are introduced back to further combine spatial classification results. For voxel
(7)
while
After global normalization across the whole scan, the eventual prior distribution
(8)
Hence, when label interacts and increased
2.3Parameter estimation
A K-Means step activates the whole procedure to generate the initial GMM intensity average and standard deviation
(9)
Here,
(10)
Normally, the EM algorithm is used here for parameter optimization [15]. The E-step obtains the posterior probability of tissue
(11)
in which
(12)
In the M-step, the derivative of Eq. (9) is assigned to be 0, and the (
(13)
The iteration terminates as the absolute value of successive log-likelihood in Eq. (9) is no larger than 0.001.
3.Experiments, results, and discussion
3.1Data
Table 1
Detailed information of the experimental dataset
| Dataset | Scan | Dimension | Voxel spacing (mm | Slice | Slice thickness (mm) |
|---|---|---|---|---|---|
| BrainWeb | 15 | 181 | 1.0 | 20–181 | [1.00, 9.00] |
| IBSR | 18 | 256 | [0.84, 1.00] | 256 | [0.84, 1.00] |
| MRBrainS13 | 5 | 240 | 0.96 | 48 | 3.0 |
Three public benchmark datasets, explained in Table 1, were used for evaluation:
• Synthetic MR scans from BrainWeb database hosted by McGill University [16]. A subset of 15 scans were randomly selected, including {1.0, 3.0, 5.0, 7.0, 9.0} mm slice thickness, {0%, 1%, 3%, 5%, 7%, 9%} noise level and {0%, 20%, 40%} bias field level. The annotation is also publicly available.
• The Internet Brain Segmentation Repository (IBSR) [17]. IBSR is affiliated to the NeuroImaging Tools and Resources Collaboratory and consists of 18 real clinical MR volumes associated with expert manual segmentation.
• The 2013 MICCAI MR Brain Segmentation Workshop (MRBrainS13) [18]. The organizers from Utrecht University launched a set of real multi-modal brain scans and ground-truth, in which the 5 training scans were selected for this study. Only T1-weighted data was adopted, and multiple labels were merged into CSF, GM and WM tissues in this section.
3.2Experiment scheme
The whole pipeline was implemented using C
(14)
Three experiments were designed to show the practical performance of the proposed method. Experiment 1 focused on the classification accuracy compared with a set of state-of-the-art GMM-based methods. Experiment 2 reviewed the robustness performance for validating the robustness under different imaging defects and protocols. Experiment 3 demonstrated the acceleration ability.
3.3Experiment 1
Table 2 shows the accuracy comparison summary of the proposed method versus three other methods, which are: Method A based on [7], Method B based on [8] and Method C based on [9]. Individual tissue is listed with the average and standard deviation of
Table 2
Accuracy comparison of the proposed method with existing methods
| BrainWeb | IBSR | MRBrainS13 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CSF | GM | WM | CSF | GM | WM | CSF | GM | WM | |
| Method A | 77.30 | 82.44 | 83.98 | 40.01 | 84.90 | 84.54 | 62.80 | 77.70 | 78.20 |
| Method B | 80.29 | 82.80 | 83.89 | 62.07 | 84.68 | 84.85 | 68.03 | 80.09 | 82.76 |
| Method C | 79.65 | 83.50 | 82.73 | 64.72 | 85.21 | 86.61 | 85.03 | 82.09 | 81.78 |
| The proposed method | 81.13 | 85.28 | 85.46 | 66.41 | 85.35 | 85.63 | 87.42 | 80.57 | 83.17 |
Each method was capable of obtaining an acceptable result, but there are still some differences. The proposed method feedbacked better performance over the other methods as bold font indication. Only a tiny
Figure 1.
Example classification results: (from left to right) original MR slice, result of Method A, B, C, the proposed method, and the ground truth. The three rows indicate example BrainWeb (3 mm thickness, 1% imaging noise and 20% bias field level), IBSR, and MRBrainS13 slice, respectively. CSF, GM and WM are displayed by dark gray, light gray and white color.
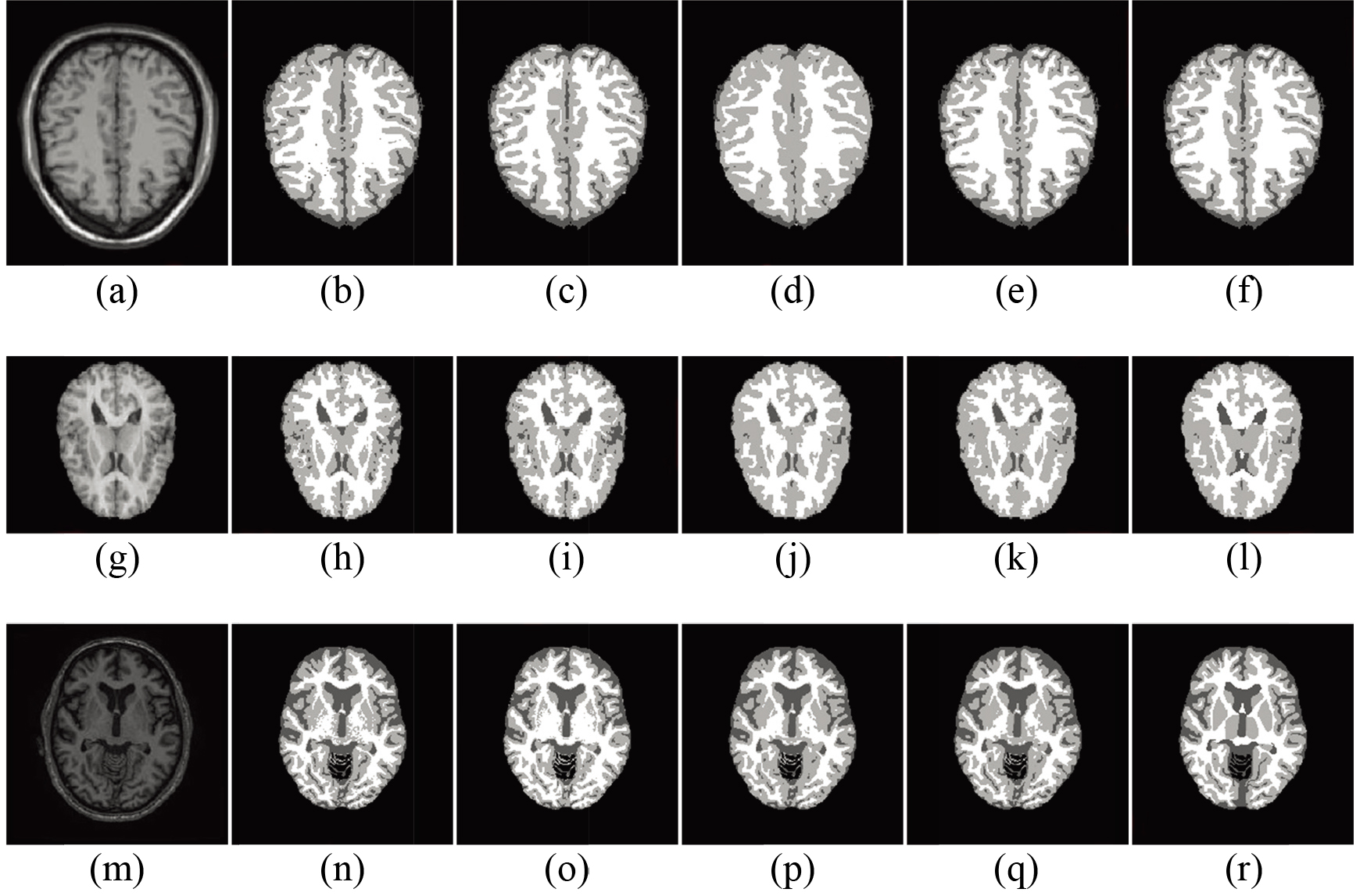
Figure 1 shows the example slices with respect to MR scan and ground truth. For BrainWeb subset, 4 compared methods revealed similar results referred to the ground truth. The presented work obtained minor advantage: no isolated tissue label was found by Method A due to imaging noises; Method B and C both detected less CSF than the proposed one when facing with intensity bias field. In IBSR comparison, tissue bias field again increased classification difficulty. Visually, Method A and B failed to precisely distinguish CSF from GM. The proposed method apparently outperformed Method C with better results, especially at the putamina and thalami. For MRBrainS13, Method B could not identify putamen and globus pallidus GM. Method A and C had improved performance but suffered from considerable label noise or thalami GM mis-labeling.
The first GMM iteration of the proposed pipeline is activated from a “simple” input generated by KMeans and tissue prior probability hypothesis, which is further detailed in Subsection 3.4 for final parameter initialization choice discussion. The pipeline is driven forward along the optimization path under global and local constraints. The spatial information employs a combination of posterior term for global constraint characterization and prior term for local characterization in Eq. (8). Definitely, an important balance should be carried out to combine both constraints for optimal classification in the meanwhile. Since both isolated tissue patches and biased boundaries by imaging defects result in label junction areas, indicating higher local label complexity in tissue neighborhood. Therefore, the voxel entropy in Eq. (6) is chosen as a ratio weighting after value normalization across the entire volume. The entropy guarantees the potential to approach classification optima gradually. The desired
3.4Experiment 2
The diversity of device vendors, scanning protocols, and imaging defects results in various intensity distributions and noise levels. This prevents GMM from maximizing its advantages when attempting to use one universal parameter initialization with high fitness. To evaluate independence on parameter choice, the pipelines were implemented 10 times with different KMeans centers and initial model prior probability in Eq. (2) by a pseudo-random function.
Figure 2.
Robustness performance under different parameter initialization choices.
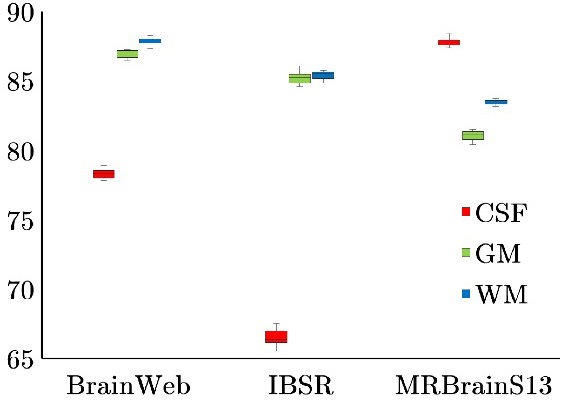
Tissue
Previous literature reported that multiple parameters in either KMeans or GMM led to entirely biased solution domains, and contrast significantly over arbitrary initializations [6]. Moreover, improper spatial information further trapped GMM by local optima in parameter estimation. On the contrary, the proposed method had distinct advantages in its robustness to initialization stage. That owes to the whole spatial information which focuses on performance improvement near tissue boundaries. The tissue label of each voxel cannot be simply determined unless the assembly weightings of its 26-neighbors are all taken into account. No matter how different parameter choices have been input, when label diversity is found, the entropy term will gradually find the solution that could lead to relatively consistent tissue boundaries. Only a few more iterations would take place to convergent results after continuous corrections.
For easy explanation, the eventual implementation set the KMeans centers as the 1/6, 3/6, and 5/6 fractional section of brain intensity range, and, arbitrary voxel tissue prior probability as 1/3. This was finally used in Experiment 1.
3.5Experiment 3
The execution time (in second) of tested methods is listed in Table 3. It is worth mentioning that the proposed method achieved the highest computation efficiency in this test compared with other GMM-based methods.
Table 3
Time-cost comparison of the proposed method with existing methods
| BrainWeb | IBSR | MRBrainS13 | |
|---|---|---|---|
| Method A | 150.3 | 183.4 | 145.0 |
| Method B | 410.6 | 430.1 | 473.9 |
| Method C | 21.3 | 23.8 | 22.3 |
| The proposed method | 13.8 | 12.0 | 11.3 |
Originally, less attention had been paid to cope with the time cost, which commonly came from complicated calculation of the prior probability in Eq. (2). Authors tended to compromise to higher accuracy performance as the statement in [9]. Other techniques, such as parallel computing, might improve pipeline efficiency. In this work, the dedicated acceleration term in Eq. (12) enables a remarkably faster convergence of GMM iterations. It should be emphasized here that unlike additional items summed in log-likelihood in Eq. (9), this term is combined directly as weighting enhancement together with the E-step to obtain tissue posterior probability. That still contributes to a stable EM format, and does not bring in other burdens in parameter estimation procedures. Besides, the proposed entropy also decreases complex probability evaluation inside uniform label regions. The nature of entropy has more influence on the correction of mis-classified tissues.
To sum up, the proposed method yields more competitive performance. The key point is the entropy-enhanced spatial term to balance the ratio of prior and posterior probability. The posterior one dominates when various tissue label occurs in voxel neighborhood, and the prior one dominates in uniform labels. Hence, the effect of spatial information improves the resulting quality around tissue boundaries, where mis-classification is more likely to take place.
4.Conclusion
This paper elaborates on an improved GMM-based tissue classification method from T1-weighted MR brain scans. The core of the method is a spatial information enhanced scheme, using a combination of entropy-weighted tissue prior probability and posterior probability. This entropy weighting, deriving from neighborhood classification results and image spatial resolution, contributes to an optimal ratio to incorporate both probability terms. The major highlights of the entropy are its “lite” and robust implementation attributes, which realize a range of reductions of computing burdens and parameter tuning steps. The term mainly influences the area next to biased tissue boundary and imaging noise, and continuously drives the whole pipeline to finalize uniform classification results. Experimental tests were carried out on publicly available datasets with tissue reference. Compared with a number of state-of-the-art GMM-based methods, the proposed method achieved high accuracy and acceleration performance, and especially excellent robustness to any choice of initialization parameters.
Conflict of interest
None to report.
References
[1] | Lorio S, Lutti A, Kherif F, Ruef A, Dukart J, Chowdhury R, Frackowiak RS, Ashburner J, Helms G, Weiskopf N, Draganski B. Disentangling in vivo the effects of iron content and atrophy on the ageing human brain. Neuroimage. (2014) ; 103: : 280-9. |
[2] | Varnum MM, Ikezu T. The classification of microglial activation phenotypes on neurodegeneration and regeneration in Alzheimer’s disease brain. Archivum Immunologiae et Therapiae Experimentalis. (2012) ; 60: (4): 251-66. |
[3] | Zhao M, Lin HY, Yang CH, Hsu CY, Pan JS, Lin MJ. Automatic threshold level set model applied on MRI image segmentation of brain tissue. Applied Mathematics & Information Sciences. (2015) ; 9: (4): 1971-80. |
[4] | Cremers D, Rousson M, Deriche R. A review of statistical approaches to level set segmentation: Integrating color, texture, motion and shape. International journal of computer vision. (2007) ; 72: (2): 195-215. |
[5] | Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: A review. Neurocomputing. (2016) ; 187: : 27-48. |
[6] | Balafar MA, Ramli AR, Saripan MI, Mashohor S. Review of brain MRI image segmentation methods. Artificial Intelligence Review. (2010) ; 33: (3): 261-74. |
[7] | Guerrout EH, Ait-Aoudia S, Michelucci D, Mahiou R. Hidden Markov random field model and Broyden-Fletcher-Goldfarb-Shanno algorithm for brain image segmentation. Journal of Experimental & Theoretical Artificial Intelligence. (2018) ; 30: (3): 415-27. |
[8] | Azimbagirad M, Simozo FH, Senra Filho AC, Junior LO. Tsallis-entropy segmentation through MRF and Alzheimer anatomic reference for brain magnetic resonance parcellation. Magnetic Resonance Imaging. (2020) ; 65: : 136-45. |
[9] | Ji Z, Huang Y, Xia Y, Zheng Y. A robust modified Gaussian mixture model with rough set for image segmentation. Neurocomputing. (2017) ; 266: : 550-65. |
[10] | Rosen KH, Krithivasan K. Discrete mathematics and its applications: with combinatorics and graph theory: Tata McGraw-Hill Education; (2012) . |
[11] | Gauvain JL, Lee CH. Maximum a posteriori estimation for multivariate Gaussian mixture observations of Markov chains. IEEE Transactions on Speech and Audio Processing. (1994) ; 2: (2): 291-8. |
[12] | Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans on Medcal Imaging. (2001) ; 20: : 45-57. |
[13] | Shannon CE. A note on the concept of entropy. Bell System Technical Journal. (1948) ; 27: (3): 379-423. |
[14] | Krishna K, Murty MN. Genetic K-means algorithm. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics). (1999) ; 29: (3): 433-9. |
[15] | Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society: Series B (Methodological). (1977) ; 39: (1): 1-22. |
[16] | BrainWeb, http://brainweb.bic.mni.mcgill.ca/brainweb. |
[17] | IBSR: Tool/Resource Info – NITRC, https://www.nitrc.org/projects/ibsr. |
[18] | The MRBrainS13 Workshop, https://mrbrains13.isi.uu.nl/. |
[19] | Dice LR. Measures of the amount of ecologic association between species. Ecology. (1945) ; 26: (3): 297-302. |




