Brachionus rotifers as a model for investigating dietary and metabolic regulators of aging
Abstract
Because every species has unique attributes relevant to understanding specific aspects of aging, using a diversity of study systems and a comparative biology approach for aging research has the potential to lead to novel discoveries applicable to human health. Monogonont rotifers, a standard model for studies of aquatic ecology, evolutionary biology, and ecotoxicology, have also been used to study lifespan and healthspan for nearly a century. However, because much of this work has been published in the ecology and evolutionary biology literature, it may not be known to the biomedical research community. In this review, we provide an overview of Brachionus rotifers as a model to investigate nutritional and metabolic regulators of aging, with a focus on recent studies of dietary and metabolic pathway manipulation. Rotifers are microscopic, aquatic invertebrates with many advantages as a system for studying aging, including a two-week lifespan, easy laboratory culture, direct development without a larval stage, sexual and asexual reproduction, easy delivery of pharmaceuticals in liquid culture, and transparency allowing imaging of cellular morphology and processes. Rotifers have greater gene homology with humans than do established invertebrate models for aging, and thus rotifers may be used to investigate novel genetic mechanisms relevant to human lifespan and healthspan. The research on caloric restriction; dietary, pharmaceutical, and genetic interventions; and transcriptomics of aging using rotifers provide insights into the metabolic regulators of lifespan and health and suggest future directions for aging research. Capitalizing on the unique biology of Brachionus rotifers, referencing the vast existing literature about the influence of diet and drugs on rotifer lifespan and health, continuing the development of genetic tools for rotifers, and growing the rotifer research community will lead to new discoveries a better understanding of the biology of aging.
1Introduction
Our understanding of many basic biological processes, and the development of several revolutionary biomedical technologies, resulted from research on “non-model” organisms, particularly aquatic taxa. For example, the discovery of ionic transmission of action potentials in the nervous system came from research on the giant squid axon by Hodgkin, Huxley, and Eccles [1]. Work in the large, transparent oocytes of sea urchins and clams led to descriptions of regulation of the cell cycle by cyclins [2] and of the ubiquitin-proteasome system for protein degradation [3, 4]. Kinesins, the ATP-hydrolysis fueled molecular motors critical for trafficking a variety of cargo in cells, were first identified in the squid giant axon [5]. Green fluorescent protein (GFP), originally isolated from the marine jelly, Aequorea victoria, is now commonly used as a biological marker for imaging of cellular processes [6–8]. Taq polymerase, which launched the PCR and sequencing revolution, was isolated from the thermophilic hot spring bacterium, Thermus aquaticus [9].
The winnowing of animal models for biomedical research to only a few species— in particular Saccromyces cerviscea, Caenorhabditis elegans, Drosophila melanogaster, and Mus musculus— has occurred primarily over the last three decades, largely due to strong leaders and the concerted efforts of their respective research communities. Work in these select cell and animal models continues to lead to significant advances in understanding basic biological processes and to innovation in translational medicine.
Increasingly, however, the research community is moving beyond a one-size-fits-all approach and is rediscovering that different taxa have unique traits that make them useful for addressing particular biological questions [10]. As pointed out by August Krogh [11], “For such a large number of problems there will be some animal of choice or a few such animals on which it can be most conveniently studied.” That is, no single organism is perfect for answering all questions in biology; the unique traits of different taxa may be leveraged to interrogate specific phenomena. Frequently, mechanisms or therapies identified to improve health or increase lifespan in a single model species have not successfully translated to humans in clinical trials. Indeed, the mechanisms found to be conserved across a wide range of species are those most likely to be relevant to human biology. Thus, a comparative biology approach embracing a variety of taxa may be required for efficient and effective translation of basic research to human treatments [10, 12, 13].
Study of the biology of aging is a rapidly growing field within biomedicine. Understanding the molecular drivers of aging, the causative role of aging in disease, the evolutionary basis of aging, and finding therapies that improve health in late age and extend lifespan are all of interest. Much of what we know about the biology of aging and interventions to aging has been revealed in model organisms. Here too, research on aging in a diversity of study systems is likely to bolster our understanding of the mechanisms of aging and the successful application of therapies to humans.
2Rotifers
With the goal diversifying the animal study systems relevant to human health, several research groups are developing rotifers as a modern model for the study of aging. Rotifers are aquatic, microscopic, invertebrate animals. More than 2000 species have been identified in four main groups: the monogononts (literally “single gonad”); the bdelloids (from the Greek, bdella, for “leech-like”), a lineage with degenerate tetraploid genomes, in which all individuals are females and reproduction appears to be entirely asexual; the seisonids, marine parasites of invertebrates; and the acanthocephala, spiny-headed parasitic worms [14]. Rotifers inhabit nearly all freshwater habitats, including ephemeral environments like vernal ponds, desert pools, and puddles, as well as coastal estuaries and inland salt lakes. Though different species have incredibly diverse morphologies, ranging from worm-like, to colonial, to round and armored, most rotifers are less than 1 mm long. Similar in complexity to C. elegans, rotifers have approximately 1000 cells, digestive and secretory organs, muscles, structures for swimming and crawling, nervous systems with centralized brains and peripheral sensory structures, and reproductive systems including ovaries and testes (Fig. 1).
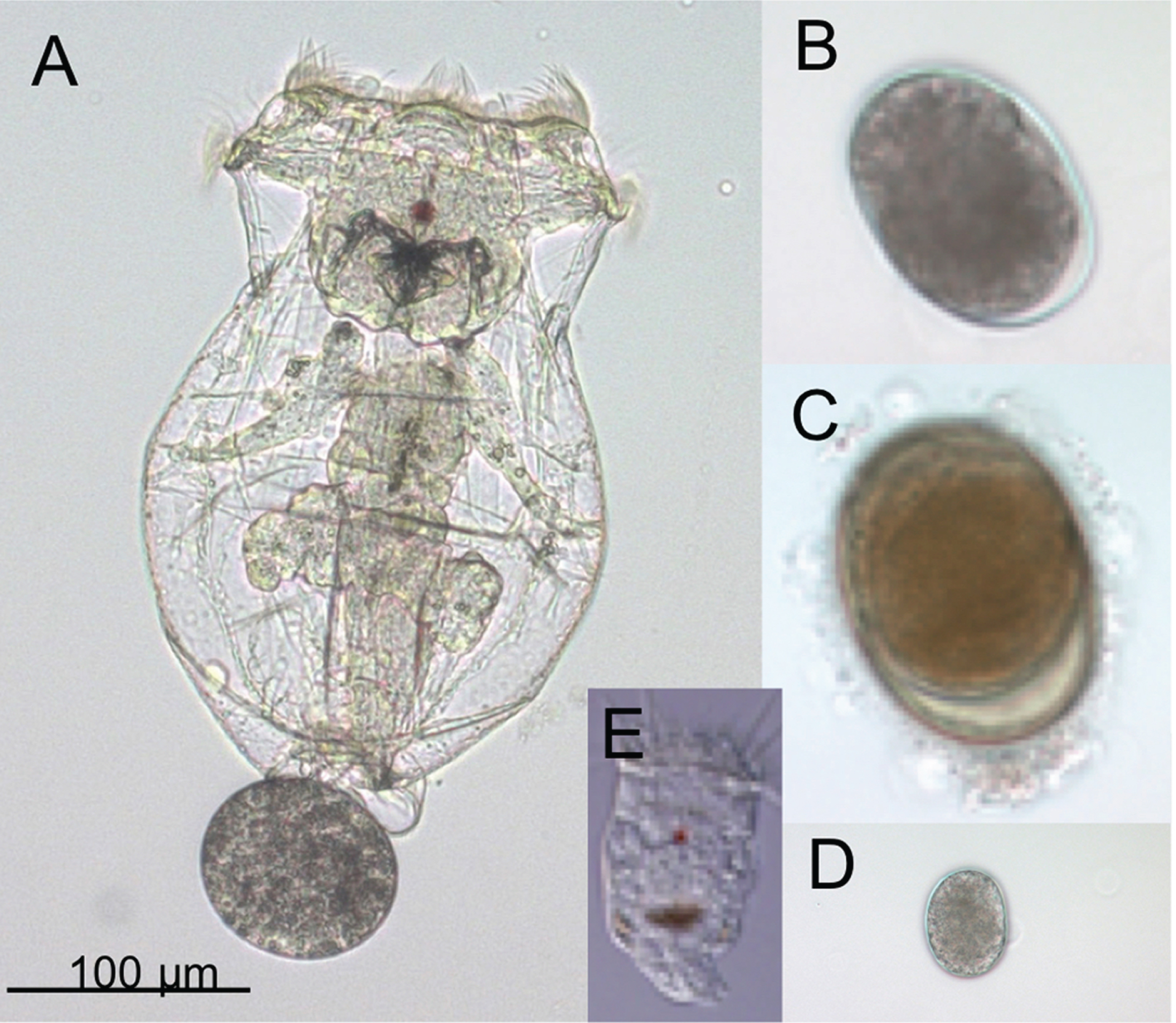
Fig. 1
Adults and eggs of the rotifer Brachionus manjavacas. (A) Female B. manjavacas carrying one egg. Females have apical cilia for swimming and feeding, a red eyespot used for sensing light; and digestive, reproductive, muscle, and nervous systems visible through a transparent body wall. (B) Amictic egg, produced asexually via mitosis, which will hatch into an asexual female. (C) Dormant resting egg, the product of sexual reproduction. (D) Small egg that will hatch into a male; (E) Male, with cilia for swimming and red eyespot, but no mouth or gut. Males are one-third the size of females and are haploid.
Monogonont rotifers have been used in studies of aging for over a century and continue to be a standard model for studies of aquatic ecology, evolutionary biology, and ecotoxicology. Additionally, because rotifers are a primary first food for larval fish in aquaculture, a great deal of research has been directed at fatty acid, protein, and micronutrient manipulations of rotifer diet to improve fish feed quality. Much of the experimental research has focused on manipulations of life history, including lifespan, reproduction, and aspects of fitness relevant to aging. Because most of this work has been published in the ecology and evolutionary biology literature, however, it may not be known to biomedically-oriented biology of aging scientists.
Our goal is to bring relevant studies to the attention of biomedical and geroscience researchers. In this review, we provide an overview of the use of Brachionus rotifers, with occasional examples from other rotifer species, to investigate nutritional and metabolic regulators of aging, with a focus on recent studies of dietary and metabolic pathway manipulation.
2.1Brachionus rotifers
Brachionus is a genus of monogonont rotifers with advantages as a model for aging similar to those of other short-lived invertebrate models: a short lifespan of two weeks; the ability to scale experi-ments from single individuals to large populations; transparency, allowing imaging of cellular morphology and processes; and genomic, transcriptomic, and gene expression manipulation tools [15, 16]. Additionally, Brachionus spp. have a number of unique advantages as invertebrate models of aging. They have direct development without a larval stage or metamorphosis which simplifies the execution and analysis of lifespan assays. Asexual reproduction allows experimentation on isogenic lines, and inducible sexual reproduction permits outcrossing and genetics. (Fig. 2). Because they swim in liquid medium and lack a resistant cuticle, pharmaceuticals are easily delivered to rotifers [17, 18].
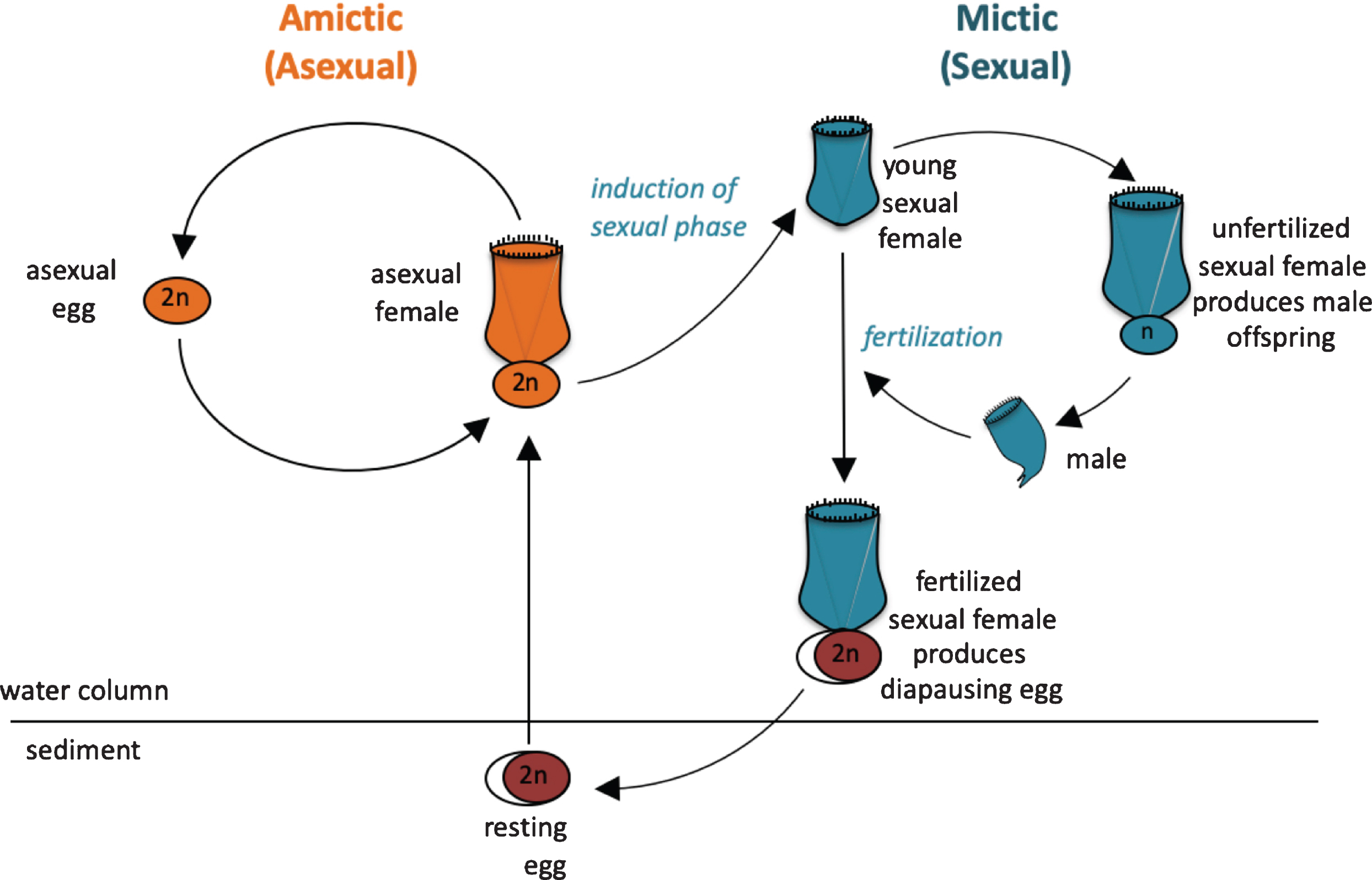
Fig. 2
Life cycle of the monogonont rotifer Brachionus manjavacas. (A) Left, the asexual (amictic) cycle, in which a female produces clonal diploid eggs by mitosis. Right, the sexual (mictic) cycle, in which crowding conditions prompt a portion of females in the population to become mictic, producing haploid gametes via meiosis. If haploid gametes are not fertilized, they hatch into diminutive haploid males. Fertilized gametes develop into dormant resting eggs, able to desiccate and overwinter in the sediments. Upon hatching, resting egg restore the asexual cycle.
The established invertebrate model systems, C. elegans and D. melanogaster, are closely related evo-lutionarily; both fall within the superphylum Ecdy-sozoa. Using rotifers, which are part of the distantly-related Spiralia, as an additional model thus adds evolutionary breadth to studies of aging [12, 19]. Both C. elegans and D. melanogaster have undergone extensive gene loss since they shared a common ancestor with humans, and thus are missing many aging-related genes found in humans [20, 21]. In contrast, many other invertebrates, including Brachionus rotifers, have retained hundreds of human homologs not found in C. elegans and D. melanogaster [15, 20, 21]. The B. manjavacas transcriptome was found to have homologs to 614 and 910 human transcripts that are absent in D. melanogaster and C. elegans genomes. respectively. Approximately 25% of these genes were significantly differentially expressed with aging in B. manjavacas [20]. Non-ecdysozoan animal models may thus reveal new genes relevant to human aging that will not be identified in established invertebrate models.
Brachionus provides a useful model system in which to study intergenerational and transgenerational effects on aging. Like humans, but unlike established invertebrate models, Brachionus rotifers make a large investment in individual offspring, with asexual females producing 25– 30 progeny sequentially over a week-long period. Sexually reproducing females produce half that number of offspring. Eggs are large and, beginning at the one-cell stage, are carried externally by the female for most of the 20 – 24-hour development period. Neonates are half to two-thirds the size of their mothers at the time of hatching and develop directly, without a larval stage or metamorphosis. These features allow easy collection of specific developmental stages and age-synchronized cohorts for lifespan studies.
2.2Experimental methods in brachionus
Lifespan, fecundity, and health in Brachionus can be measured using simple methods. Brachionus are typically grown at 15°C – 25°C in artificial seawater with salinity ranging from 0 – 20 ppt. Rotifer cultures are fed unicellular algae, bacteria, or yeast grown in the laboratory, or they may be reared on commercially available dried or frozen food. Lifetable assays are started by vortexing or blending reproductive, asexual females to collect externally-brooded eggs. Asexual eggs are hatched over 5 – 16 hours to create an age-synchronized, isogenic cohort (Fig. 3). Alternatively, cohorts may be established by rehydrating desiccated, dormant resting eggs over 24 hours, resulting in highly coordinated hatching. Because resting eggs are produced via sexual reproduction, these experimental populations are inbred, but not clonal. These alternate starting points provide flexibility for addressing questions requiring either isogenic or genetically variable populations.
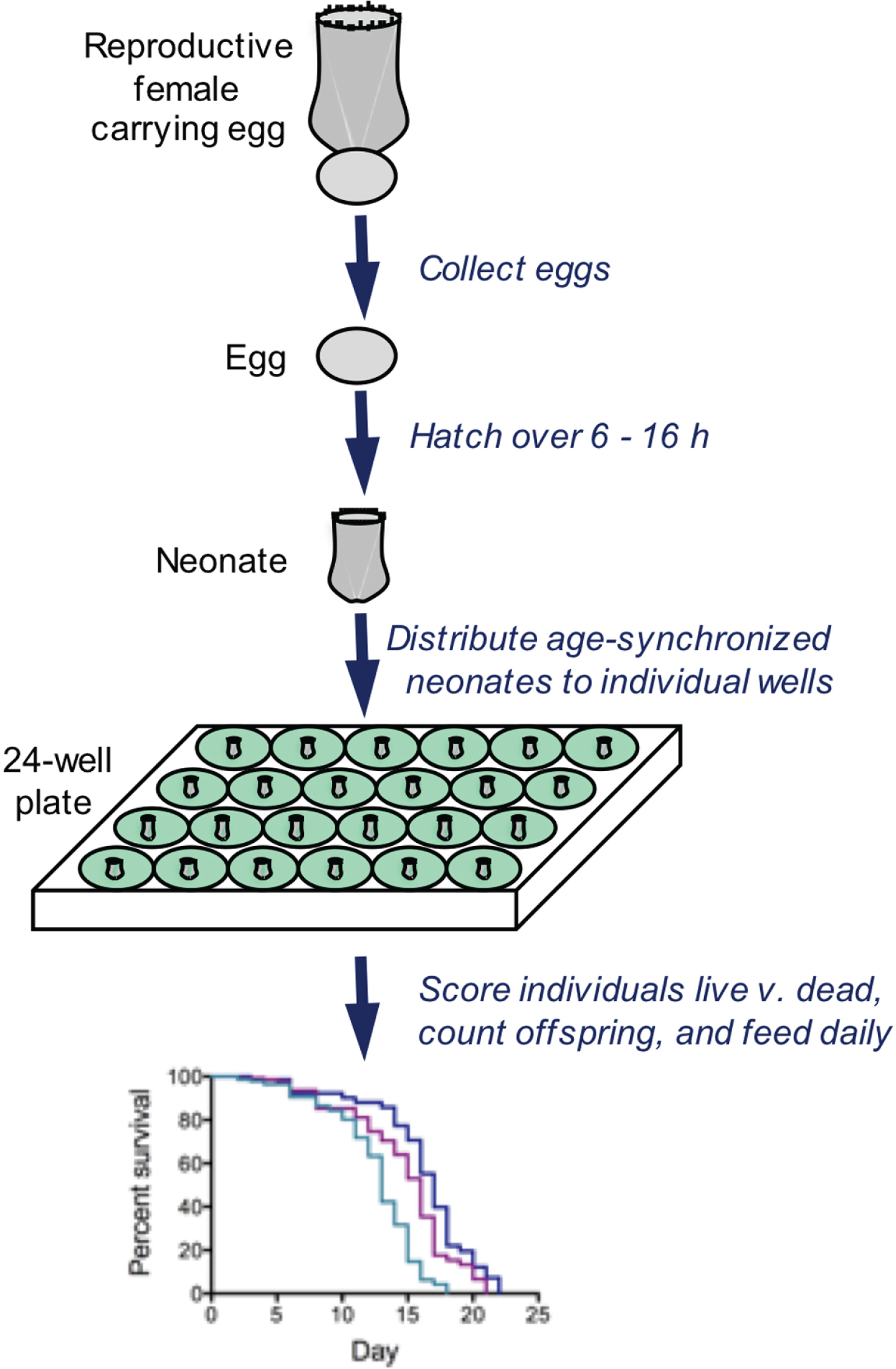
Fig. 3
Typical methods for analyzing survivorship and fecundity of Brachionus. Eggs are collected and hatched over a short time window, resulting in an age-synchronized cohort. Neonates are distributed to wells containing 1 ml of seawater, algae as food, and drugs or other treatment. Every 24 h, all individuals are observed by dissecting microscope; each rotifer is fed, scored as live or dead, assessed for reproductive status, and the number of offspring is counted. These methods allows individual-level measurements of lifespan and reproduction with high replication.
Lifespan is typically measured by daily observation of rotifers isolated into artificial seawater and food in well plates, and death is characterized by the cessation of swimming, and of movement in the cilia, muscles, and the mastax (jaw). These methods permit tracking of lifespan and fecundity in individuals, rather than in populations. Health is measured using sensory, motility, and stress resistance assays [Fig. 4; 16, 18]. 5-fluoro-deoxyuridine (FUDR) may be used to prevent egg hatching, decreasing the daily labor required to separate offspring from experimental animals [18]. However, as in C. elegans, FUDR extends life span by approximately 25% in Brachionus rotifers and suggesting off-target effects that may interfere with assessment of phenotypes and mechanisms [22].
2.3Brachionus aging phenotype
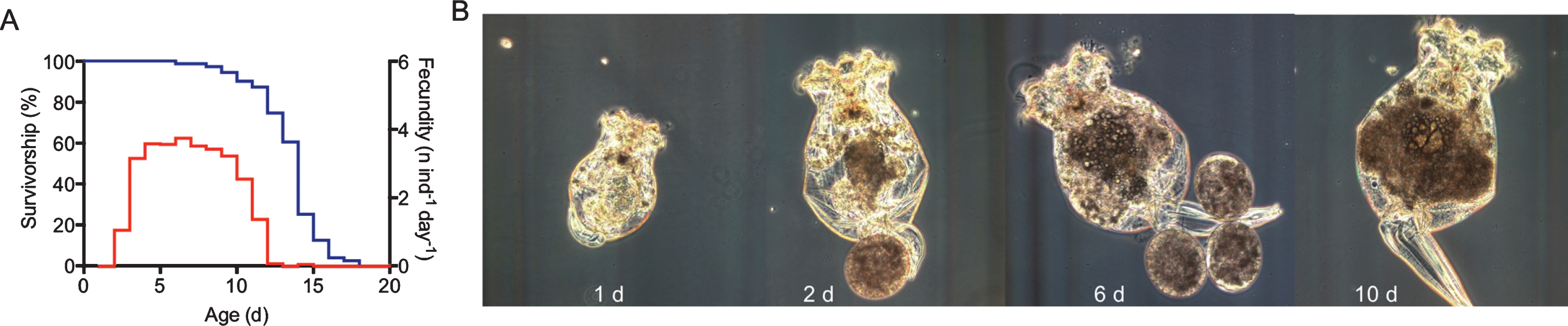
The aging phenotype of Brachionus rotifers recapitulates that seen in humans and other animal models. Rotifers have a pre-reproductive period of 1 – 2 days, a peak in reproduction early in life, followed by a decline and a short post-reproductive period before death (Fig. 5). With age, rotifers increase in size (despite being eutelic, with no somatic cell division post-hatching), swimming speed peaks in midlife and then slows, phototaxis (the propensity to swim toward a light source) declines, and many animals develop an opaque pigmentation [Fig. 4, Fig. 5; 15, 19]. Preliminary evidence suggests that, as in D. melanogaster and C. elegans, integrity of the gut epithelium is lost in late age [23, 24]; food and dyes can escape the gut and invade the tissues in the hours or days before death (personal observation).
Fig. 4
Measures of changes in health with age in asexual, female Brachionus manjavacas (K. Gribble, unpublished data). As rotifers age, (A) swimming speed declines (n = 30), (B) behavior changes from primarily swimming in a straight line or in large circles to predominantly stationary or twitching movements with no forward motion (n = 30), (C) phototaxis (the tendency to swim toward a light source) declines (n = 10/replicate, 5 replicates per age), and (D) survival 24 hours after exposure to heat stress declines by 75% (n = 48 per age).
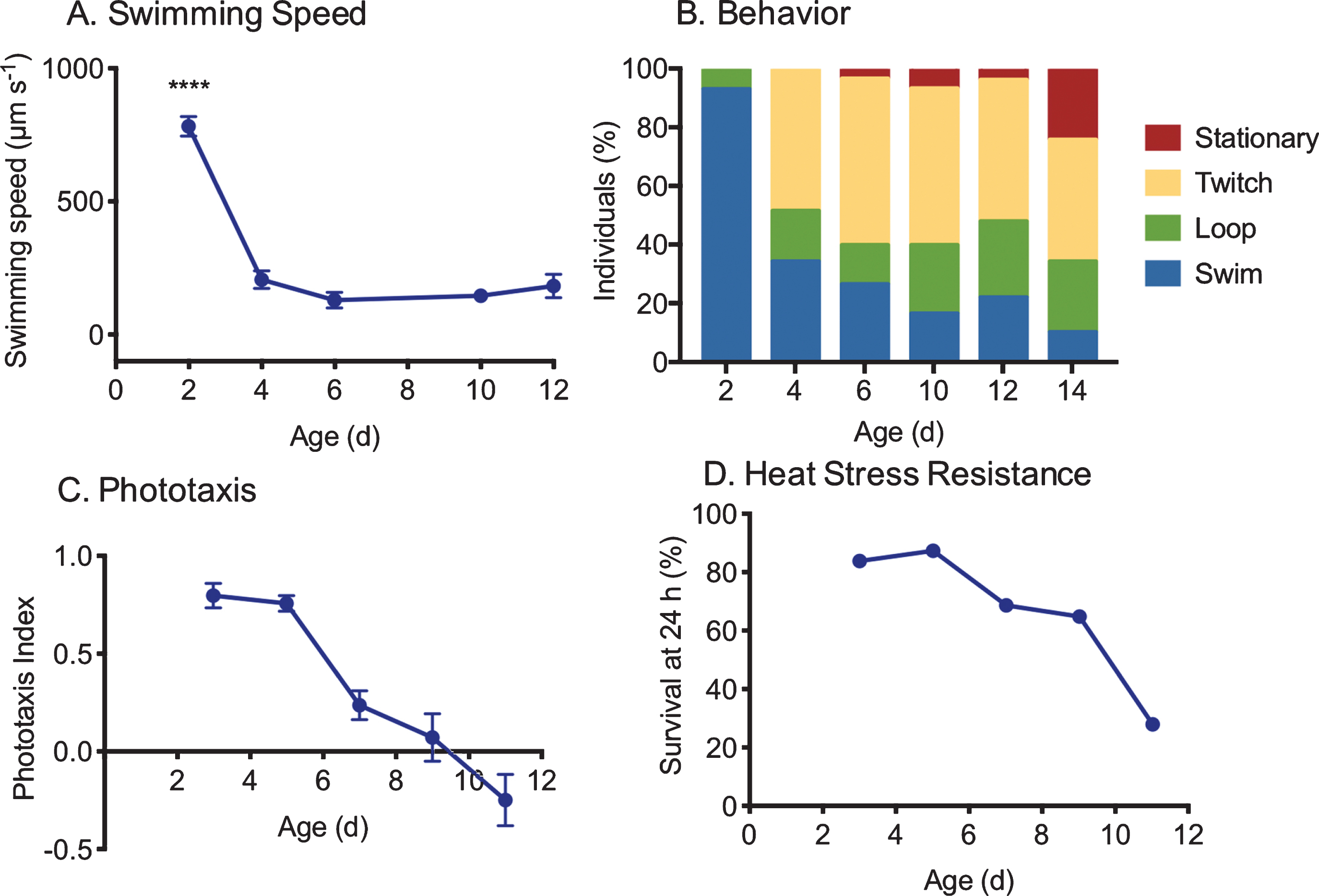
Fig. 5
Aging phenotype of asexual, female Brachionus manjavacas. (A) Typical survivorship (blue) and fecundity (red) profiles for B. manjavacas fed ad libitum. Median life span is approximately 2 weeks for asexual females. Reproduction begins after about 2 days; reproduction peaks at 5 days old. The post-reproductive period is typically 1– 3 days long. (C) Morphology changes over the life span of B. manjavacas. In late age, individuals increase in size, stop producing eggs, and the tail-like foot often drags behind the animal instead of being tucked against the body.
The transcriptomic signature of aging in B. manjavacas is similar to that observed in humans and other animal models [25]. Post-reproductive B. man-javacas females show decreased expression of pathways involved in proteostasis, mitochondrial function, oxidative phosphorylation, and calcium signaling [20, 26, 27]. Multiple metabolic pathways are down regulated in late age, including fatty acid, pentose phosphate, porphyrin and ether lipid metabolisms; glycolysis; and the TCA cycle. Additionally, signaling pathways with roles in nutrient sensing and metabolism, including insulin signaling, TOR, and adipocytokine pathways are down regulated in post reproductive females. Interestingly, expression of several transcription factors and kinases that act as key regulators and integrators of metabolism, including JNK, INSR, IGF, IGFALS, and TOR, increase slightly in late age. Together, these data suggest a dysregulation of multiple metabolic functions in late life. Thus, these pathways and genes provide targets for possible interventions to aging, though considerable work remains to be done to establish causality of mechanisms correlated with aging in rotifers.
3Metabolic manipulation of lifespan and health in rotifers
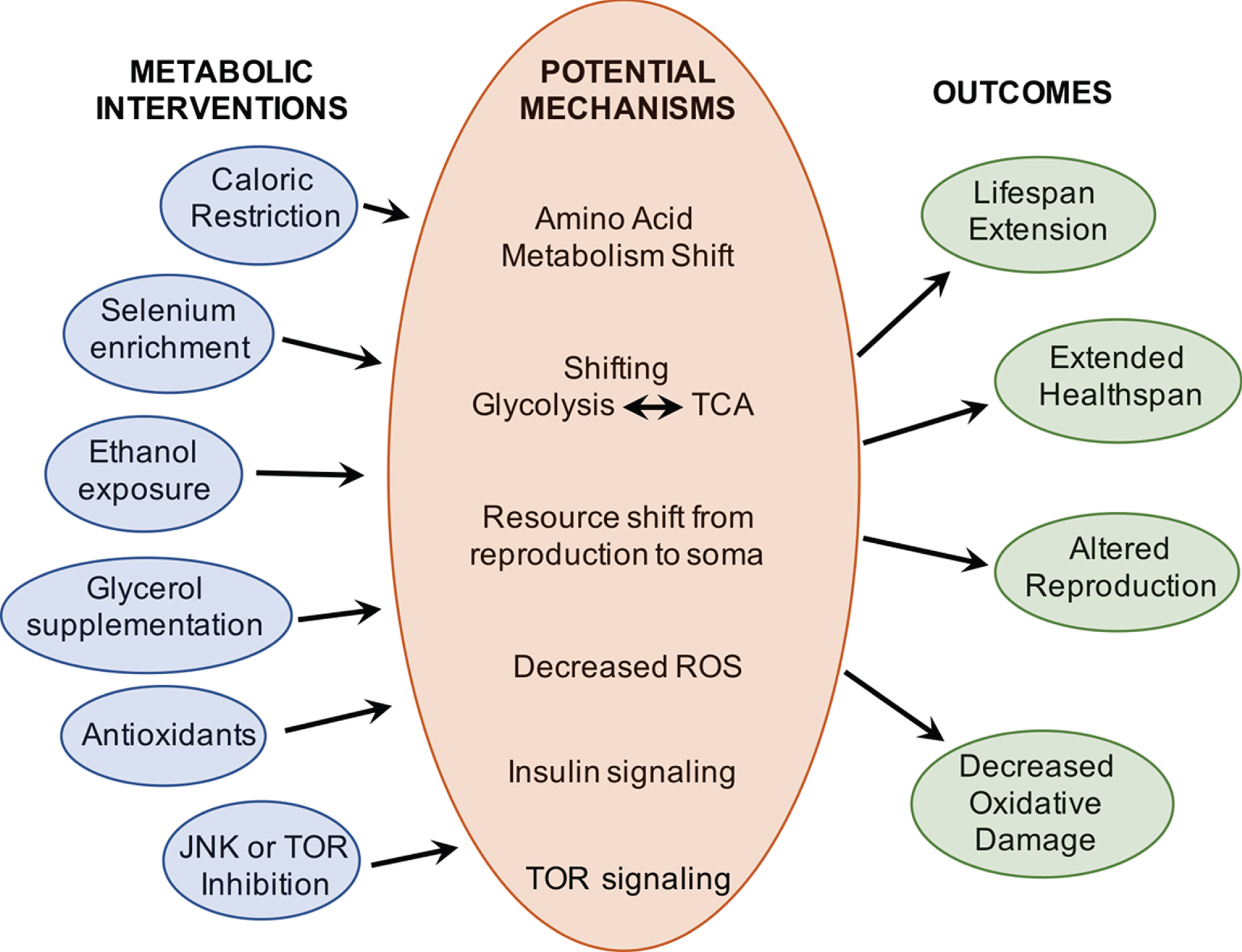
Interventions to aging in Brachionus rotifers have primarily targeted dietary and metabolic treatments. Endpoint measures have included lifespan, reproduction, gene expression changes, and proxies for healthspan (Fig. 6).
Fig. 6
A wide range of metabolic interventions have been shown to extend lifespan and improve health in Brachionus rotifers. Work remains to be done to determine the molecular mechanisms by which these interventions alter aging.
3.1Dietary interventions
3.1.1Caloric restriction
As is the case for most animal models of aging, caloric restriction (CR) has been extensively studied in Brachionus and non-Brachionus rotifers. A reduction in food intake is achieved through any of several methods, including dilution of single-cell algae food to a low concentration (chronic CR), alternating full food conditions with starvation on time scales ranging from hours to days (intermittent fasting), or restricting food for short periods of life (age-specific CR or fasting). Depending on the rotifer strain and the level of CR, these varied regimens extend median lifespan from 10% – 100% [28– 34]. The greatest delays in aging occur under intermittent fasting; the most extreme lifespan extension occurred in B. plicatilis that was fed for only 1 h/day, resulting in a doubling of lifespan [29, 31]. Fasting periods ranging from 24 – 36 h increased lifespan relative to a 12 h fasted control, while fasting 48 – 72 h decreased lifespan in B. calyciflorus [33]. Fecundity typically declines under CR, suggesting a trade-off in resource allocation between reproduction and somatic maintenance under food limiting conditions [35– 37].
Age-specific dietary restriction or starvation in Brachionus shows that early-life food limitation significantly extends lifespan [32]. Late-life CR also increases lifespan, though to a lesser extent. Interestingly, the longer rotifers were fed in early life, and the more offspring they produced before the onset of starvation, the shorter their lifespan. Rotifers fed for only one or two days before starvation nearly doubled lifespan relative to continuously fed controls, but those fed for three days or more had a 50% shorter lifespan than controls [32]. In this case, it is difficult to separate the effects of reproduction from the effects of feeding duration and age at onset of starvation.
These results suggest that CR need not be applied throughout life to achieve benefit. However, the greater benefit of early life intervention – often just as large as intervention throughout life – suggests that early life events disproportionately influence aging trajectories. While a primary focus in geroscience has been to determine which therapies will be effective when implemented late (for obvious reasons, as there are logistical, ethical, and biological issues with treating aging in the very young), the field may benefit from a better understanding of the early life mechanisms that dictate late-life aging phenotypes.
3.1.2Dietary manipulation
Manipulating specific dietary components or micronutrients also extends lifespan in Brachionus. Feeding rotifers different species of phytoplankton (which vary in their nutrient composition) results in large differences in lifespan, reproduction, and population growth rates. In a comparison of three different algae diets in individual and all pairwise combinations, B. plicatilis median lifespan ranged from 2.5 – 10.5 d. These lifespan differences were not correlated (either positively or negatively) with changes in body size or number of offspring, suggesting there is not a direct trade-off between lifespan, reproduction, and/or body size, but rather that restriction or addition of specific dietary components may lead to shifts in life history [38]. Mean lifespan of B. manjavacas ranged from 10.3 – 15 days when fed equal quantities of one of four different species of algae [16]. When fed ad libitum on one of five different algae species, Brachionus koreanus median lifespan ranged from 6 – 10 days and maximum lifespan ranged 6.5 – 11 days, differences of 40% [39]. These different diets led to significant changes in fatty acid composition and in expression of lipid metabolism genes in the rotifers, though the role of these shifts in directly determining lifespan is unclear [39]. As expected, when food is not limiting there was no correlation or trade-off between reproduction and lifespan [39, 40]. Interestingly, an investigation of B. calyciflorus and B. patulus fed two different algae singly or in combination demonstrated that the difference in lifespan and reproduction was as great between the two rotifer species on the same diet as for the same species fed different diets [40], emphasizing the significant variability in lifespan and response to dietary changes among species, even when very closely related.
Changes in growth conditions of food cultures lead to large differences in rotifer lifespan and reproduction. Changing the media renewal rate for algae cultures, thereby altering nutrient levels, light exposure, and growth rate, shifts the algae’s biochemical composition [41, 42]. For two different algae species, increasing media renewal rate from 10% to 40% or 50% led to a doubling of protein content, decreased carbohydrate content by half, and left relative lipid content unchanged [41, 42]. Rotifers fed high media replacement rate algae (with higher protein: carbohydrate) have significantly longer lifespan and higher reproduction rates than those fed low replacement rate algae, even when total food levels are the same [16].
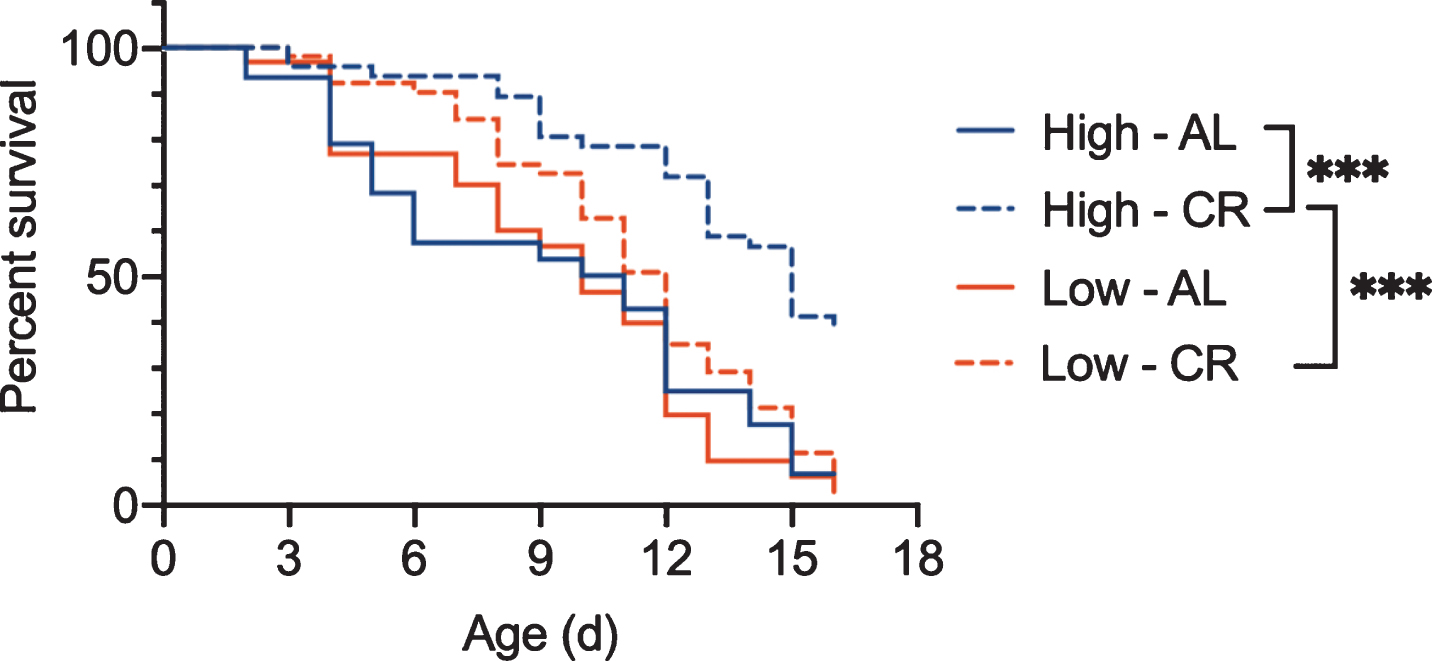
Similarly, using the methods described in Bock et al., 2019 [43], our work demonstrates that rotifers fed Tetraselmis suecica grown under high light intensity have both a longer lifespan under ad libitum fed conditions and a greater response to CR than those fed low-light algae (Fig. 7). Even algae culture temperature affects rotifer aging. Mean and maximum lifespan of B. manjavacas are reduced by 18% and 31% respectively when fed algae grown at 30 – 34 °C instead of 25 – 28 °C [44]. These results suggest that unquantified differences in culture conditions for the bacteria or yeast provided as food for other invertebrate models of aging may lead to variability between experiments, and as shown in rotifers, could change the response to dietary or other interventions to aging.
Fig. 7
Survivorship of asexual female Brachionus manjavacas fed algae that was grown under high light (Hi) or low light (Low) conditions. Rotifers were fed under ad libitum (AL) or CR. Though there was no difference in lifespan for rotifers fed high-light or low-light algae for under ad libitum conditions, the response to CR was significantly different depending on the algae growth conditions. B. manjavacas fed low light algae did not have increased lifespan under CR. n = 48 per treatment. *** p < 0.0001, Mantel-Cox test.
Manipulation of specific nutrients levels in algae used as food also changes rotifer aging. Feeding B. plicatilis with selenium-enriched Chlorella algae led to a 37% increase in median lifespan and a 40% increase in maximum lifespan relative to growth on non-enriched Chlorella [45]. However, other metabolites, proteins, carbohydrates, and fatty acids are likely to have also changed in response to selenium enrichment in these experiments, possibly contributing to the lifespan effects in unknown ways.
Understanding how specific nutrients change health and lifespan can provide insight into mechanisms of aging and provide new targets for thera-pies. In humans, plant-based and low-protein diets are associated with lower all-cause mortality and improved insulin signaling compared with animal-based diets [46– 49]. Human and mouse studies suggest this effect may be caused by lower total protein or by differences in amino acid composition that change mTOR and metabolic signaling [50, 51]. The changes in the total or relative amounts of specific nutrients in different algae diets, and the role of these changes in determining rotifer lifespan, have not been characterized in most previous studies; many of these experiments were focused on ecological and population outcomes, rather than mechanisms of aging. Given the ease of the experiments and ability to collect large amounts of biomass for analysis, Brachionus rotifers provide a tractable system for future investigation of the metabolic and lifespan responses to specific nutrients or nutrient combinations using integrated lifetable, proteomic, metabolomic, and transcriptomic approaches.
4Molecular mechanisms of caloric restriction in brachionus
Because the genomes of Brachionus species have been sequenced relatively recently [52, 53], characterization of the molecular mechanisms of CR-mediated lifespan extension in rotifers is not as extensive as in established cell and animal models. Most studies of molecular mechanisms have been descriptive, correlating changes in gene expression with changes in aging [54– 57]. Still, this work demonstrates the potential conservation of CR-mediated lifespan extension mechanisms between Brachionus and and other models. In the future, gene knockdown, editing, or transgenesis experiments will allow testing of hypothesized mechanisms and discrimination between causation and correlation.
Multiple lines of investigation suggest that lifespan extension via dietary restriction or nutrient manipulation in rotifers is associated with a shift in metabolism between glycolysis and the TCA cycle. Glycerol supplementation led to a 40 – 50% extension of median lifespan; improved resistance to oxidative, heat, osmotic, and starvation stress; and increased swimming speed in late age in Brachionus [58]. It is hypothesized that glycerol shifts metabolism from glycolysis to aerobic respiration by increasing the availability of pyruvate for incorporation into the oxidative phosphorylation pathway and additionally acts as a chemical chaperone to protect cells from detrimental protein aggregation [58].
Lifespan extension upon exposure to ethanol also suggests that a switch from glycolysis to the TCA cycle modulates aging in B. plicatilis [59]. Lifelong exposure to 0.1% ethanol increased median lifespan by 20%, boosted fecundity by 30%, and increased resistance to hydrogen peroxide induced oxidative stress [59]. The suggested mechanism for these effects is the conversion of ethanol to acetyl-coA in the TCA cycle and conversion of excess acetyl-CoA to fatty acids, which activates glucogenesis and decreases reliance on glycolysis. A similar mechanism was found to drive lifespan extension in ethanol-exposed C. elegans [60]. Alternatively, ethanol may change feeding behavior or alter the microbiome, and thus indirectly influence aging via metabolic pathways. Metabolomic analyses of ethanol-exposed animals compared with controls could help to discriminate between hypotheses and clarify the mechanism for ethanol-induced lifespan extension.
In contrast, the observation that intermittent fasting increases survival under hypoxia suggests that CR induces anaerobic respiration in Brachionus [61]. Both treatments–intermittent fasting and population density induced hypoxia–are concomitant with increases in expression of the glycolytic enzymes glyceraldehyde 3-phosphate dehydrogenase (GAPDH), enolase (ENO), and phosphoglycerate mutase (PGM) [61]. This metabolic switch is thought to contribute to population stability when natural populations reach high density (greater than 100 animals/ml at times in some lake systems), simultaneously depleting food and oxygen in the water column. These hypotheses are supported by a recent study showing that both early-life and life-long hypoxia significantly extend lifespan in B. manjavacas [62].
Gene expression studies suggest that CR upregulates genes involved in oxidative stress response and DNA repair in Brachionus, including MnSOD [54, 63], catalase [64], HSP70, and GRP94 [65]. Heat stress also increases MnSOD expression [63], and UV exposure upregulates most heat shock proteins [66], confirming that these proteins are generally involved in ROS-related stress responses. It remains to be tested whether stress response genes like SOD, catalases, and HSPs contribute to lifespan extension under CR in Brachionus, or if their increased expression is a by-product of stress due to limiting food.
A suppression subtractive hybridization study between fully fed and intermittently fasted (feeding 3 hr/day) B. plicatilis identified 38 differentially expressed genes, as well as 54 unknown ESTs [55]. Because subtraction was done in only one direction, only genes upregulated by intermittent fasting were revealed; down regulated genes were not identified. These upregulated genes fell into six gene ontology functional groups: cell structure, transport and division, DNA synthesis, metabolism, transcription, and RNA biosynthesis. Changes in expression of cell structure, DNA synthesis, and cell cycle genes are likely attributable to the declines in reproduction associated with intermittent fasting, given that cell division occurs only in the germline in Brachionus, as is also the case in C. elegans. Some of these genes, including BCCIP, pole, Pols, and CDT1 have additional roles in DNA repair, and thus may indicate enhanced genome stability under intermittent fasting. Upregulation of glycogen phosphorylase (Glase), galactose-4-epimerase (Gale), B-galactosidase (Glb), and succinate dehydrogenase complex subunit D (SDHD) suggest a shift in metabolism, possibly a shift from the TCA cycle to glycolysis under CR. Increases in IPS2 and peptidylglycine-hydroxylating monooxygenase (Phm) show differences in amino acid synthesis and protein post-translational modification under intermittent fasting [55]. Functional testing of these genes in lifespan extension under intermittent fasting remains to be done.
In these experiments, the level of CR imposed was severe, and reproduction greatly depressed, so it is difficult to determine whether genes identified are involved in increasing lifespan or simply reflect a decline in reproduction. Gene knockdown experiments, transcriptomic analyses of mild CR-treated rotifers without a significant decrease in reproduction, or comparisons between responder and non-responder strains of the same species would clarify the molecular mechanisms involved in diet-mediated lifespan extension.
Feeding selenium-enriched Chlorella algae to B. plicatilis increases median lifespan by 37%, maximum lifespan by 50%, and reproduction by 100% [45]. Selenium has been associated with anti-cancer, anti-aging, and antioxidant activity in humans [67]. In ecological systems, low doses have beneficial effects on reproduction in invertebrates and fish [68], but at higher doses selenium acts as a toxin [69].
Comparative genome-wide proteogenomic analysis (combined RNA-Seq and proteomics) revealed that, upon selenium enrichment, differentially ex-pressed proteins associated with lifespan extension were primarily involved in ROS defense, protection from lipid peroxidation, thioredoxin reductase-mediated selenocompound metabolism, glycolysis, and amino acid metabolism. Results suggested a shift from the TCA cycle to glycolysis; this was associated with decreased ROS levels. Indeed, antioxidant enzymes GPX and CAT increased, and lipid peroxidation and protein carbonylation were lower in Se-algae fed rotifers. AMPK expression was lower in these rotifers, suggesting changes in mTOR pathway energy metabolism. Additionally, selenium enrichment increased expression of transaminase, hydroxymethyltransferase, and adenosyltransferase; these enzymes are involved in metabolism of B-alanine, arginine, proline, glycine, serine, threonine, histidine, and tryptophan, and in the degradation of valine, leucine, isoleucine, and lysine. Together, these results suggest a shift to glycolysis and improved amino acid recycling may contribute to lifespan extension. Given that amino acid metabolism and degradation decrease in elderly humans, upregulation of these processes by Se enrichment could provide benefits in aging and should be explored further [45].
5Metabolic pathways regulating lifespan in rotifers
The roles of major metabolic pathways in aging appear to be conserved between rotifers and established invertebrate and vertebrate animal models. In B. manjavacas, signaling pathways with roles in metabolism, nutrient sensing, and oxidative phosphorylation – including insulin, TOR, and adipocytokine signaling – are down-regulated in post-reproductive females [20]. In general, metabolic pathways are downregulated in senescent females, but the expression of key aging-related metabolic genes, including JNK, INSR, IGF, IGFALS, and TOR, increases slightly in senescence. RNAi knockdown of these genes is known to increase lifespan in rotifers and other animals [22, 70, 71]. Other components of the insulin signaling and TOR pathways, particularly calcium-signaling genes, have decreased expression late in life [20]. Together, these data suggest a late-age breakdown in coordination of metabolic function. As in other organisms, studies in rotifers suggest that interventions targeting these pathways can extend healthspan and lifespan [72, 73].
5.1Insulin signaling
The mechanisms of insulin signaling appear to be largely conserved between rotifers, other animal model systems, and humans. Aqueous extracts from B. plicatilis cultures have insulin/insulin-like growth factor activity. When extracts were added to culture media of rat L6 myoblasts, phosphorylation of MAPK/ERK and Akt substrates increased. Insulin/IGF-1 activity was increased in extracts from fed rotifers relative to starved rotifers suggesting that insulin/IFG-like activity was stimulated upon feeding [74].
Pharmaceutical inhibition of PI3-kinase extends lifespan by 30% in B. plicatilis, suggesting involvement of insulin/IGF-1 signaling via PI3-kinase in regulating rotifer lifespan [56]. The inhibitor used, LY294002, also extends lifespan in C. elegans via age-1 (PI3-kinase) suggesting conservation of this mechanism of aging across diverse animals [75]. Although LY294002 does not inhibit the activity of PI4-k, PKC, PKA, or MAPK, it may influence the activity of other PI3-k-like kinases, so it is unclear if lifespan extension is specifically controlled by PI3-kinase inhibition in Brachionus.
5.2TOR signaling
TOR and JNK inhibition both increase lifespan in rotifers, consistent with findings from C. elegans and Drosophila. Rapamycin, which suppresses TOR, increases lifespan and decreases reproduction in a dose dependent manner in B. calyciflorus and B. manjavacas [22, 76]. Rapamycin additionally conferred increased resistance to starvation stress, but not to UV, oxidative, osmotic, or heat stressors, indicating ROS-mediated stressors are not affected by decreases in TOR signaling [22].
Pharmaceutical inhibition of JNK extended me-dian lifespan by 35% and maximum lifespan by 37%, but reduced lifetime reproduction by 77% [22]. Inhibition of JNK activity increased resistance to starvation and osmotic stressors, but not UV, oxidative, or heat stressors, suggesting JNK is not involved in mediating ROS production or damage. As measured by MitoTracker, mitochondrial activity increased upon JNK inhibition, further suggesting that JNK-mediated lifespan extension is not affected via ROS reduction [22]. RNAi knockdown of TOR and JNK produced significant lifespan extension similar to chemical inhibition. Although treatment with a combination of rapamycin and JNK inhibitors extended lifespan more than either treatment alone, RNAi knockdown of TOR and JNK did not produced this additive effect, suggesting that the pharmaceuticals may not target only the JNK and TOR genes [22].
6Comparative biology of aging using the brachionus species complex
The Brachionus species complex is comprised of 15 morphologically similar but genotypically diverse species, providing a unique opportunity to use a comparative biology approach to study aging. The genetic distance of the common barcoding gene, COI, ranges from 0.3 to 13 % within species and from 12 to 23% between species [77, 78]. This spectrum of relatedness is correlated with a gradient of reproductive compatibility among species, with more closely related species having a greater degree of mate recognition and reproductive success. The ability to interbreed between strains and subspecies allows outcrossing and traditional genetic techniques to identify genes involved in aging.
Each Brachionus species comprises a diversity of strains; these strains evolved in varied environments where they were subject to different diet, temperature, salinity, predation, hydroperiodicity, and pathogen selective pressures, leading to natural genetic and phenotypic variability. These different strains have diverse aging phenotypes and responses to aging interventions and thus function as “natural mutants” that can be used in comparative investigations to identify mechanisms driving aging and interventions [28, 79, 80]. Decades of work by dozens of research groups have characterized shifts in lifespan, health, and reproduction in response to altered environmental conditions and have revealed significant variability in aging plasticity among Brachionus strains. Lifespan ranges from a few days to a few weeks depending on the strain, and responses to interventions assumed to be conserved among distantly related taxa actually vary greatly among strains. This diversity can be leveraged experimentally to provide insights into the genetic and molecular controls on aging and therapies to aging.
6.1Caloric restriction
Comparisons between strains or species of Brachionus show that the response to CR depends on genetic background, with different strains exhibiting increased, decreased, or no change in lifespan under CR or intermittent fasting diets [28, 81]. Similar results were found in 41 recombinant inbred mouse strains [82]. Comparative genomics and transcriptomics of strains with diverse responses to the same CR treatments will yield insights into the pathways specifically involved in lifespan extension versus those differentially regulated in a non-adaptive stress response.
Even within a strain there may be variability in the response to CR. In B. manjavacas, different reproductive modes of the same strain have varied responses to CR. Median lifespan of asexual females is 14% longer than that of sexual females under ad libitum conditions. However, under intermittent fasting, sexual females increase median lifespan by 71%, while asexual females increase lifespan by 50% [29]. These sexual and asexual females are genetically identical, suggesting that as-yet undefined epigenetic factors determine lifespan and modulate the response to interventions. Males in Brachionus do not feed, and thus are not used to study sex-specific differences in the response to CR.
6.2Pharmaceuticals, gene knockdown, and low temperature
Responses to a range of other treatments, including low temperature [80, 83, 84] and pharmaceuticals [58], range from positive to negative among Brachionus genotypes. In a survey of 11 closely-related strains, growth at low temperature led to changes in lifespan ranging from a 6% decrease to a 100% increase, depending on the strain [80]. These results suggest active genetic control of lifespan extension under low temperature, rather than a passive thermo-dynamic response that lowers metabolism and thereby slows the accumulation of ROS-related cellular damage, as has been hypothesized. Pharmaceutical treatment and RNAi knockdown suggests the involvement of TRP channels in mediating low-temperature lifespan extension in Brachionus, a mechanism conserved in C. elegans [84, 85]. Within the B. calyciflorus species complex, two mitochondrial DNA lineages that exhibit differences in thermotolerance demonstrate differential expression of lipid and xenobiotic metabolism genes, suggesting divergent adaptive selection and metabolic control of temperature-regulation of aging [86]. Thus, it seems that understanding how selective evolutionary pressures act on different populations may be important for predicting how interventions to aging will affect different genotypes.
The diversity of well-characterized rotifer strains in culture provides a valuable resource for investigating the biology of aging. Comparative genomics, transcriptomics, and metabolomics can be used to contrast the positive and negative responders to interventions, in order to identify genes and pathways specifically involved in lifespan extension versus those involved only in stress responses or shifts in the allocation of resources. Mechanisms identified as conserved across a range of species have a higher likelihood of success for translation to human health.
7Challenges and future directions
The wealth of prior studies, genetic homology with humans, experimental tractability, and growing genetic tools poise rotifers to be a powerful model for the study of aging. Establishing a new, genetically tractable model system for the biology of aging is not without challenges, however. Building a rotifer aging research community will be key to determining the success of Brachionus as a model system. Currently, only a handful of laboratories use Brachionus rotifers as their primary model for studying aging. Most rotifer biologists have focused on questions of ecology, evolution, taxonomy, and utility in aquaculture. With a few exceptions [e.g., 87, 88, 89], the use of rotifers to address biomedical questions is relatively recent. Collaboration between rotifer biologists and those with expertise in aging, genetics, cell, and molecular biology of other systems will be key to expanding the use of this valuable system.
Currently, the genetic and genomic tools for rotifers are limited, though considerable progress has been made in the development of these techniques. Sequenced genomes, transcriptomes, and RNAi allow genetic analysis and manipulation [20, 52, 53, 90, 91]. Transgenics and genome editing methods such as CRISPR/Cas9 are lacking for Brachionus, however. The continued growth of rapid, low-cost sequencing, transcriptome manipulation, genome editing, and imaging technologies enables use of diverse species to investigate both novel and long-standing questions in aging. Establishing these tools for rotifers will allow the model to be used more widely.
Rotifers are valuable for studying questions that are not easily addressed in other short-lived invertebrate models for aging. For example, because they have retained hundreds of human gene homologs that are absent from the genomes of C. elegans and D. melanogaster, Brachionus spp. may be uniquely used to investigate the role of these human-relevant genes in aging and as potential targets for interventions [20]. As members of the Gnathaferia clade, rotifers are evolutionarily distantly related to C. elegans and D. melanogaster, which are both Ecdysozoans [92]. Thus, results from rotifers provide phylogenetic breadth to comparative biology studies, which may help to answer questions about the evolution of aging and evolutionary conservation of the mechanisms of aging. Because rotifers swim in liquid medium, they may be used for easy screening of pharmaceuticals as potential therapies [17]. In addition, rotifers’ complex and easily observed behaviors, in combination with a centralized brain, a simplified peripheral nervous system, highly conserved neurotransmitter function, and tractable labelling and imaging of nervous system morphology and function, will permit these simple invertebrates to be used for studies of the neurobiology of aging and metabolic therapies for age-related neurodegeneration. By using the unique biology of rotifers to answer novel questions that cannot be fully addressed in other organisms, we can capitalize on Brachionus as an additional tool to advance progress on our understanding of the biology of aging.
Conflict of interest
The author has no conflict of interest to report.
Funding
This work was supported by NIH/NIA K01AG049049, NIH/NIA R21AG067034, and NSF-CAREER 1942606.
References
[1] | Schwiening CJ . A brief historical perspective: Hodgkin and Huxley. Journal of Physiology. (2012) ;590: (11):2571–5. |
[2] | Evans T , Rosenthal ET , Youngblom J , Distel D , Hunt T . Cyclin: A protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33. |
[3] | Hershko A , Heller H , Elias S , Ciechanover A . Componenets of ubiquitin-protein ligase system: Resolution, affinity purification, and role in protein breakdown. The Journal of Biological Chemistry. (1983) ;258: (13):8206–14. |
[4] | Hershko A , Leshinsky E , Ganoth D , Heller H . ATP-dependent degradation of ubiquitin-protein conjugates. Proceedings of the National Acadamy of Sciences, USA. (1984) ;81: :1619–23. |
[5] | Vale RD , Reese TS , Sheetz MP . Identification of a Novel Force-Generating Protein, Kinesin, Involved in Microtubule-Based Motility. Cell. (1985) ;42: :39–50. |
[6] | Morise H , Shimomura O , Johnson FH , Winant J . Intermolecular energy transfer in the bioluminescent system of Aequorea. Biochemistry. (1974) ;13: (12):2656–62. |
[7] | Shimomura O , Johnson FH , Saiga Y . Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. Journal of Cellular and Comparative Physiology. (1962) ;59: (3):223–39. |
[8] | Ormö M , Cubitt AB , Kallio K , Gross LA , Tsien RY , Remington SJ . Crystal Structure of the Aequorea victoria Green Fluorescent Protein. Science. (1996) ;273: :1392–5. |
[9] | Saiki RK , Bugawan TL , Horn GT , Mullis KB , Erlich HA . Analysis of enzymatically amplified β-globin and HLA-DQα DNA with allele-specific oligonucleotide probes. Nature. (1986) ;324: (6093):163–6. |
[10] | Russell JJ , Theriot JA , Sood P , Marshall WF , Landweber LF , Fritz-Laylin L , et al. Non-model model organisms. BMC Biol. (2017) ;15: (1):55. |
[11] | Krogh A . The Progress of Physiology. The American Journal of Physiology. (1929) ;90: (2):243–51. |
[12] | Austad SN . Is there a role for new invertebrate models for aging research? Journal of Gerontology. (2009) ;64A: (2):192–4. |
[13] | Gladfelter AS . How nontraditional model systems can save us. Mol Biol Cell. (2015) ;26: (21):3687–9. |
[14] | Fontaneto D , De Smet WH . Rotifera. In: Schmidt-Rhaesa A, editor. Handbook of Zoology, Gastrotricha and Gnathifera. Handbook of Zoology/ Handbuch der Zoologie. A Natural History of the Phyla of the Animal Kingdom / Eine Naturgeschichte der Stämme des Tierreiches. 3. Berlin, Boston: De Gruyter; 2015. p. 217-300. |
[15] | Gribble KE , Snell TW . Rotifers as a model for the biology of aging. In: Ram JL, Conn PM, editors. Conn’s Handbook of Models for Human Aging. 2nd ed. London, U.K.: Academic Press; 2018. p. 483-95. |
[16] | Snell TW . Rotifers as models for the biology of aging. International Review of Hydrobiology. (2014) ;99: (1-2):84–95. |
[17] | Snell TW , Johnston RK , Srinivasan B , Zhou H , Gao M , Skolnick J . Repurposing FDA-approved drugs for anti-aging therapies. Biogerontology. (2016) ;17: (5-6):907–20. |
[18] | Snell TW , Fields AM , Johnston RK . Antioxidants can extend lifespan of Brachionus manjavacas (Rotifera), but only in a few combinations. Biogerontology. (2012) ;13: (3):261–75. |
[19] | Snell TW , Johnston RK , Gribble KE , Mark Welch DB . Rotifers as experimental tools to investigate aging Invertebrate Reproduction and Development. (2015) ;59: (1):5–10. |
[20] | Gribble KE , Mark Welch DB . Genome-wide transcriptomics of aging in the rotifer Brachionus manjavacas, an emerging model system. BMC Genomics. (2017) ;18: (1):217. |
[21] | Kortschak RD , Samuel G , Saint R , Miller DJ . EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Current Biology. (2003) ;13: (24):2190–5. |
[22] | Snell TW , Johnston RK , Rabeneck B , Zipperer C , Teat S . Joint inhibition of TOR and JNK pathways interacts to extend the lifespan of Brachionus manjavacas (Rotifera). Experimental Gerontology. (2014) ;52: :55–69. |
[23] | McGee MD , Weber D , Day N , Vitelli C , Crippen D , Herndon LA , et al. Loss of intestinal nuclei and intestinal integrity in aging C. elegans. Aging Cell. (2011) ;10: (4):699–710. |
[24] | Jasper H . Exploring the physiology and pathology of aging in the intestine of Drosophila melanogaster . Invertebr Reprod Dev. (2015) ;59: (sup1):51–8. |
[25] | López-Otín C , Blasco MA , Partridge L , Serrano M , Kroemer G . The hallmarks of aging. Cell. (2013) ;153: :1194–217. |
[26] | Yang J , Dong S , Jiang Q , Kuang T , Huang W , Yang J . Changes in expression of manganese superoxide dismutase, copper and zinc superoxide dismutase and catalase in Brachionus calyciflorus during the aging process. PLoS One. (2013) ;8: (2):e57186. |
[27] | Yang J , Mu Y , Dong S , Jiang Q , Yang J . Changes in the expression of four heat shock proteins during the aging process in Brachionus calyciflorus (Rotifera). Cell Stress Chaperones. (2014) ;19: :33–52. |
[28] | Gribble KE , Kaido O , Jarvis G , Mark Welch DB . Patterns of intraspecific variability in the response to caloric restriction. Experimental Gerontology. (2014) ;51: :28–37. |
[29] | Gribble KE , Mark Welch DB . Life-span extension by caloric restriction is determined by type and level of food reduction and by reproductive mode in Brachionus manjavacas (Rotifera). Journals of Gerontology Series A: Biological Sciences. (2013) ;68: (4):349–58. |
[30] | Kaneko G , Yoshinaga T , Yanagawa Y , Ozaki Y , Tsukamoto K , Watabe S . Calorie restriction-induced maternal longevity is transmitted to their daughters in a rotifer. Functional Ecology. (2011) ;25: (1):209–16. |
[31] | Yoshinaga T , Hagiwara A , Tsukamoto K . Effect of periodical starvation on the life history of Brachionus plicatilis O. F. Müller (Rotifera): a possible strategy for population stability. Journal of Experimental Marine Biology and Ecology. (2000) ;253: :253–60. |
[32] | Yoshinaga T , Hagiwara A , Tsukamoto K . Life history response and age-specific tolerance to starvation in Bachi-onus plicatilis O. F. Müller (Rotifera). Journal of Experimental Marine Biology and Ecology. (2003) ;287: :261–71. |
[33] | Ozdemir N . The effect of caloric restriction on the life span and reproduction of fresh water rotifer (Brachionus calyciflorus). Journal of Animal and Veterinary Advances. (2009) ;8: (4):669–73. |
[34] | Oo AKS , Kaneko G , Hirayama M , Kinoshita S , Watabe S . Identification of genes differentially expressed by calorie restriction in the rotifer (Brachionus plicatilis). Journal of Comparative Physiology B. (2010) ;180: :105–16. |
[35] | Kirkwood TBL , Shanley DP . Food restriction, evolution and aging. Mechanisms of Aging and Development. (2005) ;126: :1011–6. |
[36] | Kirkwood TBL . Evolution of ageing. Mechanisms of Aging and Development. (2002) ;123: :737–45. |
[37] | Kirkwood TBL . Evolution of aging. Nature. (1977) ;270: :301–4. |
[38] | Korstad J , Olsen Y , Vadstein O . Life history characteristics of Brachionus plicatilis (Rotifera) fed different algae. Hydrobiologia. (1989) ;186/187: :43–50. |
[39] | Lee M-C , Park JC , Yoon D-S , Choi H , Shin K-H , Kim H-J , et al. Lipid metabolism modulation by five different food types in the monogonont marine rotifer Brachionus koreanus . Aquaculture. (2019) ;503: :596–601. |
[40] | Flores-Burgos J , Sarma SSS , Nandini S . Effect of Single Species or Mixed Algal (Chlorella vulgaris and Scenedesmus acutus) Diets on the Life Table Demography of Brachionus calyciflorus and Brachionus patulus (Rotifera: Brachionidae). Acta hydrochimica et hydrobiologica. (2005) ;33: (6):614–21. |
[41] | Ferreira M , Seixas P , Coutinho P , Fabregas J , Otero A . Effect of the nutritional status of semi-continuous microalgal cultures on the productivity and biochemical composition of Brachionus plicatilis . Mar Biotechnol (NY). (2011) ;13: (6):1074–85. |
[42] | Fabregas J , Otero A , Morales E , Cordero B , Patino M . Tetraselmis suecica cultured in different nutrient concentrations varies in nutritional value to Artemia . Aquaculture. (1996) ;143: :197–204. |
[43] | Bock MJ , Jarvis GC , Corey EL , Stone EE , Gribble KE . Maternal age alters offspring lifespan, fitness, and lifespan extension under caloric restriction. Scientific Reports. (2019) ;9: (3138). |
[44] | Renaud SM , Thinh L-V , Lambrinidis G , Parry DL . Effect of temperature on growth, chemical composition and fatty acid composition of tropical Australian microalgae grown in batch cultures. Aquaculture. (2002) ;211: :195–214. |
[45] | Sun X , Cui Y , Wang Q , Tang S , Cao X , Luo H , et al. Proteogenomic Analyses Revealed Favorable Metabolism Pattern Alterations in Rotifer Brachionus plicatilis Fed with Selenium-rich Chlorella . J Agric Food Chem. (2018) ;66: (26):6699–707. |
[46] | Song M , Fung TT , Hu FB , Willett WC , Longo VD , Chan AT , et al. Association of Animal and Plant Protein Intake With All-Cause and Cause-Specific Mortality. JAMA Internal Medicine. (2016) ;176: (10):1453–63. |
[47] | Dinu M , Abbate R , Gensini GF , Casini A , Sofi F . Vegetarian, vegan diets and multiple health outcomes: A systematic review with meta-analysis of observational studies. Critical Reviews in Food Science and Nutrition. (2017) ;57: (17):3640–9. |
[48] | Orlich MJ , Singh PN , Sabaté J , Fan J , Sveen L , Bennett H , et al. Vegetarian Dietary Patterns and the Risk of Colorectal Cancers. JAMA Internal Medicine. (2015) ;175: (5):767–76. |
[49] | Fontana L , Villareal DT , Das SK , Smith SR , Meydani SN , Pittas AG , et al. Effects of 2-year calorie restriction on circulating levels of IGF-1, IGF-binding proteins and cortisol in nonobese men and women: a randomized clinical trial. Aging Cell. (2016) ;15: (1):22–7. |
[50] | Cummings NE , Williams EM , Kasza I , Konon EN , Schaid MD , Schmidt BA , et al. Restoration of metabolic health by decreased consumption of branched-chain amino acids. J Physiol. (2018) ;596: (4):623–45. |
[51] | Green CL , Lamming DW . Regulation of metabolic health by essential dietary amino acids. Mechanisms of Ageing and Development. (2019) ;177: :186–200. |
[52] | Blommaert J , Riss S , Hecox-Lea B , Mark Welch DB , Stelzer CP . Small, but surprisingly repetitive genomes: transposon expansion and not polyploidy has driven a doubling in genome size in a metazoan species complex. BMC Genomics. (2019) ;20: (1):466. |
[53] | Kim HS , Lee BY , Han J , Jeong CB , Hwang DS , Lee MC , et al. The genome of the freshwater monogonont rotifer Brachionus calyciflorus . Molecular Ecology Resources. (2018) ;18: (3):646–55. |
[54] | Kaneko G , Yoshinaga T , Yanagawa Y , Kinoshita S , Tsukamoto K , Watabe S . Molecular Characterization of Mn-superoxide Dismutase and Gene Expression Studies in Dietary Restricted Brachionus plicatilis Rotifers. Hydrobiologia. (2005) ;546: (1):117–23. |
[55] | Oo AKS . Screening of Calorie Restriction-induced Genes in the Rotifer Brachionus plicatilis. Jour Myan Acad Arts & Sc. (2013) ;11: (4):163–81. |
[56] | Yoshinaga T , Kaneko G , Kinoshita S , Furukawa S , Tsukamoto K , Watabe S . Insulin-like Growth Factor Signaling Pathway Involved in Regulating Longevity of Rotifers. Hydrobiologia. (2005) ;546: (1):347–52. |
[57] | Lee MC , Park JC , Yoon DS , Han J , Kang S , Kamizono S , et al. Aging extension and modifications of lipid metabolism in the monogonont rotifer Brachionus koreanus under chronic caloric restriction. Sci Rep. (2018) ;8: (1):1741. |
[58] | Snell TW , Johnston RK . Glycerol extends lifespan of Brachionus manjavacas (Rotifera) and protects against stressors. Exp Gerontol. (2014) ;57: :47–56. |
[59] | Udo T , Guissou PS , Ushio H , Kaneko G . Ethanol extends lifespan of the rotifer Brachionus plicatilis. Hydrobiologia. 2018. |
[60] | Patananan AN , Budenholzer LM , Eskin A , Torres ER , Clarke SG . Ethanol-induced differential gene expression and acetyl-CoA metabolism in a longevity model of the nematode Caenorhabditis elegans . Experimental Gerontology. (2015) ;61: :20–30. |
[61] | Ozaki Y , Kaneko G , Yanagawa Y , Watabe S . Calorie restriction in the rotifer Brachionus plicatilis enhances hypoxia tolerance in association with the increased mRNA levels of glycolytic enzymes. Hydrobiologia. (2010) ;649: :267–77. |
[62] | Snell TW , Johnston RK , Jones BL . Hypoxia extends lifespan of Brachionus manjavacas (Rotifera). Limnetica. (2019) ;38: (1):159–66. |
[63] | Yang J , Dong S , Zhu H , Jiang Q , Yang J . Molecular and expression analysis of manganese superoxide dismutase (Mn-SOD) gene under temperature and starvation stress in rotifer Brachionus calyciflorus . Mol Biol Rep. (2013) ;40: (4):2927–37. |
[64] | Kailasam M , Kaneko G , Oo AKS , Ozaki Y , Thirunavukk-arasu AR , Watabe S . Effects of calorie restriction on the expression of manganese superoxide dismutase and catalase under oxidative stress conditions in the rotifer Brachionus plicatilis . Fisheries Science. (2011) ;77: :403–9. |
[65] | Yoshinaga T , Kinoshita S , Kaneko G , Watabe S , Tsukamoto K . Gene expression pattern during population growth of the rotifer Brachionus plicatilis . Fisheries Science. (2002) ;68: (1):793–6. |
[66] | Kim R-o , Rhee J-S , Won E-J , Lee K-W , Kang C-M , Lee Y-M , et al. Ultraviolet B retards growth, induces oxidative stress, and modulates DNA repair-related gene and heat shock protein gene expression in the monogonont rotifer, Brachionus sp. Aquatic Toxicology. (2011) ;101: :529–39. |
[67] | Rayman MP . The importance of selenium to human health. The Lancet. (2000) ;356: (9225):233–41. |
[68] | Kim H-J , Nakamura K , Hagiwara A . Dietary effect of selenium-fortified Chlorella vulgaris on reproduction of Brachionus plicatilis species complex (Rotifera: Monogononta). International Review of Hydrobiology. (2014) ;99: (1-2):161–5. |
[69] | Hamilton SJ . Review of selenium toxicity in the aquatic food chain. Sci Total Environ. (2004) ;326: (1-3):1–31. |
[70] | Stanfel MN , Shamieh LS , Kaeberlein M , Kennedy BK . The TOR pathway comes of age. Biochim Biophys Acta. (2009) ;1790: (10):1067–74. |
[71] | Guarente L , Kenyon C . Genetic pathways that regulate ageing in model organisms. Nature. (2000) ;408: :255–62. |
[72] | de Magalhães J , Curado J , Church GM . Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. (2009) ;27: (7):875–81. |
[73] | López-Otín C , Galluzzi L , Freije José MP , Madeo F , Kroemer G . Metabolic Control of Longevity. Cell. (2016) ;166: (4):802–21. |
[74] | Ozaki Y , Kaneko G , Hakuno F , Takahashi S-I , Watabe S . Insulin/insulin-like growth factor-like activity in the aqueous extracts of the rotifer Brachionus plicatilis . Fisheries Science. (2012) ;79: (1):47–53. |
[75] | Babar P , Adamson C , Walker GA , Walker DW , Lithgow GJ . Pl3-kinase inhibition induces dauer formation, thermotolerance and longevity in C. elegans. Neurobiology of Aging. (1999) ;20: :513–9. |
[76] | Xu H , Liu L , Su Y , Liang Y , Yang J . Effects of rapamycin on life span and on expression of TOR and S6K in Brachionus calyciflorus (Rotifera). Aquatic Biology. (2017) ;26: :49–56. |
[77] | Mills S , Alcántara-Rodríguez A , Ciros-Pérez J , Gómez A , Hagiwara A , Galindo KH , et al. Fifteen species in one: deciphering the Brachionus plicatilis species complex (Rotifera, Monogononta) through DNA taxonomy. Hydrobiologia. 2016. |
[78] | Gómez A , Serra M , Carvalho GR , Lunt DH . Speciation in ancient cryptic species complexes: Evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera). Evolution. (2002) ;56: :1431–44. |
[79] | Mark Welch DB . The Potential of Comparative Biology to Reveal Mechanisms of Aging in Rotifers. In: Ram JL, Conn PM, editors. Conn’s Handbook ofModels for Human Aging. 2nd ed. London, United Kingdom: Academic Press; 2018. p. 497-505. |
[80] | Gribble KE , Moran BM , Jones S , Corey EL , Mark Welch DB . Congeneric variability in lifespan extension and onset of senescence suggest active regulation of aging in response to low temperature. Experimental Gerontology. (2018) ;114: :99–106. |
[81] | Kirk KL . Dietary restriction and aging: comparative tests of evolutionary hypotheses. The Journal of Gerontology. (2001) ;56A: (3):B123–B9. |
[82] | Liao C-Y , Rikke BA , Johnson TE , Diaz V , Nelson JF . Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. (2010) ;9: :92–5. |
[83] | Seifert LI , de Castro F , Marquart A , Gaedke U , Weithoff G , Vos M . Heated relations: temperature-mediated shifts in consumption across trophic levels. PLoS One. (2014) ;9: (5):e95046. |
[84] | Johnston RK , Snell TW . Moderately lower temperatures greatly extend the lifespan of Brachionus manjavacas (Rotifera): Thermodynamics or gene regulation? Experimental Gerontology. (2016) ;78: :12–22. |
[85] | Xiao R , Zhang B , Dong Y , Gong J , Xu T , Liu J , et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. (2013) ;152: (4):806–17. |
[86] | Paraskevopoulou S , Dennis AB , Weithoff G , Hartmann S , Tiedemann R . Within species expressed genetic variability and gene expression response to different temperatures in the rotifer Brachionus calyciflorus sensu stricto. PLoS One. (2019) ;14: (9):e0223134. |
[87] | Snell TW , King CE . Lifespan and fecundity patterns in rotifers: the cost of reproduction. Evolution. (1977) ;31: (4):882–90. |
[88] | Lansing AI . Increase of cortical calcium with age in the cells of a rotifer, Euchlanis dilatata, a planarian, Phagocata sp., and the toad, Bufo fowleri, as shown by the microincineration technique. Biological Bulletin. (1942) ;82: :392–400. |
[89] | Lansing AI . A transmissible, cumulative and reversible factor in aging. Journal of Gerontology. (1947) ;2: (3):228–39. |
[90] | Snell TW , Shearer TL , Smith HA . Exposure to dsRNA produces RNA interference in Brachionus manjavacas (Rotifera). Marine Biotechnology. (2011) ;13: (2):264–74. |
[91] | Franch-Gras L , Hahn C , Garcia-Roger EM , Carmona MJ , Serra M , Gomez A . Genomic signatures of local adaptation to the degree of environmental predictability in rotifers. Sci Rep. (2018) ;8: (1):16051. |
[92] | Marletaz F , Peijnenburg K , Goto T , Satoh N , Rokhsar DS . A New Spiralian Phylogeny Places the Enigmatic Arrow Worms among Gnathiferans. Curr Biol. (2019) ;29: (2):312–8 e3. |




