Reuse of the FoodOn ontology in a knowledge base of food composition data
Abstract
We describe our work to integrate the FoodOn ontology with our knowledge base of food composition data, WikiFCD. WikiFCD is knowledge base of structured data related to food composition and food items. With a goal to reuse FoodOn identifiers for food items, we imported a subset of the FoodOn ontology into the WikiFCD knowledge base. We aligned the import via a shared use of NCBI taxon identifiers for the taxon names of the plants from which the food items are derived. Reusing FoodOn benefits WikiFCD by allowing us to leverage the food item groupings that FoodOn contains. This integration also has potential future benefits for the FoodOn community due to the fact that WikiFCD provides food composition data at the food item level, and that WikiFCD is mapped to Wikidata and contains a SPARQL endpoint that supports federated queries. Federated queries across WikiFCD and Wikidata allow us to ask questions about food items that benefit from the cross-domain information of Wikidata, greatly increasing the breadth of possible data combinations.
1.Introduction
Regional and cultural variations in food consumption often reflect the availability of food sources. Even in our current, more global, food system where transportation of food sources is possible, great variety in food consumption patterns exists. Food composition tables (FCTs) are often created at the national level, and reflect the food consumption practices per country. WikiFCD is a global food composition database that brings together national food composition data sets from many sources in a single, web-based system.11
Food tracking applications reuse web-based data related to food composition. Food tracking via applications is a popular way to record food intake [5]. Food Data Central, the food composition database of the United States Department of Agriculture, has more food composition data for processed foods than for unprocessed foods. People who want to monitor their nutritional intake need to be able to find the foods that they eat in food tracking applications. If the underlying data sets do not contain data for a food item, those foods will not be available in food tracking applications, and people will not be able to record them. Some people in the United States have been consuming an increasing number of ultra-processed foods in the last two decades [15]. However, others are consuming plant-based foods [12]. Food composition data sets from countries other than the United States contain data for many whole foods and minimally-processed foods. We want to ensure that food composition data for as many whole foods and minimally-processed foods as possible are available via WikiFCD.
Our work to gather food composition data from published sources, and provide access to them on the web, creates the conditions for easier reuse. Our aim is to provide access to these data so that a greater breadth of human food items can be reused in food tracking applications, data sets, and research. Integrating FoodOn identifiers into this system will increase discoverability of the data, as FoodOn is a widely used ontology.
2.WikiFCD
WikiFCD is a knowledge base containing multiple food composition data sets [28]. The inspiration for WikiFCD is to have a web-based knowledge base that would provide access to food composition data for food items sourced from all over the world. Our aim is to center data related to unprocessed and minimally-processed food items, and to work toward providing data for foods from a large number of culturally-specific cuisines.
We implemented this knowledge base in an instance of Wikibase, an extension to the Mediawiki software. Mediawiki and the Wikibase extension are published under a free software license that encourages reuse. This is the same software that is used for Wikidata, a large public knowledge base. We chose Wikibase because it supports web editing by multiple users, and has both a SPARQL endpoint as well as an application programming interface (API). We also chose Wikibase so that it would be easier to donate this data back to Wikidata at some point in the future if the Wikidata community wants this level of detail about food composition.
The data model for WikiFCD consists of items and properties. Each item has a unique identifier. Statements made up of properties and values can be added to items to express information. Statements can also contain qualifiers, which are also properties, which scope statements in some way. For example, in Fig. 1, we see five statements. For the property water, we see the value of 80 grams. This statement is modified by the qualifier ‘per’ with a value of ‘100 grams’ to indicate that this measurement is based on a sample of 100 grams of the food item.
Fig. 1.
Item for Serviceberry in WikiFCD showing several statements.
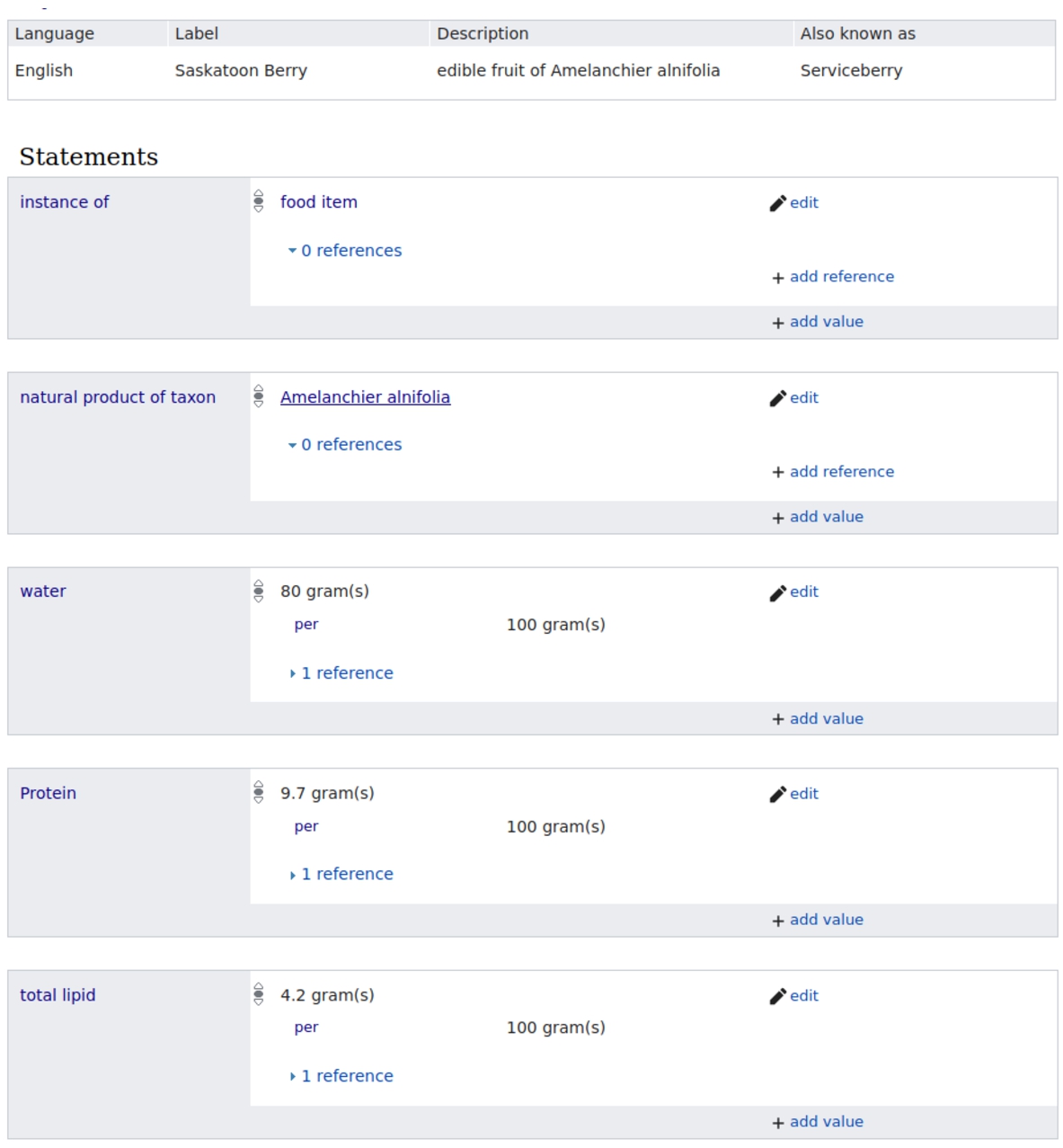
We have created three hundred fourteen properties in WikiFCD.22 Two hundred of these properties express nutrient values. We also have properties that express aspects of the publication process, how foods are related to species of plants, and where data were sourced.
The nutrient properties were created as we found nutrients expressed in the source FCTs we consulted. In this way, we are able to communicate the values as we observed them in our data sources. We then created items that represent the subject of the property. For example, Property 20 ‘Copper’ is linked to an item Q567680 for ‘Copper’ through the use of Property 304 ‘subject item of this property’. The item Q567680 for ‘Copper’ is mapped to the Wikidata item for ‘Copper’. This mapping allows us to reuse data about copper the element that is stored in Wikidata. It also allows us to bring together properties that partially overlap. The names of these properties are consistent with the nutrient names provided in the food composition tables we referenced.
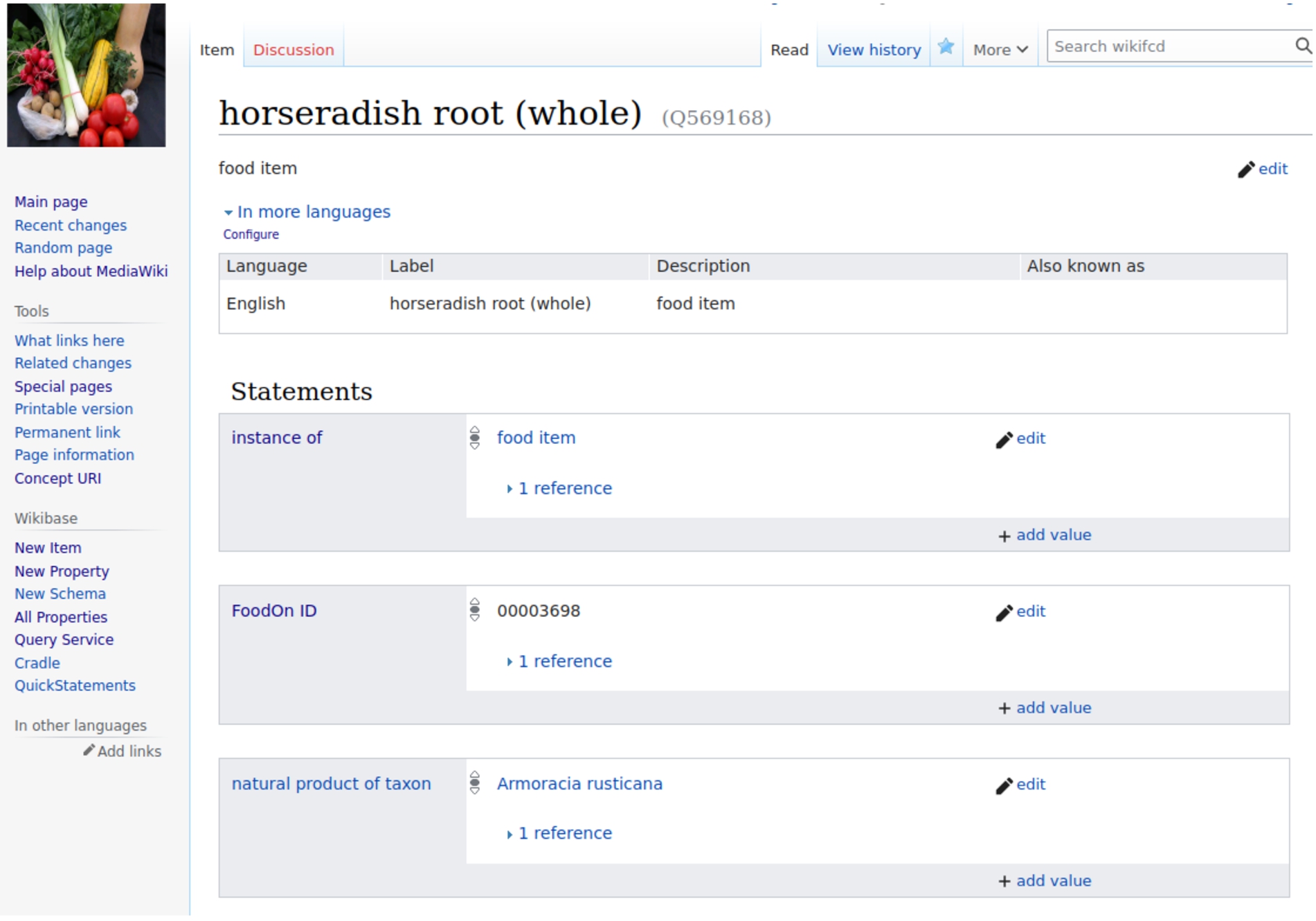
In order to connect food items in WikiFCD to the food items in FoodOn, we created a property for FoodOn identifiers, Property 309. This allows us to write queries such as https://tinyurl.com/264kt6mf which returns all foods with a FoodOn identifier and the labels of those foods. In Fig. 2, we see the WikiFCD item for ‘horseradish root (whole)’ and the second statement contains the FoodOn identifier 00003698.
Fig. 2.
Item for horseradish root (whole) with a statement containing the FoodOn identifier.
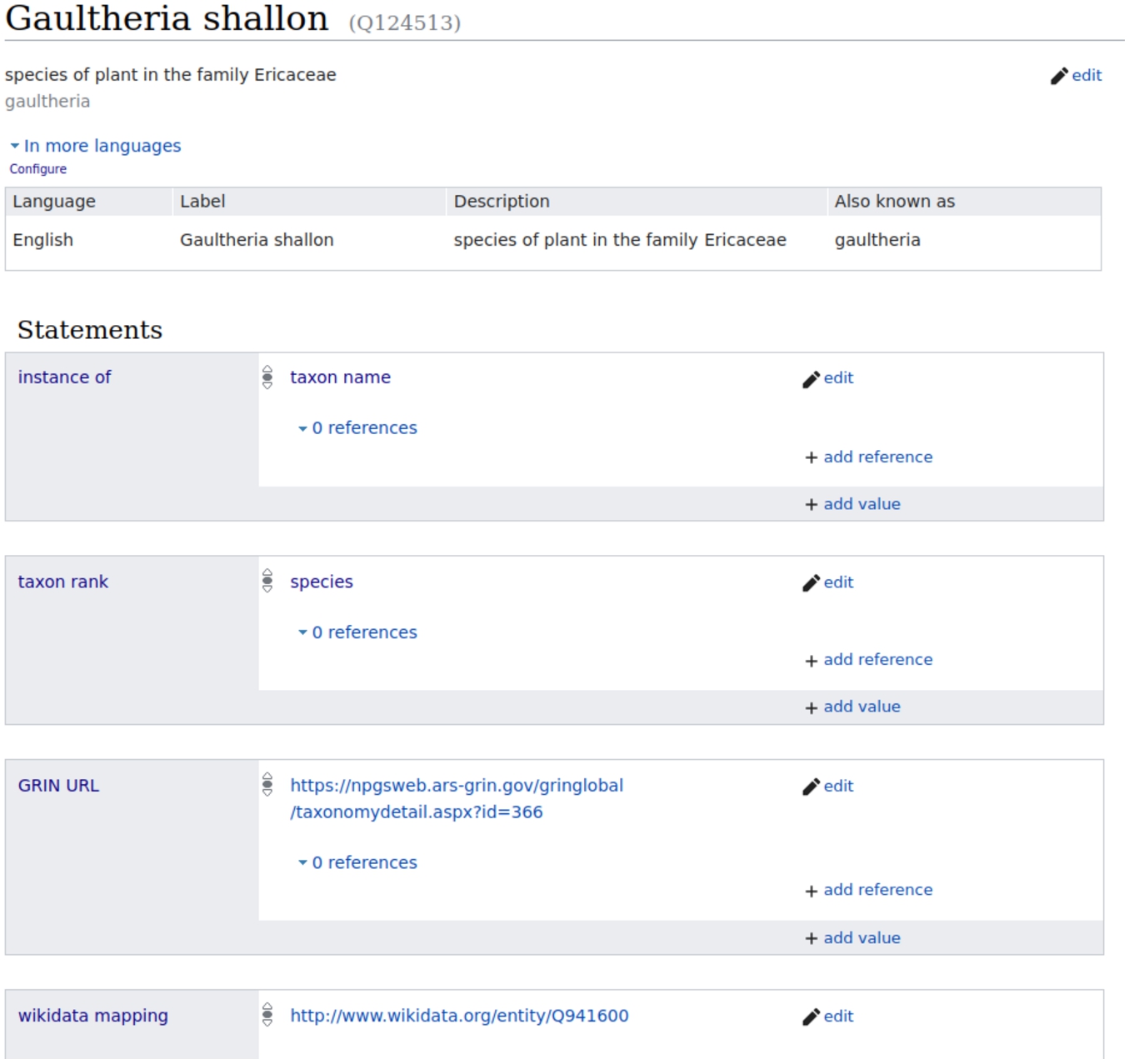
We created direct mappings to corresponding Wikidata items for several classes in WikiFCD. The classes we mapped are: Taxon names, languages, countries, and source item (nutrient) for nutrient properties. In Fig. 3 we see a screenshot of the item for Gaultheria shallon in WikiFCD, and the fourth property is the Wikidata mapping.
Fig. 3.
Item for Gaultheria shallon in WikiFCD showing Wikidata mapping.
We reused a subset of Wikidata covering human languages, creating items for each language in WikiFCD, and creating a mapping back to Wikidata. We added these items so that we could use them to indicate the languages that food names are expressed in from food composition tables. In this way, we can identify the source languages for different food names for the same food item. Here is a query for all human languages listed in WikiFCD and their mappings to the corresponding Wikidata items: https://tinyurl.com/29lajjgx.
Many food composition tables are created by national organizations and describe food items relevant to individual countries. Thus it is important to be able to indicate what country published a food composition data set. We used the same subset process to create items for countries in WikiFCD. We brought this subset in from Wikidata in order to be able to use these country items to indicate where food composition tables were published. Following the same process, we reused the subset of Wikidata items that describe chemical compounds and chemical elements that are considered nutrients in the human diet. These mappings allow us to connect the properties in WikiFCD that represent these nutrients with additional information about them from Wikidata. Nutrient properties and their related chemical items can be seen in this query: https://tinyurl.com/2bmmqhhb.
We have imported data from Food Data Central, as well as the national food composition tables from Vietnam, Laos, Cambodia, Thailand, Malawi, Uganda and Nigeria. This means we have multiple records for many food items. For example, searching the WikiFCD system for ‘brussels sprouts’ returns two hundred eighteen results. This search is matching the string ‘brussels sprouts’ with the label text of each food item. Prior to integrating FoodOn into WikiFCD, this was our only way to group these similar records. National food composition tables may provide data about what plant species a food item is derived from, or provide a category such as ‘vegetables and grains’, but these are not standardized data fields across FCTs, thus there is variability in the reporting of these data. To provide information about food groupings, we decided to integrate FoodOn into the structure of WikiFCD. Currently we need to query FoodOn externally to find these food groupings, we have not yet integrated them into WikiFCD. We plan to add food grouping statements to WikiFCD after completing the process of adding relevant FoodOn identifiers to the food items from our FCT sources, which we discuss in [29].
2.1.Data provenance and references
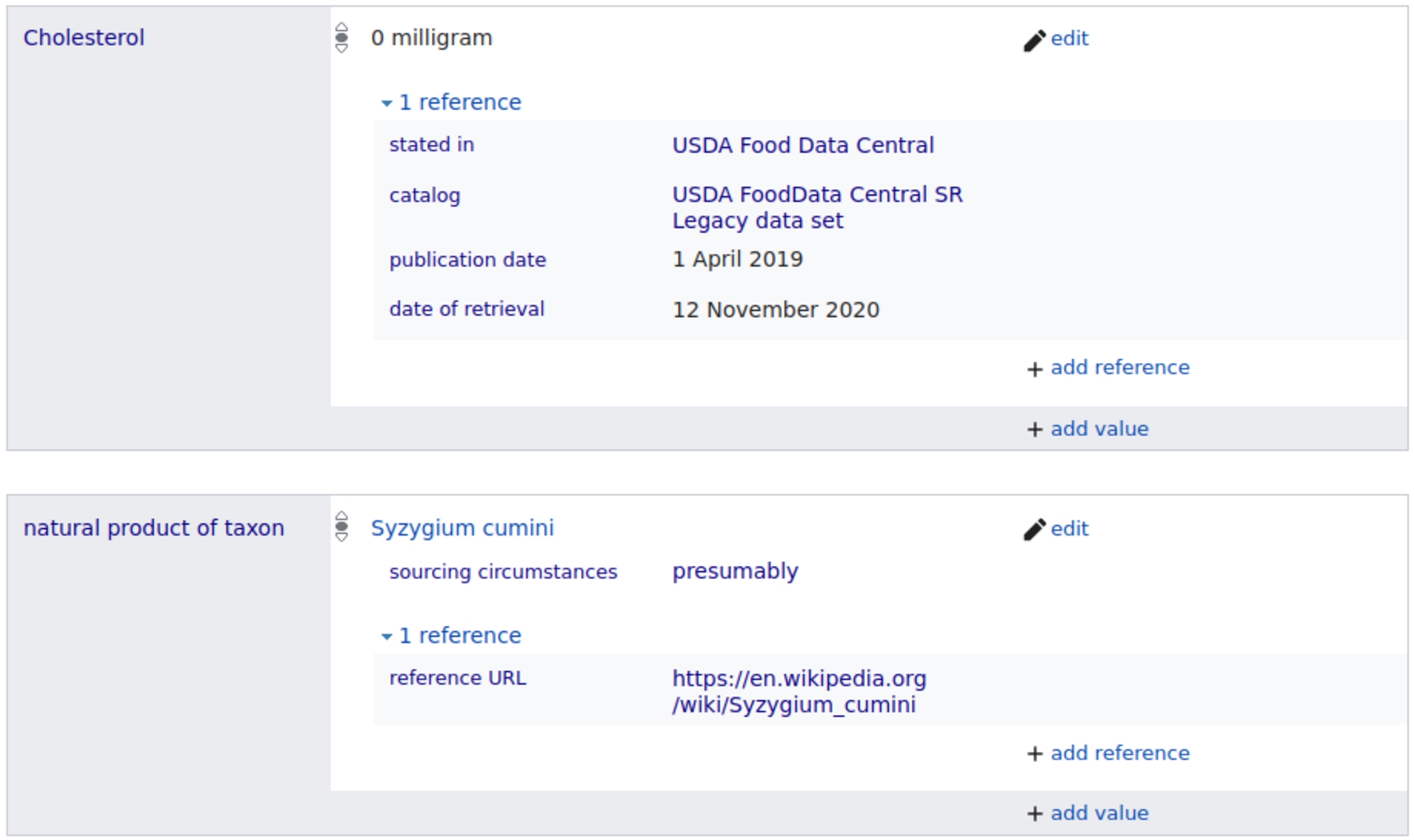
Accurate information about the provenance of information is important for a system that brings together data from many sources. WikiFCD supports references at the level of the statement. For example, the WikiFCD page for ‘Java-plum’,33 contains references to Food Data Central, as seen in Fig. 4. This ensures that each statement can be traced back to the source in which it was found. We also have a property ‘sourcing circumstances’ which can be used with a set of qualifiers (presumably, misprint, miscalculation, approximately, contradiction) to indicate additional information about the claim. In Fig. 4, we see that the statement containing the property ‘natural product of taxon’ is sourced to English Wikipedia, and that this statement contains the qualifier ‘sourcing circumstances’ with a value of ‘presumably’. This statement communicates that human judgement was used to connect ‘Java-plum’ with the taxon ‘Syzygium cumini’. This allows us to add statements to an item while maintaining provenance of the statements. In August of 2020 when we imported data from Food Data Central, it did not include taxon information. Currently Food Data Central does include taxon information for foods in the Foundation Foods catalog thanks to their collaboration with FoodOn.
Fig. 4.
Item for Java-plum in WikiFCD showing references.
Some FCTs provide taxon information for food items. There are already several hundred food items in WikiFCD where the source taxon for a food item can be referenced to a specific publication, as seen in this query: https://tinyurl.com/2bqyz5u3. Applying references to each statement ensures that when results are returned via SPARQL, we can quickly identify the source of the information. The reference structure supported by the Wikibase software has been effective for Wikidata [24]. Building on our experiences with the Wikidata system, we work to add references to as many statements as a possible in WikiFCD. People who reuse this data will be able to see, per statement, where the data originated and make decisions on whether or not it is relevant for their use case.
3.FoodOn
FoodOn is an ontology that describes foods and the organisms from which they are derived [7]. By making use of the FoodOn ontology, we can bring together food items across diverse FCT sources. FoodOn is a member of the Open Biological and Biomedical Ontology (OBO) Foundry, and is actively curated by an expert community [7]. The OBO Foundry provides tools, such as the Dashboard and automated tests, to member onotologies which help improve the quality of each individual ontology [14].
FoodOn builds on the work of the LanguaL thesaurus, reusing their product description facets to describe foods in terms of their plant or animal origins, product types, etc. [7]. Reusing the terms from LanguaL, but providing benefits of a machine-readable ontological structure, FoodOn improved the usability of LanguaL.
FoodOn’s design includes reuse of sixteen OBO Foundry ontologies such as that of the National Center for Biotechnology Information (NCBI) for taxon identification, The Environment Ontology (ENVO), Chemical Entities of Biological Interest (ChEBI), and the Disease Ontology (DO). FoodOn’s reuse of these ontologies allows us to benefit from all of the work that has gone into integrating these resources.
The Joint Food Ontology Working group, that curators of FoodOn participate in, is a regular meeting of experts from different domains with an interest in food data [5]. The discussions that take place in this working group lead to prioritization of work activities for the FoodOn curators. The fact that FoodOn is actively curated, and still being extended, allows the WikiFCD team to learn about the needs and data usage of the gathered domain experts. These discussions inform our development priorities and our data modeling activities.
4.Mapping FoodOn identifiers to food items in WikiFCD
The fact that FoodOn is a consortium-driven standard ontology for food description means that many stakeholders are ensuring that FoodOn has broad coverage to meet the needs of diverse reusers. This also increases the awareness of, and reuse of FoodOn, making it a widely-recognized ontology. We wanted to reuse as much of FoodOn as possible in the creation of WikiFCD. Rather than attempting to design and implement our own classifications for food types, we decided that FoodOn would meet our needs for food item groupings and that matching the food items in WikiFCD to FoodOn identifiers would make our data more intelligible to a wider community.
Inspired by the work of the Su Lab to integrate the Disease Ontology to Wikidata [21], we worked from the FoodOn Owl file to identify the subsets of FoodOn to import into WikiFCD. We used the ROBOT tool44 to extract subsets of the FoodOn ontology for import into the WikiFCD knowledge base.
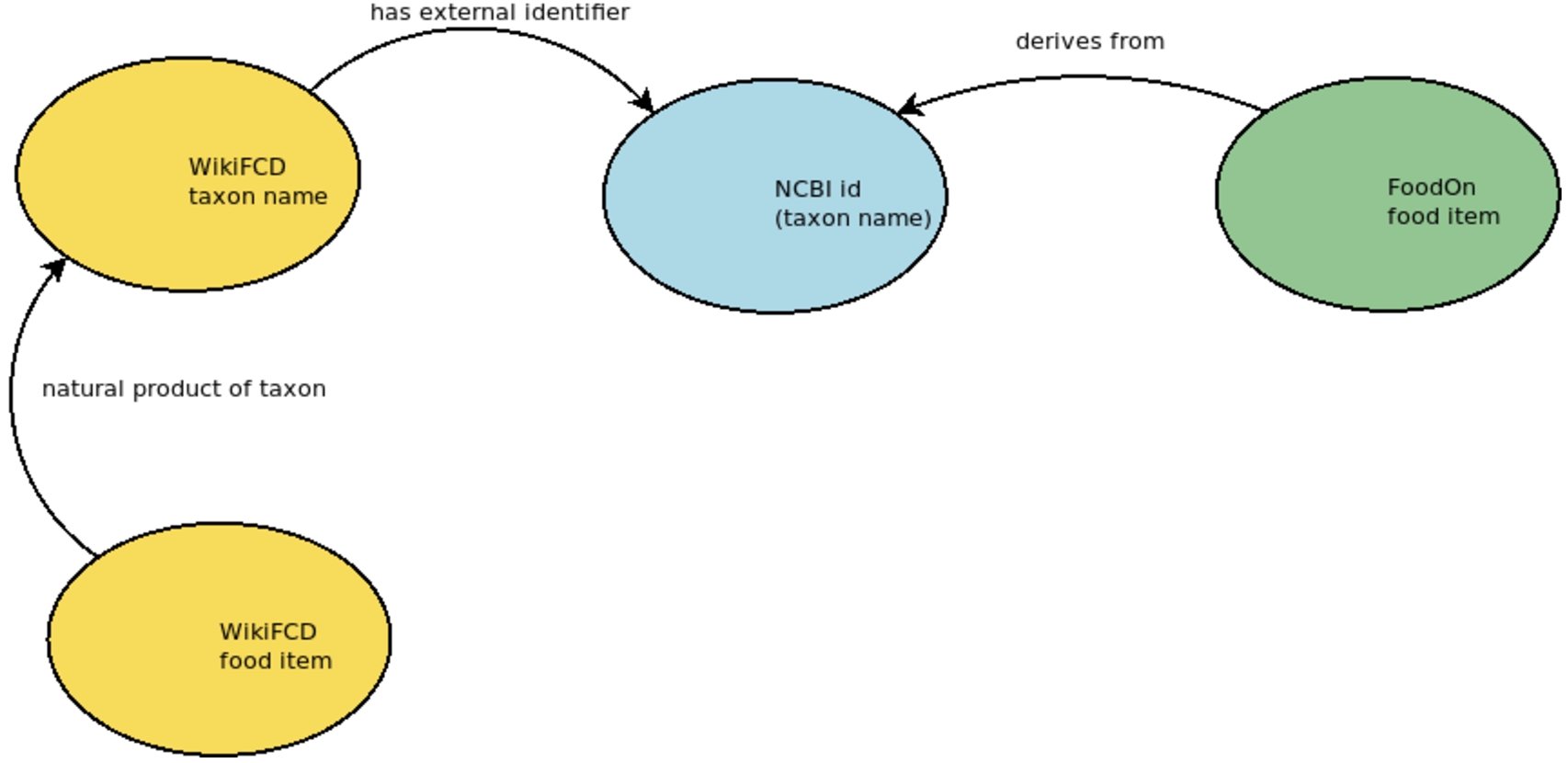
FoodOn reuses NCBI taxon identifiers for taxon names from which foods are derived. For example, ‘mango (whole)’ derives from Mangifera indica. Mangifera indica has an NCBI id ‘29780’. WikiFCD also has items for many taxa. We created statements expressing NCBI identifiers on our taxon name items by extracting a subset of Wikidata, as described above. We then extracted data from FoodOn for all foods that have a FoodOn identifier that are derived from something with an NCBI identifier, which allowed us to align food items in WikiFCD and FoodOn as seen in Fig. 5. We used this data to create statements in WikiFCD expressing these relationships and sourcing them to FoodOn. This was our first automated import of a large set of food items from FoodOn into WikiFCD.
Fig. 5.
NCBI taxon identifiers are used to align food items in WikiFCD and FoodOn.
After producing CSV files of subsets of FoodOn, we used WikidataIntegrator (WDI) to create bots to populate WikiFCD with relevant information from FoodOn. WDI is a Python library for interacting with data from Wikidata [31]. WDI was created by the Su Lab of Scripps Research Institute and published under an open-source software license via GitHub.55
5.Benefits of reusing FoodOn in WikiFCD
Reusing FoodOn provides several advantages that improve the utility of WikiFCD. FoodOn provides named groupings of food items. For example, an apple is a pomaceous fruit food product. Without FoodOn the food items in WikiFCD would not have additional information about food categories. Food categories such as ‘brassicas’ or ‘vegetables’ are widely recognized, and we wanted to be able to make use of these groupings in the future as we design interfaces that will support reuse of food composition data in applications.
FoodOn identifiers have been harmonized with terms from domains that require precision in food reference. The reuse of domain specific terminology across OBO Foundry Ontologies is exemplified by the Genomic Epidemiology Ontology’s reuse of FoodOn food terms for the purpose of investigating and tracking infectious disease outbreaks resulting from contaminated food. Reuse of terms avoids the need to apply more error-prone methods of data harmonization: either mapping food related ontology terms from different ontologies or agency databases, or free-text matching of food terms across data sets [10]. This alignment work is necessary to ensure that when food items are mentioned in the description of outbreaks and the related laboratory analyses and epidemiological research, all parties can be confident of which food items are under discussion. Complex data related to food microbiology are needed at the time of disease outbreak [17]. Reducing ambiguity about which foods are being discussed can help reduce mis-identifications.
We designed our data model so that each sourced entry of food composition data would be a new item in the knowledge base. This means that we have multiple entries for many food items. The ability to map these items to FoodOn identifiers helps us group items that describe the same food as well as items that describe similar foods. Using FoodOn terms to organize the food items in WikiFCD allows us to indicate helpful information about the part of the plant that is being used. Before integrating FoodOn the only way to find this information would be within the label of the food item which is not as helpful as a machine-readable property with an unambiguous value.
FoodOn contains terms related to processes that are very helpful for providing additional information about how foods are prepared. The method of processing used in food preparation can change the nutritional composition of the food item. Prior to integrating FoodOn the only way that processing information was indicated was in the label, and then only inconsistently. Reusing FoodOn allows us to indicate this as a property of the food item which allows for more effective search and retrieval.
FoodOn provides cross-references to databases. Providing FoodOn identifiers on food items from WikiFCD makes our data more reusable by others familiar with their identifiers. We want others to reuse data from WikiFCD, thus by providing as many links out to related resources, we hope to increase the value of this system.
6.Validating WikiFCD data with ShapeExpressions
Our open contribution policy for WikiFCD means that we welcome people beyond the members of our project team to edit WikiFCD. We have established data models for food items, food composition tables, scholarly publications, and taxon names. We use ShEx (ShapeExpressions) [2], a formal data modeling and validation language for RDF, to represent these data models and store them in the schema namespace of WikiFCD. Wikibase has had a ShEx-based schema namespace since 2018 [30]. Using WikiFCD’s schema namespace allows us to communicate our expectations for properties and their allowed values to our contributors. These ShEx schemas will also reduce work for anyone looking to combine data from our knowledge graph with other data sets. For example, if researchers would like to explore our data, rather than writing exploratory SPARQL queries to find out what data can be found and the details of our data models, they can simply review our ShEx schemas to quickly understand our data models. ShEx can be used to validate RDF graphs for conformance to a schema. This allows us to create forms for data contributors that will ensure data consistency. Data contributors will not need to familiarize themselves with our data models, the form-based contribution interaction will guide curation. Our ShEx schemas will also be useful when integrating additional RDF data sets as the project matures. When we encounter new RDF data sources we can explore them with the use of our ShEx schemas to determine where they overlap with our existing data models. We will also be able to extend our schemas as the need for greater expressivity or complexity arises.
We created a schema for food items with a FoodOn identifier. This schema represents what properties and qualifiers are expected for food items of this type. For example, all food items with a FoodOn identifier must also have a statement containing property 73 “natural product of taxon”, the value for which must be a taxon with an NCBI identifier. This corresponds to the structure of FoodOn itself in which foods with FoodOn identifiers are members of a class that have an owl Restriction of “derives from” and owl someValuesFrom a term from NCBI. Here is a query for the WikiFCD SPARQL endpoint for food items with a FoodOn identifier and the name of the taxon from which they are a natural product: https://tinyurl.com/297uvk7m.
Validating entity data against a schema allows us to create lists of items that are not currently in conformance with a schema and need human review. This will help us maintain our data models, while encouraging the participation of newcomers to the project as we will be able to generate conformance reports periodically and look for non-conformant data.
7.Using WikiFCD data in food tracking applications
Tools such as food frequency questionnaires and dietary recall logs are often used to collect self-reported data about food intake [11,25]. Digital food diaries are increasingly being used for the recording of food intake [5]. [3,6,8,9,13,19,20,22,23,27,32]. These food diaries rely on data about food composition. If a user of a digital food diary does not find a food in the database, they will not be able to create an accurate record of their nutritional intake.
Food tracking applications make use of food composition data sets available in convenient formats. By making FCTs that were previously available only as CSV or PDF files available in the web-based WikiFCD system, people can now reuse these data via the application programming interface (API) or the SPARQL endpoint.
Some food tracking applications provide data from multiple sources [16]. Our goal is to make WikiFCD data exports available for reuse in food tracking applications to increase the breadth of foods from international sources which can be more easily tracked. Many of the FCTs we have added to WikiFCD so far contain food composition data for vegetables and other plant-derived foods which are not included in Food Data Central. We are emphasizing these additions so that a greater number of plant-derived foods will be available to people who record their food intake. Providing data so that more people can find relevant food composition information for the foods they eat will increase accuracy and lead to greater satisfaction with applications. People who follow specific diets, such as vegetarians and vegans, will be able to find food composition information for more plant-based foods.
8.Future work
Some authors of food composition tables include taxon information along with food items. When authors make this information available, it is possible to use those taxa to map the food items to an appropriate FoodOn identifier because FoodOn contains relationships between taxa and food items. If authors do not include this information we need to make an educated guess as to which FoodOn identifier should be assigned to the food item. Our workflow will continue to require mapping food items to FoodOn identifiers as we ingest each additional food composition table.
There are many additional types of information we would like to import from FoodOn into WikiFCD. Now that we have established a backbone of correspondences between taxon items in WikiFCD and FoodOn identifiers for derived foods, we can bring in additional information from FoodOn more easily. For example FoodOn contains information about plant parts. This would be very useful information in WikiFCD that would allow us to express with greater precision which part of the plant is being described. For example, we have food composition data for mung beans as well as mung bean sprouts. Currently they are only distinguished by label in WikiFCD. We would like to be able to indicate this with a dedicated property mapped to FoodOn’s structure for describing plant and animal parts.
We may consider creating a multilingual index of food names that we could contribute to the FoodOn curation team for review. This would allow FoodOn to increase the number of labels it contains in many languages. Multilinguality is an important consideration in semantic web technologies intended for global audiences [4]. There are three pathways to consider as sources of multilingual data for potential reuse by FoodOn: FCTs, Wikidata, and interwiki links. For food items that are sourced from food composition tables there are often food names from a language spoken in the country publishing the FCT. All of these food names are sourced, and would be excellent candidates for consideration by the FoodOn curation team. The second source would be Wikidata itself. This pathway would use the NCBI mappings shared by WikiFCD and Wikidata and then check for food items in Wikidata that are natural products of those taxa (represented by Wikidata Property 1582) and then check the items to see if Wikidata Property 1843 ‘taxon common name’ has values, and for which languages. The third pathway is to use the NCBI mappings in WikiFCD and Wikidata, and then check for food items in Wikidata that are natural products of those taxa (Wikidata Property 1582) and then check for pages for those items in different language versions of the Wikipedias and collect the page names. The second and third options are likely to produce noisier data because there are some taxa for which multiple food items are natural products, and thus would require an additional review stage. There is also the drawback that the common names from pathway options two and three are often not connected to a reference, and thus can’t be verified. However, the number of common names available in these two options is larger than that of the first option, and covers hundreds of human languages. Thus, pathway options two and three could be useful in certain language contexts where communities would have no other local-language option for aligning their data to FoodOn.
WikiFCD could provide inspiration for additional terms to be added to FoodOn. The fact that WikiFCD has data about relationships between food items and taxon sources of the food items, and that those taxa are mapped to their corresponding NCBI identifiers, candidate food items could be transferred from WikiFCD to the FoodOn curation team for consideration. If the FoodOn curation team wants to consider adding a new database cross-reference to FoodOn, it is possible that the WikiFCD mappings to Wikidata may prove useful. For example, for species that do not have a NCBI identifier, Wikidata could provide mappings to GBIF, Wormbase, Flybase, etc. At the time of this writing, Wikidata has more than 9,000 external identifiers. Of course many of these are not relevant to FoodOn, but dozens of them pertain to food or biological domains, meaning the potential for harvesting database cross-references for FoodOn could be valuable.
Food composition tables provide information about macronutrients in food items. The chemical constituents of many food items that provide benefits to human health extend beyond nutrients profiled in FCTs [1]. Wikidata contains information about many different chemical compounds found in plants. We could explore creating profiles for different food items derived from plants the provide overviews of secondary metabolites such as flavonoids, terpenoids and alkaloids. As bioactive components of food items have been shown to regulate gene expression, having a more complete profile of chemical constituents of food items is necessary to understand how diet contributes to health [18].
The mappings we maintain between WikiFCD and Wikidata help us to keep our data in alignment with data in Wikidata. This is an important first step if we would like to contribute this data to Wikidata at some point in the future. We plan to continue adding food composition tables to this knowledge base. We aim to eventually include every national-level FCT that has been published within the last twenty years. We also hope to partner with teams working to develop new or updated FCTs so that they can publish data directly to the web via WikiFCD.
9.Conclusion
Reusing FoodOn in WikiFCD saves us time attempting to design our own ad-hoc structure for categorizing foods. Beyond saving time, reuse also increases the utility of WikiFCD, as FoodOn elements are recognizable to an existing audience of bioinformaticians and food data experts. The curation decisions that have gone into the design of FoodOn are based on deliberation among domain experts. This means that we benefit from the knowledge of the entire FoodOn curation team.
Taking food composition tables from CSV, Excel, or PDF formats and importing them into a knowledge base allows food data to become part of the semantic web. Mapping properties and items to Wikidata and FoodOn integrates food data with many other data sets and web resources. Having these data in a knowledge base allows us to compare food composition values from different sources and regions for the same food item.
People with access to supermarkets may be able to purchase foods imported from many regions of the world due to the increase of international food transportation. In this more global food system, the range of food choices is broad. Food choices impact human health [26]. Data about the nutritional value of food is available in food composition tables and databases. Bringing these data together in a publicly-available knowledge base is one strategy to increase access to nutritional information to a wider audience.
We hope that these data are reused by researchers, application developers, health practitioners and people who are interested in tracking their own dietary intake. Applying technologies of the semantic web to the challenge of bringing many disparate data sets together in a single system allow us to provide a SPARQL endpoint for querying this data, making it easier to ask questions of these data in combination with other RDF data on the web. As data relating to the nutritional composition of foods is fractured across national data sets, scholarly publications, and technical reports, semantic web technologies are a solution for more easily working with these data.
Notes
Acknowledgements
We would like to thank Adam Shoreland for creating wbstack and Rhizome for funding wbstack. Thank you to Wikimedia Deutschland for providing the Wikibase.cloud platform. Thank you to the Su Lab at Scripps Research Institute for creating WikidataIntegrator and making it available under an open license. Thank you to Andra Waagmeester for maintaining and creating new features for WikidataIntegrator. Thank you to Eric Prud’hommeaux for assistance developing queries. Thank you to the FoodOn curators for creating the ontology. Thank you everyone in the Wikidata community for contributing data and structuring the knowledge base.
References
[1] | A.-L. Barabási, G. Menichetti and J. Loscalzo, The unmapped chemical complexity of our diet, Nature Food 1: (1) ((2020) ), 33–37. doi:10.1038/s43016-019-0005-1. |
[2] | I. Boneva, J.E. Labra Gayo, S. Hym, E.G. Prud’hommeau, H.R. Solbrig and S. Staworko, Validating RDF with shape expressions, CoRR, 2014, arXiv:1404.1270. |
[3] | M.L. Bracken and B.M. Waite, Self-efficacy and nutrition-related goal achievement of MyFitnessPal users, Health Education & Behavior 47: (5) ((2020) ), 677–681. doi:10.1177/1090198120936261. |
[4] | C. Caracciolo, M. Sini and J. Keizer, Requirements for the treatment of multilinguality in ontologies within FAO, 2007, http://hdl.handle.net/10760/15660. |
[5] | L. Chan, N. Vasilevsky, A. Thessen, J. McMurry and M. Haendel, The landscape of nutri-informatics: A review of current resources and challenges for integrative nutrition research, Database 2021: ((2021) ), baab003, ISSN 1758-0463. doi:10.1093/database/baab003. |
[6] | J. Chen, W. Berkman, M. Bardouh, C.Y.K. Ng and M. Allman-Farinelli, The use of a food logging app in the naturalistic setting fails to provide accurate measurements of nutrients and poses usability challenges, Nutrition 57: ((2019) ), 208–216. doi:10.1016/j.nut.2018.05.003. |
[7] | D.M. Dooley, E.J. Griffiths, G.S. Gosal, P.L. Buttigieg, R. Hoehndorf, M.C. Lange, L.M. Schriml, F.S. Brinkman and W.W. Hsiao, FoodOn: A harmonized food ontology to increase global food traceability, quality control and data integration, npj Science of Food 2: (1) ((2018) ), 1–10. doi:10.1038/s41538-018-0032-6. |
[8] | E. Dunford, H. Trevena, C. Goodsell, K.H. Ng, J. Webster, A. Millis, S. Goldstein, O. Hugueniot and B. Neal, FoodSwitch: A mobile phone app to enable consumers to make healthier food choices and crowdsourcing of national food composition data, JMIR mHealth and uHealth 2: (3) ((2014) ), 37. doi:10.2196/mhealth.3230. |
[9] | C. Evenepoel, E. Clevers, L. Deroover, W. Van Loo, C. Matthys and K. Verbeke, Accuracy of nutrient calculations using the consumer-focused online app MyFitnessPal: Validation study, Journal of Medical Internet research 22: (10) ((2020) ), 18237. doi:10.2196/18237. |
[10] | E. Griffiths, D. Dooley, M. Graham, G. Van Domselaar, F.S.L. Brinkman and W.W.L. Hsiao, Context is everything: Harmonization of critical food microbiology descriptors and metadata for improved food safety and surveillance, Frontiers in Microbiology 8: ((2017) ), 1068, ISSN 1664-302X. doi:10.3389/fmicb.2017.01068. |
[11] | M. Hatami, F. Vahid, M.E. Akbari, M. Sadeghi, F. Ameri, H. Eini-Zeinab, Y. Jamshidi-Naeini and S.H. Davoodi, The vitamins involved in one-carbon metabolisms are associated with reduced risk of breast cancer in overall and subtypes, International Journal for Vitamin and Nutrition Research ((2019) ). doi:10.1024/0300-9831/a000501. |
[12] | E.C. Hemler and F.B. Hu, Plant-based diets for personal, population, and planetary health, Advances in Nutrition 10: (Supplement_4) ((2019) ), 275–283. doi:10.1093/advances/nmy117. |
[13] | C. Höchsmann, N. Fearnbach, J.L. Dorling, T.L. Fazzino, C.A. Myers, J.W. Apolzan and C.K. Martin, Preference, expected burden, and willingness to use digital and traditional methods to assess food and alcohol intake, Nutrients 13: (10) ((2021) ), ISSN 2072-6643. doi:10.3390/nu13103340. |
[14] | R. Jackson, N. Matentzoglu, J.A. Overton, R. Vita, J.P. Balhoff, P.L. Buttigieg, S. Carbon, M. Courtot, A.D. Diehl, D.M. Dooley, W.D. Duncan, N.L. Harris, M.A. Haendel, S.E. Lewis, D.A. Natale, D. Osumi-Sutherland, A. Ruttenberg, L.M. Schriml, B. Smith, C.J. Stoeckert Jr., N.A. Vasilevsky, R.L. Walls, J. Zheng, C.J. Mungall and B. Peters, OBO foundry in 2021: Operationalizing open data principles to evaluate ontologies, Database 2021: ((2021) ), baab069, ISSN 1758-0463. doi:10.1093/database/baab069. |
[15] | F. Juul, N. Parekh, E. Martinez-Steele, C.A. Monteiro and V.W. Chang, Ultra-processed food consumption among US adults from 2001 to 2018, The American Journal of Clinical Nutrition ((2021) ), nqab305, ISSN 0002-9165. doi:10.1093/ajcn/nqab305. |
[16] | R. Kunkel, Privatization of government information as primitive accumulation, Journal of Radical Librarianship 6: ((2020) ). doi:10.7282/t3-ejh3-2r26. |
[17] | D. Lambert, A. Pightling, E. Griffiths, G. Van Domselaar, P. Evans, S. Berthelet, D. Craig, P.S. Chandry, R. Stones, F. Brinkman, A. Angers-Loustau, J. Kreysa, W. Tong and B. Blais, Baseline practices for the application of genomic data supporting regulatory food safety, Journal of AOAC INTERNATIONAL 100: (3) ((2019) ), 721–731, ISSN 1060-3271. doi:10.5740/jaoacint.16-0269. |
[18] | J. Mierziak, K. Kostyn, A. Boba, M. Czemplik, A. Kulma and W. Wojtasik, Influence of the bioactive diet components on the gene expression regulation, Nutrients 13: (11) ((2021) ), ISSN 2072-6643, https://www.mdpi.com/2072-6643/13/11/3673. doi:10.3390/nu13113673. |
[19] | C.L. Millar, A. Cohen, S.P. Juraschek, A. Foley, M. Shtivelman, K.J. Mukamal and S. Sahni, The feasibility of using computrition software for nutrition research – a pilot study, Nutrients 13: (2) ((2021) ), ISSN 2072-6643, https://www.mdpi.com/2072-6643/13/2/329. doi:10.3390/nu13020329. |
[20] | L. Mistura, F.J. Comendador Azcarraga, L. D’Addezio, D. Martone and A. Turrini, An Italian case study for assessing nutrient intake through nutrition-related mobile apps, Nutrients 13: (9) ((2021) ), ISSN 2072-6643, https://www.mdpi.com/2072-6643/13/9/3073. doi:10.3390/nu13093073. |
[21] | E. Mitraka, A. Waagmeester, S. Burgstaller-Muehlbacher, L.M. Schriml, A.I. Su and B.M. Good, Wikidata: A platform for data integration and dissemination for the life sciences and beyond, bioRxiv ((2015) ), 031971. doi:10.1101/031971. |
[22] | K. Murakami, N. Shinozaki, S. Masayasu and M.B.E. Livingstone, Web-based personalized nutrition system for delivering dietary feedback based on behavior change techniques: Development and pilot study among dietitians, Nutrients 13: (10) ((2021) ), ISSN 2072-6643, https://www.mdpi.com/2072-6643/13/10/3391. doi:10.3390/nu13103391. |
[23] | M. Ocké, C. Dinnissen, A. Stafleu, J. de Vries and C. van Rossum, Relative validity of MijnEetmeter: A food diary app for self-monitoring of dietary intake, Nutrients 13: (4) ((2021) ), ISSN 2072-6643, https://www.mdpi.com/2072-6643/13/4/1135. doi:10.3390/nu13041135. |
[24] | A. Piscopo, L.-A. Kaffee, C. Phethean and E. Simperl, Provenance information in a collaborative knowledge graph: An evaluation of Wikidata external references, in: International Semantic Web Conference, Springer, (2017) , pp. 542–558. doi: 10.1007/978-3-319-68288-4_32. |
[25] | J.-S. Shim, K. Oh and H.C. Kim, Dietary assessment methods in epidemiologic studies, Epidemiology and Health 36: ((2014) ). doi:10.4178/epih/e2014009. |
[26] | K.S. Stylianou, V.L. Fulgoni and O. Jolliet, Small targeted dietary changes can yield substantial gains for human health and the environment, Nature Food 2: (8) ((2021) ), 616–627. doi:10.1038/s43016-021-00343-4. |
[27] | V. Teixeira, S.M. Voci, R.S. Mendes-Netto and D.G. da Silva, The relative validity of a food record using the smartphone application MyFitnessPal, Nutrition & Dietetics 75: (2) ((2018) ), 219–225. doi:10.1111/1747-0080.12401. |
[28] | K. Thornton, K. Seals-Nutt and M. Matsuzaki, Introducing WikiFCD: Many food composition tables in a single knowledge base, in: CEUR Workshop Proceedings, Vol. 2969: , CEUR-WS, (2021) . http://ceur-ws.org/Vol-2969/paper4-IFOW.pdf. |
[29] | K. Thornton, K. Seals-Nutt and M. Matsuzaki, Matching Food Items with FoodOn Identifiers in a Knowledge Base, (2022) , CEUR-WS. http://ceur-ws.org/Vol-3249/paper1-IFOW.pdf. |
[30] | K. Thornton, H. Solbrig, G.S. Stupp, J.E.L. Gayo, D. Mietchen, E. Prud’Hommeaux and A. Waagmeester, Using shape expressions (ShEx) to share RDF data models and to guide curation with rigorous validation, in: European Semantic Web Conference, Springer, (2019) , pp. 606–620. doi:10.1007/978-3-030-21348-0_39. |
[31] | A. Waagmeester, G. Stupp, S. Burgstaller-Muehlbacher, B.M. Good, M. Griffith, O.L. Griffith, K. Hanspers, H. Hermjakob, T.S. Hudson, K. Hybiske, S.M. Keating, M. Manske, M. Mayers, D. Mietchen, E. Mitraka, A.R. Pico, T. Putman, A. Riutta, N. Queralt-Rosinach, L.M. Schriml, T. Shafee, D. Slenter, R. Stephan, K. Thornton, G. Tsueng, R. Tu, S. Ul-Hasan, E. Willighagen, C. Wu and A.I. Su, Science forum: Wikidata as a knowledge graph for the life sciences, eLife 9: ((2020) ), 52614, ISSN 2050-084X. doi:10.7554/eLife.52614. |
[32] | N. Zmora and E. Elinav, Harnessing SmartPhones to personalize nutrition in a time of global pandemic, Nutrients 13: (2) ((2021) ), ISSN 2072-6643, https://www.mdpi.com/2072-6643/13/2/422. doi:10.3390/nu13020422. |




