InteractOA: Showcasing the representation of knowledge from scientific literature in Wikidata
Abstract
Knowledge generated during the scientific process is still mostly stored in the form of scholarly articles. This lack of machine-readability hampers efforts to find, query, and reuse such findings efficiently and contributes to today’s information overload. While attempts have been made to semantify journal articles, widespread adoption of such approaches is still a long way off. One way to demonstrate the usefulness of such approaches to the scientific community is by showcasing the use of freely available, open-access knowledge graphs such as Wikidata as sustainable storage and representation solutions. Here we present an example from the life sciences in which knowledge items from scholarly literature are represented in Wikidata, linked to their exact position in open-access articles. In this way, they become part of a rich knowledge graph while maintaining clear ties to their origins. As example entities, we chose small regulatory RNAs (sRNAs) that play an important role in bacterial and archaeal gene regulation. These post-transcriptional regulators can influence the activities of multiple genes in various manners, forming complex interaction networks. We stored the information on sRNA molecule interaction taken from open-access articles in Wikidata and built an intuitive web interface called InteractOA, which makes it easy to visualize, edit, and query information. The tool also links information on small RNAs to their reference articles from PubMed Central on the statement level. InteractOA encourages researchers to contribute, save, and curate their own similar findings. InteractOA is hosted at https://interactoa.zbmed.de and its code is available under a permissive open source licence. In principle, the approach presented here can be applied to any other field of research.
1.Introduction
In this work, we address the challenge of precisely referencing knowledge items to their exact locations in open access scholarly publications using the Wikidata knowledge graph. Furthermore, we exemplify structuring unstructured data within literature text, and enhance data accessibility and reusability with dynamic querying and visualization.
While the pace of data generation is continuously increasing, the translation of data into actionable insights remains a significant challenge. This transformation happens along four stages: Data, Information, Knowledge, and Wisdom (DIKW) [8]. Data, in its raw form, lays the foundation as pieces of facts. Information arises from organizing and structuring this data, while knowledge is harvested from the compilation of information, pattern recognition, and drawing insights, ultimately paving the way to wisdom. However, this process suffers from obstacles beyond just data processing. For instance, a significant portion of data and information remains unstructured, residing in non-machine-readable formats such as scholarly articles, thereby impeding its reuse. It is essential to have the data structured and organized in a predefined manner for facilitating efficient querying and analysis, allowing for transitions along the DIKW continuum. The absence of structure often represents information accessibility barriers, hampering the transformation of data into knowledge and wisdom. Moreover, accessibility barriers extend beyond the absence of structuring, like access blocked by paywalls, lack of centralization in data collection, or even data loss in scattered, decaying databases [41].
In response to these multifaceted challenges, openly accessible Knowledge Graphs have emerged as a robust solution, providing a dynamic and interconnected framework for representing and utilizing data across various domains. They can facilitate the structuring of complex relationships between heterogeneous data points, making them particularly suitable for transforming data, information, and knowledge into a format that can be holistically understood and efficiently retrieved. Moreover, Knowledge Graphs are instrumental in discovering new insights, thereby advancing the DIKW progression. In this landscape, Wikidata is one of several platforms, offering an open, collaborative environment for constructing and leveraging Knowledge Graphs. Its commitment to openness, scalability, and long-term preservation renders it an invaluable asset to the global research community. Wikidata represents a showcase of the potential of Knowledge Graphs to overcome traditional barriers in data management and to foster the advancement from data to wisdom.
2.Background and related work
2.1.Knowledge graphs
The term “Knowledge graph” was coined by Google in 2012.11 Since then, knowledge graphs have attracted growing attention from researchers due to their robustness in many areas of science, besides application in numerous other fields [12]. Although several attempts have been made to define a knowledge graph, there is still no single, agreed-upon definition of what it entails [14]. As the name implies, a knowledge graph, also known as a semantic network, is a means of storing knowledge in a graph-based model. It is a structured data model that represents a network of real-world entities, such as objects or concepts, and illustrates the relationships between them. This information is usually stored in a graph database and can be visualized in a graph structure. Knowledge graphs can be viewed as a network of nodes and edges, where nodes represent the entities and edges represent the relationships. They can store vast amounts of heterogeneous data in a structured manner and handle complex relationships, making them well-suited for applications in various domains and research areas [18].
The Resource Description Framework (RDF) [4] is one model for implementing knowledge graph storage. The RDF specification22 highlights the simplicity of the RDF data model, which is represented at its fundamental level in the form of subject-predicate-object triples. These triples can describe anything in a flexible and extensible way. To query these RDF triple stores, a query language known as SPARQL, which is similar to SQL, is used to retrieve and manipulate data stored in an RDF format.
There are several open source and proprietary Knowledge Graph systems available that play significant roles in data storage, management, and retrieval across various domains. For example, Apache Jena,33 Neo4J,44 and Virtuoso,55 are among the leading platforms that leverage the capabilities of Knowledge Graphs. Apache Jena utilizes RDF and triple stores for storage, and SPARQL querying. Unlike Apache Jena, Neo4J does not primarily focus on RDF data and SPARQL queries but instead uses its own query language, Cypher. This language makes Neo4J particularly effective in applications where relationships between data points are highly interconnected and need to be analyzed efficiently using machine learning and artificial intelligence. Virtuoso, on the other hand, is a universal server that supports RDF and other data models, in addition to relational database management systems.
2.2.Wikidata
Wikidata is one of the prime examples of knowledge graphs, and it combines two of their potential benefits: openness (as the content is published under the Creative Commons Zero licence66) and ease of editability [35,36]. It is based on the Wikibase software, which allows RDF exports [7], and its content can be queried via the SPARQL endpoint known as the Wikidata Query Service. Wikidata is maintained by the Wikimedia Foundation, which aims to make world’s knowledge available for anyone to use and extend collaboratively. A number of projects are underway to analyse and increase the quality of Wikidata [31]. It is regarded as a key source of identifiers [34] and has tremendous potential which remains largely untapped [23].
2.3.Insufficient management of data, information, and knowledge in the life sciences
Data, information, and knowledge are being generated at an ever-increasing pace in the field of biology. This is due in large part to the widespread availability of high-throughput platforms in biological research, which can generate vast amounts of data, for example, on genes, proteins, and other biological entities and their interactions. The ability to collect, organise, and analyse this data – and the information and knowledge derived from it – is crucial for future biological and biomedical research. While the FAIR principles are now widely acknowledged as a useful framework for managing data [40], a large fraction of the knowledge generated on the basis of this data continues to be stored in unstructured formats such as scholarly articles, which have formed the core of knowledge management in research for centuries. Moreover, a significant proportion of these articles are not available in machine-readable formats (e.g. PDF), which in turn hinders the downstream information mining. Even the machine-readable formats (e.g. in HTML and XML) continue to lack the semantic enrichment. Attempts have been made to address this [9,32], but these have been largely ignored by the publishing industry and scholarly community. Recent projects such as the Open Research Knowledge Graph (ORKG) [2] have tried to overcome this hurdle by offering a semantic database to represent the research findings of publications in a separate machine-actionable form but are referencing only on the article level and not to exact passages. Furthermore, domain-specific biological knowledge graphs instances with SPARQL endpoints are also available, such as UniProt [6], which is dedicated to storing information on proteins, and IntAct [19] for molecular interactions – both also referencing only on the article level.
Alongside their unstructured representation in academic literature, data, information, and knowledge are also stored in biological databases. Even though these databases represent a valuable resource for larger or more specialised research communities, their development and maintenance are often limited to the lifetime of the respective research project. There are numerous cases of valuable databases that were created as part of such projects but are no longer accessible. Examples of databases that cannot be accessed via their published URLs at the time of writing this article include BSRD [20] and sRNAdb [26], which are databases for bacterial small RNAs, as well as AANT [13], a database for amino acid-nucleotide interactions, and cpnDB [11], a database for bacterial Chaperonin sequences. Several studies have examined this issue. Their results show that more than 30% of bioinformatics web services published over the last 23 years are currently unavailable, with lack of maintenance being the primary cause of this decay [1,21,25,41]. Additionally, restrictive licensing can make it difficult for researchers to access and (re-)use these resources, further hindering scientific advancement.
Another challenging aspect of knowledge management in research is the granularity of references. Claims are cited on the level of full articles, which makes it very time-consuming for readers to find the exact location of a particular statement in the referenced source. This poses a major obstacle to the verification and contextualisation of such references by readers.
2.4.Wikidata as a knowledge-graph solution for biological data, information, and knowledge
In the life sciences and in other research fields, knowledge graphs such as Wikidata are used to represent and integrate information from a variety of sources, including genomic data, literature, and experimental results [3,37,38]. Knowledge graphs can be employed to model complex biological systems and processes, and to facilitate data mining and analysis. This may include the representation and integration of genomic data such as genes, proteins, and pathways. Furthermore, they can be used to model the interactions of biomedical entities, for example linking genes associated with antibiotic resistance in a pathogen [42].
Widespread use of knowledge graphs in the life sciences would facilitate the discovery of new biological insights and relationships through data mining and visualisation, and improve the interpretation and understanding of biological data through the integration of diverse data sources. One example of the implementation of knowledge graphs for biological data is the Clinical Knowledge Graph (CKG) [29]. The CKG aims to assist in the delivery of personalised medical treatment by using machine-learning methods to mine data from heterogeneous domains. Wikidata pushes these capabilities further by facilitating the sharing and reuse of biological data through the use of standardised data formats and open-access solutions. Further examples of existing solutions include WikiGenomes [28], an openly editable knowledge graph for genomic annotations that is geared towards the molecular biology community, ChlamBase [27], which is a central access point for genomic and proteomic information specifically for the Chlamydia research community, and WikiPathways [22], an open, collaborative, community-based platform dedicated to the curation of biological pathways. A number of articles have been published that encourage the use of Wikidata-based solutions in biology, such as the Gene Wiki initiative [5]
2.5.Small RNA regulatory networks
In bacteria and archaea, gene expression is controlled by a variety of regulators. One class of regulators is the small RNAs (also known as non-coding RNAs) [39], which are not translated into proteins to perform their regulatory functions. This class of RNAs is responsible for vital regulatory roles in gene expression. These small RNAs are often expressed in their hundreds to control cellular functions such as the response to environmental changes, and each small RNA can influence the activity of multiple targets of proteins or messenger RNAs. Small RNAs regulate their targets through various mechanisms [16,33] such as down-regulation (by disrupting mRNA translation through base-pairing to the ribosomal binding sites) and up-regulation (by inhibiting mRNA degradation). Despite their importance in all known bacterial and archaea species, knowledge of small RNAs is comparatively limited, and they often fail to be included in the creation of holistic models in systems biology.
The genomic locations of small RNAs are first identified through experiments and annotated in genomic reference sequences; then, the interactions between these annotated small RNAs and genes are computationally predicted and experimentally confirmed. The numerous interactions at the cellular level that occur under different environmental conditions between different regulators, such as small RNAs and their target genes, can be represented as a network. This network consists of nodes such as regulators and gene targets, with the edges between these nodes representing the identified interactions. Information on these interactions can be represented and visualised in network graphs to facilitate understanding of their complexity. Typically, researchers report their findings on bacterial small RNA regulatory networks in various, usually unstructured formats that lack a consistent standard. This data is presented in the main body of articles, in supplementary materials such as spreadsheets, or in flat files. For a limited number of species, data is manually compiled by experts into web-based databases such as RegulonDB [15]. Without a uniform format for reporting such information, it is difficult to gain a comprehensive understanding of these regulatory networks.
3.Approach and implementation
3.1.InteractOA as a Wikidata-based application for small RNA regulatory networks
With these needs and technological foundation in mind, we modelled small RNA interactions in Wikidata and built an application called InteractOA (OA for Open Access as it builds on Open Access scholarly articles) that presents the data. The application features a graphical interface that allows users to query Wikidata for a chosen organism and displays the network of interactions or citation information at the statement level for all referenced interactions. The semantic nature of Wikidata, combined with its openness, can help to overcome many challenges in the management of data, information, and knowledge in gene regulation research. Therefore, Wikidata was chosen as the platform to implement this solution to compile extracted scholarly knowledge for the unique combination of the following features, 1. It has an easy-to-use web interface. 2. It has a permissive licence (CC0). 3. It already has a large corpus of data (including literature metadata) to which other data can be connected. 4. The availability of a SPARQL endpoint for querying data. 5. A platform to create applications building upon the data (Toolforge77). 6. A strong (not only scientific) community backed by a sustainable organisation (Wikimedia Foundation).
The Wikidata-based approach of linking biological entities in order to represent regulatory networks offers several benefits in terms of openness and sustainability. Since intracellular interactions are often complex and numerous, it is important to use a digital representation approach that is scalable, extensible, easily queryable, and usable. Wikidata possesses all of these properties.
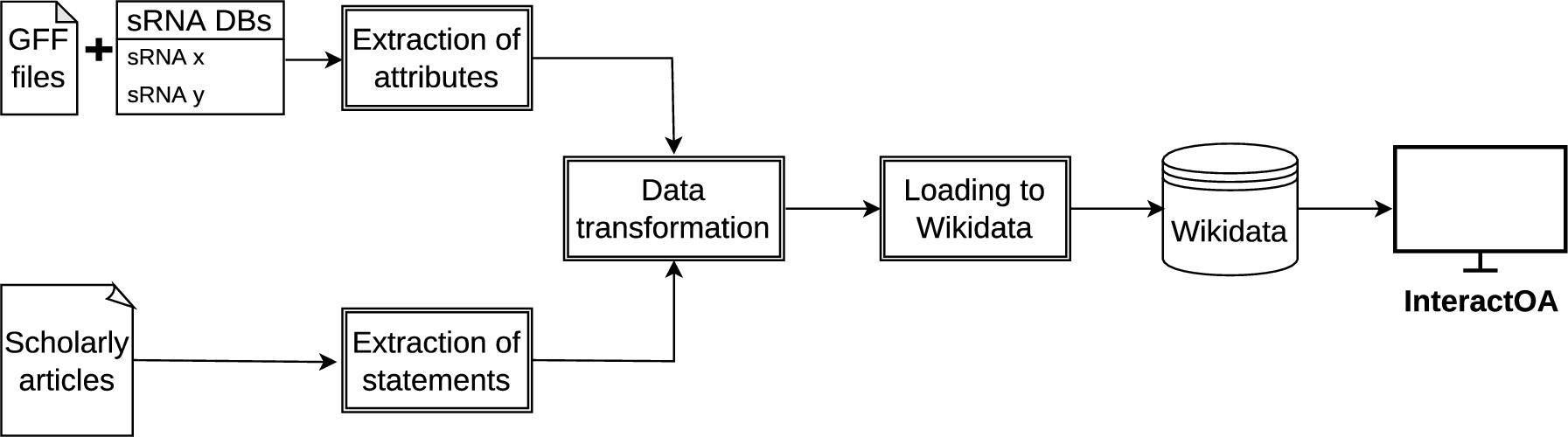
Fig. 1.
Extract-transform-load workflow used for integrating the data into Wikidata and presentation by InteractOA.
3.2.Modelling small RNA interaction data in Wikidata
Table 1
Properties list used in the modelling of the annotations, interactions, and citations of small RNA interactions
| ID | Name | Usage/Description |
| P703 | Found in taxon | Links entities of annotations to a certain Wikidata item that represents an organism at the strain level |
| P31 | Instance of | Assigns GFF entry type e.g. gene or ncRNA to a Wikidata item |
| P644 | Genomic start | Assigns the genomic start location of an annotation entry to a Wikidata item |
| P645 | Genomic end | Assigns the genomic end location of an annotation entry to a Wikidata item |
| P2548 | Strand orientation | Assigns the genomic strand of an annotation entry to a Wikidata item |
| P688 | Encodes | Links two Wikidata items of a gene to its product, e.g. protein, or RNA |
| P702 | Encoded by | Vice versa of P688 |
| P361 | Part of | Describes a partial product of a gene, like genes that splice to multiple mRNAs |
| P527 | Has part(s) | Vice versa of P361 |
| P351 | Entrez Gene ID | Assigns NCBI Entrez identifier for an annotation entry |
| P2249 | RefSeq genome ID | Assigns NCBI RefSeq identifier qualifier the start, end, strand of an annotation entry |
| P2393 | NCBI locus tag | Assigns NCBI identifier for an annotation locus tag |
| P637 | RefSeq protein ID | Assigns NCBI identifier for protein GFF entry |
| P128 | Regulates (molecular biology) | Links 2 annotation entities based on the interaction of any type if type is unspecified |
| P3777 | Antisense inhibitor of | Similar to P128, when interaction type is known as inhibition by antisensing |
| P3771 | Activator of | Similar to P128, when interaction type is known as up-regulation by activation |
| P3774 | Blocker of | Similar to P128, when interaction type is known as mRNA translation blocking |
| P3773 | Antagonist of | Similar to P128, when interaction type is known as antagonizing |
| P3772 | Agonist of | Similar to P128, when interaction type is known as agonizing |
| P248 | Stated in | Adds reference about an interaction claim, which is a Wikidata item that represents a scholarly article |
| P1683 | Quotation | Adds quote statement to the interaction claim reference |
| P932 | PMCID | Adds PubMed Central identifier for the reference article of an interaction claim |
Table 2
Items list used in the modelling of the annotations, interactions, and citations of small RNA interactions
| ID | Name | Usage/Description |
| Q427087 | Non-coding RNA | Class of RNA that is not translated into proteins |
| Q423832 | Antisense RNA | RNA molecules hybridizing to complementary sequences in either RNA or DNA, altering the function of the latter |
| Q201448 | Transfer RNA | Adaptor molecule composed of RNA |
| Q285904 | Transfer-messenger RNA | Bifunctional RNA that has properties of a tRNA and an mRNA |
| Q424665 | Signal recognition particle | Protein-RNA complex facilitating translocation of proteins across membranes |
| Q1012651 | Ribonuclease P activity | Catalysis of the endonucleolytic cleavage of RNA, removing 5’ extra nucleotides from tRNA precursor. |
| Q11053 | RNA | Family of large biological molecules |
| Q7187 | Gene | Basic physical and functional unit of heredity |
| Q22809680 | Forward strand | Forward oriented strand in a double-stranded DNA molecule |
| Q22809711 | Reverse strand | Reverse oriented strand in a double-stranded DNA molecule |
| Q215980 | Ribosomal RNA | Ribosome RNA molecule, essential for protein synthesis in all living organisms |
| Q277338 | Pseudogene | Functionless relative of a gene |
Typically, data related to genomes and genes including small RNAs are stored in text files such as GFF3 (General Feature Format)88 files. These files can be obtained from public repositories like NCBI RefSeq [24]. Each line within a GFF file contains details about a single annotation, including the gene’s location on the genome, its type, function, and other attributes. To model small RNAs and their interactions, we translated each gene annotation in these GFF files into Wikidata items. These items were then interconnected using specific properties to represent the interactions. Selected, widely used bacterial strains were used for this modelling process due to the abundance of their small RNA interaction data, including Escherichia coli strain K-12 substr. MG1655 (NCBI genome assembly GCF_000005845.2,99 RefSeq accession NC_000913.31010). For the processing and integration of the data, an ETL (extract-transform-load) workflow was established (see Fig. 1). The first step was to obtain genomic annotation data for these organisms from the NCBI RefSeq repository. Next, a Python tool was developed to automate the process of importing annotation records from GFF files and linking them to Wikidata entries using defined IDs corresponding to specific strains. To do this, the tool extracts information from each annotation record, generates Wikidata item definitions (including name, synonyms, and description), and handles communication with Wikidata’s API to create new non-redundant Wikidata items. These newly created Wikidata items were then expanded by importing the remaining annotation data, including entry type (e.g., protein or RNA), genomic location, and external gene identifiers. This process was based on selected Wikidata items and properties defined for the biology domain, which were collected from Wikidata’s listings for life sciences.1111 See Tables 1 and 2 for the properties list and the items list used here.
Next, Wikidata items representing RNAs were linked to model the interactions. To do this, interaction data for numerous small RNAs was manually obtained from research articles, RegulonDB [15], and the Staphylococcal Regulatory RNA Database (SRD) [30]. For each of the selected interactions taken from the databases, the underlying statements were manually extracted from the corresponding article and the statements collected in a dedicated file. The compiled information from this manual curation as well as the data automatically extracted from the GFF files was imported to Wikidata using a dedicated Python tool which first extracts the interaction from input files and then maps the names of interaction partners in the parsed file to the respective pre-imported annotations in Wikidata. The item of the pre-imported annotation is retrieved by the tool using a query template to get the corresponding item’s ID. After that, the tool links the Wikidata item representing RNAs by using the selected properties. For example, the property “antisense inhibitor of” (P3777) was used to link the interaction between sRNA omrA (Item ID Q50419343) and gene csgF (Item ID Q23087296). For an example of such a link, see Fig. 2. If the type of interaction has no corresponding property in Wikidata, the tool falls back to the more generic property (P128 regulates). To showcase the implementation until the point of writing this article, 776 small RNA annotations were imported and 253 connections between small RNAs and their targets were contributed specific to 3 species: Escherichia coli, Vibrio Cholerae, and Staphylococcus aureus. Both Python tools used to import the data were built upon Wikidataintegrator1212 [37] and pywikibot to facilitate interaction with the Wikidata API during querying and importing. They are available on GitHub,1313,1414 and they are archived in Zenodo.1515
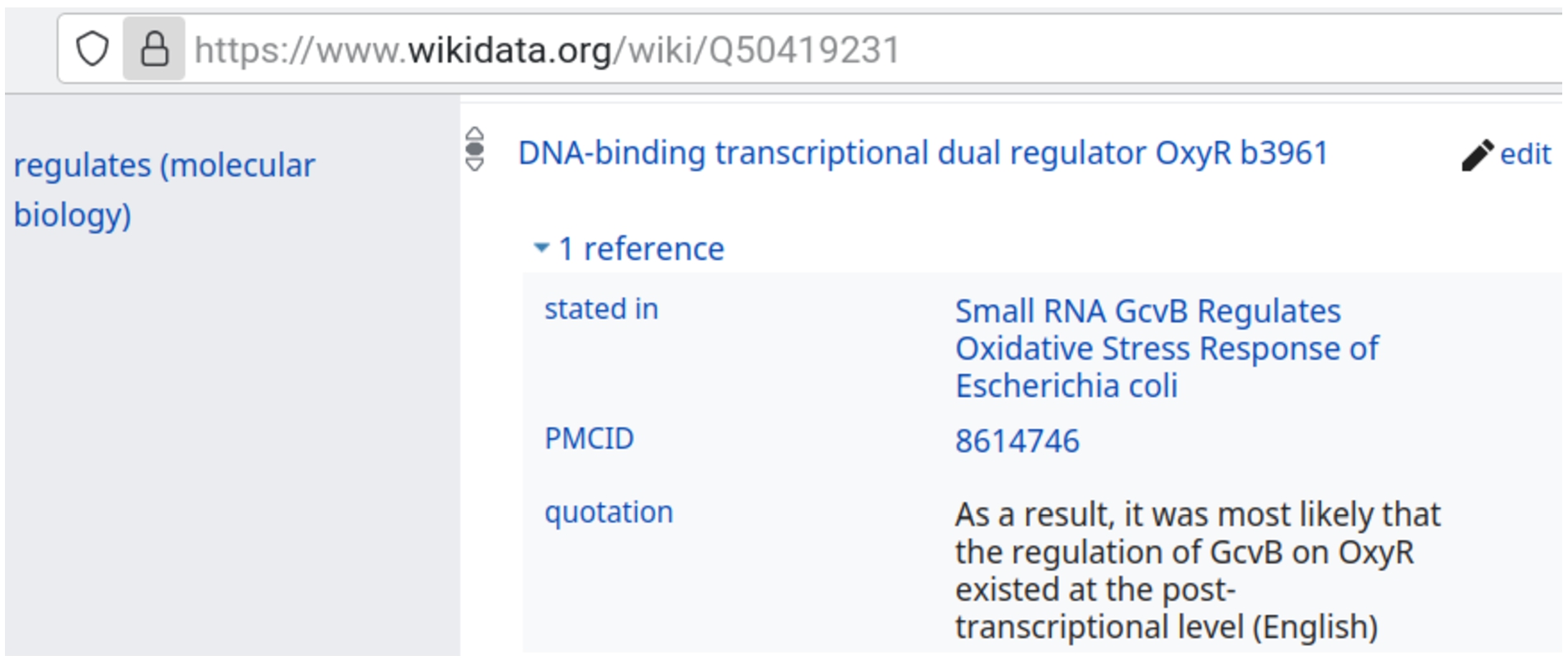
3.3.Statement-level citations stored in Wikidata
A significant proportion of the sRNA interactions modelled using the method described above appear in open-access articles or other articles freely accessible on PubMed Central.1616 For several of these interactions, the statements describing the specific interactions were extracted manually. The Wikidata items of the source article, the PubMed Central ID, and statements (using the “quotation” property P1683) were added as interaction properties. For example, as shown in Fig. 2, the property “stated in” (P248) was used with the Wikidata item that corresponds to a journal article entitled “Small RNA GcvB Regulates Oxidative Stress Response of Escherichia coli” [17] (Q115652789) in order to link the interaction to the article in which it was mentioned. Moreover, the PubMed Central ID of this article was linked with the property “PMCID” (P932), and the statement mentioning the interaction was also linked using the “quotation” property (P1683). This method of storing source statements of sRNA interactions in Wikidata makes it easy to check and correct the features of interaction models.
3.4.InteractOA as a front end to relevant Wikidata items
The web front end InteractOA was developed to facilitate interactive exploration of the data stored in Wikidata, as described above. InteractOA is implemented in Python and the Flask web framework.1717 Its code is available on GitHub1818 and its releases are archived at Zenodo.1919 Wikidata features a query service2020 that enables users to enter SPARQL queries. The service generates tables as well as various types of interactive visualisations, including bar charts and in this case, most importantly, network plots. The web front end uses these Wikidata capabilities to visualise the regulatory interaction as an interactive network plot (see Fig. 3 for an example). The interface allows users to customise their queries using filters and to search using keywords without requiring any technical knowledge. Once the user has selected the desired filters and keywords, InteractOA sends the generated SPARQL query to the Wikidata Query Service and displays the results. The full landscape of small RNAs is displayed if no filters or keywords were used. Figure 3 shows an example network of three small RNAs and their interactions with other protein-coding genes based on a limited set of locus tag IDs as keywords.
Fig. 3.
Screenshot of a network visualization for 3 small RNAs (orange labels) and their interactions with their gene targets (white labels).
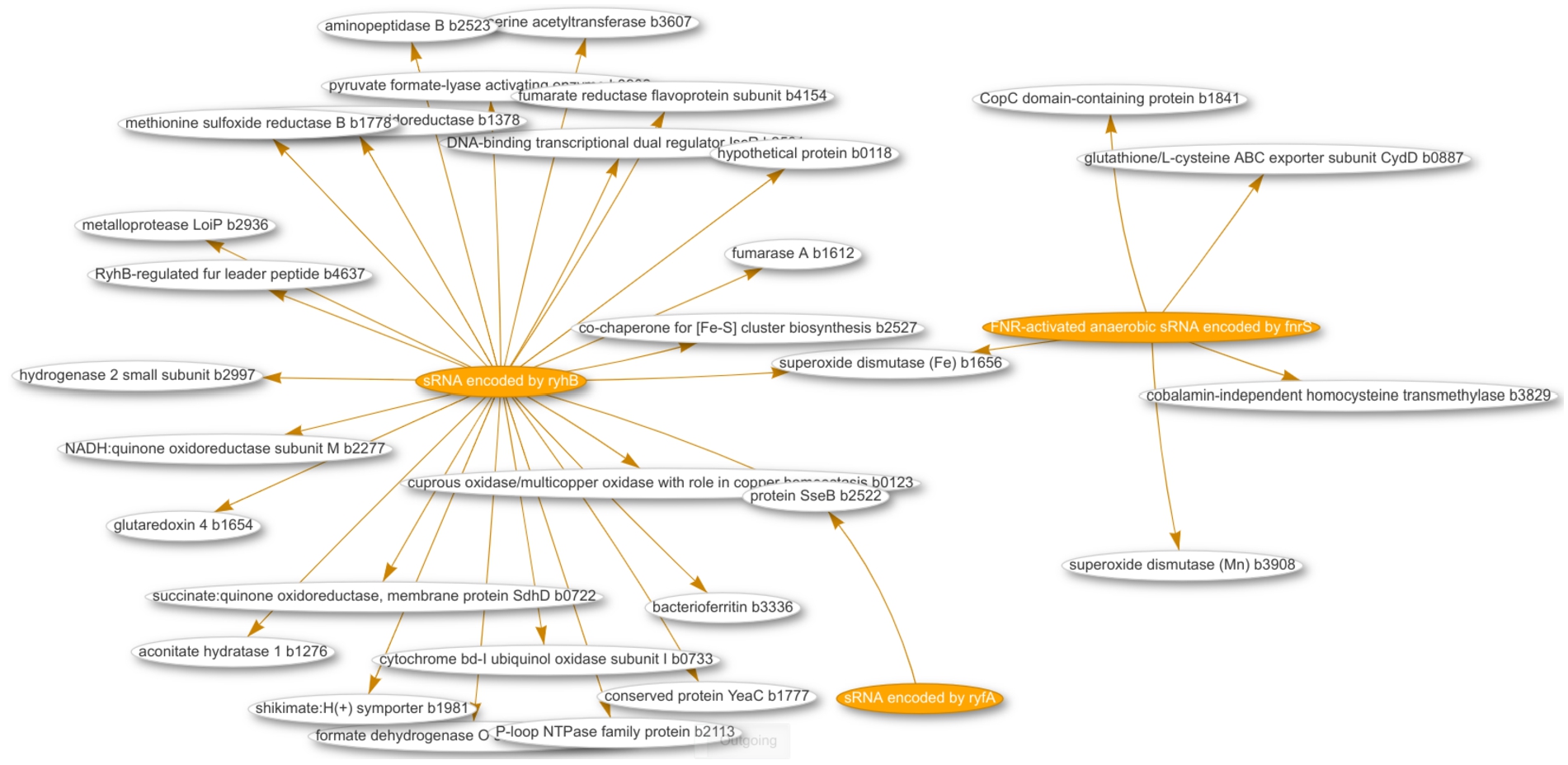
Fig. 4.
Screenshot of all available statement level citations for a selected organism, which can be filtered with keywords.
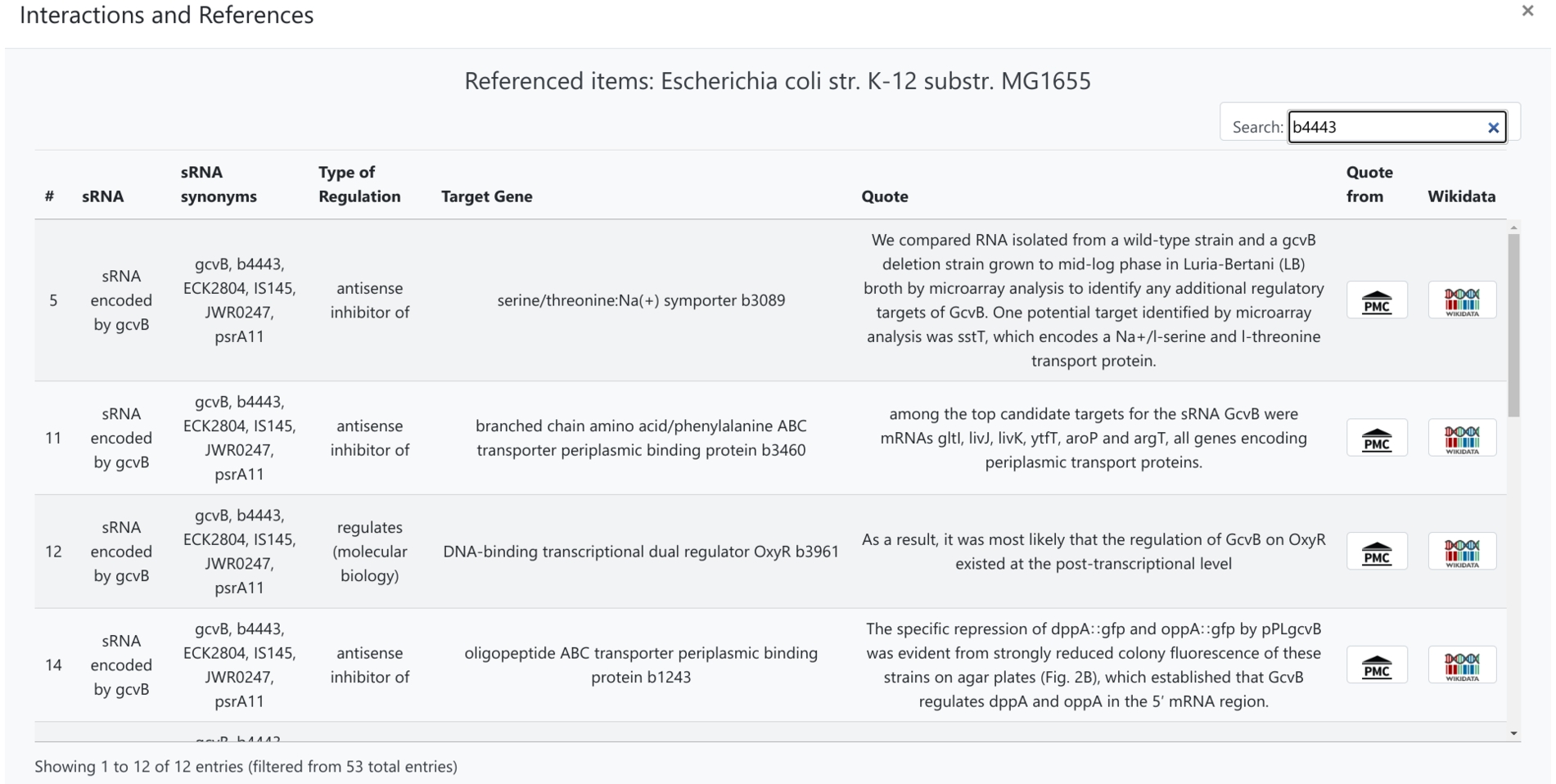
Additionally, InteractOA provides a tabular view for all the extracted statements in Wikidata for a strain selected by the user and presents them in a searchable table (see Fig. 4 for an example). Its search function can filter results by several criteria, for example by partners of interactions or by type of interaction. This solution enables users to combine multiple statements from several studies at once, which aims to shorten the time needed to consult previous research on individual small RNAs. Moreover, the user can open the scholarly article from which the statement originated, with the statement itself highlighted (see Fig. 5 for an example).
Fig. 5.
The highlighted text in an article [17] is a statement level citation example for a claim about small RNA interaction.
![The highlighted text in an article [17] is a statement level citation example for a claim about small RNA interaction.](https://content.iospress.com:443/media/sw-prepress/sw--1--1-sw243685/sw--1-sw243685-g005.jpg)
InteractOA functions as an interface between the user and the Wikidata endpoint, with no data storage taking place on the platform itself. Essentially, InteractOA operates as a middle layer, designed to facilitate dynamic data querying based on predefined SPARQL query templates. These templates contain parameters that correspond to specific fields and filters on the front end. When a user selects an organism and applies filters, the app processes these inputs and tailors the query from the template accordingly. In the case of visualising small RNA networks, the specific query is channelled via URL parameters to be executed on Wikidata’s query service. An iframe component on the webpage then employs this URL to display the results in the form of a network plot. For the highlighting of statements on small RNA interactions in scholarly articles, the app employs the user’s selected organism to pass it to the corresponding parameter in the query template. The Python module WikidataIntegrator2121 [37] is then utilized to execute the query, and the results are included in a tabular view which is then presented to the client. Before transferring the results to the user’s web browser, they are enriched with links to the Wikidata items of the sRNAs as well as links to the corresponding article hosted at PubMed Central (PMC) where the selected statements can be highlighted. This colouring works by retrieving the HTML content of the PMC’s articles to the server’s memory based on the PMC ID and wrapping the content to be highlighted with marking tags, which is then returned to the user’s browser and rendered there with the changed background colour. This process allows for aiding users in identifying and focusing on the relevant information.
4.Discussion
Nowadays, much of the research data on which scholarly articles are based is deposited in a structured format in dedicated repositories, yet the actual insights and knowledge derived from this research are often only accessible in an unstructured format within the confines of the article text. In this work, we have presented a solution to this problem based on the open-source Wikidata knowledge graph. As shown here, Wikidata provides a structured way to store knowledge generated within specific fields of research, for example by interleaving items of biological entities with bibliographic information while also providing links to the exact statements that are the source of each knowledge item in the corresponding open-access articles. In this work, we have showcased this approach by modelling the regulatory networks of small RNAs in bacteria and built a dedicated web tool that makes it easy to explore and visualise the data stored in Wikidata. The chosen approach also includes granular referencing of knowledge sources. Storing the data in Wikidata ensures its long-term availability while opening up access to a large tool chain for imports, queries, and visualisation. Moreover, it facilitates links to other relevant entities modelled in Wikidata.
We consider this tool to be a valuable method to our own research field, and we intend to apply it to further topics. Currently, the steps required to extract statements from research articles are carried out manually. As the quantity of such manually curated article excerpts grows, we aim to train language models to assist with the human curators’ extraction work. This text-mining-based approach will build upon related work conducted in our research group (Halder et al., in preparation). For this named-entity recognition (NER) of sRNA taken from databases followed by the extraction of potential interaction statements combined with a language model will generate the foundation for manual curation of the data. The curation will be conducted by collaborating microbiologist with expertise in certain bacterial species, especially regarding their regulatory networks. For the curation, a web-based tool currently under development (also Halder et al., in preparation) will be used. The curated data will then be integrated into Wikidata as described above and by that can be explored in InteractOA. The compiled data will also be used to improve the underlying language model and make the automatic extraction better. Besides this, there is an opportunity to directly incorporate other cellular interactions such as regulatory proteins, protein-protein interactions, and cellular sensing.
Despite the numerous useful features provided by Wikidata and its ecosystem of tools, there are challenges that should be considered when choosing a similar approach. Wikidata’s API is comparatively slow, which makes the ingestion of larger data sets very time-consuming. Similarly, SPARQL queries are limited by the constraints of the Wikidata Query Service. This latter issue could be solved by working with full data dumps provided by Wikidata. Besides these technical issues, there is inevitably a risk of lower quality, or even significant vandalism, that comes with choosing an openly-editable form of data storage in which anybody can add, remove, or modify entries. Thanks to versioning, problematic edits do not pose a critical risk, however, and an interface to curate new edits could further address this issue.
One additional concern pertains to the availability of item classes and properties within Wikidata. The implemented model of InteractOA applies Wikidata’s currently existing properties to the topic of small RNAs and their interaction partners. These item classes and properties, despite their sufficiency for the current implementation, are limited to relatively high-level descriptions such as “regulates” and “antisense inhibitor of”. To further improve the model, more item classes and properties would need to be agreed upon for Wikidata, for example specifying if the regulation is positive or negative for the “regulates” property. There is also a need for further options similar to the “antisense inhibitor of” property and for other interaction types such as “promoter of”, “cis-acting”, or “trans-acting”. Fortunately, there is currently an ongoing discussion on how to extend the pool of Wikidata properties for biological data.2222
Additionally, it has to be considered that currently only a fraction of the scholarly literature is covered by Wikidata2323 [10] (estimated 22.5 Million of 389 Million articles listed in Google Scholar) and that our suggested approach might have technical limitation in terms of scalability considering the current technical setup of Wikidata. In case this approach would be applied on a large scale to numerous fields and millions of articles, the capacity and performance of Wikidata might not be sufficient. A solution for this would be domain-specific Wikibase instances. This approach could also be helpful to reduce the friction of introducing new item classes and properties which is usually time-consuming as a consensus has to be found with all community members participating in the discussion and the overall state of the ontology be considered. Still, for the time being, a solution built on Wikidata lowers the access barriers to explore this approach by other research communities.
The here presented approach and its implementation stands at the intersection of Wikidata and biological scholars’ communities to bridge the gap between them, and to bring the advantages of Wikidata to the forefront, promoting its use among biological scholars. Based on the showcase given by this, the prospective users would be encouraged to contribute, curate, and save their findings as a result of highlighting Wikidata as a readily achievable centralised solution for the preservation of their discoveries, as well a platform for promoting and citing their work. The introduced statement-level citation in InteractOA as a feature that could potentially simplify the process of locating and comparing information due to the amassed collective knowledge. Despite the challenges, we are convinced that the approach showcased in this project can be applied to numerous other communities, even those that require more complex data models. We hope the example provided here will motivate other research communities to make knowledge and its sources available in a more structured fashion.
Notes
Acknowledgements
This work was supported by the Bundesministerium für Bildung und Forschung (BMBF, grant number 16OA031Z).
References
[1] | T.K. Attwood, B. Agit and L.B.M. Ellis, Longevity of Biological Databases, EMBnet. Journal 21(0) ((2015) ). doi:10.14806/ej.21.0.803. |
[2] | S. Auer, A. Oelen, M. Haris, M. Stocker, J. D’Souza, K.E. Farfar, L. Vogt, M. Prinz, V. Wiens and M.Y. Jaradeh, Improving access to scientific literature with knowledge graphs, Bibliothek Forschung und Praxis 44: (3) ((2020) ), 516–529. doi:10.1515/bfp-2020-2042. |
[3] | S. Bonner, I.P. Barrett, C. Ye, R. Swiers, O. Engkvist, A. Bender, C.T. Hoyt and W.L. Hamilton, A review of biomedical datasets relating to drug discovery: A knowledge graph perspective, Briefings in Bioinformatics 23: (6) ((2022) ), bbac404. doi:10.1093/bib/bbac404. |
[4] | D. Brickley, R.V. Guha and B. McBride, RDF schema 1.1. W3C recommendation, World Wide Web Consortium 2 ((2014) ). |
[5] | S. Burgstaller-Muehlbacher, A. Waagmeester, E. Mitraka, J. Turner, T. Putman, J. Leong, C. Naik, P. Pavlidis, L. Schriml, B.M. Good and A.I. Su, Wikidata as a semantic framework for the Gene Wiki initiative, Database 2016: ((2016) ), baw015. doi:10.1093/database/baw015. |
[6] | T.U. Consortium, UniProt: The universal protein knowledgebase, Nucleic Acids Research 46: (5) ((2018) ), 2699. doi:10.1093/nar/gky092. |
[7] | F. Erxleben, M. Günther, M. Krötzsch, J. Mendez and D. Vrandečić, Introducing Wikidata to the Linked Data Web, in: The Semantic Web – ISWC 2014, Springer International Publishing, (2014) , pp. 50–65. doi:10.1007/978-3-319-11964-9_4. |
[8] | M.H. Frické, Data-Information-Knowledge-Wisdom (DIKW) pyramid, framework, continuum, in: Encyclopedia of Big Data, Springer International Publishing, Cham, Switzerland, (2017) , pp. 1–4. ISBN 9783319320014. doi:10.1007/978-3-319-32001-4_331-1. |
[9] | A. Garcia, F. Lopez, L. Garcia, O. Giraldo, V. Bucheli and M. Dumontier, Biotea: Semantics for pubmed central, PeerJ 6: ((2018) ), e4201. doi:10.7717/peerj.4201. |
[10] | M. Gusenbauer, Google Scholar to overshadow them all? Comparing the sizes of 12 academic search engines and bibliographic databases, Scientometrics 118: (1) ((2018) ), 177–214. doi:10.1007/s11192-018-2958-5. |
[11] | J.E. Hill, S.L. Penny, K.G. Crowell, S.H. Goh and S.M. Hemmingsen, cpnDB: A chaperonin sequence database, Genome Research 14: (8) ((2004) ), 1669–1675. doi:10.1101/gr.2649204. |
[12] | P. Hitzler, A review of the semantic web field, Communications of the ACM 64: (2) ((2021) ), 76–83. doi:10.1145/3397512. |
[13] | M.M. Hoffman, AANT: The amino acid-nucleotide interaction database, Nucleic Acids Research 32: (90001) ((2004) ), D174–D181. doi:10.1093/nar/gkh128. |
[14] | A. Hogan, E. Blomqvist, M. Cochez, C. D’amato, G.D. Melo, C. Gutierrez, S. Kirrane, J.E.L. Gayo, R. Navigli, S. Neumaier, A.-C.N. Ngomo, A. Polleres, S.M. Rashid, A. Rula, L. Schmelzeisen, J. Sequeda, S. Staab and A. Zimmermann, Knowledge Graphs, ACM Computing Surveys 54: (4) ((2021) ), 1–37. doi:10.1145/3447772. |
[15] | A. Huerta, RegulonDB: A database on transcriptional regulation in Escherichia coli, Nucleic Acids Research 26: (1) ((1998) ), 55–59. doi:10.1093/nar/26.1.55. |
[16] | M.G. Jørgensen, J.S. Pettersen and B.H. Kallipolitis, sRNA-mediated control in bacteria: An increasing diversity of regulatory mechanisms, Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms 1863: (5) ((2020) ), 194504. doi:10.1016/j.bbagrm.2020.194504. |
[17] | X. Ju, X. Fang, Y. Xiao, B. Li, R. Shi, C. Wei and C. You, Small RNA GcvB regulates oxidative stress response of escherichia coli, Antioxidants 10: (11) ((2021) ), 1774. doi:10.3390/antiox10111774. |
[18] | M. Kejriwal, Knowledge graphs: A practical review of the research landscape, Information 13: (4) ((2022) ), 161. doi:10.3390/info13040161. |
[19] | S. Kerrien, Y. Alam-Faruque, B. Aranda, I. Bancarz, A. Bridge, C. Derow, E. Dimmer, M. Feuermann, A. Friedrichsen, R. Huntley, C. Kohler, J. Khadake, C. Leroy, A. Liban, C. Lieftink, L. Montecchi-Palazzi, S. Orchard, J. Risse, K. Robbe, B. Roechert, D. Thorneycroft, Y. Zhang, R. Apweiler and H. Hermjakob, IntAct–open source resource for molecular interaction data, Nucleic Acids Research 35: (Database) ((2007) ), D561–D565. doi:10.1093/nar/gkl958. |
[20] | L. Li, D. Huang, M.K. Cheung, W. Nong, Q. Huang and H.S. Kwan, BSRD: A repository for bacterial small regulatory RNA, Nucleic Acids Research 41: (D1) ((2012) ), D233–D238. doi:10.1093/nar/gks1264. |
[21] | S. Mangul, T. Mosqueiro, R.J. Abdill, D. Duong, K. Mitchell, V. Sarwal, B. Hill, J. Brito, R.J. Littman, B. Statz, A.K.-M. Lam, G. Dayama, L. Grieneisen, L.S. Martin, J. Flint, E. Eskin and R. Blekhman, Challenges and recommendations to improve the installability and archival stability of omics computational tools, PLOS Biology 17: (6) ((2019) ), e3000333. doi:10.1371/journal.pbio.3000333. |
[22] | M. Martens, A. Ammar, A. Riutta, A. Waagmeester, D.N. Slenter, K. Hanspers, R.A. Miller, D. Digles, E.N. Lopes, F. Ehrhart, L.J. Dupuis, L.A. Winckers, S.L. Coort, E.L. Willighagen, C.T. Evelo, A.R. Pico and M. Kutmon, WikiPathways: Connecting communities, Nucleic Acids Research 49: (D1) ((2020) ), D613–D621. doi:10.1093/nar/gkaa1024. |
[23] | M. Mora-Cantallops, S. Sánchez-Alonso and E. García-Barriocanal, A systematic literature review on Wikidata, Data Technologies and Applications 53: (3) ((2019) ), 250–268. doi:10.1108/dta-12-2018-0110. |
[24] | N.A. O’Leary, M.W. Wright, J.R. Brister, S. Ciufo, D. Haddad, R. McVeigh, B. Rajput, B. Robbertse, B. Smith-White, D. Ako-Adjei, A. Astashyn, A. Badretdin, Y. Bao, O. Blinkova, V. Brover, V. Chetvernin, J. Choi, E. Cox, O. Ermolaeva, C.M. Farrell, T. Goldfarb, T. Gupta, D. Haft, E. Hatcher, W. Hlavina, V.S. Joardar, V.K. Kodali, W. Li, D. Maglott, P. Masterson, K.M. McGarvey, M.R. Murphy, K. O’Neill, S. Pujar, S.H. Rangwala, D. Rausch, L.D. Riddick, C. Schoch, A. Shkeda, S.S. Storz, H. Sun, F. Thibaud-Nissen, I. Tolstoy, R.E. Tully, A.R. Vatsan, C. Wallin, D. Webb, W. Wu, M.J. Landrum, A. Kimchi, T. Tatusova, M. DiCuccio, P. Kitts, T.D. Murphy and K.D. Pruitt, Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation, Nucleic Acids Research 44: (D1) ((2015) ), D733–D745. doi:10.1093/nar/gkv1189. |
[25] | Á. Ősz, L.S. Pongor, D. Szirmai and B. Győrffy, A snapshot of 3649 web-based services published between 1994 and 2017 shows a decrease in availability after 2 years, Briefings in Bioinformatics 20: (3) ((2017) ), 1004–1010. doi:10.1093/bib/bbx159. |
[26] | J. Pischimarov, C. Kuenne, A. Billion, J. Hemberger, F. Cemič, T. Chakraborty and T. Hain, sRNAdb: A small non-coding RNA database for gram-positive bacteria, BMC Genomics 13(1) ((2012) ). doi:10.1186/1471-2164-13-384. |
[27] | T. Putman, K. Hybiske, D. Jow, C. Afrasiabi, S. Lelong, M.A. Cano, G.S. Stupp, A. Waagmeester, B.M. Good, C. Wu and A.I. Su, ChlamBase: a curated model organism database for the Chlamydia research community, Database 2019 ((2019) ). doi:10.1093/database/baz041. |
[28] | T.E. Putman, S. Lelong, S. Burgstaller-Muehlbacher, A. Waagmeester, C. Diesh, N. Dunn, M. Munoz-Torres, G.S. Stupp, C. Wu, A.I. Su and B.M. Good, WikiGenomes: an open web application for community consumption and curation of gene annotation data in Wikidata, Database 2017 ((2017) ). doi:10.1093/database/bax025. |
[29] | A. Santos, A.R. Colaço, A.B. Nielsen, L. Niu, M. Strauss, P.E. Geyer, F. Coscia, N.J.W. Albrechtsen, F. Mundt, L.J. Jensen and M. Mann, A knowledge graph to interpret clinical proteomics data, Nature Biotechnology 40: (5) ((2022) ), 692–702. doi:10.1038/s41587-021-01145-6. |
[30] | M. Sassi, Y. Augagneur, T. Mauro, L. Ivain, S. Chabelskaya, M. Hallier, O. Sallou and B. Felden, SRD: A staphylococcus regulatory RNA database, RNA 21: (5) ((2015) ), 1005–1017. doi:10.1261/rna.049346.114. |
[31] | K. Shenoy, F. Ilievski, D. Garijo, D. Schwabe and P. Szekely, A study of the quality of Wikidata, Journal of Web Semantics 72: ((2022) ), 100679. doi:10.1016/j.websem.2021.100679. |
[32] | D. Shotton, K. Portwin, G. Klyne and A. Miles, Adventures in semantic publishing: Exemplar semantic enhancements of a research article, PLoS Computational Biology 5: (4) ((2009) ), e1000361. doi:10.1371/journal.pcbi.1000361. |
[33] | G. Storz, S. Altuvia and K.M. Wassarman, An abundance of RNA regulators, Annual Review of Biochemistry 74: (1) ((2005) ), 199–217. doi:10.1146/annurev.biochem.74.082803.133136. |
[34] | T.V. Veen, Wikidata – from “an” identifier to “the” identifier, Information Technology and Libraries 38: (2) ((2019) ), 72–81. doi:10.6017/ital.v38i2.10886. |
[35] | D. Vrandečić, Wikidata: A new platform for collaborative data collection, in: WWW ’12: Proceedings of the 21st International Conference on World Wide Web, ACM, Association for Computing Machinery, New York, United States, (2012) . doi:10.1145/2187980.2188242. |
[36] | D. Vrandečić and M. Krötzsch, Wikidata: A free collaborative knowledgebase, Communications of the ACM 57: (10) ((2014) ), 78–85. doi:10.1145/2629489. |
[37] | A. Waagmeester, G. Stupp, S. Burgstaller-Muehlbacher, B.M. Good, M. Griffith, O.L. Griffith, K. Hanspers, H. Hermjakob, T.S. Hudson, K. Hybiske, S.M. Keating, M. Manske, M. Mayers, D. Mietchen, E. Mitraka, A.R. Pico, T. Putman, A. Riutta, N. Queralt-Rosinach, L.M. Schriml, T. Shafee, D. Slenter, R. Stephan, K. Thornton, G. Tsueng, R. Tu, S. Ul-Hasan, E. Willighagen, C. Wu and A.I. Su, Wikidata as a knowledge graph for the life sciences, eLife 9 ((2020) ). doi:10.7554/elife.52614. |
[38] | A. Waagmeester, E.L. Willighagen, A.I. Su, M. Kutmon, J.E.L. Gayo, D. Fernández-Álvarez, Q. Groom, P.J. Schaap, L.M. Verhagen and J.J. Koehorst, A protocol for adding knowledge to Wikidata: Aligning resources on human coronaviruses, BMC Biology 19(1) ((2021) ). doi:10.1186/s12915-020-00940-y. |
[39] | E.G.H. Wagner and P. Romby, Small RNAs in bacteria and archaea, in: Advances in Genetics, Elsevier, (2015) , pp. 133–208. doi:10.1016/bs.adgen.2015.05.001. |
[40] | M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L.B. da Silva Santos, P.E. Bourne, J. Bouwman, A.J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C.T. Evelo, R. Finkers, A. Gonzalez-Beltran, A.J.G. Gray, P. Groth, C. Goble, J.S. Grethe, J. Heringa, P.A.C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S.J. Lusher, M.E. Martone, A. Mons, A.L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M.A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, The FAIR Guiding Principles for scientific data management and stewardship, Scientific Data 3(1) ((2016) ). doi:10.1038/sdata.2016.18. |
[41] | J.D. Wren, C. Georgescu, C.B. Giles and J. Hennessey, Use it or lose it: Citations predict the continued online availability of published bioinformatics resources, Nucleic Acids Research 45: (7) ((2017) ), 3627–3633. doi:10.1093/nar/gkx182. |
[42] | J. Youn, N. Rai and I. Tagkopoulos, Knowledge integration and decision support for accelerated discovery of antibiotic resistance genes, Nature Communications 13(1) ((2022) ). doi:10.1038/s41467-022-29993-z. |