Impact of long-term high dietary fat intake and regular exercise on serum TMAO and microbiome composition in female rats
Abstract
BACKGROUD:
Trimethylamine oxide (TMAO) represents a gut metabolite and it's physiological concentration is proposed to be influenced by the abundance of precursor trimethylamine (TMA)-producing microbiota in the gut.
OBJECTIVE:
Lifestyle factors, such as diet and exercise, modify the microbiome, but their long-term impact on plasma TMAO and TMA production in the gut is poorly understood.
METHODS:
Fecal microbiome composition was analyzed and correlated with TMAO serum concentrations in female Sprague-Dawley rats that received either a healthy normal or a high-fat diet (HFD) for 10 months. Half of the animals performed regular running exercise, the other half served as sedentary controls.
RESULTS:
HFD consumption induced an alteration of the fecal microbiome with increased alpha diversity on ASV levels. More importantly, HFD reduced the abundance of genera containing potential TMA producers, such as certain members of Clostridia and Lachnospiraceae. In line with this observation, serum TMAO concentrations were found to be lower with HFD and to correlate with the abundance of these genera. Regular exercise also induced changes in microbiome beta diversity but had no effect on serum TMAO in female rats.
CONCLUSION:
A diet containing high amounts of dietary fat reduced serum TMAO, probably through a reduced intestinal abundance of TMA-producing bacteria. In contrast, regular exercise altered beta diversity of the microbiome composition, but without significant effects on serum TMAO concentrations.
1Introduction
Trimethylamine oxide (TMAO) is a gut microbiota-derived metabolite that is synthesized from dietary nutrients and its synthesis is closely related to the composition of the diet and the microbiome of the gut [1]. Originally, TMAO is known as a compound that helps saltwater animals and fish to maintain an iso-osmotic state and to stabilize cellular proteins against the mounting hydrostatic pressure with increasing water depth. In addition, it serves as defrosting agent in arctic fish [2].
The plasma concentration of TMAO has been linked to a wide range of health outcomes including all-cause and cardiovascular mortality, cardiovascular disease (CVD), stroke, hypertension, renal failure, frailty, diabetes mellitus, and cancer [3–9]. Experimental studies suggest that TMAO promotes cardiovascular disease [10], endothelial dysfunction [11], and platelet hyper-reactivity and thrombosis [12]. Furthermore, it enhances cholesterol uptake into macrophages and the accumulation of foam cells in arterial walls [10]. Also renal function is impaired in the presence of elevated TMAO concentrations [6].
While many studies propose TMAO as a risk factor for multiple adverse health outcomes, others suggest the opposite. For example, beneficial or protective effects have been reported in glucose homeostasis [13, 14], carcinogenesis [15] atherosclerosis and cardiovascular function [16, 17], steatohepatitis [18], and inflammatory insults [19]. Moreover, the deleterious associations between TMAO and different cardiovascular conditions fade when corrected for renal function [20, 21]. This suggests that observational evidence for associations between TMAO and cardiovascular diseases may be due to confounders or reverse causality [21].
Circulating TMAO is primarily produced in the liver from trimethylamine (TMA). This enzymatic reaction is catalysed by flavin monooxygenases (FMOs) [22]. In humans, FMO3 is mainly responsible for the oxidation of TMA to TMAO [23]. TMA is produced by the gut microbiome, mainly from choline and carnitine which has passed the small intestine [1, 24]. Supplementing Apoe-/- mice with L-carnitine markedly enhanced TMA/TMAO production [1]. However, the genetic modification disrupts lipid handling in these mice, which impedes a direct translation to the wild-type situation. Also, only specific bacteria, so called TMA high-producers, are capable of synthesizing TMAO [25, 26]. Therefore, the composition of the gut microbiome, as well as the availability of required precursors, seem to be major determinants of the serum TMAO concentration, and may be target factors that could be modified through appropriate lifestyle interventions.
Mounting evidence indicates that the gut microbiome is influenced by multiple lifestyle factors, such as dietary composition, physical activity, smoking, alcohol consumption and others. For example, a high dietary fat intake does not only trigger obesity, but also alters intestinal microbiome diversity [27] and gut permeability through inflammation [28, 29]. Therefore, the gut microbiome appears to play a central role in establishing the characteristic phenotype of individuals that nurture themselves with a meat-based diet including a high content of saturated fat (HFD). Exercise represents another modulator of the gut microbiome [30]. Scheimann et al. have shown that exercise increases the relative abundance of Veillonella atypica, a microbe that improves athletic performance through the enzymatic conversion of lactate into propionate [31]. However, a complex interaction seems to be between dietary composition and physical activity with variable net-effects [32]. In mice, exercise has been shown to prevent weight gain despite administration of a high fat diet (HFD) [33]. This effect was accompanied with a unique composition of the gut microbiome that differed from sedentary animals fed with the same HFD. Also, the microbiome of obese rats differs from non-obese rats and exercise has been shown to induce additional alterations. [34]. Despite substantial evidence that diet and exercise modify the composition of the gut microbiome, there is insufficient knowledge on the specific responses to various dietary and exercise regimens [30, 35]. Therefore, more research is needed to explore the complex interaction between diet, exercise and net effects on disease risk.
Until today, the impact of potentially healthy lifestyle factors, such as regular exercise, on the abundance of TMA producing species in the gut and plasma TMAO concentrations has not been revealed yet. Therefore, the present study aimed to investigate the long-term effects of HFD and regular physical activity on the gut microbiome and plasma TMAO concentrations in a rat model.
2Materials and methods
2.1Animal experiments
All animal experiments were performed following permission by the Federal Ministry of Education, Science and Research GZ: 66.010/0070-V/3b/2018. One hundred twenty 4-month-old healthy female Sprague-Dawley rats with an average body weight of approximately 300 g were purchased from Janvier Labs (France) and kept in groups of three animals per cage under standardized conditions on a 12 h light and 12 h dark cycle in the institutional animal facility. Only female animals were included to avoid gender effects and to reduce the risk of dropouts due to aggressive behaviour between animals. After 1 week of acclimatization, the animals were randomly assigned to receive either a normal chow-based diet (ND; Altromin, Germany) with 3,230 kcal/kg and 11% fat or a custom-designed beef-tallow high-fat diet (HFD), rich in saturated fatty acids (SFA) with 5,150 kcal/kg and 60% fat (ssniff, Germany). The HFD composition was based on previous studies [36, 37]. Food and tap water were provided ad libitum.
The animals were divided into 5 groups of 24 each. One group of rats was sacrificed immediately after the acclimatization period and served as young adult baseline control (BL; n = 24). The remaining 96 animals were allocated to receive either ND (n = 48) or HFD (n = 48) for 10 months. For the entire duration of the study, half of the ND (exeND; n = 24) and HFD (exeHFD; n = 24)-fed rats completed five 30 min running sessions per week on consecutive days followed by 2 days of rest. Running was performed on a treadmill (Panlab, Spain) with a constant speed of 30 cm/s. The training protocol was based on previous experimental studies [38, 39]. The remaining ND and HFD animals were used as sedentary controls (coND; n = 24 and coHFD; n = 24).
Fecal samples for analysis of the microbiome were collected from the dissected descending colon from all animals directly after sacrifice. Samples were stored at -80°C until analysis. In addition, blood was collected by heart puncture into S-Monovette Serum-Gel tubes and S-Monovette Plasma-EDTA tubes (Sarstedt, Nümbrecht, Germany) under deep isoflurane anaesthesia (Forane, Abbott, Austria). Blood collections were performed in non-fasting conditions to minimize emotional stress. Serum and EDTA-plasma were obtained by centrifugation of the samples at 2000 g for 12 min at room temperature. Subsequently, serum and plasma were split and aliquots were stored at –80 °C until analysis.
2.2Analysis of blood biomarkers
Serum TMAO was measured with a high-performance liquid chromatography-tandem mass spectrometry method on a SCIEX QTRAP 6500 triple quadrupole instrument (Applied Biosystems, Framingham, MA, USA) according to Enko et al. [40]. Within-day CVs for TMAO were 5.5% (2.8μmol/L) and 2.2% (12.6μmol/L), and between-day CVs were 9.9% (2.8μmol/L) and 7.6% (12.6μmol/L).
In addition to serum, TMAO was also analysed in the two diets in order to account for dietary TMAO uptake. For this purpose, samples from both diets were extracted in ethanol containing internal standard with a disperser (IKA T 18 digital ULTRA-TURRAX, IKA, Staufen, Germany) and centrifuged. The supernatant was reduced by evaporation with nitrogen and used for analysis in the same manner as serum. Measurements were performed in triplicates and calculated to grams per diet.
2.3Microbiome analysis
DNA isolation for microbiome analysis was done on a QIAsymphony (QIAGEN, Hilden, Germany) device using the QIAsymphony DSP Virus/Pathogen Mini Kit (QIAGEN, Hilden, Germany) with a protocol adapted to stool samples. This protocol combined mechanical and enzymatic lysis with two additional washing steps. From each sample, a small amount of feces (approximately 200 mg) was dissolved in 500μl PBS buffer. 100μl of this mixture (or 100μl PBS buffer for the negative control) were added to a MagNA Lyser green bead tube (Roche, Mannheim, Germany) containing 600μl ATL buffer. Mechanical sample homogenization was performed twice for 30 s at 6500 rpm on a MagNA Lyser (Roche, Mannheim, Germany) instrument followed by a one-minute cool down after each round of mechanical homogenization. Subsequently, two enzymatic digestion steps were performed. First, the homogenates were incubated with 35μl of lysozyme (100 mg/ml) for 30 min at 37 °C followed by a second incubation for 1 hour at 68°C with 70μl of proteinase K (>600 mAU/ml, QIAGEN, Hilden, Germany). At the end of the digestion process, enzymes were inactivated by heating samples to 95 °C for 10 min. Finally, samples were cooled for 5 min on ice and subsequently centrifuged at 13,000 rpm for 5 min.
100μl of the prepared samples were analyzed with the customized program for the QIAsymphony DSP Virus/Pathogen Mini Kit (QIAGEN, Hilden, Germany), adapted with two additional washing steps to suit the requirements of the sample type. Elution volume was 110μl. Automated DNA isolation was done in batches of 24 samples with one negative control per batch comprising 100μl PBS and 600μl ATL buffer. The batches were handled according to a data collection sheet to properly process the stool samples.
For 16 S rRNA gene PCR amplification the FastStart High Fidelity PCR system, dNTPack (Roche, Mannheim, Germany), a blend of FastStart Taq DNA Polymerase and a thermostable proofreading protein, was used. PCR reactions were performed in triplicates and each 25μl PCR reaction included 2.5μl of Fast Start High Fidelity Reaction Buffer (Roche, Mannheim, Germany), 0.25μl High Fidelity Enzyme Blend (Roche, Mannheim, Germany) containing 1.25 U of enzymatic activity, 0.5μl PCR grade nucleotide mix with 10 mM dNTPs (200μM dNTP end concentration) (Roche, Mannheim, Germany), 0.4μM each of forward and reverse primer, and PCR-grade water (Roche, Mannheim, Germany). For amplification of the hypervariable region V4, modified primers according to Walters et al. [41] supplemented with Illumina adaptors for indexing PCR reaction according to Illumina’s 16 s metagenomic sequencing library preparation guide were used. The primer sequences were as follows:
MiSeqF-v4_515Fmod (TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGYCAGCMGCCGCGGTAA)
MiSeqR-v4_806Rmod (GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACNVGGGTWTCTAAT)
For amplification the following protocol was used: 3 min initial denaturation at 95 °C followed by 30 cycles of 95 °C for 45 sec, 55 °C for 45 sec, 72 °C for 1 min with a final extension step at 72°C for 7 min.
The triplicate PCR products from each sample were pooled and checked on a 1.5 % agarose gel for correct amplification. 25μl of the pooled PCR products were used for normalization with a NormPlate (Life Technologies Corporation, Frederick, MD, USA) and incubated for 60 min, following the manual’s instructions. Thereafter, 7.5μl of the normalized PCR products were used as template in a 25μl indexing PCR reaction. The reaction composition resembles the targeted PCR reaction described above, with the difference that Illumina-capable index primers were used, and the cycle number was reduced to 8. 5μl of each indexed sample were used for a second pooling and another 30μl of this pool run through a 1.5% agarose gel for purification, following the instructions of the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany).
The received final sequencing library was quantified and subjected to quality control using the QuantiFluor ONE dsDNA Dye on Quantus Fluorometer (Promega, Mannheim, Germany) and an automated electrophoresis tool for the quality control, the BioAnalyzer 2100 (Agilent, Waldbronn, Germany). Finally, 9 pM libraries containing 20% PhiX Control DNA (Illumina, Eindhoven, Netherlands) were sequenced on a MiSeq desktop sequencer (Illumina, Eindhoven, Netherlands) using the MiSeq Reagent v3 (600cycles) Kit (Illumina, Eindhoven, Netherlands) according to the manufacturer’s instructions. FASTQ raw reads were used for subsequent data analysis.
2.4Statistical analyses
Weight and TMAO data are presented as means ± standard deviations. One-way ANOVA followed by Tukey’s honest significant difference (HSD) correction for multiple testing was used to explore differences of normally distributed variables between multiple groups. Two-way ANOVA followed by Tukey's HSD post hoc test was performed to analyse the impact of diet and exercise on the dependent variables. A p value of <0.05 was considered statistically significant. Analyses were performed by explorative data analyses using SPSS for Windows (IBM©SPSS©Statistics, version 25).
16 S rRNA gene analysis was done on a local Galaxy instance using QIIME2. A SILVA database (version 138) was used for taxonomic classification. Abundance data, metadata, and taxonomy files were analysed with R (”phyloseq”, “vegan”, “tidyverse”) and Calypso V8.84. TMAO serum concentrations were included in the metadata file. Data were filtered as follows: samples with less than 200 sequence reads were removed. Sequences from Cyanobacteria/chloroplasts were removed. Then the data were normalized via total sum scaling and subsequent square root transformation (Hellinger transformation).
Alpha diversity was determined by Shannon Index, and richness, beta diversity with Principal coordinate analysis (PCoA) based on Bray-Curtis dissimilarities, and Redundancy analysis (RDA). Further core microbiome and phylum distribution, as well as linear discriminant analysis effect size (LEfSE) of genera between diet groups were determined. Significance of different abundancies between study groups was determined by 2-way ANOVA. Linear regression analysis was done by point-by-point correlation of TMAO and Shannon index values or abundances of single genera respectively. Significance was calculated with Spearman's correlation coefficient.
3Results
3.1Characterization of animals
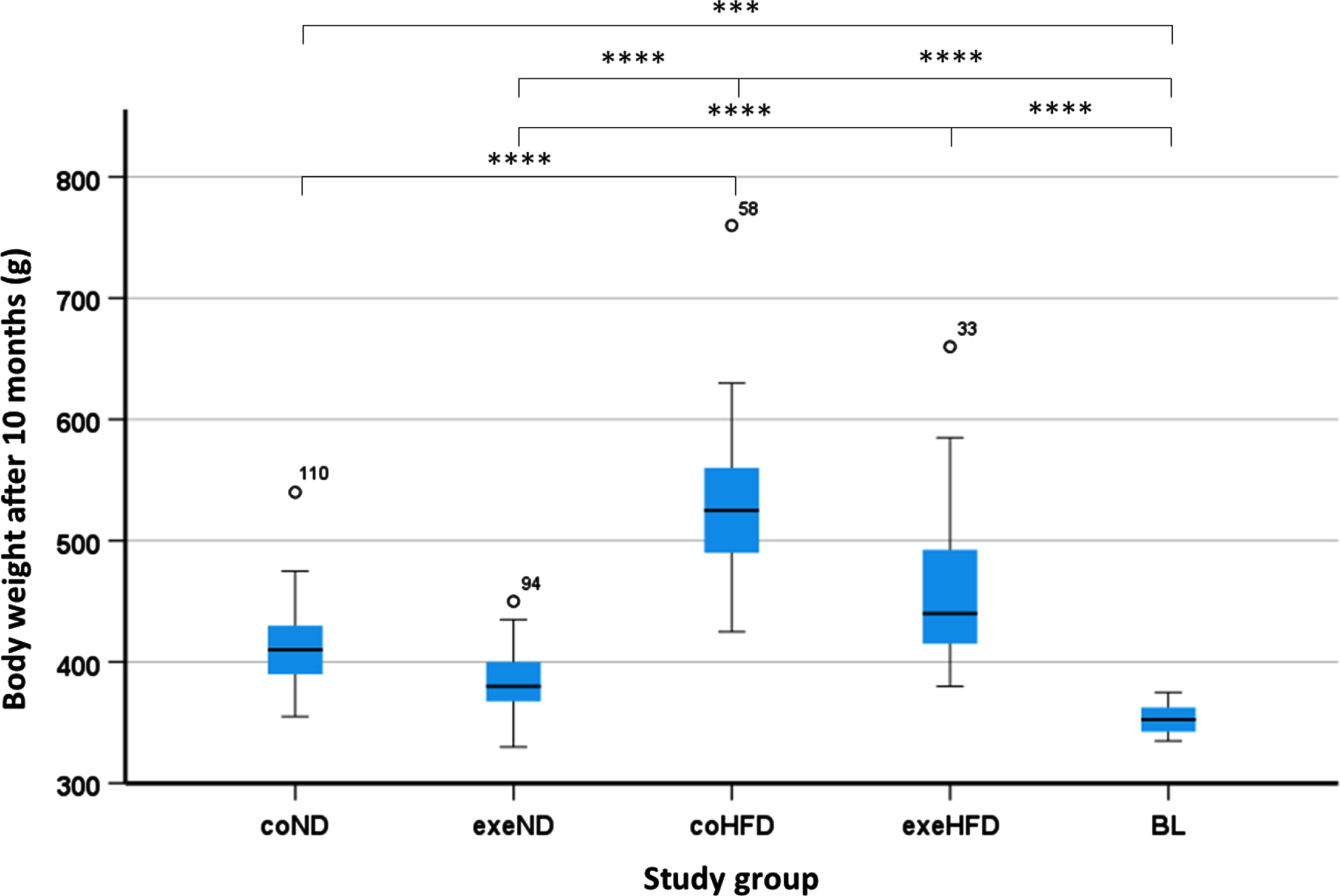
Of the 120 rats included in this study, 6 animals were culled before the study ended due to conspicuous benign tumors (n = 3) or other various health issues (n = 3). Additional 10 animals completed the study but were excluded from statistical analysis because of missing data points. Consequently, the final analysis included a total of 104 animals that were distributed as follows: coND = 22, exeND = 20, coHFD = 21, exeHFD = 21 and BL = 20. At study end, the HFD animals showed a significantly higher body weight compared to ND animals (Fig. 1). Both exercise and diet, but not their combination, had a significant impact on the body weight (Table S1). A thorough characterization of the animals, including food consumption, organ weight, lipid metabolism, and oxidative stress, has been published previously [42, 43].
Fig. 1
Body weight from all study groups. Box and Whisker Blots showing the body weight. The impact of the study conditions was analysed with the 2-way ANOVA followed by Tukey’s HSD post hoc test. Differences among all study groups was assessed by one-way ANOVA followed by Tukey’s HSD correction for multiple tests. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
3.2Serum TMAO analysis
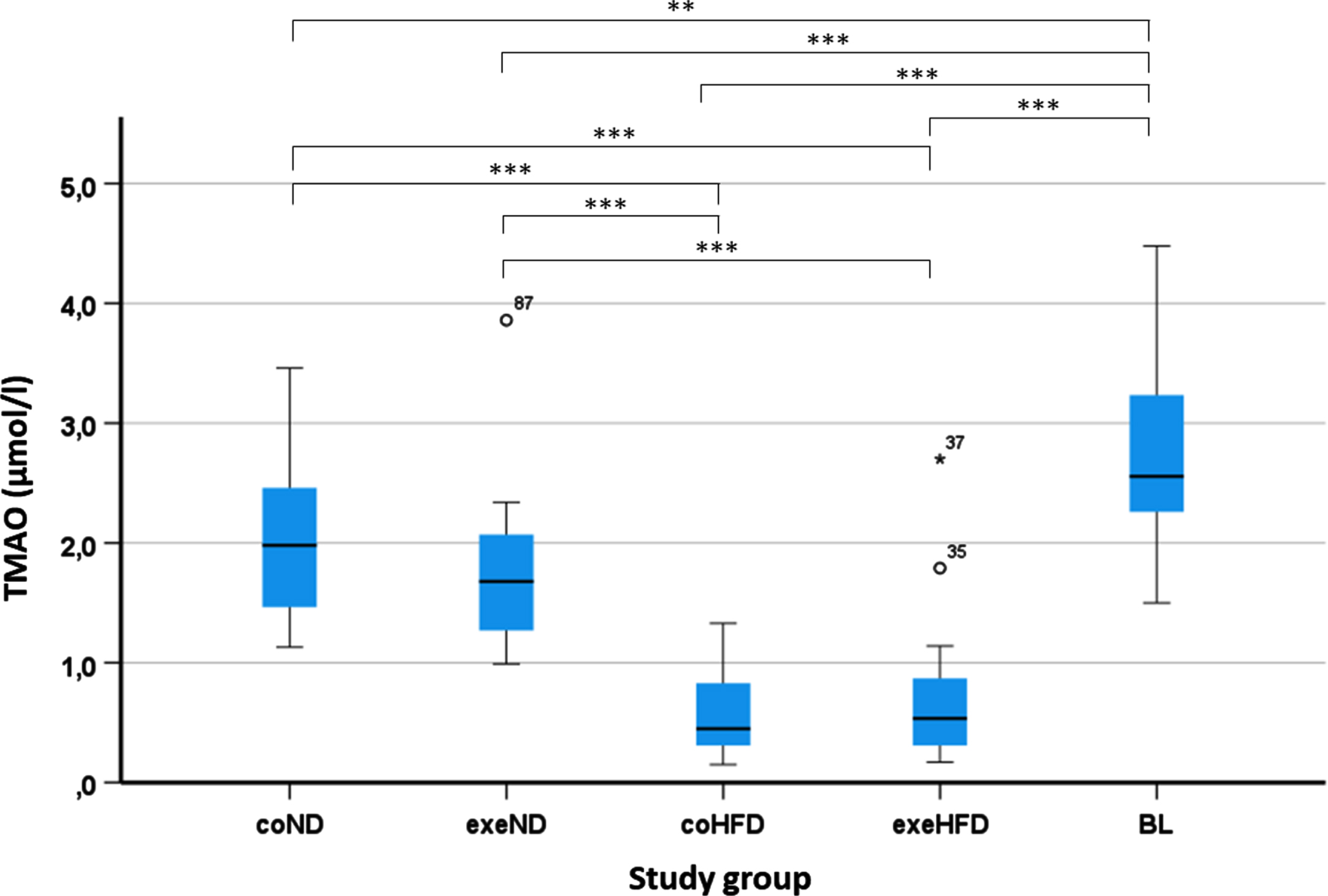
The serum concentrations of TMAO varied markedly among animals ranging from 0.15 to 4.48μmol/L. Young adult rats in the BL group showed the highest TMAO concentrations (2.75μmol/l±0.17). In aged adult rats that received ND, mean TMAO concentrations were 2.04μmol/l±0.14 (coND) and 1.74μmol/l±0.13 (exeND), respectively. Administration of HFD had a pronounced impact on serum TMAO with markedly lower concentrations in the two HFD groups, with means of 0.60μmol/l±0.08 (coHFD) or 0.69μmol/l±0.13 (exeHFD) respectively (Fig. 2, Table S2). In contrast to diet, exercise, or diet and exercise combined, had no significant influence on serum TMAO (Table S3).
Fig. 2
Determination of TMAO concentrations from rat serum from all study groups. Blood was collected and serum prepared from all groups at the end of the runtime and from the young adult group (BL). TMAO serum levels were determined by HPLC/MS. The impact of the study conditions was analysed with the 2-way ANOVA followed by Tukey’s HSD post hoc test. Differences among all study groups was assessed by one-way ANOVA followed by Tukey’s HSD correction for multiple tests. *p<0.05; **p<0.01; ***p<0.001. Multiple comparisons of TMAO concentration between all groups are shown in Table S2.
3.3Microbiome analysis
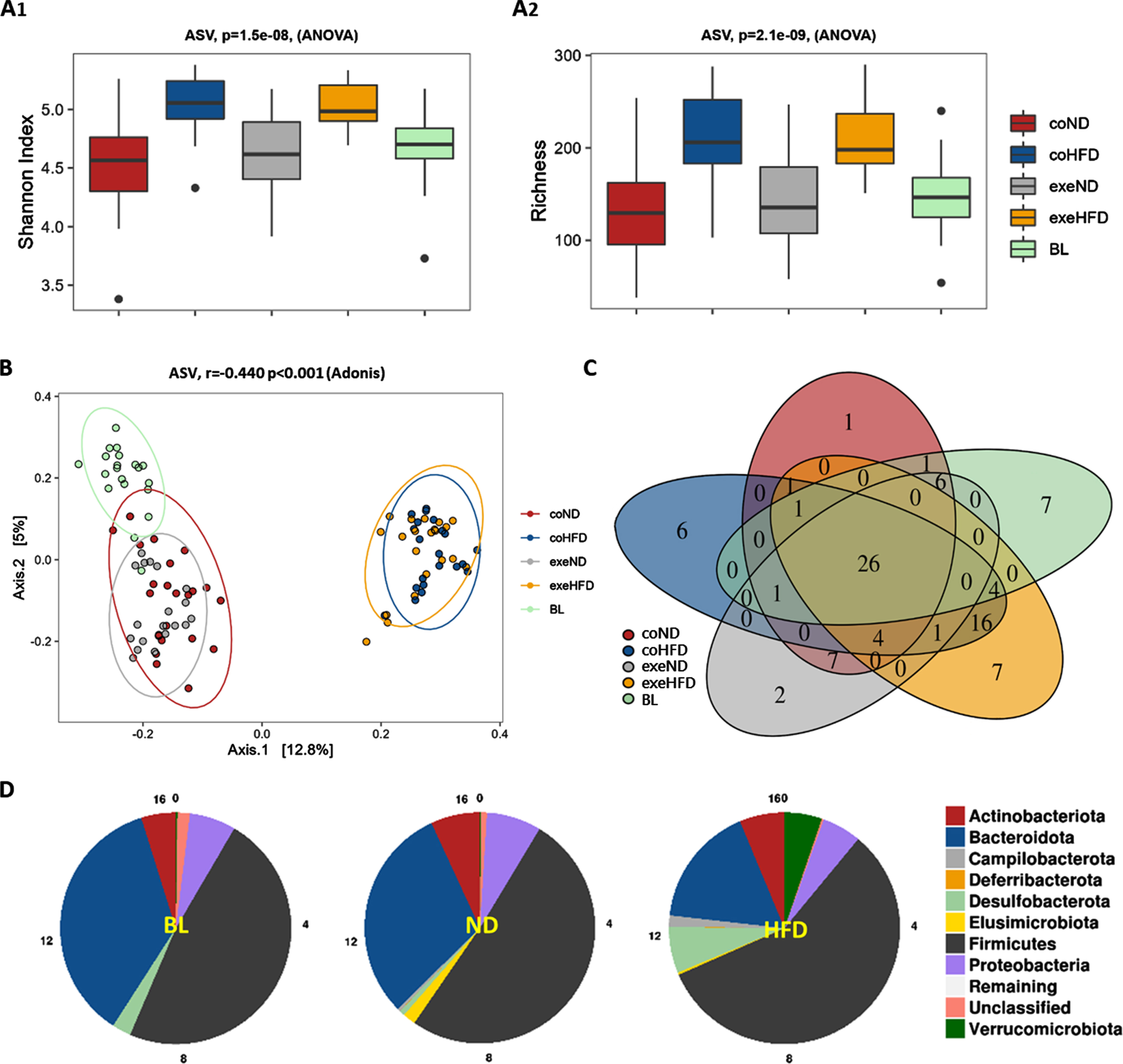
Bacterial alpha diversity was analyzed at ASV levels. Shannon Index was found to be higher in aged adult rats on ND compared to their young counterparts implying increased diversity. When compared to the respective controls on ND, Shannon index and different taxa (richness) showed an increased bacterial diversity in coHFD and exeHFD rats. Exercise had no significant effect on alpha diversity, regardless of diet (Fig. 3A). Principal component analysis, a qualitative representation of beta diversity, showed a clear separation of the microbiome from ND and HFD animals at the ASV level (Fig. 3B). This indicates substantial dissimilarity of the individual taxa in stool samples upon long-term feeding with ND or HFD (r = 0.440, p = 0.001, Adonis model). In addition, core microbiome analysis of all groups at the genus level revealed more unique genera in the HFD fed rats than in aged rats on ND (29 HFD vs. 10 ND) (Fig. 3C). Finally, phylum distribution showed that ten months of HFD administration changed the composition of the microbiome with more Firmicutes, Desulvobacteroidetes, Verrucomicrobiota, and less Bacteroidetes than in ND animals (Fig. 3D). However, no obvious differences were found when the diet groups were further separated into exercise and control groups (Figure S1).
Fig. 3
Microbiome alpha and beta diversity analysis from feces of all rat study groups. (A) Shannon Index (A1) and richness (A2) for alpha diversity at ASV level. Significance of differences was calculated with the 2-way ANOVA. (B) Principal coordinate analysis (PCoA) for beta diversity at the ASV level. Significance of differences was calculated with Adonis model (Bray-Curtis). (C) Venn diagram of core microbiome depicting unique and shared taxa on genus level between all study groups. (D) Diet-based phylum distribution independent of exercise.
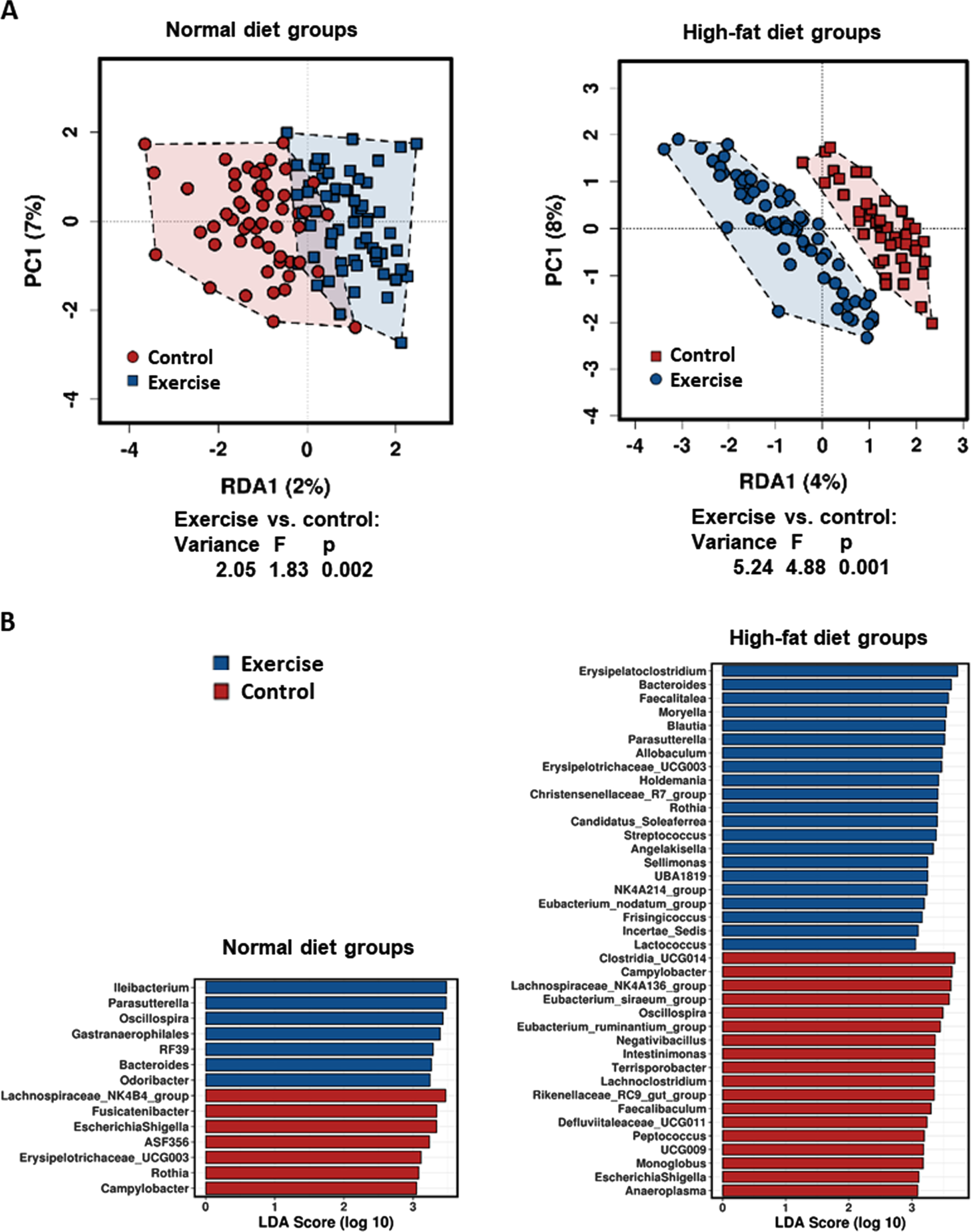
For the specific analysis of the impact of long-term exposure to HFD and exercise on microbiome diversity, only the long-term study groups were compared in the subsequent statistical analyses. These analyses demonstrate a stronger influence of exercise on the fecal microbiome composition upon feeding with HFD rather than ND. Redundancy analyses (RDA) on genus level showed a greater impact of exercise on beta diversity in HFD (Variance = 5.24, F = 4.88, p = 0.001) than in ND rats (Variance = 2.05, F = 1.83, p = 0.002; Fig. 4A). In addition, linear discriminant analysis effect size analysis (LEfSE) at the genus level identified more genera with differences when comparing exeHFD and coHFD rats than exeND and coND rats (Fig. 4B). Both, RDA and LefSE analysis including all groups are shown in Figure S2.
Fig. 4
Influence of high-fat diet and exercise on abundancy differences of taxa on genus level between the long-term study groups. (A) Exercise-related variances within the diet groups are shown by Redundancy analysis (RDA). (B) Linear discriminant analysis effect size analysis (LEfSE) showing the bacterial genera that most likely explain the differences between exercising and control animals fed either normal diet or high-fat diet.
3.4Microbiome diversity and serum TMAO
Next, the relationship between serum TMAO concentrations and microbiome diversity from all groups was explored. A linear regression with point-by-point correlation (Fig. 5A) shows that the Shannon index for alpha diversity at the genus level increased significantly with lower TMAO concentrations (r=–0.507, p < 0.001), and a clustering of the HFD fed animals (Fig. 5A, blue and yellow colour codes represent coHFD and exeHFD groups). Furthermore, serum TMAO was positively correlated with beta diversity at the genus level as shown by the Adonis model in the PCoA (r = 0.171, p < 0.001, Fig. 5B).
Fig. 5
TMAO levels in relation to microbiome diversity and possible TMA producing taxa. Serum TMAO levels from the study groups determined at the end of the trial were integrated into the microbiome metadata set. (A) Linear regression analysis of TMAO serum concentrations of all groups with alpha diversity’s Shannon Index. (B) Relation of TMAO with beta diversity of all groups shown by PCoA; significance of differences was calculated with Adonis model (Bray-Curtis). ND, normal diet groups; HFD, high-fat diet groups; NA, not available. (C) Taxa at the genus level with different abundancies between HFD and ND groups. Significance of differences in abundancies between study groups was calculated with the 2-way ANOVA. *p<0.05; **p<0.01; ***p<0.001. (D) Linear regression analysis of TMAO concentrations with single taxa abundances on genus level suggests them to be possible TMA-producing genera.
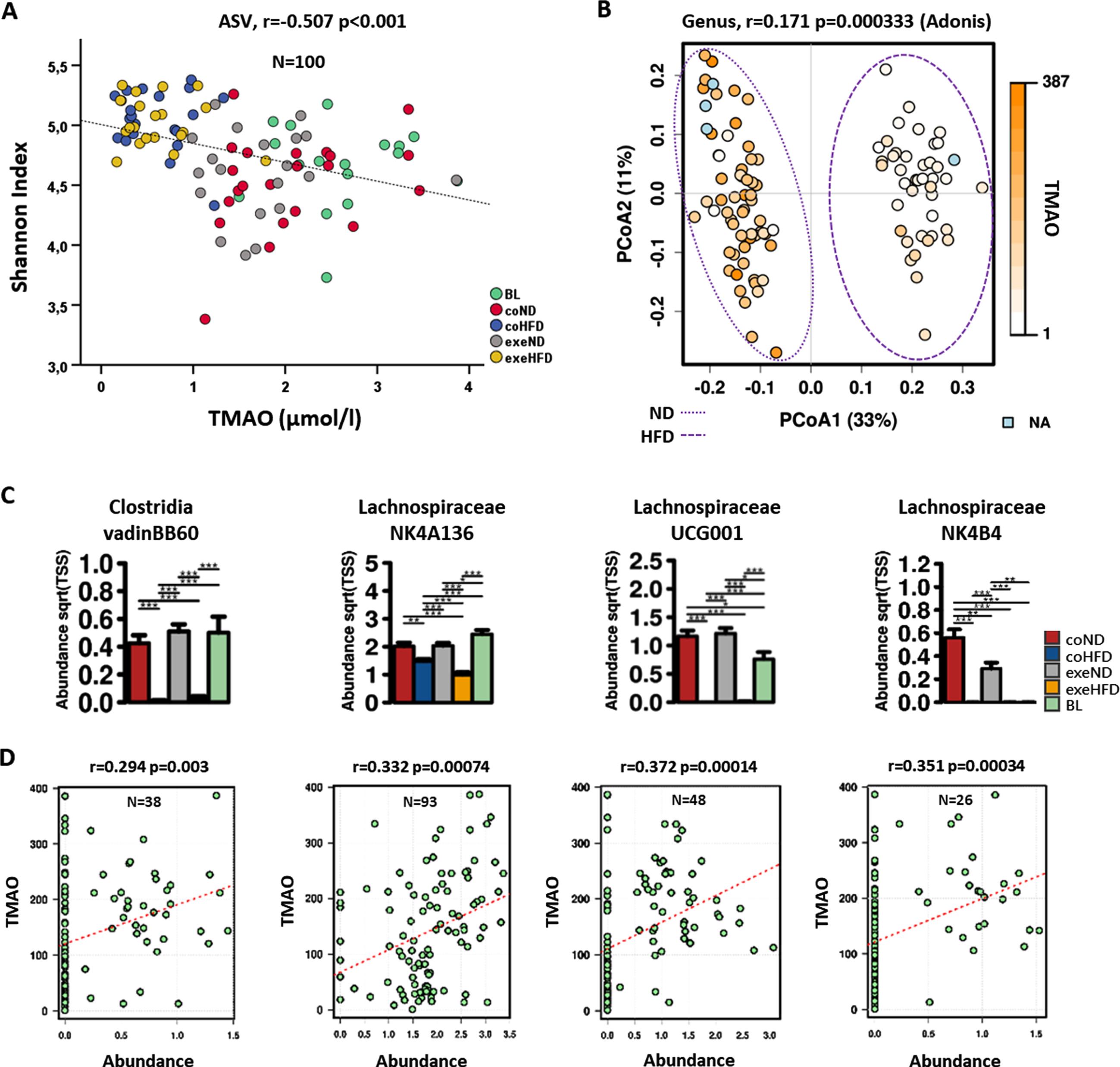
The two-way ANOVA revealed a profound reduction of Clostridia vadinB660 in the microbiome of HFD-fed rats when compared to ND controls (p < 0.001). In addition, Lachnospiraceae NK4A136, UCG001, and NK4B4 were significantly less abundant in these animals (Fig. 5C). Clostridia and Lachnospiraceae groups are known to contain high producers of TMA, the precursor for hepatic TMAO synthesis [25, 44]. The relative abundance of all four shown genera of the microbiome significantly positively correlated with TMAO serum concentrations (Fig. 5D).
4Discussion
The present study provides evidence for a relationship between dietary composition, diversity of the gastrointestinal microbiome, and the serum TMAO concentration, a potential risk factor for CVD. Long-term consumption of high amounts of nutritional fat induced pronounced alterations of the gut microbiome with an increased alpha diversity at the genus level and opposite effects at the phylum level. In particular, HFD reduced the abundance of genera that possibly contain high producers of TMA, the precursor for hepatic TMAO synthesis. In line with this observation, serum TMAO concentrations were directly correlated with the abundance of these genera. In contrast, regular moderate running exercise also correlated with changes in microbiome beta diversity but had no effect on serum TMAO.
There is robust evidence that serum TMAO can be increased through the intake of TMAO-containing food items, such as saltwater fish and other sea products [45]. In addition, the consumption of L-carnitine and choline-rich food, such as red meat and eggs, can lead to higher serum TMAO concentrations [1, 46]. Both compounds are metabolized by the gut microbiome to TMA, the substrate for hepatic TMAO synthesis, which is catalysed by FMO-3 in the liver [23]. Only a few studies have investigated the effect of other meat (e.g., sausages, bacon) and dairy (milk, yogurt, cheese) products on serum TMAO with mixed results [45]. On this background, low TMAO concentrations in our HFD-fed animals may seem not as surprising as first thought. Exploratory analysis of the TMAO content in both diets revealed quantities that were found to be different (Table S4) but can explain no more than 5% of the measured TMAO serum concentrations (Table S4 appendix). Furthermore, the used beef-tallow-based HFD is rich in saturated fat, but poor in carnitine. Also, the choline content of the HFD was actually higher than in the ND (920 vs. 535 mg/kg), which argues against dietary choline as a relevant driver of serum TMAO in our study. Comparable to this observation, an increase in plasma choline concentration was observed in a human study with an intake of three eggs per day, whereas plasma TMAO concentration did not change [46]. Together, these facts suggest that dietary TMAO and its precursors carnitines and choline had no significant influence on the serum TMAO concentrations of the animals in our study.
In addition to carnitine and choline, other nutritional TMA(O) precursors, such as betaine, dimethylglycine, crono-betaine, μ-butyrobetaine and ergothioneine should also be mentioned. Some of these dietary compounds may have an effect on TMA and TMAO blood concentrations, depending on the intestinal microflora in the colon [47, 48]. Besides other sources, the TMA(O) precursors proline betaine and glycine betaine were found in many flowering plants and cereals [49, 50]. Therefore, possible dietary effects involving other TMA(O) precursors or dietary sources cannot be ruled out completely.
The fecal microbiome of both HFD groups showed greater diversity at ASV (and genus) levels than the respective ND groups. Although unexpected, this result is in line with a recent meta-analysis of murine studies that revealed inconsistent effects of different HFD types on microbiome composition and diversity [51]. The authors argue that such inconsistencies could be the result of different animal strains and variable dietary composition. Furthermore, technical factors, such as sequencing depth and clustering/inference algorithms, could also contribute to the heterogeneity of previous studies. Concluding, one can speculate that the used HFD, which was enriched in triglycerides but not in cholesterol and simultaneously necessarily reduced in carbohydrates, also caused beneficial effects, finally leading to a higher alpha diversity and lower TMAO concentrations compared to the groups on normal diet. In line with this consideration, we found evidence that HFD-related alterations of the microbiome may have lowered intestinal TMA synthesis. More importantly, the abundance of genera with possible TMA high producers, such as Clostridia vadinB660, Lachnospiraceae NK4A136, UCG001, and NK4B4, was markedly lower in HFD animals than in ND animals. In addition, the serum TMAO concentrations were significantly correlated with the abundance of these genera.
A relationship between TMAO, Lachnospiraceae, and Clostridia in healthy men has been described before by Cho et al. [44]. Participants with a higher abundance of potential TMA high-producers in their microbiome had higher urinary TMAO concentrations after consuming a choline-rich meal. Nevertheless, there were also reports of an inverse relationship between TMAO and Lachnospiraceae [52]. A very recent work by Cai et al. identified a Lachnochlostridium sp. as TMA high-producer [53]. In our study, we did not find a direct correlation between Lachnoclostridium genera and TMAO serum concentrations, although we found a high abundancy of Lachnoclostridium in the baseline group (Figure S2B), the study group with the highest TMAO serum concentrations on average.
Our results support a previous study from Crawford et al. demonstrating a significant alteration of the gut microbiome in male Sprague-Dawley rats that were fed for six weeks with a 60% HFD [54]. This change in microbiome composition was accompanied by adverse gastrointestinal effects and increased inflammation. Also human studies found clear differences in the community structure of microbiota from the distal guts of lean and obese adolescents, but also that their microbiomes exhibited partial functional redundancy [55]. Further, Wang et al. reported a higher microbiome diversity in mice upon HFD feeding [56]. However, their HFD was not only rich in fat but also contained a substantial amount of fibres. In fact, the authors suggested that the increased fibre intake rather than high amounts of fat were responsible for the observed increases in gut microbiome diversity, short-chain fatty acids, and energy expenditure. The importance of dietary fibres’ content is further supported by Dalby et al. showing major alterations of the murine gut microbiome after six weeks of feeding with refined diets that were either high or low in fat [57]. Recent publications also reported an influence of dietary fibres’ content on serum TMAO and microbiome diversity [56, 58]. In our study, HFD and ND had a similar fibres’ content (5 g vs. 5.4 g/100 g dry weight). Therefore, the observed microbiome changes in the HFD animals are more probably caused by the changed ratio of saturated fat to carbohydrates. From this it can be speculated that a HFD which contains primarily saturated lipids and a reduced carbohydrate amount may increase dyslipidaemia-driven diseases while lowering TMAO at the same time.
In the same cohort we also investigated cardiovascular parameters recently [59]. The observed opposing effects may explain the rather small and inconsistent findings in cardiac remodelling, myocardial telomere length and telomerase-reverse transcriptase expression in the hearts of our rats.
The present results further support previous evidence of an exercise-induced alteration of the gut microbiome [31, 60, 61]. In these studies, exercise-related changes in the microbiome have been linked to gastrointestinal barrier function, physical performance, and the adaptation of skeletal muscle to exercise. In our study, effects worth mentioning were seen for the genera of Parasutterella and Bacteroides. Both taxa were higher in exercising animals than in the respective controls. Interestingly, these exercise-related effects were more pronounced in HFD than in ND animals (Fig. 4B and S3). In line with our results, 12 weeks of brisk walking exercise has also been shown to increase Bacteroides in healthy elderly women [62]. However, this change in microbiome composition was not accompanied by significant improvements in glycaemic control or lipid metabolism, but with only 17 participants in the exercise group, statistical power was rather low. A marked increase in the intestinal abundance of Bacteroides has also been demonstrated in rats after 12 weeks of resistance training [63]. In contrast, top endurance athletes with high training volumes and intensity have been found with a lower abundance of Bacteroides compared to inactive controls [64]. In a review of the existing literature, Cerdà et al. reported mixed results regarding the impact of exercise on Bacteroides [30]. In addition to effects on Bacteroides, here we showed for the first time a marked exercise-induced increase in Parasutterella, which was even stronger than that observed for Bacteroides. While alterations of Bacteroides and Parasutterella in response to exercise are interesting, both genera are not known to be involved in TMAO metabolism. Also other potential metabolic and pathophysiological implications of such changes are poorly understood and require further research.
This study has some strengths and limitations that should be considered when interpreting the results. To our knowledge, this is the first long-term in vivo study that investigated the effects of a HFD and moderate aerobic exercise on serum TMAO and fecal microbiome composition. With 20-22 animals per group, this study is well powered and statistically valid, which creates substantial credibility to the results. Measurement of TMAO was performed with a validated LC-MS/MS method ensuring accurate and precise results. It should also be mentioned that the alpha diversity of the microbiome was stable over time as shown by intermediate analyses of stool samples at 4 and 8 months (Figure S4). Impaired renal function as a potential confounder can be ruled out based on comparable creatinine levels that have been derived from semi-quantitative metabolomic studies (Figure S5).
As limitations may be considered the use of only female rats and the different basis of the two diets (Figure S6), with the HFD containing no cholesterol and being enriched only in triglycerides, and necessarily correspondingly reduced in carbohydrate, so that it may be considered more “artificial”. However, the dietary amount of TMAO was very low so substantial variability in the intake of TMAO and its precursors is unlikely, and the amounts of dietary fibre were comparable between the two diets. Despite a comparable fibre content, fibre composition may have varied between the two diets as they were obtained from different providers. To mention, the long-term influence of different high caloric diets should be investigated in future trials. Another limitation is the lack of baseline stool and blood samples, which impedes longitudinal comparisons. This shortcoming is, at least partially, mitigated by the inclusion of a group of young adult rats (BL group) that was purchased together with the other animals, but sacrificed directly after acclimatization. Also, associations between stool and serum results should be interpreted with caution as the microbiome may vary along the gastrointestinal tract [65] and the rather transient microbiome in stool may differ from the more resident bacteria that are found in close proximity to the gut mucosa. Finally, additional analysis of serum TMA and hepatic FMO3 activity would have been useful for the interpretation of TMAO results but appropriate methods were not available. Nevertheless, this should be considered in future investigations.
5Conclusions
High amounts of dietary fat that are consumed over an extended period of time strongly reduced serum TMAO. This effect was accompanied by a profound alteration in microbiome composition with a reduced abundance of possible TMA-producing bacteria, such as Clostridia and Lachnospiraceae. Regular moderate endurance exercise did alter beta diversity of the microbiome composition but had no effect on serum TMAO and TMA-producing bacteria in the gut. Future studies should investigate the impact of diet-related differences in serum TMAO and, besides its controversial associations with CVD, upcoming investigations would be needed to explore its possible benefits more in depth.
Acknowledgments
The authors thank Lisa Wink for 16 S rRNA gene analysis and taxonomic classification and Gerd Hörl for graphical improvement.
Funding
This research received no external funding.
Author contributions
Conception: Gunter Almer and Markus Herrmann; Performance of work: Gunter Almer, Maria Donatella Semeraro, Hans-Jürgen Gruber, Andreas Meinitzer, Angela Horvath. and Giovanny Rodriguez Blanco; Interpretation of data: Gunter Almer, Angela Horvath, Christine Moissl-Eichinger, Dietmar Enko and Markus Herrmann; Resources: Gunter Almer, Andreas Meinitzer, Birgit Gallé and Markus Herrmann; Writing the article: Gunter Almer, Angela Horvath and Markus Herrmann; Writing—Review & Editing: Maria Donatella Semeraro, Dietmar Enko, Christine Moissl-Eichinger, Holger Till and Markus Herrmann.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary data
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/NHA-220198.
References
[1] | Koeth RA , Wang Z , Levison BS , Buffa JA , Org E , Sheehy BT , et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. (2013) ;19: (5):576–85. |
[2] | Yancey PH . Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. (2005) ;208: (Pt 15):2819–30. |
[3] | Fretts AM , Hazen SL , Jensen P , Budoff M , Sitlani CM , Wang M , et al. Association of Trimethylamine N-Oxide and Metabolites With Mortality in Older Adults. JAMA Netw Open. (2022) ;5: (5):e2213242. |
[4] | Gencer B , Li XS , Gurmu Y , Bonaca MP , Morrow DA , Cohen M , et al. Gut Microbiota-Dependent Trimethylamine N-oxide and Cardiovascular Outcomes in Patients With Prior Myocardial Infarction: A Nested Case Control Study From the PEGASUS-TIMI 54 Trial. J Am Heart Assoc. (2020) ;9: (10):e015331. |
[5] | Lee Y , Nemet I , Wang Z , Lai HTM , de Oliveira Otto MC , Lemaitre RN , et al. Longitudinal Plasma Measures of Trimethylamine N-Oxide and Risk of Atherosclerotic Cardiovascular Disease Events in Community-Based Older Adults. J Am Heart Assoc. (2021) ;10: (17):e020646. |
[6] | Tang WH , Wang Z , Kennedy DJ , Wu Y , Buffa JA , Agatisa-Boyle B , et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. (2015) ;116: (3):448–55. |
[7] | Xu KY , Xia GH , Lu JQ , Chen MX , Zhen X , Wang S , et al. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci Rep. (2017) ;7: (1):1445. |
[8] | He W , Luo Y , Liu JP , Sun N , Guo D , Cui LL , et al. Trimethylamine N-Oxide, a Gut Microbiota-Dependent Metabolite, is Associated with Frailty in Older Adults with Cardiovascular Disease. Clin Interv Aging. (2020) ;15: :1809–20. |
[9] | Li D , Lu Y , Yuan S , Cai X , He Y , Chen J , et al. Gut microbiota-derived metabolite trimethylamine-N-oxide and multiple health outcomes: an umbrella review and updated meta-analysis. Am J Clin Nutr. (2022) ;116: (1):230–43. |
[10] | Wang Z , Klipfell E , Bennett BJ , Koeth R , Levison BS , Dugar B , et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. (2011) ;472: (7341):57–63. |
[11] | Li T , Gua C , Wu B , Chen Y . Increased circulating trimethylamine N-oxide contributes to endothelial dysfunction in a rat model of chronic kidney disease. Biochem Biophys Res Commun. (2018) ;495: (2):2071–7. |
[12] | Zhu W , Gregory JC , Org E , Buffa JA , Gupta N , Wang Z , et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell. (2016) ;165: (1):111–24. |
[13] | Papandreou C , Bullo M , Zheng Y , Ruiz-Canela M , Yu E , Guasch-Ferre M , et al. Plasma trimethylamine-N-oxide and related metabolites are associated with type 2 diabetes risk in the Prevencion con Dieta Mediterranea (PREDIMED) trial. Am J Clin Nutr. (2018) ;108: (1):163–73. |
[14] | Dumas ME , Rothwell AR , Hoyles L , Aranias T , Chilloux J , Calderari S , et al. Microbial-Host Co-metabolites Are Prodromal Markers Predicting Phenotypic Heterogeneity in Behavior, Obesity, and Impaired Glucose Tolerance. Cell Rep. (2017) ;20: (1):136–48. |
[15] | Georgescauld F , Mocan I , Lacombe ML , Lascu I . Rescue of the neuroblastoma mutant of the human nucleoside diphosphate kinase A/nm23-H1 by the natural osmolyte trimethylamine-N-oxide. FEBS Lett. (2009) ;583: (4):820–4. |
[16] | Huc T , Drapala A , Gawrys M , Konop M , Bielinska K , Zaorska E , et al. Chronic, low-dose TMAO treatment reduces diastolic dysfunction and heart fibrosis in hypertensive rats. Am J Physiol Heart Circ Physiol. (2018) ;315: (6):H1805–H20. |
[17] | Collins HL , Drazul-Schrader D , Sulpizio AC , Koster PD , Williamson Y , Adelman SJ , et al. L-Carnitine intake and high trimethylamine N-oxide plasma levels correlate with low aortic lesions in ApoE(-/-) transgenic mice expressing CETP. Atherosclerosis. (2016) ;244: :29–37. |
[18] | Zhao ZH , Xin FZ , Zhou D , Xue YQ , Liu XL , Yang RX , et al. Trimethylamine N-oxide attenuates high-fat high-cholesterol diet-induced steatohepatitis by reducing hepatic cholesterol overload in rats. World J Gastroenterol. (2019) ;25: (20):2450–62. |
[19] | Hoyles L , Pontifex MG , Rodriguez-Ramiro I , Anis-Alavi MA , Jelane KS , Snelling T , et al. Regulation of blood-brain barrier integrity by microbiome-associated methylamines and cognition by trimethylamine N-oxide. Microbiome. (2021) ;9: (1):235. |
[20] | Winther SA , Ollgaard JC , Tofte N , Tarnow L , Wang Z , Ahluwalia TS , et al. Utility of Plasma Concentration of Trimethylamine N-Oxide in Predicting Cardiovascular and Renal Complications in Individuals With Type 1 Diabetes. Diabetes Care. (2019) ;42: (8):1512–20. |
[21] | Jia J , Dou P , Gao M , Kong X , Li C , Liu Z , et al. Assessment of Causal Direction Between Gut Microbiota-Dependent Metabolites and Cardiometabolic Health: A Bidirectional Mendelian Randomization Analysis. Diabetes. (2019) ;68: (9):1747–55. |
[22] | Veeravalli S , Karu K , Scott F , Fennema D , Phillips IR , Shephard EA . Effect of Flavin-Containing Monooxygenase Genotype, Mouse Strain, and Gender on Trimethylamine N-oxide Production, Plasma Cholesterol Concentration, and an Index of Atherosclerosis. Drug Metab Dispos. (2018) ;46: (1):20–5. |
[23] | Warrier M , Shih DM , Burrows AC , Ferguson D , Gromovsky AD , Brown AL , et al. The TMAO-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep. (2015) ;10: (3):326–38. |
[24] | Bin-Jumah MN , Gilani SJ , Hosawi S , Al-Abbasi FA , Zeyadi M , Imam SS , et al. Pathobiological Relationship of Excessive Dietary Intake of Choline/L-Carnitine: A TMAO Precursor-Associated Aggravation in Heart Failure in Sarcopenic Patients. Nutrients. (2021) ;13: (10). |
[25] | Jameson E , Quareshy M , Chen Y . Methodological considerations for the identification of choline and carnitine-degrading bacteria in the gut. Methods. (2018) ;149: :42–8. |
[26] | Rath S , Rud T , Pieper DH , Vital M . Potential TMA-Producing Bacteria Are Ubiquitously Found in Mammalia. Front Microbiol. (2019) ;10: :2966. |
[27] | Vangay P , Johnson AJ , Ward TL , Al-Ghalith GA , Shields-Cutler RR , Hillmann BM , et al. US Immigration Westernizes the Human Gut Microbiome. Cell. (2018) ;175: (4):962–72 e10. |
[28] | de La Serre CB , Ellis CL , Lee J , Hartman AL , Rutledge JC , Raybould HE . Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. (2010) ;299: (2):G440–8. |
[29] | Ding S , Chi MM , Scull BP , Rigby R , Schwerbrock NM , Magness S , et al. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS One. (2010) ;5: (8):e12191. |
[30] | Cerda B , Perez M , Perez-Santiago JD , Tornero-Aguilera JF , Gonzalez-Soltero R , Larrosa M . Gut Microbiota Modification: Another Piece in the Puzzle of the Benefits of Physical Exercise in Health? Front Physiol. (2016) ;7: :51. |
[31] | Scheiman J , Luber JM , Chavkin TA , MacDonald T , Tung A , Pham LD , et al. Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat Med. (2019) ;25: (7):1104–9. |
[32] | Clarke SF , Murphy EF , O’Sullivan O , Lucey AJ , Humphreys M , Hogan A , et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. (2014) ;63: (12):1913–20. |
[33] | Evans CC , LePard KJ , Kwak JW , Stancukas MC , Laskowski S , Dougherty J , et al. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. (2014) ;9: (3):e92193. |
[34] | Petriz BA , Castro AP , Almeida JA , Gomes CP , Fernandes GR , Kruger RH , et al. Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genomics. (2014) ;15: :511. |
[35] | Campbell SC , Wisniewski PJ , Noji M , McGuinness LR , Haggblom MM , Lightfoot SA , et al. The Effect of Diet and Exercise on Intestinal Integrity and Microbial Diversity in Mice. PLoS One. (2016) ;11: (3):e0150502. |
[36] | Bhandari U , Kumar V , Khanna N , Panda BP . The effect of high-fat diet-induced obesity on cardiovascular toxicity in Wistar albino rats. Hum Exp Toxicol. (2011) ;30: (9):1313–21. |
[37] | Matsuo T , Takeuchi H , Suzuki H , Suzuki M . Body fat accumulation is greater in rats fed a beef tallow diet than in rats fed a safflower or soybean oil diet. Asia Pac J Clin Nutr. (2002) ;11: (4):302–8. |
[38] | Li FH , Sun L , Zhu M , Li T , Gao HE , Wu DS , et al. Beneficial alterations in body composition, physical performance, oxidative stress, inflammatory markers, and adipocytokines induced by long-term high-intensity interval training in an aged rat model. Exp Gerontol. (2018) ;113: :150–62. |
[39] | Rocha-Rodrigues S , Rodriguez A , Goncalves IO , Moreira A , Maciel E , Santos S , et al. Impact of physical exercise on visceral adipose tissue fatty acid profile and inflammation in response to a high-fat diet regimen. Int J Biochem Cell Biol. (2017) ;87: :114–24. |
[40] | Enko D , Zelzer S , Baranyi A , Herrmann M , Meinitzer A . Determination of Trimethylamine-N-oxide by a Simple Isocratic High-Throughput Liquid-Chromatography Tandem Mass-Spectrometry Method. Clin Lab. (2020) ;66: (9). |
[41] | 16S Metagenomic Sequencing Library Preparation. Available from: https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistrydocumentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf. |
[42] | Semeraro MD , Almer G , Kaiser M , Zelzer S , Meinitzer A , Scharnagl H , et al. The effects of long-term moderate exercise and Western-type diet on oxidative/nitrosative stress, serum lipids and cytokines in female Sprague Dawley rats. Eur J Nutr. (2022) ;61: (1):255–68. |
[43] | Semeraro MD , Almer G , Renner W , Gruber HJ , Herrmann M . Influences of Long-Term Exercise and High-Fat Diet on Age-Related Telomere Shortening in Rats. Cells. (2022) ;11: (10). |
[44] | Cho CE , Aardema NDJ , Bunnell ML , Larson DP , Aguilar SS , Bergeson JR , et al. Effect of Choline Forms and Gut Microbiota Composition on Trimethylamine-N-Oxide Response in Healthy Men. Nutrients. (2020) ;12: (8). |
[45] | Lombardo M , Aulisa G , Marcon D , Rizzo G , Tarsisano MG , Di Renzo L , et al. Association of Urinary and Plasma Levels of Trimethylamine N-Oxide (TMAO) with Foods. Nutrients. (2021) ;13: (5). |
[46] | DiMarco DM , Missimer A , Murillo AG , Lemos BS , Malysheva OV , Caudill MA , et al. Intake of up to 3 Eggs/Day Increases HDL Cholesterol and Plasma Choline While Plasma Trimethylamine-N-oxide is Unchanged in a Healthy Population. Lipids. (2017) ;52: (3):255–63. |
[47] | Wang Z , Bergeron N , Levison BS , Li XS , Chiu S , Jia X , et al. Impact of chronic dietary red meat, white meat, or non-meat protein on trimethylamine N-oxide metabolism and renal excretion in healthy men and women. Eur Heart J. (2019) ;40: (7):583–94. |
[48] | Gatarek P , Kaluzna-Czaplinska J . Trimethylamine N-oxide (TMAO) in human health. EXCLI J. (2021) ;20: :301–19. |
[49] | Hanson AD , Rathinasabapathi B , Rivoal J , Burnet M , Dillon MO , Gage DA . Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc Natl Acad Sci U S A. (1994) ;91: (1):306–10. |
[50] | Servillo L , D’Onofrio N , Giovane A , Casale R , Cautela D , Ferrari G , et al. The betaine profile of cereal flours unveils new and uncommon betaines. Food Chem. (2018) ;239: :234–41. |
[51] | Bisanz JE , Upadhyay V , Turnbaugh JA , Ly K , Turnbaugh PJ . Meta-Analysis Reveals Reproducible Gut Microbiome Alterations in Response to a High-Fat Diet. Cell Host Microbe. (2019) ;26: (2):265–72 e4. |
[52] | Arias N , Arboleya S , Allison J , Kaliszewska A , Higarza SG , Gueimonde M , et al. The Relationship between Choline Bioavailability from Diet, Intestinal Microbiota Composition, and Its Modulation of Human Diseases. Nutrients. (2020) ;12: (8). |
[53] | Cai YY , Huang FQ , Lao X , Lu Y , Gao X , Alolga RN , et al. Integrated metagenomics identifies a crucial role for trimethylamine-producing Lachnoclostridium in promoting atherosclerosis. NPJ Biofilms Microbiomes. (2022) ;8: (1):11. |
[54] | Crawford M , Whisner C , Al-Nakkash L , Sweazea KL . Six-Week High-Fat Diet Alters the Gut Microbiome and Promotes Cecal Inflammation, Endotoxin Production, and Simple Steatosis without Obesity in Male Rats. Lipids. (2019) ;54: (2-3):119–31. |
[55] | Ferrer M , Ruiz A , Lanza F , Haange SB , Oberbach A , Till H , et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol. (2013) ;15: (1):211–26. |
[56] | Wang B , Kong Q , Li X , Zhao J , Zhang H , Chen W , et al. A High-Fat Diet Increases Gut Microbiota Biodiversity and Energy Expenditure Due to Nutrient Difference. Nutrients. (2020) ;12: (10). |
[57] | Dalby MJ , Ross AW , Walker AW , Morgan PJ . Dietary Uncoupling of Gut Microbiota and Energy Harvesting from Obesity and Glucose Tolerance in Mice. Cell Rep. (2017) ;21: (6):1521–33. |
[58] | Genoni A , Christophersen CT , Lo J , Coghlan M , Boyce MC , Bird AR , et al. Long-term Paleolithic diet is associated with lower resistant starch intake, different gut microbiota composition and increased serum TMAO concentrations. Eur J Nutr. (2020) ;59: (5):1845–58. |
[59] | Semeraro MD , Beltrami AP , Kharrat F , Almer G , Sedej S , Renner W , et al. The impact of moderate endurance exercise on cardiac telomeres and cardiovascular remodeling in obese rats. Front Cardiovasc Med. (2022) ;9: :1080077. |
[60] | Motiani KK , Collado MC , Eskelinen JJ , Virtanen KA , Loyttyniemi E , Salminen S , et al. Exercise Training Modulates Gut Microbiota Profile and Improves Endotoxemia. Med Sci Sports Exerc. (2020) ;52: (1):94–104. |
[61] | Valentino TR , Vechetti IJ Jr, Mobley CB , Dungan CM , Golden L , Goh J et al. Dysbiosis of the gut microbiome impairs mouse skeletal muscle adaptation to exercise. J Physiol. (2021) ;599: (21):4845–63. |
[62] | Morita E , Yokoyama H , Imai D , Takeda R , Ota A , Kawai E , et al. Aerobic Exercise Training with Brisk Walking Increases Intestinal Bacteroides in Healthy Elderly Women. Nutrients. (2019) ;11: (4). |
[63] | Castro AP , Silva KKS , Medeiros CSA , Alves F , Araujo RC , Almeida JA . Effects of 12 weeks of resistance training on rat gut microbiota composition. J Exp Biol. (2021) ;224: (12). |
[64] | Kulecka M , Fraczek B , Mikula M , Zeber-Lubecka N , Karczmarski J , Paziewska A , et al. The composition and richness of the gut microbiota differentiate the top Polish endurance athletes from sedentary controls. Gut Microbes. (2020) ;11: (5):1374–84. |
[65] | Haange SB , Oberbach A , Schlichting N , Hugenholtz F , Smidt H , von Bergen M , et al. Metaproteome analysis and molecular genetics of rat intestinal microbiota reveals section and localization resolved species distribution and enzymatic functionalities. J Proteome Res. (2012) ;11: (11):5406–17. |




