AgreementMakerLight
Abstract
Ontology matching establishes correspondences between entities of related ontologies, with applications ranging from enabling semantic interoperability to supporting ontology and knowledge graph development. Its demand within the Semantic Web community is on the rise, as the popularity of knowledge graph supporting information systems or artificial intelligence applications continues to increase.
In this article, we showcase AgreementMakerLight (AML), an ontology matching system in continuous development since 2013, with demonstrated performance over nine editions of the Ontology Alignment Evaluation Initiative (OAEI), and a history of real-world applications across a variety of domains. We overview AML’s architecture and algorithms, its user interfaces and functionalities, its performance, and its impact.
AML has participated in more OAEI tracks since 2013 than any other matching system, has a median rank by F-measure between 1 and 2 across all tracks in every year since 2014, and a rank by run time between 3 and 4. Thus, it offers a combination of range, quality and efficiency that few matching systems can rival. Moreover, AML’s impact can be gauged by the 263 (non-self) publications that cite one or more of its papers, among which we count 34 real-world applications.
1.Introduction
Ontologies are a cornerstone of the Semantic Web, serving as knowledge models for describing data on the web, namely under knowledge graphs [4]. They are also critical for the realization of the FAIR data principles [27] as they provide the means for enabling findability, interoperability and reusability of (meta)data. Yet, ontologies represent particular points of view of their domains, reflecting the concrete application that motivated their development. Thus, distinct ontologies may overlap in domain but differ substantially in how they model that domain, with respect to terminology, granularity and/or structure. When related datasets are described using distinct but overlapping ontologies, we have a semantic interoperability problem, which can be addressed by establishing mappings between the ontologies [5].
Ontology matching is the process of finding correspondences between entities of ontologies that overlap in domain [5]. This is essential to a full realization of the Semantic Web vision, as it links entities from different ontologies while still maintaining the distributed nature of semantic resources. It is also highly relevant for ontology development, as it is facilitates the common practice of creating explicit mappings to existing ontologies in the form of cross-references, as well as the reuse and integration of existing ontologies.
Ontology matching can be performed (semi-)automatically through the use of ontology matching systems that identify potential mappings based on the various features of ontology entities. The annual Ontology Alignment Evaluation Initiative (OAEI) [21] serves as a testing ground for these systems, by providing a wide range of benchmarks and homogeneous testing conditions.
In this article, we showcase AgreementMakerLight (AML), one of the most successful ontology matching systems with respect to performance in the OAEI as well as use in real-world applications. Originally released in 2013 [9], AML has been in continuous development since, and features numerous new functionalities.
The rest of the article is organized as follows: Section 2 details the key concepts in ontology matching; Section 3 overviews AML’s architecture, algorithms and user interface; Section 4 reviews its evaluation results; Section 5 compiles its impact and its applications to real-world ontology matching problems; and Section 6 concludes the article with our perspective on the present and future of AML.
2.Background
In its traditional form, the ontology matching process takes as input two ontologies – source and target – and outputs a set of correspondences between their entities, called an alignment [5]. A correspondence establishes a directional link between two ontology entities, typically in the form of a 4-tuple
Ontology matching systems typically employ three types of algorithms: pre-processing algorithms, which load the input ontologies and extract and process the information that will be used to match them (e.g. by normalizing labels and synonyms); matching algorithms (or matchers) which exploit the information of the ontologies to generate candidate correspondences between them, resulting in preliminary alignments; and filtering algorithms (or filters) which remove candidate correspondences from preliminary alignments according to predefined criteria, to increase the quality of the final alignment. In some systems, matching and filtering are meshed together.
Matchers can be classified according to the ontology features they rely on: terminological matchers rely on lexical information (i.e. annotations such as labels and synonyms); structural matchers rely on the relations between entities; semantic matchers interpret the semantics of the ontologies, usually using a reasoner; and data matchers rely on attributive information (i.e. the data values that characterize individuals).
Filters can be classified according to the principles that guide them, including: similarity, cardinality, coherence, conservativity, and locality. Similarity filters simply filter mappings below a given confidence score threshold. Cardinality filters aim to ensure or approximate a maximum cardinality in the alignment (i.e., a maximum number of correspondences per entity), generally 1-to-1. Coherence filters, or alignment repair algorithms, remove mappings that cause unsatisfiabilities or incoherences when the ontologies are considered together with the alignment [16]. Conservativily filters exclude correspondences that would lead to novel axioms being inferred when the ontologies are considered together with the alignment, such as two classes from the source ontology being inferred as equivalent if in the alignment they are equivalent to the same class of the target ontology [14]. Thus, these filters encompass strict 1-to-1 cardinality filtering. Finally, locality filters are predicated on the assumption that correspondences should be co-located within the structure of the ontologies, and can be either high level if they look to the top branches or blocks of the ontology, or low level if they look to the direct vicinity of mappings [14]. High level locality filtering, or blocking, can be performed a priori (i.e., before full matching) in the case of very large ontologies, to reduce the dimension of the matching space.
Ontology alignments can be stored independently from the ontologies or incorporated into them, and can have varying degrees of semantic enforcing. The de facto standard is to encode them as external files using the RDF Alignment format,11 but they can also be encoded in OWL either semantically (e.g. with equivalent class or subclass axioms) or no semantics (e.g. with annotations such as cross-references).
As an example, let us consider the OAEI’s Anatomy test case: a schema matching task that consists of matching mouse anatomic structures (represented as classes) from the Mouse Adult Gross Anatomy Ontology to the corresponding human anatomic structures (again represented as classes) from a subset of the NCI Thesaurus [3]. These ontologies include respectively 2737 and 3298 classes, making this a relatively small matching problem for the biomedical domain. On the lexical front, each class is annotated with a label and some are also annotated with synonyms (11% and 29% of the classes, respectively). Structurally, the ontologies include respectively 1807 and 3761 subclass relations between their classes (not counting subclasses of ‘owl:Thing’) but importantly, also respectively 1637 and 1662 ‘part of’ relations (represented as existential restrictions on the ‘part of’ property) denoting part-whole relations between anatomic structures (e.g. the brain is part of the central nervous system). Thus, while the terminological component is at the forefront of this matching task, with many anatomic structures having an equivalent or similar between mouse and human, the structural component is also highly relevant, with the caveat that both the subclass hierarchy and the ‘part of’ hierarchy should be considered. The manually curated reference alignment for this test case contains 1516 mappings between 1497 source classes and 1509 target classes, meaning the cardinality is approximately but not strictly 1-to-1, and therefore the conservativity principle is not observed in this test case. The coherence principle, however, is observed, as there are no unsatisfiable classes when the ontologies are merged with the alignment even though they would be possible (since the NCI Thesaurus includes disjoint class axioms). Figure 1 depicts an excerpt from this test case with examples of typical mappings found by AML. We will refer to this example throughout Section 3 to illustrate AML’s algorithms.
Fig. 1.
A subset of mappings between the Mouse Adult Gross Anatomy Ontology and the NCI thesaurus captured by different AML matching algorithms.
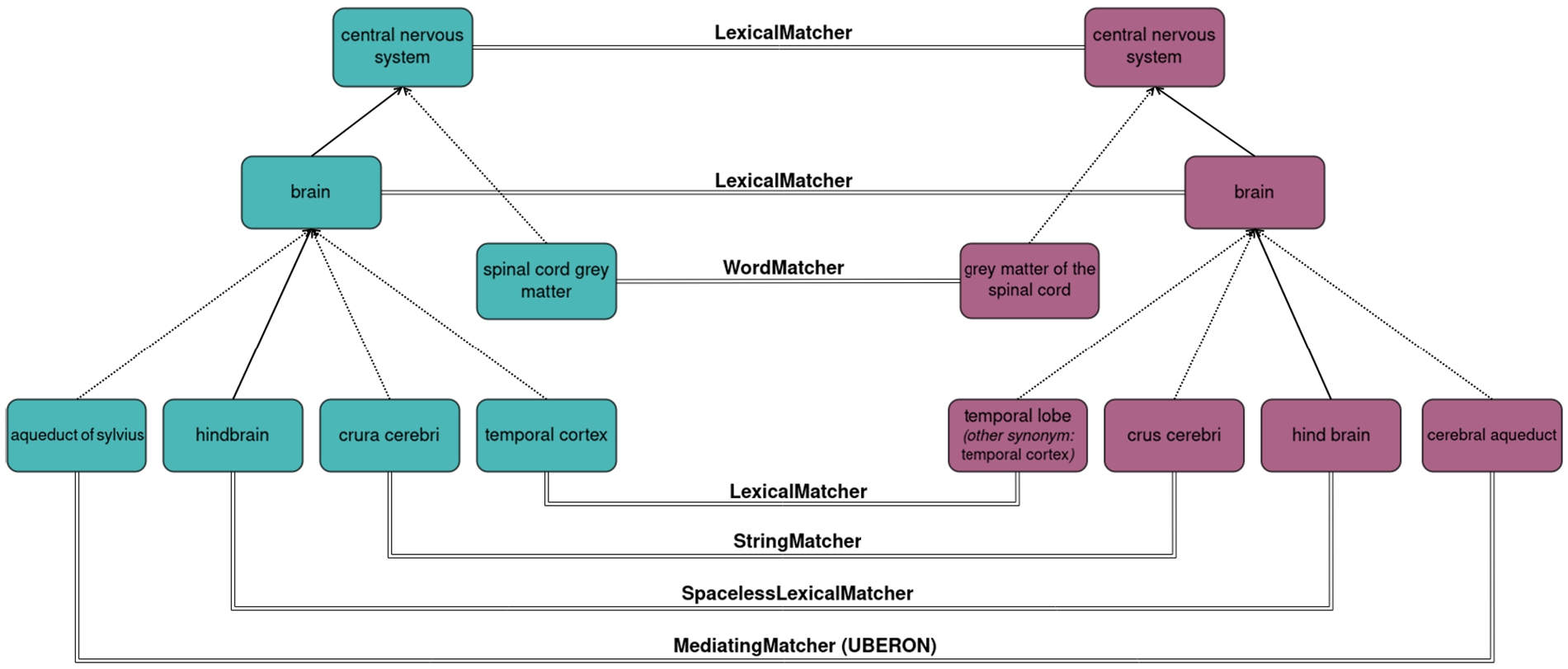
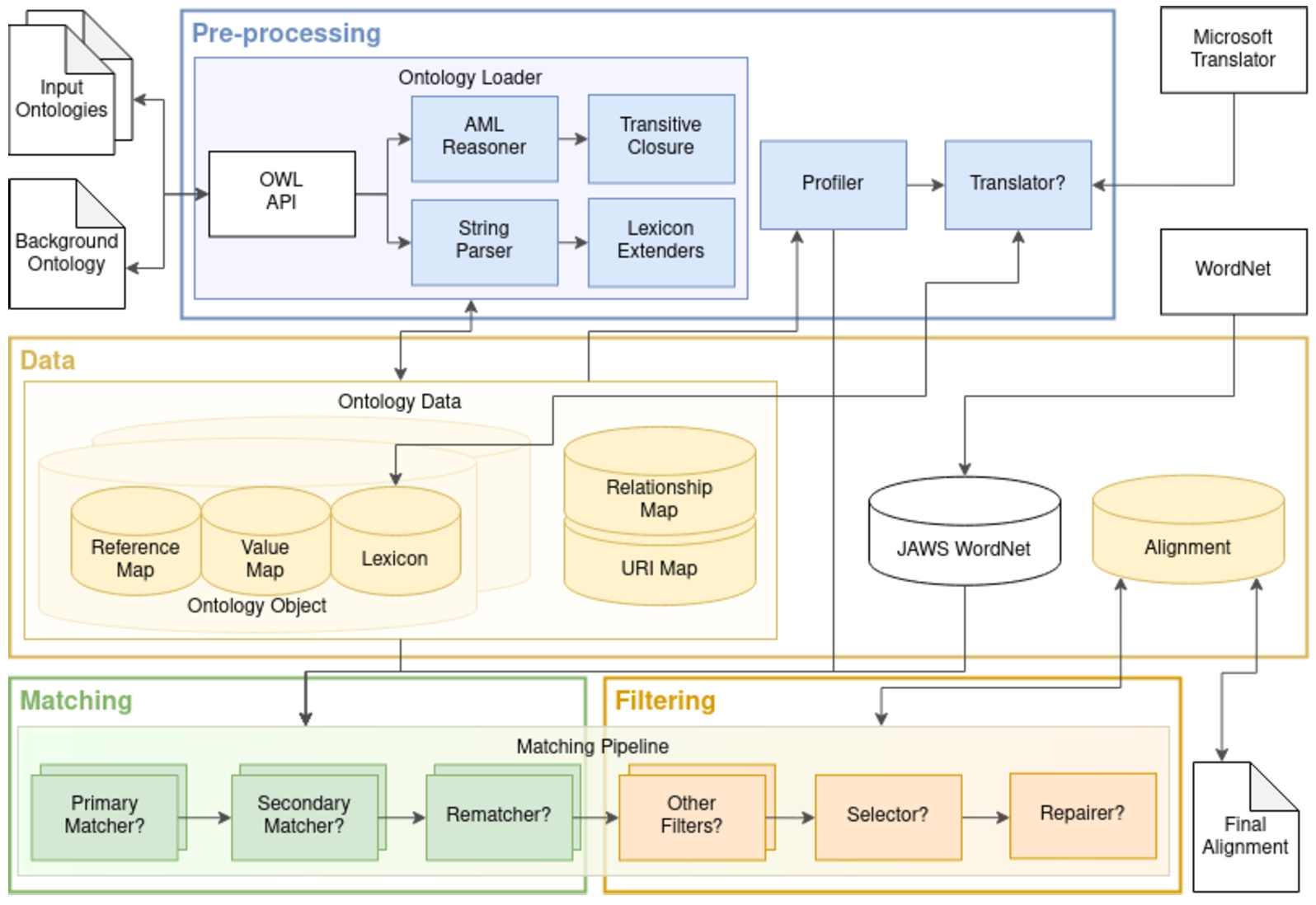
Fig. 2.
AML architecture. The pre-processing module handles the loading of ontologies and extracting all the relevant information to populate AML’s data structures. It also handles the profiling of the matching task, which configures which matching and filtering algorithms will compose AML’s matching pipeline, as well as their settings. Question marks indicate conditional steps.
3.AgreementMakerLight
AML is an ontology matching system that can perform schema and/or instance matching either automatically or interactively. It can also be used to perform repair of an existing ontology alignment and for alignment validation. AML is a Java open source project22 under the Apache License 2.0, featuring both a command line interface and a graphical user interface.
3.1.Core architecture
AML comprises four main modules: data, pre-processing, matching, and filtering (Fig. 2).
3.1.1.Data
The data module comprises AML’s data structures for representing ontology and alignment information. These are key-value tabular data structures geared to enable
– The URIMap stores the URIs and corresponding integer indexes assigned by AML to all entities of the two input ontologies.
– The RelationshipMap encompasses all semantic relations between the entities of the two input ontologies, including: subclass and equivalence relations between classes as well as class relations encoded in the ontologies through either existential or universal property restrictions (such as the part of relations displayed in our example), all of which are stored with transitive closure that also contemplates properties declared as transitive and property chains; disjoint classes; property domains and ranges; subproperty and equivalent property relations between properties; instance relations between individuals and classes; and relations between individuals.
– The Lexicon contains the lexical data of an ontology by language, including the local name of entities (when it is not an alphanumeric code), annotations of entities with lexical annotation properties (including established properties such as rdf:label, skos:prefLabel, skos:altLabel, oboInOwl:hasExactSynonym, oboInOwl:hasOtherSynonym, as well as any annotation property with local name containing ‘synonym’ or ‘SYN’ except oboInOwl:hasNarrowSynonym and oboInOwl:hasBroadSynonym which are explicitly excluded), and data values of individuals for data properties that have a local name or label ending on ‘name’ or ‘title’. All Lexicon entries are assigned one of five categories and a corresponding weight which reflects the expected precision of the property, as empirically determined from the analysis and matching of several existing ontologies [20]: local name > label > exact synonym > other synonym > formula. The latter is used for any lexical entry that AML determines not to be a word-based name, including chemical formulas, numbers, and short acronyms. In our example in Fig. 1, each class is identified by its label but the class labeled “temporal lobe” also features an other synonym “temporal cortex”. Each ontology has its own instance of Lexicon, including background knowledge ontologies.
– The ValueMap holds all attributes (i.e. data properties and their data values) for the individuals of an ontology, other than those placed in the Lexicon (i.e. name and title properties). Unlike the Lexicon, entries in the ValueMap are not normalized. Each ontology has its own instance of ValueMap.
– The ReferenceMap stores cross references and logical definitions of classes [10], which are common in biomedical ontologies. Each ontology has its own instance of ReferenceMap, including background knowledge ontologies.
– The Alignment represents an ontology alignment, storing all correspondences between entities in both a list (to enable sorting) and a key-valued table (to enable efficient search and reasoning). Each of AML’s matching algorithms will produce an instance of Alignment, and these can be combined through standard set operations – union, intersection and difference – as well as by adding only non-conflicting correspondences (i.e. those where neither entity is already present in the alignment) or only better correspondences (i.e. those with a higher confidence score than existing correspondences for either of the entities in the correspondence).
3.1.2.Pre-processing
The pre-processing module handles the loading of the input ontologies into memory, their profiling to configure the matching problem, and when necessary, their translation.
Ontology loading is mediated by the OWL API [12], which can read OWL ontologies in all official serializations, constructing an object-oriented ontology representation in Java. AML extracts from the latter all relevant information for ontology matching, populating its data structures. With respect to lexical information, all entries in the Lexicon except those detected as formulas (i.e. non-Latin-alphabet-word-based entries) are normalized by removing most non-word non-number characters, removing all diacritics, introducing spaces when case-changes or non-word characters are used as word separators, and converting all characters to lower case.
After loading, the transitive closure of class relations in the RelationshipMap is computed via the Semi-Naive algorithm [2], and two lexicon extender algorithms are applied to enrich the Lexicons of the two ontologies by generating synonyms for all lexical through the removal of all stop words (listed in AML’s StopList.txt file) and, separately, of all substrings within parenthesis.
The matching problem is then profiled by computing parameters that enable AML to decide on the matching strategy to use, including whether the ontologies need to be translated, which type(s) of entities should be matched and which matching algorithms to employ, and whether to match individuals only of the same class. The profiling parameters computed include: the number of entities in the ontologies, the proportion of entities of each type (classes, properties and individuals), the language(s) of their Lexicon entries, and classes in common between the input ontologies (in the case of instance matching problems). Default matching settings can be overridden either by declaring them in AML’s config.ini file or when running AML’s manual matcher via the graphical user interface.
Finally, when the ontologies to match do not have significant overlap between their language(s), AML performs automatic translation using Microsoft® Translator. The translation is done for each ontology, by querying Microsoft® Translator for each lexical entry (the full entry, rather than word-by-word) and translating it both into the primary language of the other ontology and into English. To improve performance, AML stores locally all translation results in dictionary files, and queries the Translator only when no stored translation is found. Users that wish to make use of this feature of AML are required to register their own instance of Microsoft® Translator and place a file with their authentication token in AML’s ‘store/’ folder.
3.1.3.Matching
The matching module encompasses AML’s matchers, which leverage the data in AML’s data structures from the input ontologies and/or external knowledge sources, to produce mappings between ontology entities. Each AML matcher can be classified in three dimensions: its matching strategy, the entity type(s) which it can match; and the matching mode(s) it supports. Additionally, matchers can be self-contained or be ensembles of other matchers.
With respect to matching strategy, matchers can be divided into pairwise and hash-based, with the former requiring an explicit pairwise comparison of the entities of the two ontologies and therefore having
Concerning entity types, matchers can apply to classes and/or properties and/or individuals, with each matcher storing the entity types it can match, and requiring a choice of entity type when called to match two ontologies. Some matchers are exclusively for a single type while others are transversal.
Matchers can implement one or more of the following matching modes: primary matcher, secondary matcher, rematcher, and lexicon extender. In primary matcher mode, the matcher performs a complete match, assessing all entities of the specified type of the two ontologies. In secondary matcher mode, an input alignment is required, and only entities (of the specified type) that are not in that alignment will be matched. Some matchers match all non-aligned entities in this mode, whereas others match only entity pairs in the vicinity of the correspondences in the input alignment, which improves both efficiency and precision. In rematcher mode, an input alignment is also required, but the matcher will only match the entities (of the specified type) that are already in correspondences in that alignment, by computing new confidence scores for the correspondence according to the metric employed by the matcher. This mode is useful for refining mappings, namely in two-stage matching algorithms where a first efficient global search determines the pairs of entities to match, then a more computationally-intensive algorithm is used to calculate the similarity of each of pair. In lexicon extender mode, instead of matching the ontologies, the algorithm extends their lexicons with additional synonyms, which can then be used by other matchers to match the ontologies. This mode is available for matchers involving background knowledge as a scalable way to enable inexact matching with string or word similarity algorithms: rather than use these algorithms to compare each input ontology with the background knowledge source, if the background knowledge source is used to enrich the input ontologies, the algorithms can be applied only between them, reducing the computational cost. Note that lexicon extenders are only considered as part of the matching stage when they involve matching the input ontologies against background knowledge sources; lexicon extenders that rely only on internal ontology information are considered part of the pre-processing stage (wherein they were listed).
The matchers currently available in AML are the following:
– LexicalMatcher: a hash-based primary matcher that finds string equivalent Lexicon entries between the ontologies and can be used for any entity type. Correspondences are scored by the product of the lexical weights of the lexical entities, so label-based correspondences such a “brain” ≡ “brain” in our example will have a higher score than the correspondences involving synonyms, such as “temporal cortex” ≡ “temporal lobe”. If several entities of the same type share the same name (e.g. if the label of one class is the synonym of another class of the same ontology) correspondences based on that name will receive a penalty to the score proportional to the number of entities that share the name, since the name is not discriminating.
– SpacelessLexicalMatcher: a matcher that is equivalent to the LexicalMatcher in all aspects except that it removes all spaces from all Lexicon entries (from clones of the Lexicons) in order to compute hash-based string equivalence ignoring spaces. This enables capturing mappings involving spaced versus agglutinated spelling variants of the same name, such as “hindbrain” ≡ “hind brain” from our example, which are fairly common, and would otherwise require computationally more intensive string matching to uncover them.
– WordMatcher: a two-staged hash-based matcher that computes name similarity through a weighted Jaccard index of the words in the Lexicon entries of ontology entities of any entity type. It can compute similarity by entity (hash-based), where all Lexicon entries of the entity are treated as a single bag of words for the Jaccard index, or by name (pairwise), where each lexical entry of the entity is treated as an independent bag of words, as well as by combining the two and taking the maximum or average. It enables capturing mappings involving spelling variants with translocated words, such as “spinal cord gray matter” ≡ “grey matter of the spinal cord” from our example. It can be used as a primary matcher, secondary matcher or rematcher.
– StringMatcher: a pairwise matcher that computes string similarity between the Lexicon entries of ontology entities of any entity type. Several traditional string similarity metrics are implemented, with the ISub metric [25] being used by default. It enables finding other minor spelling variants, such as “crura cerebri” ≡ “crus cerebri” from our example. It can be used as a primary matcher, secondary matcher or rematcher, but since it is a pairwise algorithm, use as a primary matcher will be time intensive for large ontologies. Thus its default mode for large scale class-matching problems is secondary matcher mode, which is local, meaning it will only attempt to match classes in the vicinity of classes already matched (contemplating subclass and other relations between classes); this improves both scalability and precision.
– DirectXRefMatcher: a hash-based primary matcher for classes that finds direct cross-references between the ontologies (i.e. when the cross-reference in the Reference Map of one ontology is a class in the other ontology) and shared Reference Map entries (i.e. when classes of the source and target ontologies have a cross-reference in common to a class of an external ontology, or have the same logical definition).
– MediatingMatcher: a hash-based primary matcher and lexicon extender for classes that relies on an external background knowledge ontology as a mediator, using the LexicalMatcher to compute correspondences between the mediator and both the input ontologies, and then matching each source and target class that correspond to the same mediator class. It enables finding matches that have low lexical similarity by exploiting the terminological knowledge encoded in external ontologies, such as the mapping “aqueduct of sylvius” ≡ “cerebral aqueduct” from our example, which can be found using the UBERON ontology as a mediator. The minimum score between the source-mediator correspondence and the mediator-target correspondence is used for the resulting source-target correspondence. In case the mediator class is promiscuous and would result in multiple source-target correspondences, the highest scoring is chosen if there is a single highest-scoring; or the mediator class is skipped entirely otherwise.
– MediatingXRefMatcher: a hash-based primary matcher and lexical extender for classes that relies on an external background knowledge ontology as a mediator by looking for direct cross-references between the mediator ontology and the two input ontologies, and complementing these with LexicalMatcher correspondences. As in the case of the MediatingMatcher, correspondences are made between each source and target class that correspond to the same mediator class. Promiscuous mediator classes, that have cross-references to more than one class of either of the input ontologies take a penalty to the matching score proportional to the number of classes they reference.
– WordNetMatcher: a hash-based primary matcher and lexical extender for classes that employs WordNet to extend (clones of) the Lexicons of the input ontologies with synonyms of each whole lexical entry and then finds string equivalent entries between the lexicons (analogously to the LexicalMatcher) that involve at least one WordNet synonym. It can only be used with English language Lexicon entries.
– BackgroundKnowledgeMatcher: a hash-based primary matcher for classes that is an ensemble of background knowledge algorithms. It tests the usefulness of all available knowledge sources using the MediatingXRefMatcher for background knowledge ontologies and the MediatingMatcher for external Lexicon files, as well as testing the WordNetMatcher, using the mapping gain algorithm described in [8].
– ThesaurusMatcher: a hash-based primary matcher that extends (clones of) the Lexicons of the input ontologies using synonyms inferred from lexical analysis of the entries in the Lexicons as detailed in [20] (e.g. inferring that the words ‘stomach’ and ‘gastric’ are synonymous because ‘stomach serosa’ and ‘gastric serosa’ are Lexicon entries for the same class, and then using this knowledge to generate new synonyms for all entries that include either ‘stomach’ or ‘gastric’) and then finds string equivalent entries between the ontologies that include at least one such synonym (analogously to the WordNetMatcher).
– AcronymMatcher: a pairwise matcher that aims to match an acronym and the corresponding full name in Lexicon entries of any entity type. It can be used as a primary matcher or secondary matcher, but the latter is recommended since the algorithm is evidently imprecise and should only be attempted in cases where all other lexical approaches fail to find a correspondence.
– HybridStringMatcher: a pairwise matcher that computes string similarity between Lexicon entries of any entity type through a hybrid approach combining word overlap (analogously to the WordMatcher) with WordNet synonyms (word by word, instead of whole name like the WordNetMatcher) and the ISub metric. It can be used as a primary matcher, secondary matcher or rematcher, but use as a primary matcher will be time intensive for large ontologies.
– MultiWordMatcher: a pairwise primary matcher based on the Wu-Palmer similarity [28] between two-worded Lexicon entries where one word is shared and the other is related through WordNet.
– NeighborSimilarityMatcher: a pairwise structural matcher that computes similarity between classes based on the fraction of their subclasses and superclasses that are mapped in an input alignment. It can be used as a secondary matcher or rematcher.
– BlockRematcher: a pairwise structural matcher that computes similarity between classes based on the overlap between blocks (high-level divisions) of the ontologies in an input alignment. It can only be used as a rematcher.
– InstanceBasedClassMatcher: a hash-based primary class matcher that computes similarity between classes based on the overlap between their instances, measured through a Jaccard index.
– ValueMatcher: a hash-based instance matcher that finds string equivalent literals in the ValueMaps of the two input ontologies that are values for the same data property or for matching data properties and matches the individuals that have the matching values. That is to say, it finds individuals that have the same value for the same or corresponding property. It can be used as a primary or secondary matcher.
– ValueStringMatcher: a pairwise instance matcher that employs the same algorithm as the HybridStringMatcher to measure string similarity between literals in the ValueMaps of the two input ontologies that are values for the same data property or for matching data properties and matches the individuals that have the matching values. That is to say, it finds individuals that have a similar value for the same or corresponding property. It can be used as a primary or secondary matcher.
– Value2LexiconMatcher: a pairwise instance matcher that also employs the same algorithm as the HybridStringMatcher but compares ValueMap entries of one ontology with Lexicon entries of the other (in both directions), to capture cases where one ontology uses annotation properties to describe aspects that another describes with data properties. It can be used as a primary or secondary matcher.
– ProcessMatcher: a pairwise instance matcher that employs the same algorithm as the HybridStringMatcher in a preliminary step to identify individuals with similar Lexicon entries, then propagates similarity across the individuals related to those individuals in an approach analogous to similarity flooding [17]. It was developed specifically for matching process models, where individuals are linked in a workflow. It can be used only as a primary matcher.
3.1.4.Filtering
AML’s filtering module encompasses its filters, which are responsible for producing a final high-quality alignment from the preliminary alignment produced by the matching module. AML includes two modes for filtering: filterer and flagger. The filterer mode is aimed at automatic use, as the algorithms automatically classify mappings as incorrect, whereas the flagger mode is intended for manual use, as the problematic mappings are ‘flagged’ which highlights them in the user interface for user validation. AML’s filters and their principles are the following:
– Selector: a cardinality filter that uses a greedy heuristic to select correspondences sorted by descending confidence score as long as they do not conflict with already selected correspondences, so as to produce a (near) 1-to-1 alignment. In strict mode, no conflicts are allowed; in permissive mode, conflicts where the confidence score is tied are allowed (i.e. both mappings are returned instead of choosing arbitrarily between them); in hybrid mode, all mappings above 0.7 confidence score are allowed and below that threshold selection is made as in permissive mode.
– CardinalitySelector: a cardinality filter that uses the same greedy heuristic as the Selector but allows up to n conflicts per mapping, where n is specified as an argument, with the goal of producing an n-to-n alignment.
– Repairer: a coherence filter that removes correspondences that would cause unsatisfiable classes using modularization to compute the latter efficiently and a selection heuristic to determine which correspondences to remove, as detailed in [23].
– DomainAndRangeFilterer: a coherence filter that removes property correspondences that do not have compatible domain and range declarations (i.e. the domains are not the same, equivalent, or subsumed considering existing class correspondences).
– ObsoleteFilterer: a deprecation filter that removes correspondences involving obsolete/deprecated classes.
3.2.User interfaces & functionalities
AML features both a command line interface (CLI) and a graphical user interface (GUI) [19], with the latter being called when the AML jar is executed with no arguments, and the former being called when command line arguments are given.
The CLI is intended for automatic use of AML, either to match two ontologies or to repair an existing ontology alignment, with command line options detailed in a README file. Match settings can be configured through a ‘config.ini’ file.
Fig. 3.
AML’s graphical user interface displaying the alignment panel with alignment validation functionalities. Mappings highlighted in orange were flagged automatically by AML’s filters.
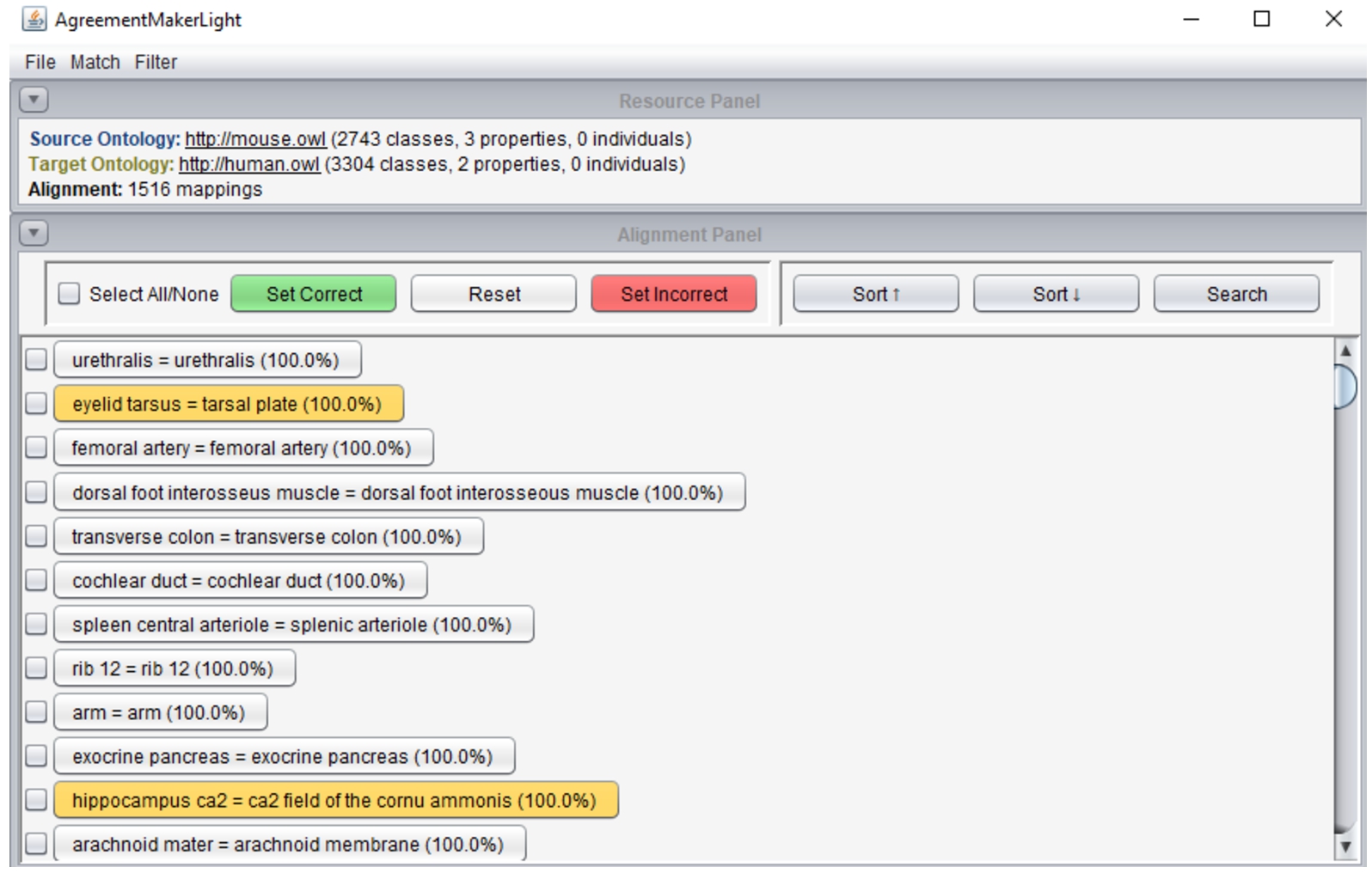
The GUI supports automatic or custom ontology matching, automatic filtering (including repair) of an input alignment, and manual alignment validation. To support the latter, AML provides two views of the alignment: a primary list view of the whole alignment, where mappings can be classified by the user with color-coding, as displayed in Fig. 3; and secondary a mapping panel with a local graph view of the alignment and information about the mapped classes, including conflicting mappings, which can be accessed by clicking on a mapping. AML’s filters can be run in flag mode, which highlights the problem-causing mappings in orange, deferring to the user the decision on which to remove. Alignments can be saved in either RDF and TSV, including information on the classification of each correspondence, which allows alignment validation to be carried out over multiple sessions.
With respect to the use of background knowledge, AML automatically checks the ‘store/knowledge’ folder for available ontologies, so users can place any ontologies they desire to use as background knowledge in this folder. By default, AML will evaluate all background knowledge sources available using the algorithm described in [8], selecting only those that are relevant for the matching problem at hand, but the list of sources to test can be manually configured either via the GUI in custom matching mode or through the ‘config.ini’ file.
4.Performance
AML’s performance in ontology matching can be assessed through its results in the OAEI, where it has entered in all editions between 2013 and 2021. Its range is evidenced by the fact that, since 2016, when AML was extended with instance matching capabilities, it participated in more distinct OAEI tracks (14) than any other matching system, spanning the variety of types and domains summarized in Table 1. It is rivaled in OAEI participation and range only by LogMap [13] which was the only other system that entered in all OAEI editions in this period, and participated in 13 distinct tracks. No other matching system participated in more than 8 distinct tracks.
Table 1
Recurring OAEI tracks in which AML participated, classified by type and domain
| Track | Type | Domain | Participations |
| Anatomy | Schema matching | Biomedical | 2013–2021 (9) |
| Large biomedical ontologies | Schema matching | Biomedical | 2013–2021 (9) |
| Disease & phenotype | Schema matching | Biomedical | 2016–2021 (6) |
| Biodiversity & ecology | Schema matching | Biomedical | 2018–2021 (4) |
| Conference | Schema matching | Conferences | 2013–2021 (9) |
| Interactive matching | Interactive schema matching | Multiple | 2013–2021 (9) |
| Multifarm | Multilingual schema matching | Conferences | 2013–2021 (9) |
| Library | Multilingual thesauri matching | Libraries | 2013–2014 (2) |
| Benchmark | Schema matching | Multiple | 2013–2016 (4) |
| Process model | Instance matching | Process models | 2016–2017 (2) |
| DOREMUS | Instance matching | Music | 2016–2017 (2) |
| SPIMBENCH | Instance matching | Creative works | 2017–2021 (5) |
| Link discovery | Link discovery & spatial matching | Path coordinates | 2017–2021 (5) |
| Knowledge graph | Schema & instance matching | Fandom wikis | 2018–2021 (4) |
Table 2
AML’s OAEI participation and results across the years
| 2013 | 2014 | 2015 | 2016 | 2017 | 2018 | 2019 | 2020 | 2021 | |
| Total tracks | 7/8 | 7/8 | 6/7 | 9/9 | 9/9 | 11/11 | 10/11 | 10/12 | 10/13 |
| Total matching tasks | 12 | 14 | 12 | 25 | 20 | 22 | 21 | 21 | 19 |
| New tasks | - | 2 | 1 | 14 | 4 | 6 | 0 | 2 | 0 |
| Median F-measure | 0.73 | 0.75 | 0.71 | 0.74 | 0.83 | 0.76 | 0.73 | 0.78 | 0.78 |
| Median rank by F-measure | 4 | 1 | 1 | 2 | 1 | 1.5 | 1 | 1.5 | 1 |
| Median rank by run time | 3 | 3.5 | 3.5 | 3 | 3 | 4 | 4 | 4 | 4 |
With respect to the quality of its results, as summarized in Table 2, AML had a median rank by F-measure across all OAEI tasks it participated in between 1 and 2 since 2014. Concerning efficiency, it had a median rank by run time between 3 and 4 in all OAEI editions. These results highlight that AML is an all-purpose ontology matching system, capable of tackling a variety of tasks efficiently and with high quality, again rivaled only by LogMap [13].
5.Impact and use
AML has also been one of the ontology matching systems with the greatest impact in research and beyond, as evidenced by the number of citations of its previous publications as well as by the several applications for which it was used.
We classified the 263 unique publications that cite one or more of AML’s previous publications33 in three dimensions: by type of publication, by type of citation, and by application domain. With respect to type of publication, we count 208 research articles, 40 theses, 7 review articles, 7 OAEI system papers and 1 book chapter. Regarding the type of citation, 123 publications merely mention AML, usually as part of the state of the art in ontology matching, 102 publications use AML as a benchmark against which a proposed algorithm is compared, 4 publications propose extensions to AML (external to the AML team) and 34 publications apply AML in real-world ontology matching problems. While 141 publications are domain agnostic, 122 focus on a specific application domain, with the distribution displayed in Fig. 4.
Fig. 4.
Domain distribution of publications citing AML.
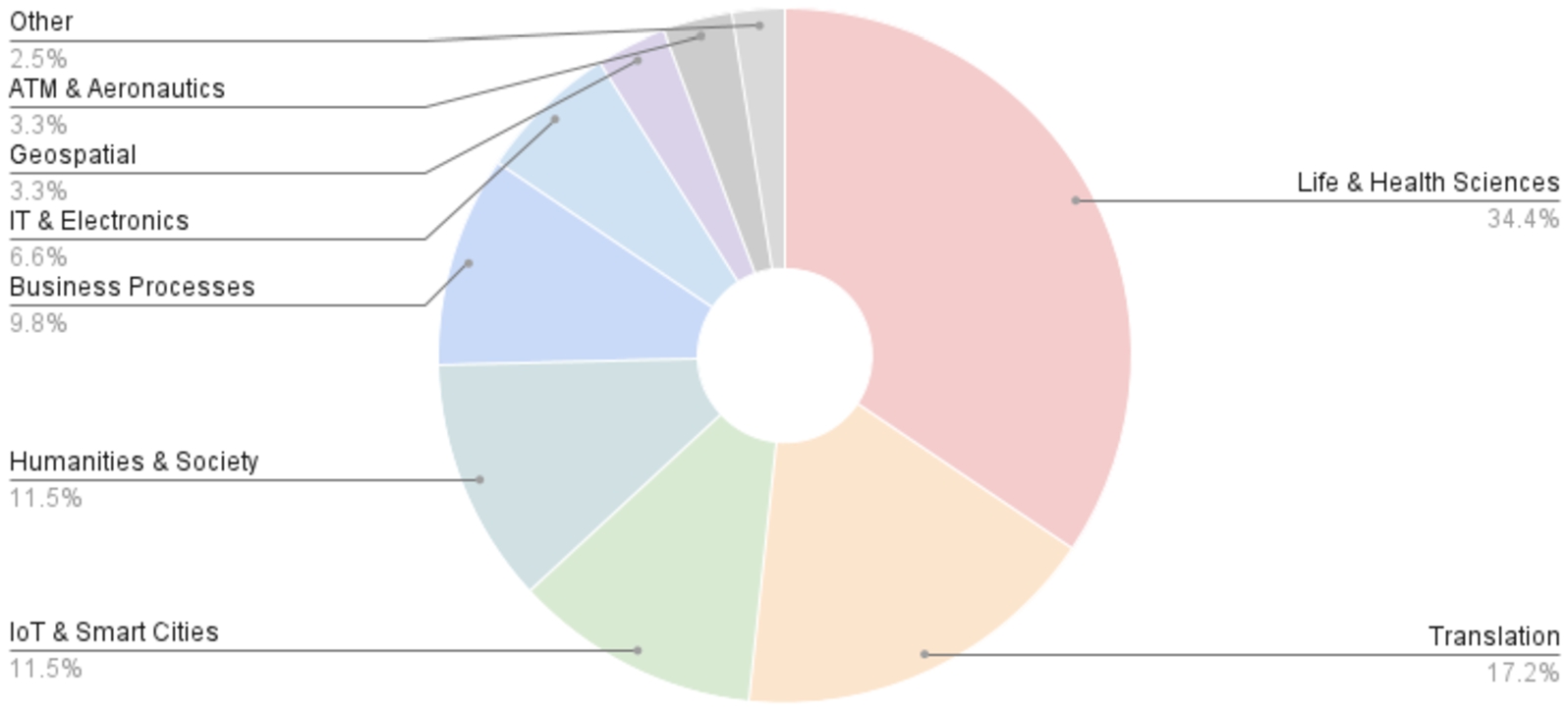
Many of the application domains under which AML is cited or applied reflect its successes in the OAEI, including the two most frequent domains, the life & health sciences and translation & cross-lingual matching, but also Business Processes and the Geospatial domain. Others, however, are domains in which we had never tested AML, including the Internet of Things (IoT) & Smart Cities, Humanities & Society, Information Technology (IT) & Electronics, and Air Traffic Management (ATM) & Aeronautics.
Among real-world problems to which AML was applied, we highlight the integration of agricultural thesauri under the Global Agricultural Concept Space for the Food and Agricultural Organization of the United Nations (FAO) [1], the creation of a knowledge graph for ecotoxicology risk assessment [18], and the alignment of ATM ontologies from the Single European Sky ATM Research (SESAR) and the National Aeronautics and Space Administration (NASA) [11].
Last but not least, further evidence of the impact and use of AML comes from the statistics of its GitHub repository, which was forked 31 times and starred 38 times.44
6.Conclusions and future work
AML is one of the most successful ontology matching systems, both in terms of its results in the OAEI and in terms of its impact in real-world applications. It evolved from a matching system focusing mainly on the biomedical domain to an all-purpose system with wide applicability, thanks to both a continuous development effort and a core architecture designed with extensibility in mind.
The future development of AML will contemplate new challenges arising in ontology matching, as we endeavor to keep it on the forefront of the field.
As the Semantic Web evolves, we are witnessing the proliferation of huge knowledge graphs supporting online information systems as well as AI applications, which will increase demand for ontology matching, both to support the construction of knowledge graphs from existing data and ontologies, and to enable interoperability between information systems relying on related knowledge graphs. These applications place additional emphasis on scalability, and may require taking ontology matching beyond the pairwise paradigm, and into holistic matching.
But by far the greatest standing challenge in ontology matching is complex matching, which requires identifying complex semantic relations between ontology entities, involving expressions such as property restrictions. Such mappings offer a solution to the problem of reconciling logical coherence with alignment completeness in ontology matching, yet detecting them automatically with reasonable accuracy is still beyond the state of the art, as our preliminary efforts have revealed [6].
Notes
1 https://moex.gitlabpages.inria.fr/alignapi/format.html, accessed April 14th 2022.
3 Citations were collected through Google Scholar on April 14th 2022, with self-citations excluded.
4 Statistics collected on April 14th 2022.
Acknowledgements
We dedicate this article to the memory of Prof. Isabel Cruz, without whom AML would not have existed. We also acknowledge the many students and collaborators who contributed to AML throughout the years.
This work was supported by: FCT through the LASIGE Research Unit (UIDB/00408/2020 and UIDP/00408/2020) and through project DeST: Deep Semantic Tagger (PTDC/CCI-BIO/28685/2017); the KATY project, funded by the European Union’s Horizon 2020 research and innovation program under grant agreement No 101017453; NSF award III-1618126; and NIGMS-NIH award R01GM125943.
References
[1] | T. Baker, B. Whitehead, R. Musker and J. Keizer, Global agricultural concept space: Lightweight semantics for pragmatic interoperability, NPJ science of food 3: (1) ((2019) ), 1–8. doi:10.1038/s41538-018-0033-5. |
[2] | F. Bancilhon, Naive evaluation of recursively defined relations, in: On Knowledge Base Management Systems, Springer, (1986) , pp. 165–178. doi:10.1007/978-1-4612-4980-1_17. |
[3] | O. Bodenreider, T.F. Hayamizu, M. Ringwald, S. De Coronado and S. Zhang, Of mice and men: Aligning mouse and human anatomies, in: AMIA Annual Symposium Proceedings, Vol. 2005: , American Medical Informatics Association, (2005) , p. 61. |
[4] | L. Ehrlinger and W. Wöß, Towards a definition of knowledge graphs, SEMANTiCS (Posters, Demos, SuCCESS) 48: (1–4) ((2016) ), 2. |
[5] | J. Euzenat and P. Shvaiko, Ontology Matching, 2nd edn, Springer-Verlag, Heidelberg (DE), (2013) . |
[6] | D. Faria, B. Lima, M.C. Silva, F.M. Couto and C. Pesquita, AML and AMLC Results for OAEI 2021, (2021) . |
[7] | D. Faria, C. Pesquita, I. Mott, C. Martins, F.M. Couto and I.F. Cruz, Tackling the challenges of matching biomedical ontologies, Journal of biomedical semantics 9: (1) ((2018) ), 1–19. doi:10.1186/s13326-017-0171-8. |
[8] | D. Faria, C. Pesquita, E. Santos, I.F. Cruz and F.M. Couto, Automatic background knowledge selection for matching biomedical ontologies, PLoS One 9: (11) ((2014) ), 111226. doi:10.1371/journal.pone.0111226. |
[9] | D. Faria, C. Pesquita, E. Santos, M. Palmonari, I.F. Cruz and F.M. Couto, The AgreementMakerLight ontology matching system, in: OTM Confederated International Conferences “On the Move to Meaningful Internet Systems”, Springer, (2013) , pp. 527–541. |
[10] | G.V. Gkoutos, C. Mungall, S. Dolken, M. Ashburner, S. Lewis, J. Hancock, P. Schofield, S. Kohler and P.N. Robinson, Entity/quality-based logical definitions for the human skeletal phenome using PATO, in: 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, IEEE, (2009) , pp. 7069–7072. doi:10.1109/IEMBS.2009.5333362. |
[11] | E. Gringinger, R.M. Keller, A. Vennesland, C.G. Schuetz and B. Neumayr, A comparative study of two complex ontologies in air traffic management, in: 2019 IEEE/AIAA 38th Digital Avionics Systems Conference (DASC), IEEE, (2019) , pp. 1–9. |
[12] | M. Horridge and S. Bechhofer, The OWL API: A Java API for OWL ontologies, Semantic Web 2: (1) ((2011) ), 11–21. doi:10.3233/SW-2011-0025. |
[13] | E. Jiménez-Ruiz and B. Cuenca Grau, Logmap: Logic-based and scalable ontology matching, in: International Semantic Web Conference, Springer, (2011) , pp. 273–288. |
[14] | E. Jiménez-Ruiz, B.C. Grau, I. Horrocks and R. Berlanga, Logic-based assessment of the compatibility of UMLS ontology sources, Journal of biomedical semantics 2: (1) ((2011) ), 1–16. doi:10.1186/2041-1480-2-1. |
[15] | I. Megdiche, O. Teste and C. Trojahn, An extensible linear approach for holistic ontology matching, in: International Semantic Web Conference, Springer, (2016) , pp. 393–410. |
[16] | C. Meilicke, Alignment incoherence in ontology matching, Universität Mannheim, 2011. |
[17] | S. Melnik, H. Garcia-Molina and E. Rahm, Similarity flooding: A versatile graph matching algorithm and its application to schema matching, in: Proceedings 18th International Conference on Data Engineering, IEEE, (2002) , pp. 117–128. doi:10.1109/ICDE.2002.994702. |
[18] | E.B. Myklebust, E. Jiménez-Ruiz, J. Chen, R. Wolf and K.E. Tollefsen, Prediction of adverse biological effects of chemicals using knowledge graph embeddings, 2021, arXiv preprint arXiv:2112.04605. |
[19] | C. Pesquita, D. Faria, E. Santos, J.-M. Neefs and F.M. Couto, Towards visualizing the alignment of large biomedical ontologies, in: International Conference on Data Integration in the Life Sciences, Springer, (2014) , pp. 104–111. |
[20] | C. Pesquita, D. Faria, C. Stroe, E. Santos, I.F. Cruz and F.M. Couto, What’s in a “nym”? Synonyms in biomedical ontology matching, in: International Semantic Web Conference (ISWC), Lecture Notes in Computer Science, Vol. 8218: , Springer, Berlin, Heidelberg, Germany, (2013) , pp. 526–541. |
[21] | N. Pour, A. Algergawy, R. Amini, D. Faria, I. Fundulaki, I. Harrow, S. Hertling, E. Jiménez-Ruiz, C. Jonquet, N. Karam et al., Results of the ontology alignment evaluation initiative 2020, in: Proceedings of the 15th International Workshop on Ontology Matching (OM 2020), Vol. 2788: , CEUR-WS, (2020) , pp. 92–138. |
[22] | D. Ritze, C. Meilicke, O. Sváb-Zamazal and H. Stuckenschmidt, A pattern-based ontology matching approach for detecting complex correspondences, in: OM, (2009) . |
[23] | E. Santos, D. Faria, C. Pesquita and F.M. Couto, Ontology alignment repair through modularization and confidence-based heuristics, PLoS ONE 10: (12) ((2015) ), 0144807. |
[24] | F. Scharffe, Correspondence patterns representation, PhD thesis, University of Innsbruck, 2009. |
[25] | G. Stoilos, G. Stamou and S. Kollias, A string metric for ontology alignment, in: International Semantic Web Conference (ISWC), Lecture Notes in Computer Science, Vol. 3729: , Springer, Berlin, Heidelberg, Germany, (2005) , pp. 624–637. |
[26] | É. Thiéblin, O. Haemmerlé, N. Hernandez and C. Trojahn, Survey on complex ontology matching, Semantic Web 11: (4) ((2020) ), 689–727. doi:10.3233/SW-190366. |
[27] | M.D. Wilkinson, M. Dumontier, I.J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L.B. da Silva Santos, P.E. Bourne et al., The FAIR guiding principles for scientific data management and stewardship, Scientific data 3: (1) ((2016) ), 1–9. |
[28] | Z. Wu and M. Palmer, Verbs semantics and lexical selection, in: Proceedings of the 32nd Annual Meeting on Association for Computational Linguistics, (1994) , pp. 133–138. doi:10.3115/981732.981751. |




