Dense deconvolution net: Multi path fusion and dense deconvolution for high resolution skin lesion segmentation
Abstract
BACKGROUND:
Dermoscopy imaging has been a routine examination approach for skin lesion diagnosis. Accurate segmentation is the first step for automatic dermoscopy image assessment.
OBJECTIVE:
The main challenges for skin lesion segmentation are numerous variations in viewpoint and scale of skin lesion region.
METHODS:
To handle these challenges, we propose a novel skin lesion segmentation network via a very deep dense deconvolution network based on dermoscopic images. Specifically, the deep dense layer and generic multi-path Deep RefineNet are combined to improve the segmentation performance. The deep representation of all available layers is aggregated to form the global feature maps using skip connection. Also, the dense deconvolution layer is leveraged to capture diverse appearance features via the contextual information. Finally, we apply the dense deconvolution layer to smooth segmentation maps and obtain final high-resolution output.
RESULTS:
Our proposed method shows the superiority over the state-of-the-art approaches based on the public available 2016 and 2017 skin lesion challenge dataset and achieves the accuracy of 96.0% and 93.9%, which obtained a 6.0% and 1.2% increase over the traditional method, respectively.
CONCLUSIONS:
By utilizing Dense Deconvolution Net, the average time for processing one testing images with our proposed framework was 0.253 s.
1.Introduction
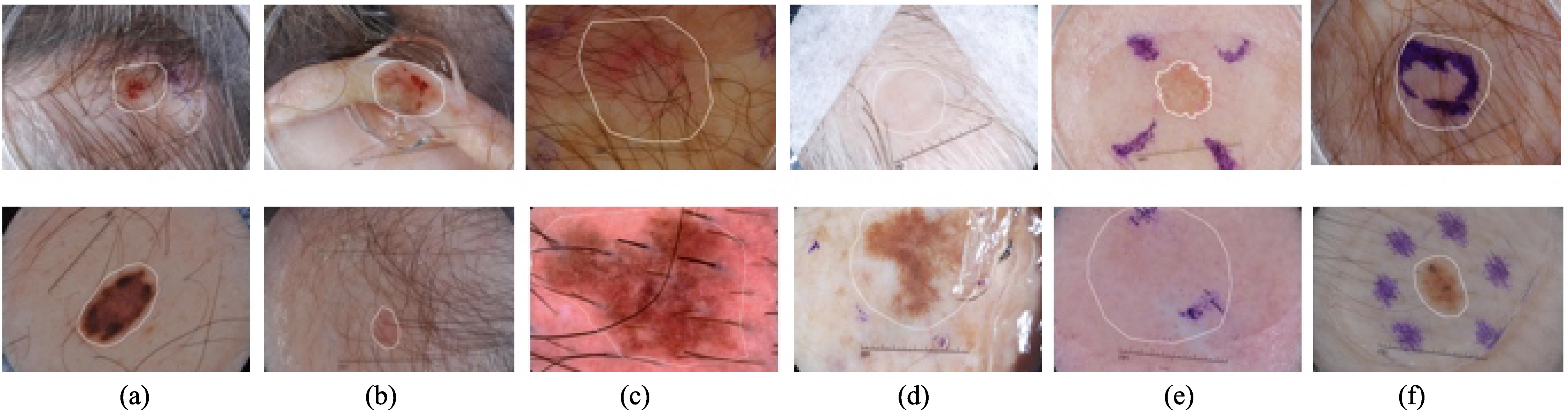
According to cancer statistics released by the American Cancer Society, melanoma increasing at a growth rate of 14% and skin cancer has a death rate of 75% [1, 2]. However, these diseases are curable if they are diagnosed timely and treated properly. Clinically, dermoscopy is adopted to assist dermatologists in classifying melanoma from nevi. The recent reports suggest that the human visual inspection only depends on ‘Ugly Duckling’ sign or experience [3]. Therefore, valid morphological features are required for the accurate segmentation. The manual segmentation process is often labor-intensive and subjective. However, clinicians who acquired the adequate levels of expertise are scarce in unprivileged countries. Otherwise, the blurry and irregular boundary degrades the segmentation accuracy. Figure 1 illustrates the main challenges for accurate diagnosis. Moreover, segmentation is critical in reducing screening errors and aiding the identification of benign and malignant melanoma. For example, deep polynomial networks can achieve more decent performance by leveraging the result of segmentation [4, 5, 6]. Automated dermoscopy image analysis is an effective way to tackle these problems.
Figure 1.
Illustration of challenges of automatic segmentation of skin lesions in dermoscopy images. The main challenge includes distinguishable inter-class, indistinguishable intra-class variations, artifacts and inherent cutaneous features in natural images. (a–c) skin lesions are covered with hairs or exploded with blood vessels; (d) air bubbles and marks occlude the skin lesions; (e–f) dye concentration downgrades the segmentation accuracy. Note that white contours indicate the skin lesions.
For automatic quantitative analysis of skin lesion, deep learning has attracted intensive attention and become a focus due to its ability to boost performance [6, 7]. Currently, the most popular deep learning method is convolutional network (CNN) [8, 9]. However, the outputs of multi-scale CNN are too coarse to fulfill the requirement of segmentation. A new type of CNN, fully convolutional network (FCN) has witnessed a great success and achieved unprecedented progress in the development of segmentation. FCN allows researchers to concentrate on network architecture design without sophisticated pre-processing and post-processing algorithms.
Meanwhile, recent studies demonstrate that increasing network depth can further boost performance due to the discriminative representations from deep layers of a network [10]. The latest generation CNN deep residual neural network (ResNet) is proposed by He et al. [11], which outperforms state-of-the-art techniques in classification task by solving vanishing/exploding gradient problem which inhibits convergence. However, ResNet has degraded segmentation accuracy due to the contradiction between classification and localization. When the network gets deeper, the spatial resolution of the feature maps (layer outputs) decreases significantly when CNN is employed in a fully convolutional way. In addition, obtaining spatial transformation invariant features for a classifier needs to discard local information. Despite the use of CNN, there are still significant differences between the result of automatic segmentation and the dermatologist’s delineation.
Many previous works have concentrated on solve this problem and obtained promising segmentation performance. The typical works include deconvolution and the conditional random field (CRF) algorithm. Deconvolution, also named as transposed convolution, has been widely used in deep-learning based method with up-sampling requirement. In encoder-decoder architecture, the coarse probability maps flow through the stacked deconvolution path to recover spatial resolution. Specifically, fully connected residual network (FCRN) adopted the encoder-decoder based network to boost performance [12]. Also, Krähenbühl and Koltun [13] adopted the CRF to integrate all pairs of pixels on an image to leverage spatial low-level information since obtaining spatial transformation invariant features through down-sampling path needs to discard local information.
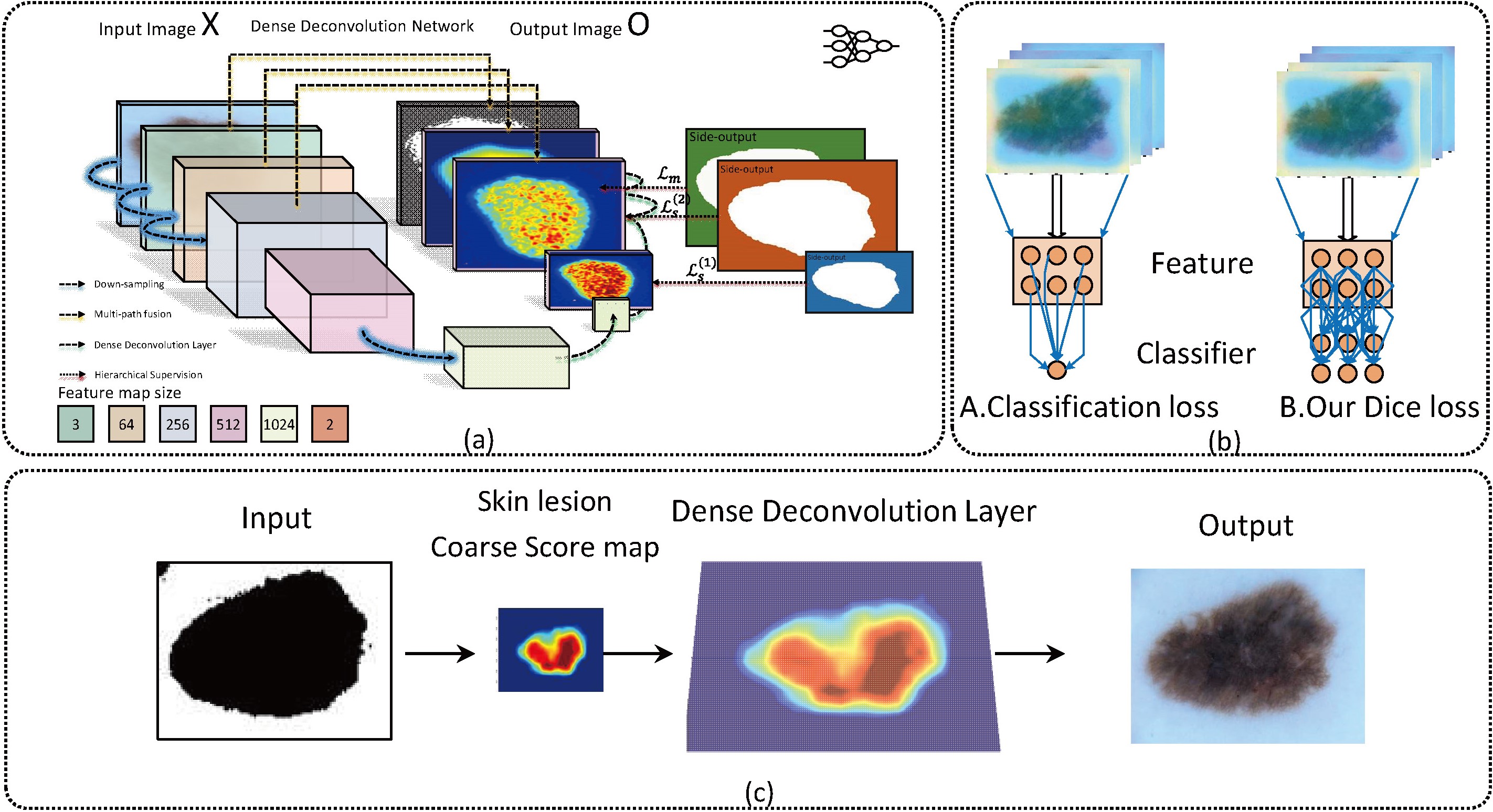
Although traditional deconvolution layers and CRF have witnessed their capability for reconstruction, they are trapped in ‘checkerboard’ problem [14, 15]. Also, the methods with the CRF cannot train the network in an end-to-end way and lack the ability of semantic interpretation. These previous works have their inherent limitations. To address the limitations, we construct a network in this paper using multi-path features for segmentation with direct relationship between the intermediate probability maps. The combination of the deep refinement network and dense deconvolution layer has been proposed for segmenting dermoscopy in this paper. Figure 2 illustrates the flowchart of our proposed method.
Figure 2.
Flowchart of the proposed network architecture for melanoma segmentation; (a) Multi-path processing to fuse various contrast information with deep supervision; (b) Comparison of Dice loss and Classification loss; (c) dense deconvolution layer further refines the contour.
2.Methodology
2.1Residual block
Our network starts with residual block inspired by ResNet. The first part of residual block contains an adaptive convolution layer via fine-tuning to maintain the weights of pretrained ResNet for our task. The main part of ResNet is the skip-connection and residual block. To train a very deep network for segmentation, we leverage a novel method named residual learning. The characteristics of residual learning are the skip connection to refine the flow of gradient. Also, a combination of multiple skip-connection structure has demonstrated an evident activity of early layers. Accordingly, the residual learning can speed up the convergence of deep network and maintain accuracy gains by substantially increasing the network depth. Moreover, we also adopt the dropout mechanism, batch normalization, and careful initialization to address the problem of gradient vanishing. A residual block with identity mapping is formulated as
(1)
where
By directly applying skip-connection, our network suffers from the drawback of the small receptive field. To tackle it, each input path is followed by two residual blocks, which are similar to that in the original ResNet. In our architecture, the filter channel for each input path is set to 512, and the remaining ones are fixed to 256 for keeping dimension balanced in our experiments.
2.2Dense deconvolution block
Since FCRN and U-Net still suffer from down-sampling and losing fine structure, a coarse-to-fine strategy is developed to recover high resolution [16, 17]. Instead of fusing all path inputs into the highest resolution feature map at once, we only integrate two features with adjacent size in each stage. This layer first performs convolutions for input adaptation, which generates feature maps of the same number, and then uses dense deconvolution layer to interpolate smaller feature maps with the largest resolution of the inputs. All feature maps are fused by summation.
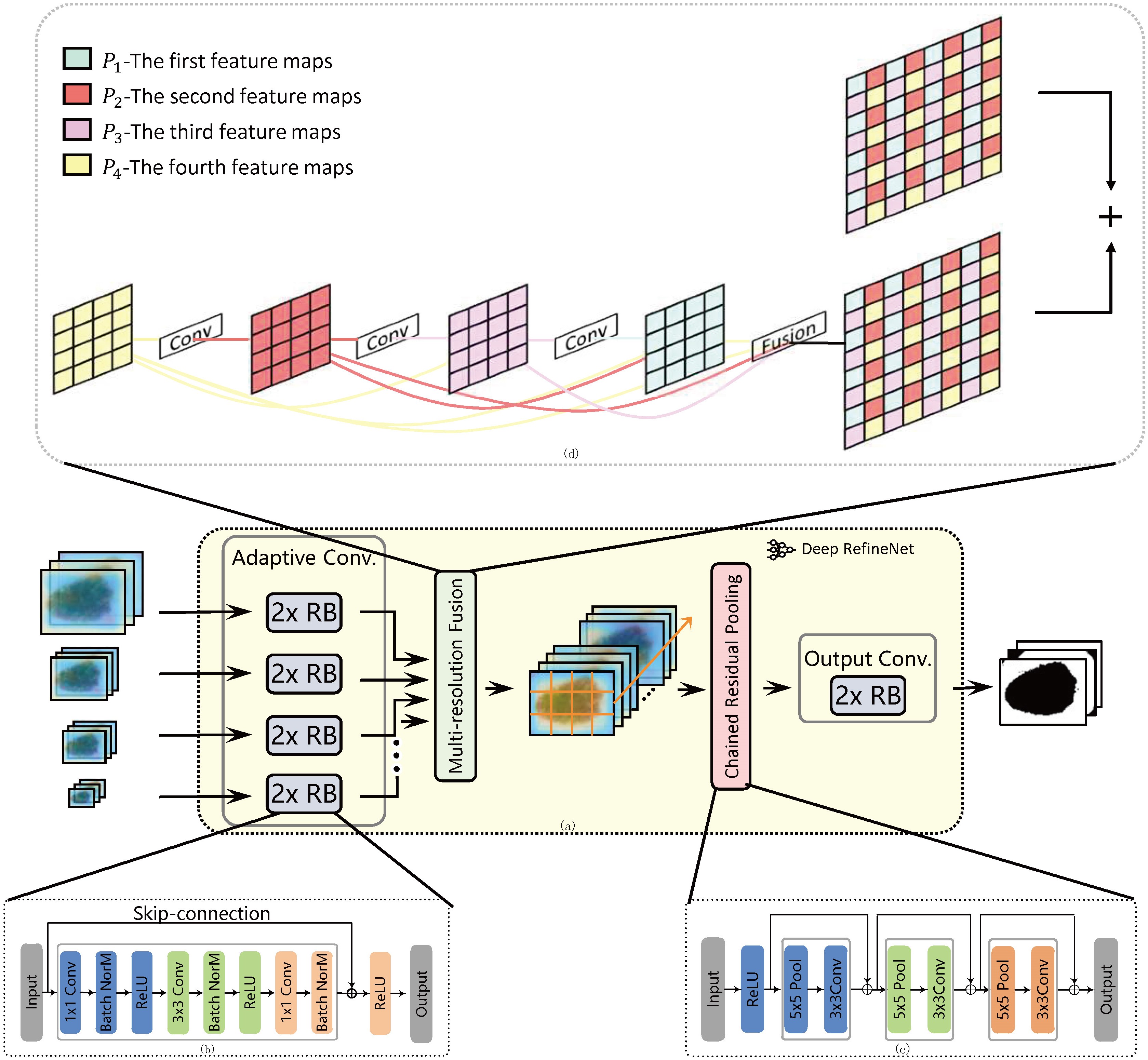
During the dense deconvolution layer, we perform the up-sampling to combine two path inputs together. In this paper, dense deconvolution layer explores a two-layer strategy named dense deconvolution layer to improve the process of multi resolution fusion, namely dense deconvolution layer and fusion layer. Dense deconvolution layer is shown in Fig. 3.
Figure 3.
Illustration of proposed method; (a) Deep RefineNet; (b) residual block; (c) chained residual pooling; (b) dense deconvolution block.
2.2.1Dense generation layer
The first step of dense deconvolution layer is to generate intermediate probability maps and add the relationship among them [18]. Note that Dense Generation Layer generates intermediate probability maps one-by-one rather than simultaneously since adjacent pixels are from different intermediate probability maps. In this method, we introduce dense connection to form any intermediate probability map from all subsequent intermediate probability maps. The relationship among intermediate probability maps not only adds dependence among intermediate probability maps, but also enhances the information flow. Therefore, there isn’t checkerboard phenomenon in the final result. Given
(2)
where
2.2.2Fusion layer
After generating four intermediate probability maps, we interweave them together to get the final probability maps Assuming that (
(3)
(4)
2.3Chained residual pooling block
After the size observation and analysis of skin lesions, there is rich contextual information. Recently, pooling can utilize global image-level feature to support our research. Feature map passes sequentially to the chained residual pooling block, and is schematically depicted in Fig. 3. Fused feature map passes sequentially one max-pooling layer and one convolution layer. Nevertheless, pooling once needs large pooling window in network for the segmentation task. The proposed chained residual pooling method concatenates pooling layers as a chain with learnable weight. Noted that ReLu is adopted to improve the pooling efficiency. During the course of training, two pooling blocks are adopted and each is with stride 1.
The output of all pooling blocks is concatenated together with the input feature map by summation of skip-connection. We employ skip-connections to facilitate gradient propagation during training. In one pooling block, each pooling operation is followed by convolutions, which serve as a weighting layer for the summation fusion. We expect that chained residual pooling can be learnable to identify the importance of pooling layers during the training course.
2.4Dense deconvolution net with hierarchical supervision
The first phase of segmentation is to obtain every pixel’s prediction. Since the output of ResNet is a probability of each class being designed for classification, a multi-prediction layer should replace the single label prediction layer. Hence, segmentation can be viewed as a dense prediction problem. The final prediction is based on feature maps from various receptive fields. The requirement of complex boundary delineation is met by local intensity information which only appears early in forward propagation. The main idea of the proposed dense deconvolution net is shown in Fig. 3.
Directly training such deep network may cause difficulty in optimization due to the issue of vanishing gradients. Motivated by the previous studies on training neural networks with deep supervision, we utilize four side-output layers in our net to supervise early layers [19]. Each side-output layer is in charge of one size of feature maps. However, explosion at high level side-output layers does not work well. By probing the outputs from the first side-output layer associated with the smallest feature maps, we cannot find any cue between the dense prediction and ground truth. The underlying reason for this is that the small skin lesions cannot be captured. Hence, we only exploit side-output layers on the last two layers. To address the above-mentioned problems, we combine all side-output layers and final-output layers, which is formulated as
(5)
(6)
where the first part is side-output loss terms and another one is main function between the predicted results and ground truth,
The skin lesions occupy only small regions, and the learning process is trapped into local minima. Therefore, we fit dermoscopy images’ characteristics by taking different loss functions into account. Therefore, our network is defined with per-pixel categorization [20]. The main loss layer is formulated as
(7)
where
3.Experiments and results
3.1Dataset and implementation
In this study, we perform the experiment to evaluate performance of our purposed method using the public challenge datasets – ISBI skin lesion segmentation dataset released in 2016 and 2017. The skin images are based on the International Skin Imaging Collaboration (ISIC) Archive, acquired from various international clinical centers’ devices. The dataset released in 2016 consists of 900 images for training and 350 images for validation, which are extended to 2000 and 600 respectively in 2017. Due to independent evaluation, the organizer excludes the ground truth of the validation part.
Our algorithm is implemented in MATLAB R2014b based on MatConvNet with a NVDIA TITAN X GPU. We utilize stochastic gradient descent (SGD) to optimize our objective function. The mini-batch involves 20 images. We adopt a weight decay of 0.0001 and a momentum of 0.9. The learning rate is 0.00001 for the first 300 epochs and 0.000001 for the next 300 epochs. Apart from the hyper-parameters set above, we incorporate the dropout layers (dropout rate is set as 0.5) in our network to prevent co-adaptation of feature detectors. When we train Deep RefineNet, we decouple our algorithm to two stages due to computational efficiency We train our network without CRF during the first step During the second step, we fix our network’s parameters. For the CRF parameters, we assume that the unary terms are fixed.
Table 1
Segmentation results of ISBI 2016 and 2017 dataset
| Network | Parameter | DC | JA | AC | Dataset | DC | JA | AC | Dataset | Time |
|---|---|---|---|---|---|---|---|---|---|---|
| FCRN [16] | 91M | 0.689 | 0.816 | 0.905 | 2016 | 0.814 | 0.721 | 0.928 | 2017 | 0.153 |
| RN | 410M | 0.890 | 0.824 | 0.941 | 2016 | 0.828 | 0.735 | 0.931 | 2017 | 0.245 |
| RN-CRF | 410M | 0.908 | 0.841 | 0.952 | 2016 | 0.830 | 0.741 | 0.934 | 2017 | 0.320 |
| RN-ML-CRF | 410M | 0.924 | 0.860 | 0.956 | 2016 | 0.843 | 0.758 | 0.938 | 2017 | 0.343 |
| RN-ML-DDL | 412M | 0.931 | 0.871 | 0.960 | 2016 | 0.845 | 0.761 | 0.939 | 2017 | 0.253 |
Table 2
Segmentation algorithm comparison based on ISBI 2016 and 2017 dataset
| Method | DC | JA | AC | Dataset | Method | DC | JA | AC | Dataset |
|---|---|---|---|---|---|---|---|---|---|
| EXB | 0.910 | 0.843 | 0.953 | 2016 | ResNet [21] | 0.842 | 0.758 | 0.934 | 2017 |
| CUMED | 0.897 | 0.829 | 0.949 | 2016 | RECOD [22] | 0.839 | 0.754 | 0.931 | 2017 |
| Mahmudur | 0.895 | 0.822 | 0.952 | 2016 | FCN [12] | 0.837 | 0.752 | 0.930 | 2017 |
| SFU-mial | 0.885 | 0.811 | 0.944 | 2016 | SMCP [23] | 0.839 | 0.749 | 0.930 | 2017 |
| UiT-Seg | 0.881 | 0.806 | 0.939 | 2016 | INESC TECNALIA [24] | 0.810 | 0.718 | 0.922 | 2017 |
| Our proposed | 0.931 | 0.871 | 0.960 | 2016 | Our proposed | 0.845 | 0.761 | 0.939 | 2017 |
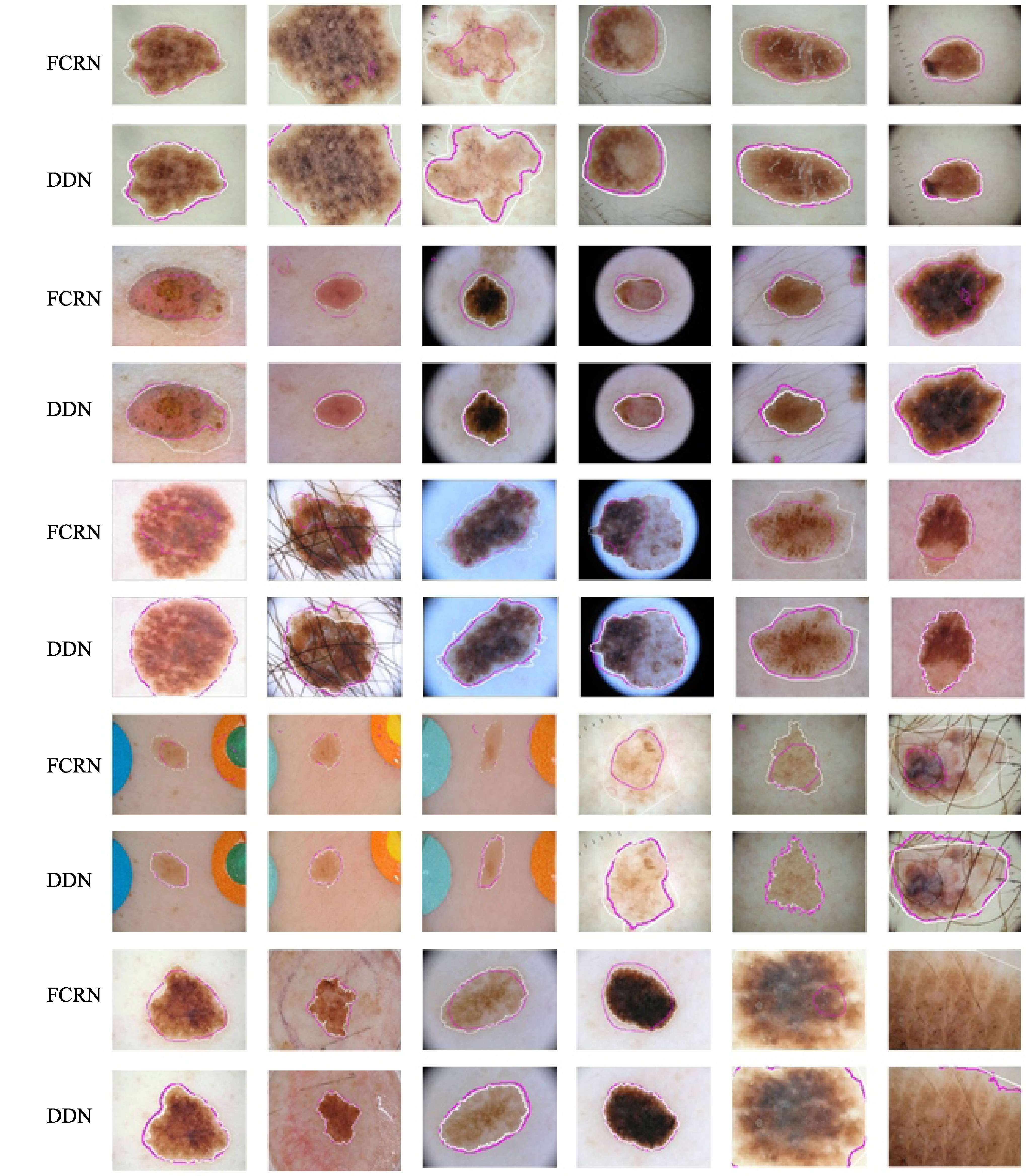
Figure 4.
Segmentation results of various methods. The pink and white contours denote the segmentation results of our method and ground truth, respectively. The upper rows are FCRN method and the bottom rows are the proposed method.
3.2Results
Due to rich parameters to learn, our network cannot train efficiently unless enough number of training images is provided. Despite the skin lesion images provided by the ISBI 2016 challenge, the segmentation performance is still greatly affected by image processing due to the huge intra-class variations. Labeling extra medical images are extremely tedious. Accordingly, data enlarging or augmentation is of great importance to boost the segmentation performance. Performing spatial transformation is one of the most effective ways to solve this problem. Motivated by this, lesion images are rotated to four degrees with (0
We apply our proposed method and evaluate our models in ISBI skin lesions datasets released in both 2016 and 2017. We use the models pretrained in ImageNet and all settings are consistent to assess our method fairly. Models are trained with and without deep supervised method, dice loss and CRF for performance comparison. The segmentation performance is evaluated based on Dice coefficient (DC), jaccard index (JA), accuracy (AC). Table 1 shows the segmentation results of various methods and the computational cost of per image at stage of validation, where ML is multi-path loss DDL is dense deconvolution network and CRF represent training with CRF. We can see that our proposed RN-ML-DDL method achieves the best result due to the multi-path information exploration and dense deconvolution layer. From the comparison, we can see that our proposed method overcomes the shortcoming of CRF method and is efficient due to end-to-end training. In addition, DDL reuses the local spatial information to make one more step in skin-lesions’ recovery, which boosts the segmentation performance. In Table 2, we provide the segmentation algorithm comparison based on ISBI 2016 and 2017 dataset. Compared with the listed methods, our proposed method gets the best segmentation performance. The main explanation is that we integrate local information and global information. Our proposed method achieved considerable improvement in terms of good DC and JA values.
Figure 4 shows segmentation results of various methods. From the comparison of the proposed automatic method and the provided doctor’s ground truth, we can see that the proposed segmentation method is consistent with ground truth. Also, our proposed method outperforms the traditional FCRN method.
4.Discussions
We propose a novel network for skin lesion segmentation, which adopt dense deconvolution layer (DDL) to reconstruct high-resolution image during the phase of decoder. By dense connections, DDL can enhance flow of information and gradients DDL throughout the network. The hierarchical supervision ensures the rapid convergence of the DDN, since gradient feedback can easily reach the early layers. Accordingly, our network possesses more discriminative feature than network without the hierarchical supervision. The prior experience that the shape of skin lesion can be approximated by the ellipse has been learned by our proposed method. However, even if it is easy to distinguish by human naked eye, it is still confused in our proposed method.
5.Conclusion
In this paper, we proposed a deep learning framework for dermoscopy image segmentation based on DDL with multi-path processing. The advantage of our proposed network based on multi-path has been fully demonstrated. Meanwhile, we performed the extensive experiments on the publicly available ISBI 2016 and 2017 challenge skin lesion datasets. Experiments showed that our proposed method outperforms state-of-the-arts methods. Our future work will focus on how to integrate texture information in our network.
Conflict of interest
None to report.
References
[1] | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Ca-a Cancer Journal for Clinicians (2017) ; 67: (1): 7. |
[2] | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2016. Ca-a Cancer Journal for Clinicians (2016) ; 66: (1): 7. |
[3] | Gaudy-Marqueste C, Wazaefi Y, Bruneu Y, Triller R, Thomas L, Pellacani G, et al. Ugly duckling sign as a major factor of efficiency in melanoma detection. Jama Dermatology (2017) ; 153: (4): 279. |
[4] | Shi J, Wu JJ, Li Y, Zhang Q, Ying SH. Histopathological image classification with color pattern random binary hashing-based PCANet and matrix-form classifier. IEEE Journal of Biomedical and Health Informatics (2017) ; 21: (5): 1327. |
[5] | Shi J, Zheng X, Li Y, Zhang Q, Ying SH. Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE Journal of Biomedical and Health Informatics (2018) ; 22: (1): 173. |
[6] | Ying SH, Wen ZJ, Shi J, Peng YX, Peng JG, Qiao H. Manifold preserving: An intrinsic approach for semi-supervised distance metric learning. IEEE Transactions on Neural Networks and Learning Systems (2017) ; (99): 1. |
[7] | Goceri E, Goceri N. Deep learning in medical image analysis: Recent advances and future trends. The Int Conferences Computer Graphics, Visualization, Computer Vision and Image Processing (2017) ; 305. |
[8] | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature (2015) ; 521: (7553): 436. |
[9] | Yu LQ, Chen H, Dou Q, Qin J, Heng PA. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Transactions on Medical Imaging (2017) ; 36: (4): 994. |
[10] | Ataer-Cansizoglu E, Akcakaya M, Orhan U, Erdogmus D. Manifold learning by preserving distance orders. Pattern Recognition Letters (2014) ; 38: : 120. |
[11] | He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (2016) ; 770. |
[12] | Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (2015) ; 3431. |
[13] | Krähenbühl P, Koltun K. Efficient inference in fully connected CRFs with Gaussian edge potentials. Advances in Neural Information Processing Systems (2012) ; 109. |
[14] | Gao H, Yuan H, Wang Z, Ji S. Pixel deconvolutional networks. arXiv preprint arXiv:1705.06820. (2017) . |
[15] | Zeiler MD, Krishnan D, Taylor GW, Fergus R. Deconvolutional networks. 2010 IEEE Computer Vision and Pattern Recognition (2010) ; 2528. |
[16] | Laina I, Rupprecht C, Belagiannis V, Tombari F, Navab N. Deeper depth prediction with fully convolutional residual networks. 3D Vision (2016) ; 239. |
[17] | Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. Medical Image Computing And Computer-Assisted Intervention, Pt III. (2015) ; 9351: : 234. |
[18] | Huang G, Liu Z, Weinberger KQ, Laurens VDM. Densely connected convolutional networks. 2016 IEEE Conference on Computer Vision and Pattern Recognition (2016) . |
[19] | Lee CY, Xie S, Gallagher P, Zhang Z, Tu Z. Deeply-supervised nets. Eprint Arxiv (2014) ; 562. |
[20] | Milletari F, Navab N, Ahmadi SA. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. 3D Vision (2016) ; 565. |
[21] | Bi L, Kim J, Ahn E, Feng D. Automatic skin lesion analysis using large-scale dermoscopy images and deep residual networks. arXiv preprint arXiv:1703.04197. (2017) . |
[22] | Menegola A, Tavares J, Fornaciali M, Li LT, Avila S, Valle E. RECOD titans at ISIC Challenge 2017. arXiv preprint arXiv:1703.04819. (2017) . |
[23] | Jahanifar M, Tajeddin NZ, Asl BM. Segmentation of lesions in dermoscopy images using saliency map and contour propagation. arXiv preprint arXiv:1703.00087. (2017) . |
[24] | Galdran A, Alvarezgila A, Meyer MI, Saratxaga CL, Araújo T, Garrote E, et al. Data-driven color augmentation techniques for deep skin image analysis. arXiv preprint arXiv:1703.03702. (2017) . |




