Color disease leaf image segmentation using NAMS superpixel algorithm
Abstract
BACKGROUND:
Disease leaf segmentation in color image is used to explore the disease shape and lesion regions. It is of great significance for pathological diagnosis and pathological research.
OBJECTIVE:
This paper proposes a superpixel algorithm using Non-symmetry and Anti-packing Model with Squares (NAMS) for color image segmentation of leaf disease.
METHODS:
First of all, the NAMS model is presented for color leaf disease image representation. The model can segment images asymmetrically and preserve the characteristics of image context. Second, NAMS based superpixel (NAMS superpixel) algorithm is proposed for clustering pixels, which can represent large homogeneous areas by super squares. By this way, the impact of complex background and the data redundancy in image segmentation can be reduced.
RESULTS:
Experimental results indicate that compared with segmenting the original image directly and manipulating by Simple Linear Iterative Clustering (SLIC) superpixel, the proposed NAMS superpixel performs more excellently in not only saving storage but also adhering to the lesion region edge.
CONCLUSIONS:
The outcome of NAMS superpixel can be regarded as a preprocess procedure for leaf disease region detection since the method can segment the image into superpixel blocks and preserve the lesion area.
1.Introduction
Plant diseases research is challenging and meaningful. Accurate detection and identification of plant diseases can not only be crucial for disease diagnosis and pathological study, but also help to mitigate the losses of economy and ecology. Plant diseases are often expressed by visual lesion on leaf surfaces, such as abnormal color changes, streaks, and spots. There exist some image processing and machine learning methods to identify these diseases [1, 2, 3] from the leaf disease images. The performance of the identification methods largely depends on the segmentation results of the diseased leaf images. Image segmentation plays an important role in further study, such as feature extraction and recognition of leaf diseases.
Many research efforts have been made on disease leaf segmentation. Wang et al. [4] proposed an adaptive thresholding algorithm to segment the target leaf and background from the leaf images. The Canny [5] and Otsu operators [6] are used to segment the image initially. Then mapping function and shape identification algorithm are used to decide the threshold for segmentation. Phadikar et al. [7] showed Fermi energy-based algorithm to segment the lesion region from the background of disease leaf image. Han segmented the disease leaf image with Simple Linear Iterative Clustering (SLIC) superpixel algorithm [8] and then extracted the superpixel features. And then they classified superpixels as healthy region or infected region with features. These methods have achieved excellent performance in certain contexts; however, most of them regard the original image as the input image to process, which may lead to large data redundancy, since the focus is the infected region. Recently, SLIC superpixel algorithm has been adopted to preprocess the image in some method, but the segmentation may not adhere to the edge of the leaf disease region.
In this paper, a new superpixel algorithm using Non-symmetry and Anti-packing Model with Squares (NAMS) for color leaf disease image segmentation is proposed. The algorithm can be as a pretreatment process for leaf disease image segmentation. The NAMS superpixel can largely reduce the data redundancy in segment process and adhere to the edge of lesion region precisely in the meantime.
The main contributions of this paper are as follows:
Firstly, a new NAMS superpixel algorithm for color leaf disease image segmentation is proposed, which can largely reduce the data redundancy in the further disease region detection, analysis and understanding. At the same time, it can segment lesion region and adhere to the region edge.
Secondly, super square is proposed to represent the image. Greedy principle is used in NAMS superpixel algorithm, which can expand the size of super square as large as possible. The benefit is that one super square can represent a large homogeneous area and improve the computing efficiency.
Thirdly, NAMS superpixel can be regarded as a preprocess procedure for leaf disease region segmentation and detection. Compared to methods that handle the images directly, it can largely reduce the data amount and improve the efficiency. Furthermore, compared to other superixel method like SLIC superpixel, the proposed NAMS superpixel can segment the disease region more precisely.
The rest of the paper is organized as follows: In Section 2, the NAMS model is introduced. Then, the details of the NAMS superpixel algorithm are presented in Section 3. The experimental results of the proposed method and other compared methods are showed in Section 4. Finally, Section 5 is the conclusion.
2.Description of NAMS
NAMS is based on the NAM which is an anti-packing problem [9], which is specially used for image pattern. In image pattern, the idea of the NAMS can be described as follows: give an image (packed pattern) and squares (predefined sub-patterns), pick up these squares (sub-patterns) from the image (packed pattern), and then represent the image (packed pattern) with the combination of the squares (sub-patterns).
Given an image
(1)
(2)
where
Non-distortion image
The NAMS model can largely preserve the characteristics of image context and adhere to the edge, since greed principle is used in the splitting process and anti-packing method is asymmetrical. On the other hand, it can also reduce the data amount of the sub-patterns to represent images.
3.NAMS Superpixel
3.1Idea of NAMS superpixel
In image segmentation, NAMS can represent images by squares, which only need to record the axis of the start point of the square, the average value of the pixels and the length. It can reduce the data amount and improve calculation efficiency. To certain extent, NAMS can be regard as a method of superpixel. However, there exist many isolate pixels which do not belong to any square. To solve the problem, a NAMS superpixel method is proposed to reduce the number of isolate pixels. If a isolate pixel or square has close value with its adjacent pixels or squares, they can be clustered as a larger pattern, called super square in this paper. It can be described as follows:
(3)
where
(4)
where arv is the average value of all the pixels of the square, and
3.2Implementation of NAMS superpixel
NAMS superpixel use NAMS algorithm to represent the color disease leaf image firstly. CIE-Lab color space is used since the Lab color model can achieve better performance for human perceptive awareness. Euclidean distance is adopted to measure the similarity of two pixels to decide whether they belong to one square or not. Given two adjacent pixels
(5)
Without splitting color image into three gray images separately, the method preserves the relevance of three color components, which can improve the segment quality and reduce the color error.
There are many isolate pixels in the NAMS image. To reduce the data amount, the pixels can be merged with adjacent homogeneous pixels or squares by the merging rule [10]. Search every pixel or square achieved by NAMS by raster scanning order, and then find its adjacent pixels or squares. If they have close value, they will be merged to one.
After the leaf disease image has been segmented by NAMS superpixel, distortion image is used to represent the original image to reduce the data amount. Then canny operator is chosen to mark the edge of the image to show the result of NAMS superpixel segmentation.
4.Experiment results and analysis
In this section, all of the experiments are performed on a Celeron microprocessor running at 2.6 GHz and 4 GB RAM. We have implemented our algorithm of NAMS superpixel for disease leaf image segmentation and compared NAMS superpixel with original image handled by canny operator and SLIC superpixel.
Table 1
Comparison of segment accuracy between SLIC and NAMS superpixel
| Lesion region | SLIC segment | NAMS segment | SLIC | NAMS | |
|---|---|---|---|---|---|
| quantity | quantity | quantity | accuracy | accuracy | |
| Evergreen | 3 | 3 | 3 | 100% | 100% |
| Wintersweet | 13 | 9 | 12 | 69% | 92% |
| Pachira macrocarpa | 10 | 5 | 8 | 50% | 80% |
| Orchid | 3 | 2 | 3 | 67% | 100% |
Figure 1.
Comparison of segmented images of SLIC and NAMS superpixel method.
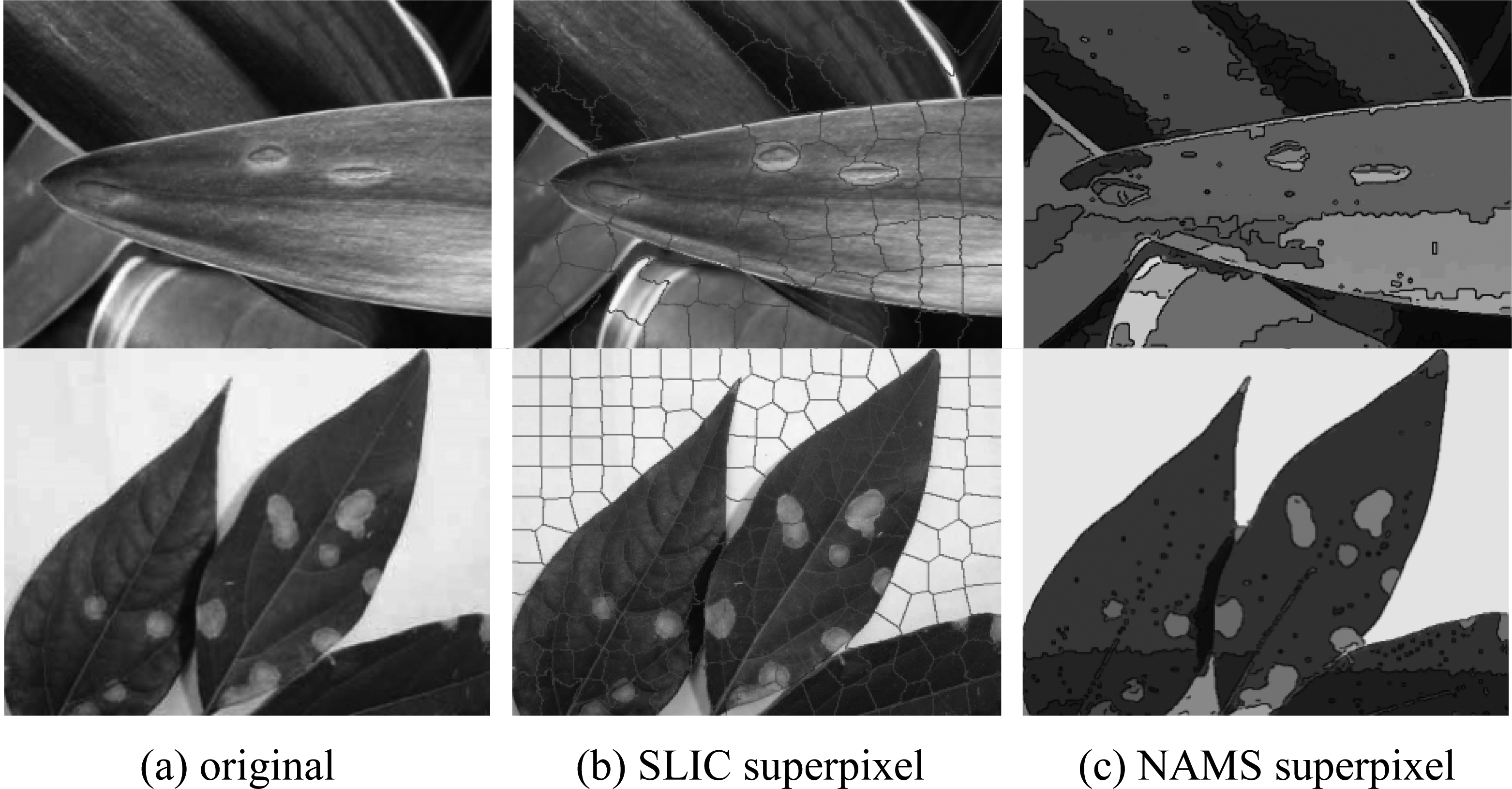
To improve the segment quality of disease leaf image, many methods preprocess the input image firstly. SLIC superpixel is one of the most popular methods. However in the leaf disease image, SLIC superpixel has shown its instability in segment the lesion region. As is presented in Fig. 1, SLIC superpixel may segment one disease region into two different region, but NAMS superpixel can adhere to the edge of disease region. Besides, large-size blank area without lesion in the image is still segmented into many small regions by SLIC superpixel. For this low context area, NAMS superpixel regards them as a whole and do not over-segment, which largely reduce the data redundancy. As it showed in Fig. 1c, the connected white region is one pattern while that in Fig. 1b is segmented to several blocks. In the proposed algorithm, isolate pixels and small patterns in NAMS image are merged into a large region. This can largely reduce the data amount.
Table 1 has figured out the segment accuracy of SLIC superpixel method and NAMS superpixel method. The lesion region quantity in Table 1 is the total number of the lesion region by manual statistics. SLIC segment quantity and NAMS segment quantity are the right lesion region marked by the algorithm respectively. If the lesion region is fully segmented as a whole, it is recorded as a right lesion region. On the contrary, if the lesion region is segmented into several parts or the edge is not adhered, it is recorded as a wrong lesion region. It is shown that SLIC superpixel cannot segment accurate lesion region stably, whereas the proposed NAMS superpixel algorithm has better performance on segmenting precisely for the disease leaf image.
5.Conclusion
Accurate segmentation of disease leaf images is crucial for disease diagnosis and pathological study. In this paper, NAMS superpixel algorithm has been proposed for clustering pixels, which can represent homogeneous area by super squares and largely improve the computing efficiency. NAMS superpixel can be regarded as a preprocess procedure for leaf lesion region detection, which can reduce the data redundancy in the following leaf lesion region segmentation and detection. Experimental results have indicated that compared with other segment method, the proposed NAMS superpixel performs more excellent in not only saving storage but also adhering to the lesion region edge. It is a highly effective algorithm for color disease leaf image segmentation.
Acknowledgments
This work was supported by Natural Science Foundation of Hubei Province, China (ZRMS20160003 46) and Natural Science Foundation of Shandong Province, China (ZR2017LF016).
Conflict of interest
None to report.
References
[1] | Chan T, Vese L. Active contours without edges. IEEE Transactions on Image Processing (2011) ; 10: (2): 266-277. |
[2] | Li BN, Chui CK, Chang S, et al. Integrating spatial fuzzy clustering with level set methods for automated medical image segmentation. Computers in Biology and Medicine (2011) ; 41: (1): 1-10. |
[3] | Bai XD, Cao ZG, Wang Y, Yu ZH, Zhang XF, Li CN. Crop segmentation from images by morphology modeling in the CIE lab color space. Computers and Electronics in Agriculture (2013) ; 99: : 21-34. |
[4] | Wang J, He J, Han Y, Ouyang C, Li D. An adaptive thresholding algorithm of field leaf image. Computers and Electronics in Agriculture (2013) ; 96: : 23-39. |
[5] | Canny J. A computational approach to edge detection. IEEE Trans Pattern Analysis and Machine Intelligence (1986) ; 8: (6): 679-698. |
[6] | Otsu N. A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man and Cybernetics (1979) ; 9: (1): 62-66. |
[7] | Phadikar S, Sil J, Das AK. Rice diseases classification using feature selection and rule generation techniques. Computers and Electronics in Agriculture (2013) ; 90: : 76-85. |
[8] | Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Susstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Transactions on Pattern Analysis and Machine Intelligence (2012) ; 34: : 2274-2282. |
[9] | Liang H, Zhao SR, Chen CB, Sarem M. The NAMlet transform: A novel image sparse representation method based on non-symmetry and anti-packing model. Signal Processing (2017) ; 137: : 251-263. |
[10] | Zheng YP, Sarem M. A fast region segmentation algorithm on compressed gray images using non-symmetry and anti-packing model and extended shading representation. Journal of Visual Communication and Image Representation (2016) ; 34: : 153-166. |



