Characterization and screening of the potential probiotic lactic acid bacteria and Bifidobacterium strains isolated of different biotopes
Abstract
The use of lactic acid bacteria (LAB) and bifidobacteria species as probiotics may help to reduce antibiotic use for therapeutic, prophylactic and growth promotion in animal husbandry. The choice of the starter cultures has a critical impact on the palatability, processability and nutritional attributes of fermented products. In our research, the aim of this study was to screen and select potent probiotic LAB and Bifidobacterium strains isolated from different niches and to evaluate their characteristic features. A total of fourteen LAB and fifty-four Bifidobacterium were isolated from four fresh cows and camel’s milk, and twenty stool samples of healthy new born infants were identified and characterized by morphology and biochemical tests in order to select most suitable strains according to their technological characteristics including probiotic proprieties, antibiotics resistance and in vitro antagonism against food-poisoning bacteria. When the results of tolerance to both gastric and bile juices are taken together it appears that between fourteen LAB and forty-five, the strain B. longum BHI 07 has significantly the highest ability to survive during gastrointestinal transit (P < 0.05). Therefore, the mixed cultures of Bf. longum strains with LAB strains were more active against pathogenic bacteria than the pure one. These results show that bifidobacteria isolated from infants may be useful for improving probiotic formulae with respect to protection against pathogenic bacteria responsible of infection.
1Introduction
Lactic acid bacteria as well as Micrococcaceae strains, which can convert fermentable carbohydrates into lactic acid [1] used as starter cultures in fermented foods such as cheese, fermented milks, butter and other fermented products [2]. They include species of the genera Carnobacterium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Oenococcus, Pediococcus, Streptococcus, Tetragenococcus, Vagococcus and Weissella [3]. The main criteria used to select microbial starters are desirable technological such as growth, acidification rate [4], organoleptic, textural, sensory and nutritional aspects, also synthesis of antimicrobial compounds [5, 6] and antifungal activity [7]. Other selection criteria of probiotic properties were required such as the adhesion to the intestinal mucosa [8, 9], the non-production of biogenic amines, antibiotic susceptibility [8] and the ability to tolerate the presence of pancreatic enzymes [10]. According to previous study probiotics are defined as live microorganisms which when administrated in adequate numbers, confer a health benefit to the host by maintaining or improving their intestinal microbial balance [11–13]. Metchnikoff [14] was the first who suggested that the longevity of the LAB-usefulness was due to their high intake. Promotion of human health via functional food is in high demand. Moreover, functional foods are currently produced mainly (but not limited) by the aid of probiotic microorganisms [15]. Consumption of probiotic bacteria via food products is an ideal way to re-establish the intestinal microflora balance for maintaining good health.
There has been much recent interest in the use of various strains of LAB as probiotics. LAB isolated from the gastrointestinal tract (GIT) of animals and humans constitute an important source of new functional bacteria, which can develop biological roles during the gastrointestinal transit (probiotics) or during food processing [16]. The natural adaptation of many LAB to the gut environment and the antimicrobial substances produced by them (organic acids and bacteriocins) has provided these organisms with a competitive advantage over other microorganisms to be used as probiotics [17, 18]. These organisms should possess the ability to cross the barriers from mouth to intestine, such as low pH in the stomach and bile in the duodenum. They should also adhere to the intestinal micelle and exhibit antagonistic activity against to harmful bacteria [19]. The health benefits attributed to probiotic bacteria in the literature can be categorized as nutritional and therapeutic [2, 20].
Most probiotic organisms are lactobacilli or bifidobacteria, which are normal inhabitants of the human colon. However, a small number of commensal LAB and bifidobacteria have been reported to be potential probiotics [21]. Borriello et al. [22] recommended that to development of novel ideal probiotics, it should be isolate from the human fecal microflora of healthy volunteers.
In many regions of African and Arabian countries [23–25] and also in Algeria, especially in environments Saharan desert, there are popular butter indigenous dairy products (lben, raib, shmen...) made from different milk sources (cow, sheep, goat or camel) witch are traditionally produced and consumed. Many studies have focused on the isolation of lactic acid bacteria and Bifidobacterium strains from fermented dairy products (raib, smen) [26, 27] and Cheese [28] traditionally fermented olives [26, 27] and fermented sausages [29].
During the last 50 years, microbial infections are a serious public health problem and physical disabilities throughout the world where they are the main cause of the high mortality rates recorded. Infections of the intestinal tract including diarrhea and gastroenteritis are common, affecting people of all ages. The alarming incidence of antibiotic resistance in bacteria becoming a major problem. One of the alternative methods is biological control using antagonistic metabolites produced by microorganisms (probiotics) including lactic acid bacteria [30]. Today, there is a growing need for selection of new strains of LAB and Bifidobacterium that not carry transferable antibiotic resistance genes and with favourable health effects on animals or man. Therefore, the main objective of this work was to perform a screening of LAB and Bifidobacterium strains previously isolated from raw cows-milk, camel-milk and healthy infant’s faeces for potential investigation as starter cultures in the production of fermented feed and foodstuffs such as fermented milk, meat and for development of probiotics.
2Materials and methods
2.1Samples
In the present study, the samples were obtained from four fresh cows and camel’s milk from various farmhouses in west and south Algeria and twenty stool samples of healthy new born infants’ aged 15 months minimum at 10 days postpartum that had no history of antibiotic treatment.
After collection, milk and feces samples were transported to the laboratory in thermoflasks containing ice and they were analyzed on arrival.
2.2Bacterial strains and culture conditions
Lactobacillus, Streptococcus, Lactococcus, Enterococcus and Pediococcus strains were isolated from milk (fourteen strains coded (S1 to S 14). While Bifidobacterium strains from infant feces (fifty-four strains) coded (BHI 1 to BHI 54).
One ml of milk-sample was aseptically added into 9 ml of sterile 0.9% NaCl solution. Homogenized and serially diluted. Lactobacillus were counted in MRS agar adjusted to pH 5.4 with sodium acetate so that the growth of other organisms could be supressed [31]. Plates were incubated under anaerobic conditions (Gas Pak System, Becton Dickinson) at 30°C for 48 to 72 h until growth was observed. Lactococcus were counted in M17 agar (Merck, Darmstadt, Germany) [32] after incubation for 48 h at 30°C. Pediococcus were counted in MRS agar. Plates were incubated under aerobic conditions at 37°C for 48 h. Streptococcus were counted in M17 agar. Plates were incubated under aerobic conditions at 42°C for 48 h. Streptococcus and Enterococcus were counted in M17 and MRS agar respectively, Plates were incubated under aerobic conditions at 44°C for 48 h. Isolation of Bifidobacteria from infant’s feces was carried out following Beeren [33], it was performed on MRS supplemented with 0.05 % cysteine-chloride (Cys-HCl, Sigma) and 2 mg/L of nalidixic acid at pH 6.8. Plates were incubated under anaerobic conditions at 37°C for 48 h to 72 h. Isolation requires strict anaerobic conditions (anaerobic jars with gas-packs). The purification is performed by successive transplanting from Petri plates containing selective medium (MRS medium supplemented with 0.05% cysteine-chloride and 2 mg/L nalidixic acid).
All LAB strains were stored at 4°C for 4 weeks in reconstituted skim milk (10 %) or frozen at – 20°C. Bifidobacteria cultures were maintained in skim milk containing 30 % glycerol, 10 % of yeast extract and 0.2 % cysteine-HCl [34], while Lactobacillus, Streptococcus and Pediococcus cultures were maintained in skim milk containing 30 % glycerol, 10 % of yeast extract. Prior to the experiment, the cultures were activated for 72 h at appropriate temperature and subcultured at least three times.
For antimicrobial activity screening, standard bacterial cultures, viz. Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 6538), Salmonella typhimurium ATCC 14028, Bacillus subtulis (ATCC 6633) and Bacillus cereus (ATCC 10876) was obtained from institute pasteur of Alger, Algeria. These pathogenic microorganisms were selected due to their role as pathogens for humans and their presence in the human gut. All the cultures were maintained as per the recommended practices. These pathogens were grown in nutrient broth and incubated at 37°C for 24 h before use.
2.3Identifications
Strains were identified using an integrated approach that included phenotypic (morphological, biochemical and physiological characterization). The morphology of the colonies and their dimension are studied after that two to three colonies from each medium were re-plated at least five times by streaking on the same medium. Colonies were then examined by Gram staining and those showing morphological characteristics.
Strains were pre-identified according to physiological and biochemical characteristics as described by Mahmoudi et al. [35] & Schillinger and Lücke [36]. Catalase, oxidase and urease test were carried out, also the fermentation of carbohydrate, production of indole, citrate on middle Kempler and Mc Kay are performed. The influence of pH was tested on strains selected at different pH (4, 5, 6.5, 8.5 and 8) and the influence of incubation temperature (25°C, 30°C, 45°C) followed by a growth test in hyper saline environment at 4 %, 6.5 % NaCl. API 50 CH and API 20 A micro-identification systems were used in this step for the identification of species of lactic acid bacteria. The purity of the bacterial isolates was also confirmed by optical microscope observation and Gram staining.
2.4Assay for functional properties of the Strains
Tolerance of isolated strains to acidic conditions, bile salts and hydrogen peroxide was tested. It’s were examined according to Chung et al. [37]; Klingberg et al. [38] with slight modifications. To investigate survival of LAB and Bifidobacterium strains under acidic condition. 1 mL of overnight bacterial culture of each culture was inoculated respectively into 5 mL of nutritive broth adjusted to pH values of 2.0, 3.0 and 6 mol.L-1 with HCl. Samples were incubated for 3 h at 37°C. Then, the number of viable LAB and Bifidobacterium cells was determined by serial 10-fold dilution in PBS, and 100μl aliquots were spread evenly on MRS or M17 agar and the residual viable population was determined after 48–72 h of incubation under aerobic or anaerobic conditions. The survival rate was calculated as the percentage of colonies grown on agar compared to the initial bacterial concentration. Each determination was conducted in triplicate.
The ability of the strains to grow in the presence of bile was determined according to the method of Vinderola and Reinheimer [39]. Each strain was inoculated (2 % v/v) into MRS or M 17 broth with 0.3, 0.5 and 1 % (w/v) of bovine bile (Sigma-Aldrich, Saint Louis, Missouri, USA). Cultures were inoculated at appropriate temperature and after 24 h; optical densities (OD) of the cultures were measured at A 620 nm every two hours during the experiment period and compared within the different concentrations of bile salts for each bacterial culture used and with a control culture (without bile salts). This was expressed as the percentage (%)±standard deviations (sd) of growth at OD 620 in the presence of bile salts compared to the control. All of the experiments were carried out in triplicate. All strains belonging to the LAB or Bifidobacterium selected for the functional properties will be the subject for later studies.
2.5Assay for technological properties of the Strains
2.5.1Acidifying activity
To determine the acidifying activity, a modified method Accolas et al. [40] was used, with using NaOH (Panreac Quimica, Espain) (N/9) in the presence of phenolphthalein indicator (Sigma-Aldrich, Germany) (1 % in alcohol) and measuring the pH by using a pH meter (HANNA Instrument, pH 211, Woonsocket, Romania) of the culture after 6, 24, and 48 h.
2.5.2Kinetics-Growth
In pure culture, the kinetics growth of LAB and Bifidobacterium strains is followed by counting on agar medium appropriate and at different time (0 h, 2 h, 4 h, 6 h...up to 72 h). The colony-forming units (CFU) were estimated. LAB and Bifidobacterium strains selected for technological properties will be the subject of assessment of antagonistic activity.
2.5.3Determination of antibiotic susceptibility testing of the strains
Antibiotic sensitivity or resistance of the selected isolates was determined by the standardized technique of diffusion of Phillips et al. [41] & Zhou et al. [42] toward 11 commercials antibiotics discs: Fusidic acide (10μg), Erythromycin (15μg), chloramphenicol (30μg), Penicillin (10 U), Gentamicin (10μg), Streptomycin (10μg), Ampicillin (10μg), Cefotaxime (30μg). All discs from Bioanalyse (France) except Amoxycillin (25μg) (Himedia, India), Oxacillin (1μg) (bioMérieux, Marcy-l’Etoile, France), Cefalaxin (30μg) Bio-rad (Marnes-la-Coquette, France).
A bacteria suspension was made by picking one or two colonies from agar plates using a sterile loop and making a suspension in sterile broth medium and left for 18 h. Using a sterile swab, the suspension was applied on the surface of Mueller-Hinton agar in sterile Petri dishes, and the strips were placed on the surface using sterile forceps. Two antibiotic reference strains Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were tested with the same technique as the control strains. The results are interpreted according to the recommendations of the French-Society-Committee of sensitivity (2007) and NCCLS (national comity of clinical laboratory standars). The result (average of 3 reading) were expressed as sensitive (S) and resistant (R), No Sensitive (NS), intermediaries (I) by precisely measuring the diameters of the clear zone appearing around the discs with using a digital caliper [43].
2.6Assay for antimicrobial activity
The ability of the supernatants from Bf. bifidum both single culture and the interaction between Bf. bifidum and the five LAB strains selected to inhibit the growth of pathogenic microorganisms was investigated using well-diffusion agar method described by Tagg et al. [44]. The antimicrobial activity was determined by measuring the diameter of the inhibition zones (clear agar) with calipers [45]. Two independent replicates were conducted for each experiment. One or two colonies from agar plates using a sterile loop and making in five mL of sterile broth medium and incubated for a night. Overnight (18 h) cultures of such organism were centrifuged at 3000×g for 20 min. The pH of the aqueous phase was adjusted to 6.5-7±0.1 with 1N NaOH and then filter-sterilized using 0.22μm filters before being subjected to the antibacterial assay. Agar was seeded with 1 % (v/v) of an overnight of pathogenic culture and poured into Petri dishes (90 mm in diameter). Welles of 6 mm were cut out of the agar and loaded with 100μL of each filter-sterilized-supernatant. The plates were left at 5°C for 2 hr, to allow migration and diffusion of the tested supernatant. Control wells filled with sterile nutritive broth and standard antibiotic solution of 100μg/ml Tetracycline (Sigma-aldrich) were performed as negative control and positive control, respectively. Plates were then incubated for 24 h under appropriate conditions for each specific pathogen.
2.7Statistical analyses
Data were statistically analyzed with IBM SPSS Statistics Base 22.0 software for Windows. One-way ANOVA analysis of variance was carried to identify significant differences between means on quantitative parameters, with the significance level at α= 0.05. T test and correlation was used to perform multiple comparisons between means. All tests were repeated at least three times to calculate the means and standard error.
3Results and discussion
3.1Results of identification of the lactic bacteria
The colonies developed on MRS, M17, Columbia and MRSc agar medium were further studied. From these, sixty-eight strains were isolated. They were appeared small, circular in shape, lenticular, more elongated or round with regular contour, whitish and cream color with varying diameter (Fig. 1A).
Fig.1
Macroscopic observation of LAB isolated (A) and purified (B) strains on Agar medium.
The result of microscopic appearance assumes that these selected isolates were Gram-positive cells with cocci or bacillus form which the grouping mode differs from species to another: shells in pairs (Entercoccus sp.) (Fig. 2E); in diplococci and tetrad (Pediococcus sp) (Fig. 2B); in clusters (Lactococcus sp.) (Fig. 2D) or in chains (Streptococcus sp.) (Fig. 2A); long bacilli in pair or chain (Lactobacillus sp.) (Fig. 2C), short-curved bacilli (Bifidobacterium sp.) (Fig. 2F).
Fig.2
LAB and Bifidobacterium strains isolated from cow’s, camel’s-milk and infants feces. (A): Streptococcus thermophilus; (B): Pediococcus cerevisiae; (C): Lactobacillus bulgaricus;(D): Lactococcus lactis; (E): Enterococcus faecalis; (F): Bifidobacterium bifidum (1000 x).
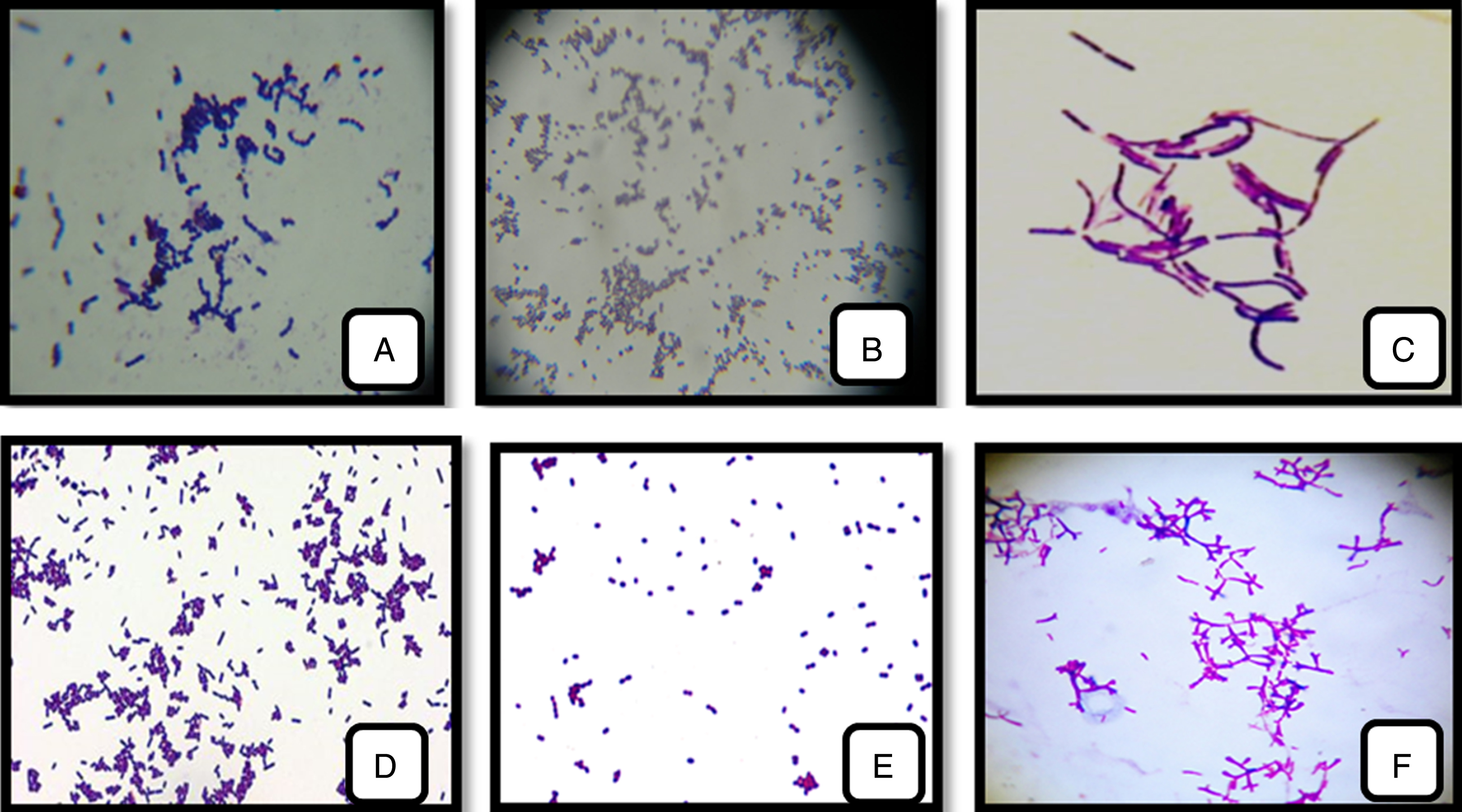
The purified strains were all Gram positive, catalase and oxidase negative, none motile, none sporulating, producing no gas from glucose. These characteristics are typical of LAB and Bifidobacterium strains.
The biochemical results of the strains isolated from milk and with reference to the studies reported by Desmazeand et al. [46] and Guirand [47] showed that the fourteen strains belong to the species quoted in the Table 1. While, the strains isolated from newborn feces were gelatinase negative, with no indole production and resisting up to 2 % of bile salt, citrate permease positive, nitrate reductase negative, no production of gas from glucose, growth in anaerobic with the exception of BHI 10, 23–25, 27–30, 36–41 (Table 2). Based on the previous tests, we assume that these selected isolates were likely to be Bifidobacterium strain. The results showed that all strains ferment some sugars (glucose, fructose, maltose and lactose).
Table 1
Biochemical and physiological characteristics of the isolated strains from fresh milks
| Biochimical Characteristics | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 |
| Catalase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Oxydase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Urease | + | + | + | + | + | + | + | + | – | – | – | – | + | + |
| Indol production | + | – | + | – | + | – | – | – | – | – | – | – | – | – |
| Mobility | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Citrat permease | – | – | – | + | – | – | + | + | – | – | – | – | + | + |
| VP | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RM | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ADH | + | + | – | + | + | + | – | – | – | – | – | + | – | – |
| LDC | – | – | – | – | – | + | + | + | – | – | – | – | + | + |
| ODC | – | – | – | – | – | + | + | + | – | – | – | – | + | + |
| Fermetation of sugars | ||||||||||||||
| Glucose | + | + | + | + | + | + | + | + | + | – | + | + | + | + |
| Lactose | + | + | + | + | – | + | + | + | + | + | + | ± | + | + |
| Saccharose | ± | + | – | + | – | + | – | ± | + | ± | + | + | ||
| Mannitol | – | + | – | + | – | – | + | + | + | + | – | + | + | + |
| Production of H2S | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
| Production of CO2 from glucose | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Resistance to bile salts (02%) | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Physical Characteristics | ||||||||||||||
| Test of thermoresistance 65°C | – | + | – | + | – | – | – | – | – | – | – | – | – | – |
| Growth at pH = 3.2 | ± | – | + | – | ± | ± | ± | ± | – | – | ± | ± | ± | ± |
| Growth at pH = 4 | + | ± | + | ± | – | + | + | + | – | – | – | + | + | + |
| Growth at pH = 6.5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at pH = 9.6 | – | ± | – | ± | – | – | – | – | – | – | – | – | – | – |
| Growth in presence of NaCl 4% | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth in presence of NaCl 6.5% | – | + | – | + | – | + | + | + | + | – | – | + | + | + |
| Growth at T 30°C | + | + | + | + | + | + | + | + | + | – | + | + | + | + |
| Growth at T 37°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at T 45°C | – | + | – | + | – | – | – | – | – | + | + | + | – | – |
| Species | Lc 01 | Ec 01 | Lc 02 | Ec 02 | Pc 01 | Pc 02 | Lb 01 | Lb 02 | Lc 03 | Sc 01 | Sc 02 | Sc 03 | Lb 03 | Lb 04 |
| Lc. Lactis | Ec. faecium | Lc. cremoris | Ec. Faecalis | Pc. cerevisiae | Pc. acidilactici | Lb. bulgaricus | Lb. Lactis | Lc. Lactis | Sc. boris | Sc. Thermophilus | Sc. agalactiae | Lb. bulgaricus | Lb. lactis |
+: positive reaction; –: negative reaction; ±: variable.
Table 2
Biochemical and physiological characteristics of the isolated strains infant’s feces
| Biochimical Characteristics | BHI 01 | BHI 02 | BHI 03 | BHI 04 | BHI 05 | BHI 06 | BHI 07 | BHI 08 | BHI 09 | BHI 10 | BHI 11 | BHI 12 | BHI 13 | BHI 14 | BHI 15 | BHI 16 | BHI 17 | BHI 18 | BHI 19 | BHI 20 |
| Catalase | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Oxydase | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Urease | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | + |
| Indol production | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Gelatinase | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Mobility | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Citrat permease | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| VP | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | + | + |
| RM | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ADH | + | + | + | + | + | + | + | + | + | + | – | – | + | – | + | – | + | + | + | + |
| LDC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + | + | – |
| ODC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | + | + | + | + |
| Production of H2S | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | + | + |
| Production of CO2 from glucose | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | + | + |
| Nitrate reductase | – | – | – | – | – | – | – | – | – | + | – | – | – | – | – | – | – | – | + | + |
| Resistance to bile salts (02%) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Physical Characteristics | ||||||||||||||||||||
| Test of thermoresistance 46.5°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at pH = 4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Growth at pH = 6.5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – |
| Growth at pH = 8.5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Growth in presence of NaCl 4% | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth in presence of NaCl 6.5% | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | ||
| Growth at T 30°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at T 37°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at T 45°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
+: positive reaction; –: negative reaction; ±: variable.
Table 2a
Biochemical and physiological characteristics of the isolated strains infant’s feces (SUITE)
| Biochimical Characteristics | BHI 21 | BHI 22 | BHI 23 | BHI 24 | BHI 25 | BHI 26 | BHI 27 | BHI 28 | BHI 29 | BHI 30 | BHI 31 | BHI 32 | BHI 33 | BHI 34 | BHI 35 | BHI 36 | BHI 37 | BHI 38 | BHI 39 | BHI 40 |
| Catalase | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Oxydase | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Urease | – | – | – | – | – | – | – | – | – | – | – | – | – | – | + | – | – | – | – | + |
| Indol production | – | – | + | + | + | – | – | + | + | + | – | – | – | – | – | + | + | + | – | – |
| Gelatinase | – | – | + | + | + | – | – | + | + | + | – | – | – | – | – | – | – | – | – | – |
| Mobility | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Citrat permease | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| VP | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RM | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ADH | + | + | + | + | + | + | – | – | – | – | + | + | ± | ± | + | + | + | + | + | + |
| LDC | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| ODC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Production of H2S | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Production of CO2 from glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Nitrate reductase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| Resistance to bile salts (02 %) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Physical Characteristics | ||||||||||||||||||||
| Test of thermoresistance 46.5°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at pH = 4 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Growth at pH = 6.5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| Growth at pH = 8.5 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Growth in presence of NaCl 4% | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth in presence of NaCl 6.5% | + | + | + | – | + | – | – | + | + | + | + | – | + | + | + | – | – | – | + | – |
| Growth at T 30°C | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Growth at T 37°C | – | – | + | – | + | – | + | – | – | – | – | – | – | – | – | – | – | – | + | + |
| Growth at T 45°C | – | – | + | – | + | – | + | – | – | – | – | – | – | – | – | – | – | – | + | + |
+: positive reaction; –: negative reaction; ±: variable.
Table 2b
Biochemical and physiological characteristics of the isolated strains infant’s feces (SUITE)
| Biochimical Characteristics | BHI 41 | BHI 42 | BHI 43 | BHI 44 | BHI 45 |
| Catalase | – | – | – | – | – |
| Oxydase | – | – | – | – | – |
| Urease | – | – | – | + | – |
| Indol production | – | – | – | – | – |
| Gelatinase | – | – | – | – | – |
| Mobility | – | – | – | – | – |
| Citrat permease | + | + | + | + | + |
| VP | + | + | + | + | + |
| RM | + | + | + | + | + |
| ADH | + | + | – | + | + |
| LDC | – | – | – | – | – |
| ODC | + | + | + | + | + |
| Production of H2S | + | + | + | + | + |
| Production of CO2 from glucose | + | + | + | + | + |
| Nitrate reductase | + | + | + | + | + |
| Resistance to bile salts (02 %) | + | + | + | + | + |
| Physical Characteristics | |||||
| Test of thermoresistance 46.5°C | + | + | + | + | + |
| Growth at pH = 4 | – | – | – | – | + |
| Growth at pH = 6.5 | + | + | + | + | + |
| Growth at pH = 8.5 | – | – | – | – | – |
| Growth in presence of NaCl 4 % | + | + | + | + | + |
| Growth in presence of NaCl 6.5 % | – | – | – | – | – |
| Growth at T 30°C | + | + | + | + | + |
| Growth at T 37°C | + | + | + | + | + |
| Growth at T 45°C | + | + | + | + | + |
+: positive reaction; –: negative reaction; ±: variable.
The identification of species has been completed by the fermentation of Carbohydrates with used of the API gallery (Tables 3 and 4) and the results were compared to the literature [48–50].
Table 3
Characters fermentation of LAB strains isolated from fresh milks samples
| Biochimical Characteristics | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 |
| Catalase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Oxydase | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Urease | + | + | + | + | + | + | + | + | – | – | – | – | + | + |
| Indol production | + | – | + | – | + | – | – | – | – | – | – | – | – | – |
| Mobility | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Citrat permease | – | – | – | + | – | – | + | + | – | – | – | – | + | + |
| VP | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| RM | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| ADH | + | + | – | + | + | + | – | – | – | – | – | + | – | – |
| LDC | – | – | – | – | – | + | + | + | – | – | – | – | + | + |
| ODC | – | – | – | – | – | + | + | + | – | – | – | – | + | + |
| Fermetation of Glucose | + | + | + | + | + | + | + | – | + | + | ||||
| Fermetation of Lactose | + | + | + | + | – | + | + | + | + | + | + | ± | + | + |
| Fermetation of Saccharose | ± | + | – | + | – | + | + | + | – | ± | + | ± | + | + |
| Fermetation of Mannitol | – | + | – | + | – | – | + | + | – | + | ||||
| Production of H2S | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
| Production of CO2 from glucose | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Resistance to bile salts (02 %) | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Physical Characteristics | ||||||||||||||
| Test of thermoresistance 65°C | – | + | – | + | – | – | – | – | – | – | – | – | – | – |
| Growth at pH = 3.2 | ± | – | + | – | ± | ± | ± | ± | – | – | ± | ± | ± | ± |
| Growth at pH = 4 | + | ± | + | ± | – | + | + | + | – | – | – | + | + | + |
| Growth at pH = 6.5 | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at pH = 9.6 | – | ± | – | ± | – | – | – | – | – | – | – | – | – | – |
| Growth in presence of NaCl 4 % | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth in presence of NaCl 6.5 % | – | + | – | + | – | + | + | + | + | – | – | + | + | + |
| Growth at T 30°C | + | + | + | + | + | + | + | + | + | – | + | + | + | + |
| Growth at T 37°C | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Growth at T 45°C | – | + | – | + | – | – | – | – | – | + | + | + | – | – |
| Species | Lc 01 | Ec 01 | Lc 02 | Ec 02 | Pc 01 | Pc 02 | Lb 01 | Lb 02 | Lc 03 | Sc 01 | Sc 02 | Sc 03 | Lb 03 | Lb 04 |
| Lc . Lactis | Ec . Faecium | Lc . cremoris | Ec . Faecalis | Pc . cerevisiae | Pc . acidilactici | Lb . bulgaricus | Lb . Lactis | Lc . Lactis | Sc . boris | Sc . Thermophilus | Sc . agalactiae | Lb . bulgaricus | Lb . lactis |
Table 4
Characters fermentation of Bifidobacterium strains isolated from infants feces
| Fermetation of sugars | BHI 01 | BHI 02 | BHI 03 | BHI 04 | BHI 05 | BHI 06 | BHI 07 | BHI 08 | BHI 09 | BHI 11 | BHI 12 | BHI 13 | BHI 14 | BHI 15 | BHI 16 | BHI 17 | BHI 18 |
| L-Arabinose | – | + | – | + | – | – | + | – | – | – | + | – | – | + | - | + | – |
| D-Ribose | + | + | + | + | + | + | – | – | + | + | + | – | + | + | + | + | + |
| Xylose | – | ± | ± | + | ± | ± | – | – | ± | – | ± | – | ± | + | ± | ± | ± |
| D-Galactose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Fructose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Mannose | + | ± | ± | ± | ± | ± | – | – | ± | + | ± | – | ± | ± | ± | ± | ± |
| L-Rhamnose | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| D-Mannitol | ± | – | – | ± | – | – | – | – | – | ± | – | – | – | ± | – | – | – |
| D-Sorbitol | ± | – | – | ± | – | – | – | – | – | ± | – | – | – | ± | – | – | – |
| Salicine | + | – | – | + | – | – | – | ± | – | + | – | – | – | + | – | – | – |
| Cellobiose | ± | – | – | + | – | – | – | ± | – | ± | – | – | – | + | – | – | – |
| Maltose | + | + | + | + | + | + | – | + | + | + | + | – | + | + | + | + | + |
| D-Lactose | + | + | + | + | + | + | + | ± | + | + | + | + | + | + | + | + | + |
| D-Melibiose | + | + | + | + | + | + | ± | + | + | + | + | ± | + | + | + | + | + |
| D-Saccharose | + | + | + | + | + | + | ± | + | + | + | + | ± | + | + | + | + | + |
| D-Trehalose | ± | – | – | ± | – | – | – | ± | – | ± | – | – | – | ± | – | – | – |
| Inuline | ± | – | ± | ± | – | ± | – | ± | ± | ± | – | – | ± | ± | ± | – | ± |
| D-Mélézitose | ± | + | – | + | + | – | – | ± | – | ± | + | – | – | + | – | + | – |
| D-Raffinose | + | + | + | + | + | + | – | + | + | + | + | – | + | + | + | + | + |
| Amidon | – | – | – | + | – | – | – | + | – | – | – | – | – | + | – | – | – |
| Gluconate | – | – | – | + | – | – | – | – | – | – | – | – | – | + | – | – | – |
| Species | breve | longum | infantis | Adolescentis | longum | infantis | bifidum | thermophilum | infantis | breve | longum | bifidum | infantis | Adolescentis | Infantis | longum | infantis |
Table 4a
| Fermetation of sugars | BHI 19 | BHI 20 | BHI 21 | BHI 22 | BHI 26 | BHI 31 | BHI 32 | BHI 33 | BHI 34 | BHI 35 | BHI 42 | BHI 43 | BHI 44 | BHI 45 |
| L-Arabinose | – | – | + | – | + | – | – | + | – | + | – | – | – | + |
| D-Ribose | + | – | + | + | + | + | – | – | + | + | – | + | + | + |
| Xylose | – | – | ± | ± | + | ± | – | + | – | ± | – | – | ± | + |
| D-Galactose | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Glucose | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Fructose | + | + | + | + | + | + | + | ± | + | + | + | + | + | + |
| D-Mannose | + | – | ± | ± | ± | ± | – | ± | + | ± | – | + | ± | ± |
| L-Rhamnose | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| D-Mannitol | ± | – | – | – | ± | – | – | – | ± | – | – | ± | – | ± |
| D-Sorbitol | ± | – | – | – | ± | – | – | – | ± | – | – | ± | – | ± |
| Salicine | + | – | – | – | + | – | – | – | + | – | – | + | – | + |
| Cellobiose | ± | – | – | – | + | – | – | – | ± | – | – | ± | – | + |
| Maltose | + | – | + | + | + | + | – | + | + | + | – | + | + | + |
| D-Lactose | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| D-Melibiose | + | ± | + | + | + | + | ± | + | + | + | ± | + | + | + |
| D-Saccharose | + | ± | + | + | + | + | ± | + | + | + | ± | + | + | + |
| D-Trehalose | ± | – | – | – | ± | – | – | – | ± | – | – | ± | – | ± |
| Inuline | ± | – | – | ± | ± | ± | – | – | ± | – | – | ± | ± | ± |
| D-Mélézitose | ± | – | + | – | + | – | – | – | ± | + | – | ± | – | + |
| D-Raffinose | + | – | + | + | + | + | – | + | + | + | – | + | + | + |
| Amidon | – | – | – | – | + | – | – | – | – | – | – | – | – | + |
| Gluconate | – | – | – | – | + | – | – | – | – | – | – | – | – | + |
| Species | breve | bifidum | longum | infantis | Adolescentis | infantis | bifidum | suis | breve | longum | bifidum* | breve | infantis | Adolescentis |
Therefore, all the various strains isolated and identified as LAB and Bifidobacterium have been screened for their acid and bile resistance.
3.2Tolerance under acidic and bile conditions
According to the suggestion of FAO/WHO [51], it is mandatory to perform a preliminary in vitro assessment before assessing the probiotic properties of bacterial strains, such as the capacity for ensure survival during passage through the gastro-intestinal tract [52–54] and have their health promoting effects as metabolically viable active cells when they arrive in the colon [55].
3.2.1Survival rate at low pH
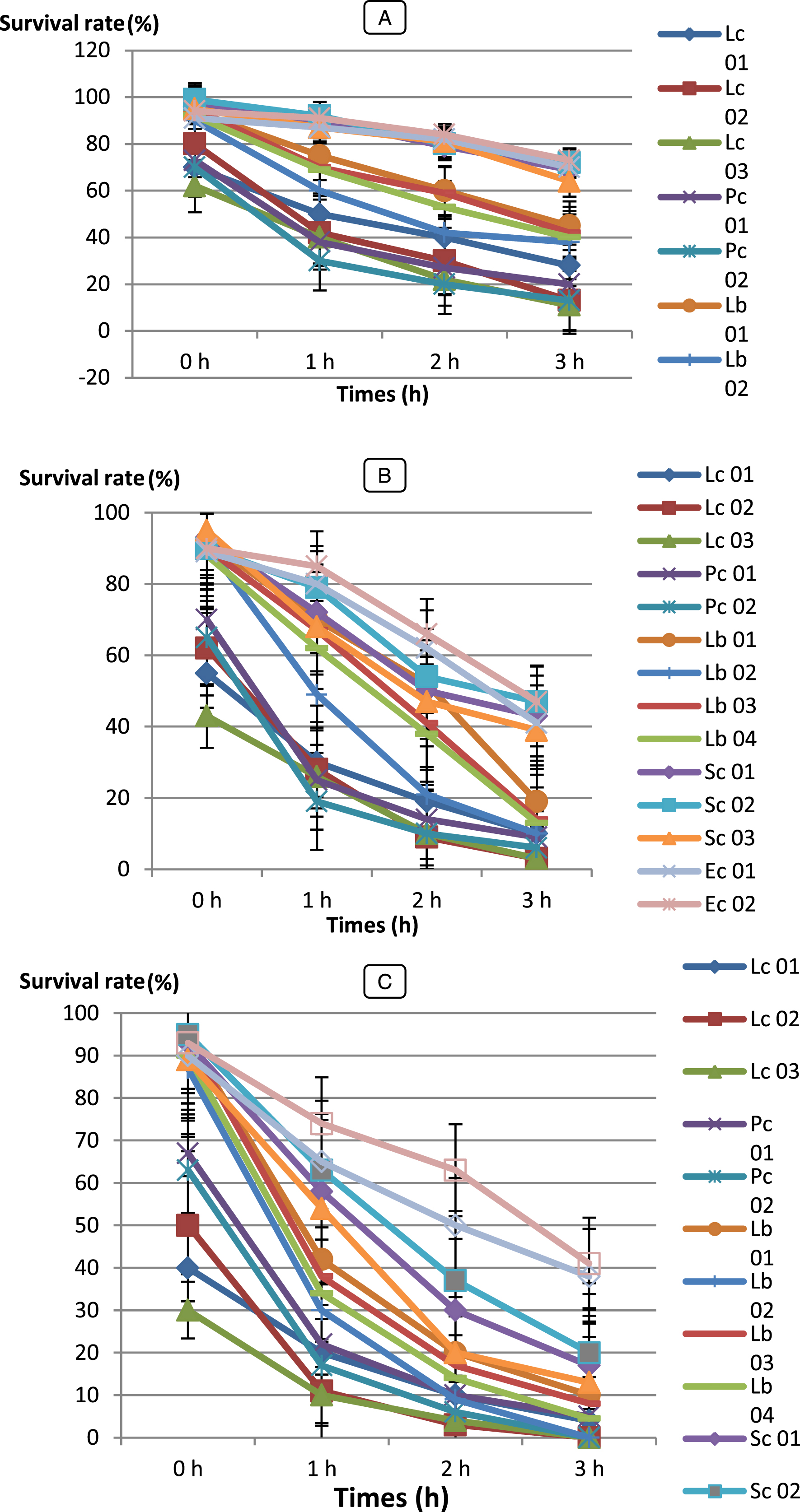
In this work, the abilities of fourteen (14) LAB strains and forty-five (45) Bifidobacterium strains to endure low pH (2.0, 3.0 and 5.0) were examined for acid tolerance, only (11) LAB and 07 strains were selected as resistant to low pH.
Survival rates of the isolates varied during incubation (Figs. 3 and 4). All LAB and Bifidobacterium strains survived at pH 3.0 and 5.0 but, BHI 21, 43, 42, 15 and 22 were the most acid sensitive strains after 4 h of incubation of all strains tested. However, at pH 2.0, Ec 01, BHI 34, 43, 32, 42, 22, 44, 16 and 9 strains were losing viability in less than 3 h of incubation, while Pc 01 and Pc 02 strains were losing viability in less than 2 h of incubation whereas, Sc 01, Sc 02, Sc 03 strains than 1 h. Kim et al. [56] reported that Lactococcus lactis subsp. cremoris and L.lactis subsp. lactis isolated from milk lost their viability at pH 2.0
Fig.3
Survival rate of Bf. longum (BHI 02, BHI 07, BHI 12, BHI 17, BHI 21, BHI 35), Bf. breve (BHI 01, BHI 11, BHI 19, BHI 34, BHI 43), Bf. bifidum (BHI 05, BHI 13, BHI 20, BHI 32, BHI 42), Bf. adolescentis (BHI 04, BHI 15, BHI 26, BHI 45); Bf. infantis (BHI 03, BHI 06, BHI 09, BHI 14, BHI 16, BHI 18, BHI 22, BHI 31, BHI 44); Bf. suis (BHI 33); Bf. thermophilum (BHI 08), at pH 2 (A); pH 3 (B); pH 5 (C) after 4 h at 37°C. Vertical bars represent standard deviations of the mean.
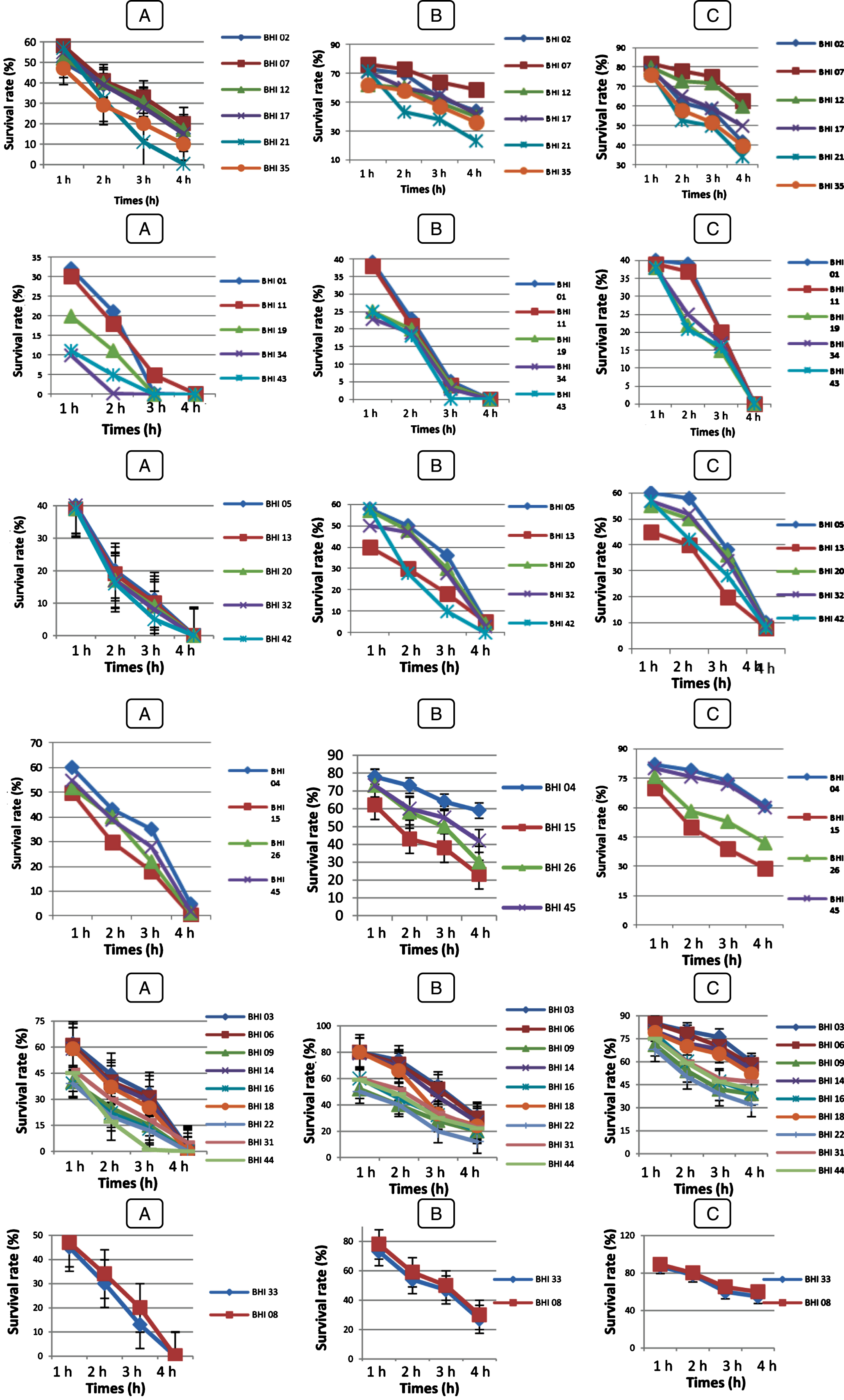
Fig.4
Survival rate of LAB strains under acidic condition at pH 2 (A); pH 3 (B); pH 5 (C) after 4 h at 37°C. Vertical bars represent standard deviations of the mean. Survival rate at bile salt
Bf. bifidum (BHI 07) showed significantly higher survival rate (SR = 63 %) after 4 h of incubation (P < 0.05) than any other isolates and followed by Bf. adolescentis (BHI 04), Bf. infantis (BHI 03), Bf. thermophilum (BHI 08), Bf. suis (BHI 33). However, LAB strains were showed higher survival rate were Lc. cremoris (Lc 03) with 73 % and followed by Pc 01, Lb 01, Sc 02 and Ec 02. When we compare our results with these reported by Xanthopoulos [57] Lactobacillus spp. isolates (L. acidophilus, L. gasseri, L. rhamnosus and L. reuteri) from infant feces, their survival ability to tolerance low pH (pH 3.0) ranged between 0.01 – 68.3 %. Our strains have shown a very good performance.
All LAB strains with the exception of Pediococcus and Streptococcus strains, Bf. bifidum BHI 07, Bf. breve BHI 01, Bf. longum BHI 05, Bf. adolescentis BHI 04, Bf. infantis BHI 03 were the most acid-tolerant strains, retaining around 20 – 50 % viability for up to 2 h at pH 2.0. Similar studies have been done by Al-Saleh [58] reported that Streptococcus thermophilus DSM 20617 and Bifidobacterium infantis DSM 20088 were the most acid sensitive of all bacterial strains tested. These strains completely lost their viability after 1.5 h at pH 2, but at pH 3, the strains improved the acid tolerance with the exception of Streptococcus thermophilus.
3.2.2Survival rate to tolerate bile salts
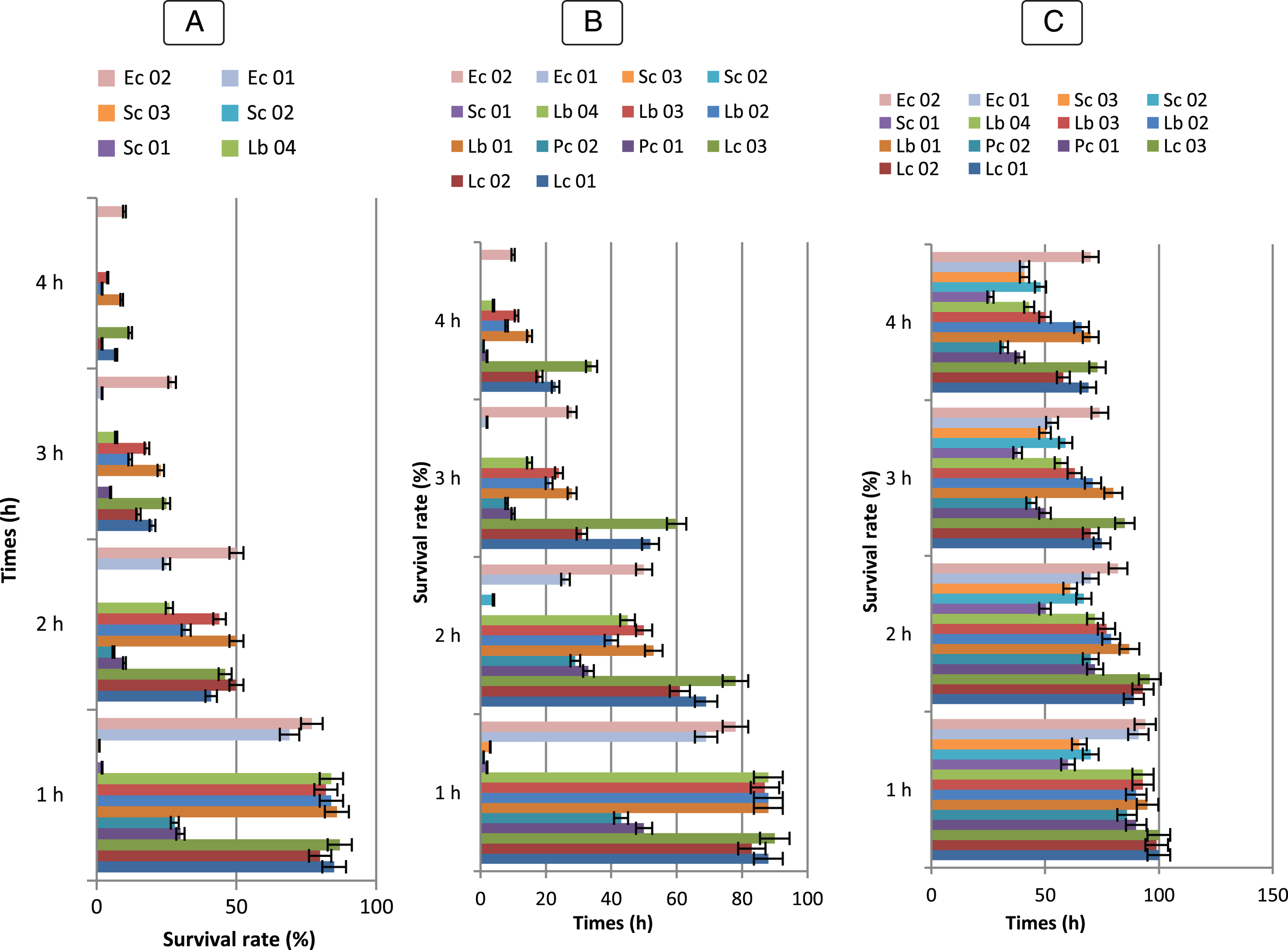
Another critical factor influencing the viability of microorganisms in the stomach is the presence of bile salt. The abilities of bile salt resistance of all LAB and Bifidobacterium strains tested for their ability to survive in gastric juice were examined. The results showed that all isolates grew at concentrations of at least 0.3 % (w/v) bile extract and the maximum value of bile resistance varied from 9 to 32 (Fig. 5 and 6). In comparison to the control culture, there was no significant difference (P > 0.05). BHI 34, 43, 20, 42 and 15 Bifidobacterium strains could not grow in 1 % after 3 h of incubation. B. bifidum strain (BHI 07) showed the maximum resistance at 1.0 % concentrations while the B. longum (BHI 02, 12, 17, 35) and B. infantis (BHI 03, 06, 14, 18) strains indicates relatively strong bile salt tolerance.
Fig.5
Survival rate of Bf. longum (BHI 02, BHI 07, BHI 12, BHI 17, BHI 21, BHI 35), Bf. breve (BHI 01, BHI 11, BHI 19, BHI 34, BHI 43), Bf. bifidum (BHI 05, BHI 13, BHI 20, BHI 32, BHI 42), Bf. adolescentis (BHI 04, BHI 15, BHI 26, BHI 45); Bf. infantis (BHI 03, BHI 06, BHI 09, BHI 14, BHI 16, BHI 18, BHI 22, BHI 31, BHI 44); Bf. suis (BHI 33); Bf. thermophilum (BHI 33), at 0,3% (A), 0,5% (B), 01% (C) of bile salts (n = 3). Vertical bars represent standard deviations of the mean.
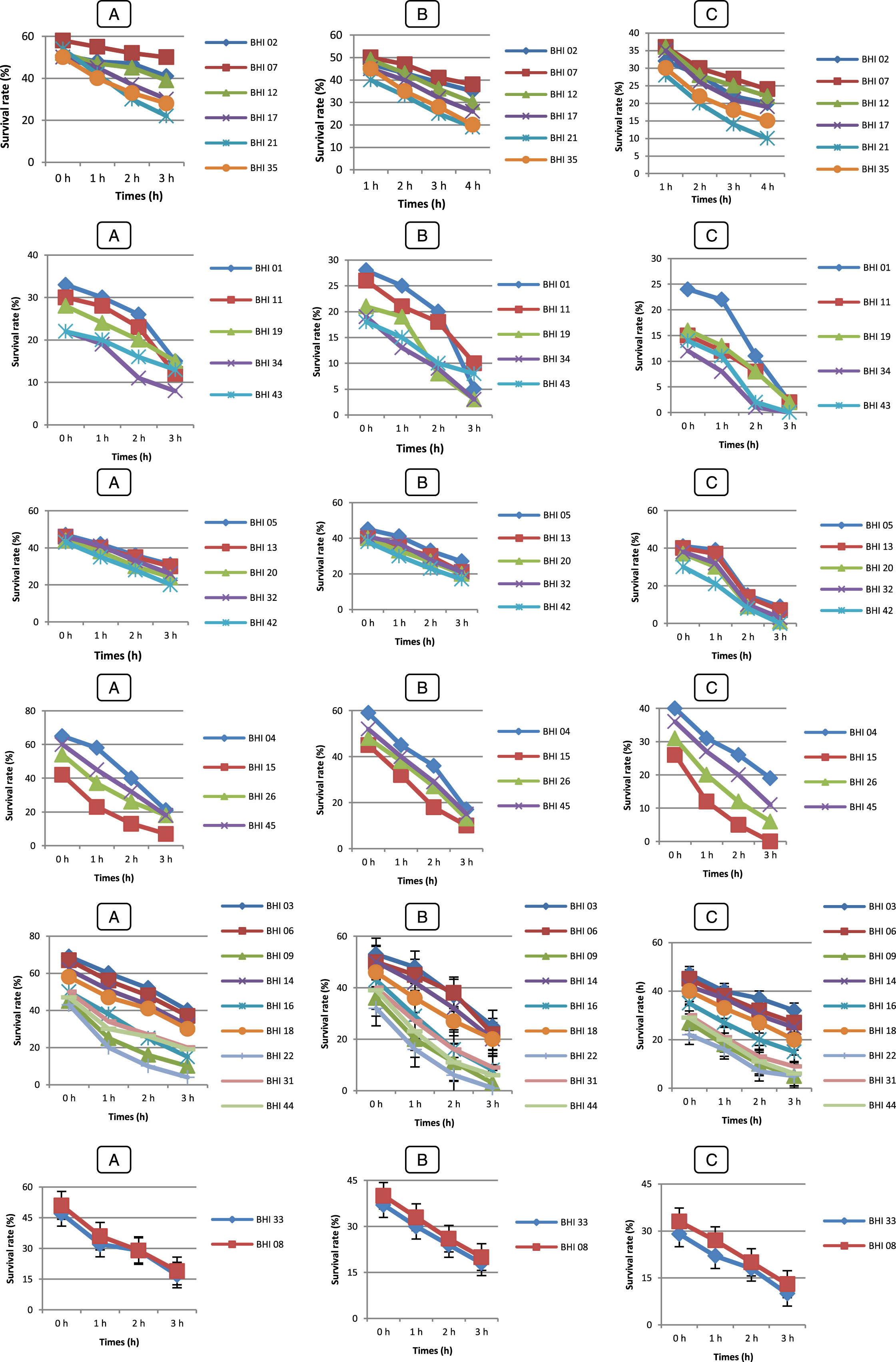
Fig.6
Survival rate tolerance at 0,3 % (A), 0,5 % (B), 01 % (C) of bile Salts of LAB strains (♦) Lc. lactis (Lc 01), (■) Lc 02, (▴) Lc 03, (◊), (×) Pc 01, (*) Pc 02, (•) Lb 01, () Lb 02, (+) Lb 03, (–) Lb 04, () Sc 01, () Sc 02, (◊) Ec 01, (□) Ec 02. Error bars show the SD of the mean of triplicate analyses.
In order to survive and establish within the human GIT, some of the desirable properties of probiotics include their ability to resist the acidity (pH 2.5 – pH 3.5) of the stomach [59] and the exposure to bile in the upper part of the intestine. Gilliland et al. [60] and Goldin et al. [61] reported that 0.3 % bile as a suitable concentration for selection of probiotics. In the present study, the exposure to pH 2 showed to be a very discriminating factor with three strains Lc 03, Lb 01 and Ec 02 out 14 LAB strains, also five B. longum (BHI 07, 12, 17, 02 and 35) out 45 strains surviving after 4 h of exposure. These strains also performed well in the acid and bile assay, suggesting their survival through the stomach and their ability to compete in the human intestine.
These results are in agreement with those of Pyoung Il Kim et al. [62] reported that large variation in the survival of the isolates in gastric juice and growth in the medium containing 0.3 % (w/v) bile and strain dependent [63]. Xanthopoulos et al. [57] reported that Lactobacillus spp. isolates (L. acidophilus, L. gasseri, L. rhamnosus and L. reuteri) from infant feces were tested for their ability to tolerance low pH (pH 3.0) and bile salt. Survival of the test strains ranged between 0.01 – 68.3 % at pH 3.0 and 10.3 – 57.4 % at 0.15 % bile salts. Another research has shown that all cells of Lactococcus lactis subsp. cremoris and L. lactis subsp. lactis lost their viability at pH 2.0 and 0.2 % bile salts [64]. When we compare our results with these results, our strains have shown a very good performance at low pH and bile salts.
Three LAB strains and 38 Bifidobactrium strains were excluded based on these criteria in the current study and only 11 LAB strains and 7 Bifidobactrium strains were selected for further examinations. When the results of tolerance to both gastric and bile juices are taken together it appears that the strain B. longum BHI 07 has significantly the highest ability to survive during gastrointestinal transit (P < 0.05). Therefore, it can be potentially used as probiotic strains.
3.3Technological properties
The ability to acidify the milk was investigated for the 17 strains selected as described above. Among 11 LAB and 7 Bifidobacterium strains examined, 5 LAB and one Bifidobacterium strains were fast acidifying isolates and produced lactic acid.
The measurement of initial and final pH shows a decrease in pH in the medium containing the lactic strains isolated. This decrease is shown in Fig. 7.
Fig.7
Acid production of the medium measured by recording the pH by LAB strains (A) and Bifidobacterium strains (B) selected for their capacity to survival during passage through the gastro-intestinal tract. Data points are shown with 02-h intervals. Determinations were carried out twice with a maximum standard deviation of±0.05. The LAB and Bifidobacterium strains were added at a level of 106 CFU/g. Error bars show the SD of the mean of triplicate analyses. (A) LAB strains:  Lc. cremoris (Lc 02),
Lc. cremoris (Lc 02),  Lc. lactis (Lc 03),
Lc. lactis (Lc 03),  Pc. cerevisiae (Pc 01),
Pc. cerevisiae (Pc 01),  Pc. acidilactici (Pc 02),
Pc. acidilactici (Pc 02),  Sc. boris (Sc 01),
Sc. boris (Sc 01),  Sc. thermophilus (Sc 02),
Sc. thermophilus (Sc 02),  Sc. agalactiae (Sc 03),
Sc. agalactiae (Sc 03),  Ec. Faecium (Ec 01),
Ec. Faecium (Ec 01),  Ec. Faecalis (Ec 02),
Ec. Faecalis (Ec 02),  Lb. bulgaricus (Lb 01),
Lb. bulgaricus (Lb 01),  Lb. lactis (Lb 02).
Lb. lactis (Lb 02).  Bf. breve (BHI 01),
Bf. breve (BHI 01),  Bf. infantis (BHI 03),
Bf. infantis (BHI 03),  Bf. Adolescentis (BHI 04),
Bf. Adolescentis (BHI 04),  Bf. bifidum (BHI 05),
Bf. bifidum (BHI 05),  Bf. Longum (BHI 07),
Bf. Longum (BHI 07),  Bf. thermophilum (BHI 08),
Bf. thermophilum (BHI 08),  Bf. suis (BHI 33).
Bf. suis (BHI 33).
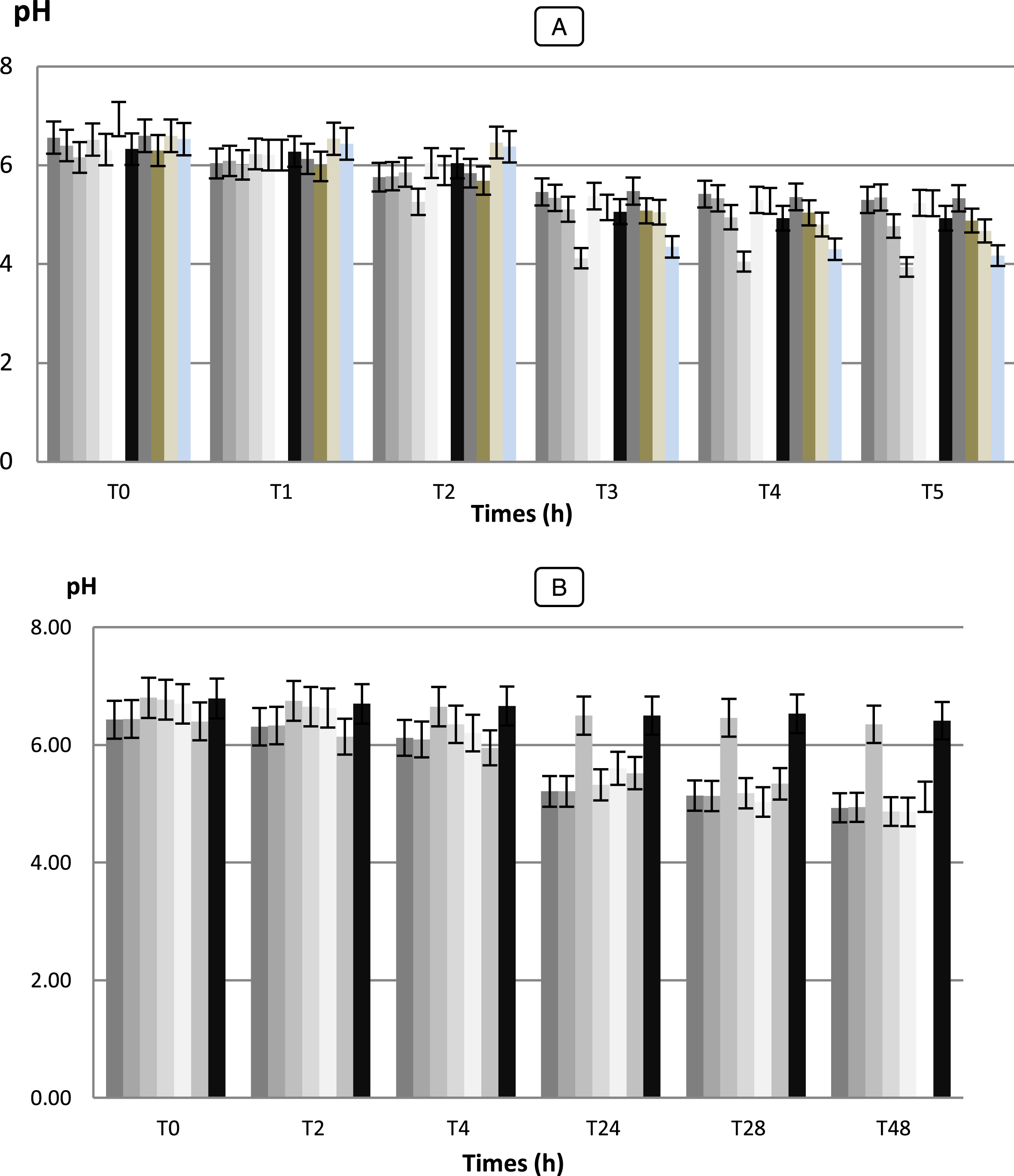
Fig.8
Growth (Log UFC/ ml) of Bifidobacterium strain (A): (■) Bf. breve (BHI 01), (♦) Bf. infantis (BHI 03), (▴) Bf. Adolescentis (BHI 04), (•) Bf. bifidum (BHI 05), (□) Bf. Longum (BHI 07), (◊) Bf. thermophilum (BHI 08), (Δ) Bf. suis (BHI 33). LAB strains (B): () Lc. cremoris (Lc 02), () Lc. lactis (Lc 03), () Pc. cerevisiae (Pc 01), () Pc. acidilactici (Pc 02), (□) Sc. boris (Sc 01), (Δ) Sc. thermophilus (Sc 02), (◊) Sc. agalactiae (Sc 03), (▴) Ec. Faecium (Ec 01), (□) Ec. Faecalis (Ec 02), (♦) Lb. bulgaricus (Lb 01), (■) Lb. lactis (Lb 02), in skim milk at 37°C. Vertical bars represent standard deviations of the mean.
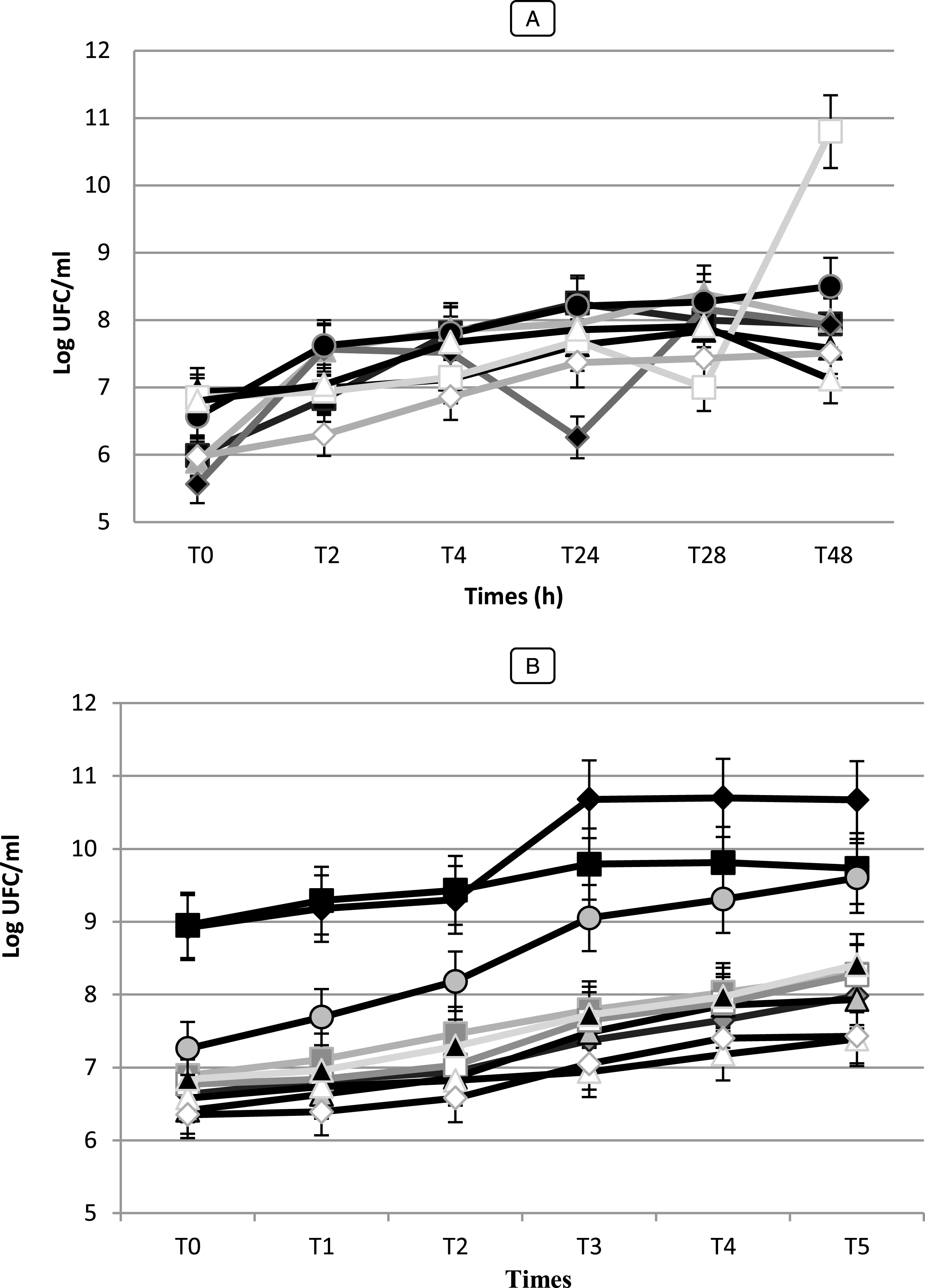
The values of acidity found show variations more or less important between the different species (p < 0.05). They vary between 1.5 and 2.8 g of lactic acid/ 100 mL to T0, after 6 h the acidity starts to gradually increase (p < 0.05) and varied between 1.9 and 3.8 g of lactic acid/ 100 mL. After 48 h of incubation, the pH of the culture medium decreased to values lower than 3.94 and the acidity reached values to 10 g of lactic acid/ 100 mL for the strain Lb 02. While, the strain Pc 02 was presented significantly the highest acidifying capacity with Δ pH = 2.58 (p < 0.05) following by Lb 02 (Δ pH = 2.36) (data not shown). While Sc. thermophilus (Sc 02) and Sc. agalactiae (SC 03) strains were the most weakly acidifying with Δ pH = 1.05 and 1.08, respectively developing an acidity of 3.4 and 3.1 g of lactic acid/ 100 mL. For the seven (7) Bifidobacterium strains, during fermentation and after 48 h of incubation, the pH values changed from 6.41 to 4.86 with Δ pH = 0.38 and 1.84, respectively (data not shown). The strains Bf. longum (BHI 05) and Bf. bifidum (BHI 07) performed the fastest acidification with Δ pH = 1.90 and 1.84, respectively and reached the highest level of viable counts with μ= 6.58 Log (UFC/mL) while Bf. suis (BHI 33) showed the lowest growth with μ= 0.53 h-1 (data not shown). A slower acidification was observed for Bf. adolescentis and Bf. suis (BHI 04, 33) reaching pH 6.35 and 6.41 after 48 h of fermentation. Our results showed that there is a positive correlation (r = 0.98 for Bf. bifidum strain, r = 0.970 for Bf. adolescentis strains) between the Acidifying capacity of the strains correlation is significant at the 0.05 level.
The results obtained in this study with regard to the technological properties of LAB and Bifidobacterium strains were shown a very important with those obtained by Herreros et al [28] observed that L. lactis subsp. lactis strains were those which showed the highest acidifying capacity, developing an acidity of 0.40 g.100 mL-1 lactic acid after 6 h and 0.65 – 0.70 g.100 mL-1 after 12 h.
3.4Resistance of LAB to antibiotic
The cultures were demonstrated sensitive (S) and resistant (R) by observing the inhibitory zone against tested antibiotics taking into consideration the clinical break points presented by the FEEDAP panel [65]. The results of antibiotic susceptibility testing are shown in Table 5. The isolates were analyzed by the diffusion method in Muller-Hinton agar for eleven antibiotics.
Table 5
Antibiotic sensitivity test of selected LAB and Bifidobacterium isolates
| Inhibition zone | |||||||||||
| Symbol μg/disc | AM 10μg | FA 10μg | E 15μg | OX 1μg | C 30μg | P 10 U | FOX 30μg | AX 25μg | CN 10μg | S 10μg | CE 30μg |
| Lc. lactis | R | R | R | R | R | R | R | R | S | S | S |
| Pc. acidilactici | R | R | R | R | I | R | R | R | S | S | I |
| Sc. boris | R | R | R | R | R | R | R | R | S | S | I |
| Ec. faecalis | S | I | I | R | I | R | R | R | S | S | S |
| Lb. L actis | S | S | S | S | I | I | I | S | S | S | S |
| Bf. breve | S | S | S | R | S | S | R | S | S | S | I |
| Bf. infantis | S | S | S | S | S | S | S | S | S | S | S |
| Bf. adolescentis | S | S | S | S | S | S | R | S | S | S | I |
| Bf. bifidum | S | S | S | R | S | R | R | I | I | S | I |
| Bf. longum | S | S | S | S | S | S | S | S | S | S | S |
| Bf. thermophilus | S | S | S | R | S | R | S | S | S | S | I |
| Bf. suis | S | S | S | S | S | R | S | S | S | S | S |
| Escherichia coli ATCC 25922 | S | – | – | S | – | I | S | S | S | S | |
| Staphylococcus aureus ATCC 25923 | S | S | S | I | I | I | S | S | S | S | |
R: resistan ce, I: intermediate, and S: sensitive; AM, Ampicillin; FA, Fusidic Acide; E, Erythromycin; OX, Oxacillin; C, Chloramphinicol; P, Penicillin; FOX, Cefoxitin; AX, Amoxicillin, CN, Gentamicin, S, Streptomycin; CE, Cefotaxime.
In the present study, it was found that the Bifidobacterium isolates were sensitive to ampicillin, fusidic acide, erythromycin, oxacillin, chloramphinicol, penicillin, amoxicillin, gentamicin, streptomycin and cefotaxime, with the exception of Bf. breve, Bf. adolescentis strains were resistant to Cefoxitin whereas Bf. longum, Bf. thermophilus, Bf. suis were resistant to Oxacillin. While Bf. bifidum strain was resistant to three antibiotics (oxacillin, penicillin, cefoxitin) and also it has been intermediate to three antibiotics (amoxicillin, gentamicin, cefotaxime). Recent studies have reported that, among the different Bifidobacterium species, B. breve possesses resistant levels to several antibiotics that are higher than those of other Bifidobacterium species [66].
Previous studies document that Bifidobacterium were highly resistant to kanamycin, gentamicin, fusidic acid, streptomycin, polymyxin B, nalidixic acid, paromomycin, neomycin and susceptible to penicillin G [67–71], metronidazole, tetracycline, cephalotin, aztreonam, bacitracin, rifampicin and bacitracin [72, 73].
Although, Lc. Lactis, Pc. Acidilactici, Sc. boris strains were resistant to eight antibiotics tested (ampicillin, fusidic acide, erythromycin, oxacillin, chloramphinicol, penicillin, amoxicillin) but it’s have been sensitive to the three antibiotics (gentamicin, streptomycin and cefotaxime). Delgado et al. [74] reported that Lactobacillus, Lactococcus and Leuconostoc species show resistance to high levels of cefoxitin (MIC≥30μg.ml-1) whereas, Bifidobacterium species are very sensitive. However, resistant strains to these agents have also been identified [74, 75] and several genes providing such resistance have been studied; e.g., a chloramphenicolresistance cat gene has been found in Lactobacillus reuteri and Lactobacillus plantarum [77]; different erythromycin-resistance genes (erm) which have been found in many species [78, 79] and a number of tetracycline resistance genes – tet (K, M, O, Q, S, W, 36) [80]. While, more studies reported that Lc. lactis have been shown resistant to chloramphenicol, clindamycin, streptomycin, erythromycin and tetracycline [81–83]. It was found to contain at least three different plasmid-encoded antibiotic resistance determinants (for tetracycline [tet (S)], chloramphenicol and streptomycin) [84] and tet (M) gene [85].
The knowledge of genetic mobility of antibiotic resistance genes constitutes an important task that should be explored. In fact, one of the essential definitions of a safe micro- organism that can be added to food preparation (e.g., B. lactis, B. breve, and B. bifidum probiotic strains) should be the absence of mobile antibiotic-resistance genes. Although it is clear that simultaneous application of susceptible probiotics with oral antibiotics is generally unreasonable, in the case of microbial infection it might be possible to use these resistant probiotics in combination with appropriate antibiotics [86].
Bifidobacteria are increasingly employed in the production of functional foods, only a few studies [69, 87] have been conducted in order to define the resistance/sensitivity to antibiotics of bifidobacteria. Antibiotic resistance is not a common trait of dairy and intestinal LAB and Bf. species. However, several strains were considered to be resistant to some antibiotics, and were thought to harbour acquired resistance genes [88]. European Commission, it can be concluded that the isolates do not carry any resistant genes and can be safely be used as starter cultures. The resistance gene reservoir hypothesis suggests that beneficial and commensal bacterial populations may play a role in the transfer of antibiotic resistance to pathogenic and opportunistic bacteria [89, 90].
The large numbers of LAB and Bifidobacteria in fermented products and in the GIT help in the appearance of different resistant mechanisms via mutation. In addition, LAB can also acquire resistances from other bacteria in the environment. Once a LAB becomes resistant, the determinant is amplified and may be transmitted to another host. Therefore, checking for signs of transferable antibiotic resistance in starter strains and bacteria used as feed and food additives is essential; quite apart from the need for their complete physiological and technological characterization [8]. Thought susceptibility levels are species dependent. LAB and bifidobacteria have probably acquired most antibiotic resistances by conjugation. Therefore, strains intended to be used in feed and food systems should be systematically monitored for resistances in order to avoid their inclusion in starters and probiotics. The widespread use of LAB and bifidobacteria in fermented foods and dairy products has a long history of safety, and it is generally assumed that the risk of infection from ingested bacteria is very low [91, 92]. At the same time, the use of LAB and bifidobacteria species as probiotics may help to reduce antibiotic use for therapeutic, prophylactic and growth promotion in animal husbandry [93].
3.5In vitro inhibition of pathogen growth
Antimicrobial activity is a very important criterion for selection of the potential-antagonists-starter and probiotic-culture against harmful bacteria. The inhibition ability of supernatants from selected strain BHI 07 Bf. bifidum and five LAB selected strains (Lc. Lactis, Pc. Acidilactici, Sc. boris, Ec. Faecalis, Lb. Lactis) for their acid and bile resistance, performed the fastest acidification and reached the highest level of viable counts was determined against 4 pathogens, E. coli ATCC 25922, Staphyloccus aureus ATCC 6538, Bacillus cereus ATCC 10876, Salmonella typhimurium ATCC 14028 by the agar diffusion test to evaluate their antagonistic activity against potentially bacterial pathogens.
The inhibition of pathogenic bacteria, causing diarrhea or other diseases in the human intestine, is a desirable property for probiotic bacteria [74]. The positive control used (tetracycline) produced inhibition zones for all pathogens with diameters ranging from 14 to 29 mm (data not shown).
All the LAB and Bifidobacterium isolates exhibited wide spectrum of antibacterial activity against the pathogenic tested strains (E. coli ATCC 25922, Staphyloccus aureus ATCC 6538, Bacillus cereus ATCC 10876) but toward the pathogen strain Salmonella typhimurium ATCC 14012, Bf. bifidum, Lc. Lactis, Sc. boris, Ec. faecalis in single trials and Bf. bifidum in mixed culture with Sc. boris. Strains showing a clear zone (Fig. 9) of lateral extension greater than 2 mm are considered as producers of antibacterial substances [94]. This inhibition could be due to the production of inhibitory substances, such as organic acids, bacteriocins or peroxide hydroxide [95].
Fig.9
Agar-well diffusion showing inhibition zones from antimicrobial activity of supernatants- LAB and Bf. bifidum BHI 07 grown alone or in combination with lactic acid bacteria grown in skim milk either alone or in combination with LAB: Lc. Lactis, Pc. Acidilactici, Sc. Boris, Ec. Faecalis, Lb. Lactis, on A) E. coli, B) S. aureus, C) Bacillus cereus, D) Salmonella typhimurium. Analyses were carried out using Muller-Hinton agar. Clear area is zone of inhibition.
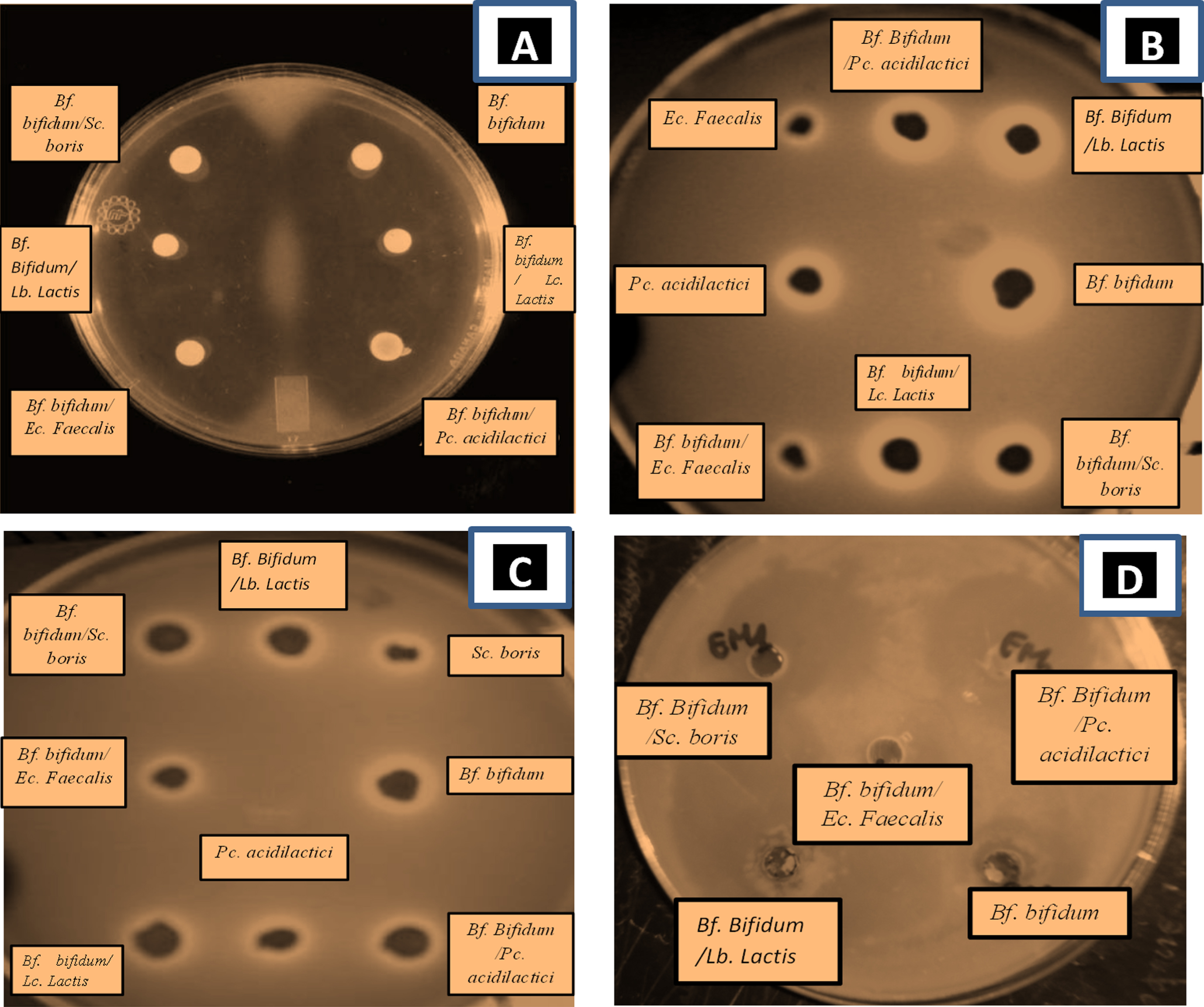
E. coli ATCC 25922 strain was the most sensitive to the presence of Bf. bifidum in mixed culture with Sc. boris showing an inhibition of 35±0.83 mm following by Bf. bifidum in mixed trial with Lc. lactis (31±1.12 mm) but Bf. bifidum strain on single trials showing somewhat low inhibition effect (25±1.83 mm). Sc. boris in single trials (9±0.8 mm) (Table 6).
Table 6
In vitro inhibition of the growth of pathogens by Bf. bifidum and LAB strains tested. Results display the diameter of the inhibition zones (mm)
| Strains | Diameter (mm) of inhibition zones | |||
| E. coli ATCC 25922 | Staphyloccus aureus ATCC 6538 | Bacillus cereus ATCC 10876 | Salmonella typhimurium ATCC 14028 | |
| Single cultures | ||||
| Bf. bifidum | 25±1.83 | 24±1.23 | 21±1.49 | 1.2±0.45 |
| Lc. Lactis | 17.5±0.9 | 19±0.09 | 13±1.36 | 0.8±0.5 |
| Pc. acidilactici | 21±1.2 | 23±0.05 | 19±0.02 | 3.78±0.14 |
| Sc. boris | 09±0.8 | 14±1.03 | 10±0.09 | 0.3±0.91 |
| Ec. Faecalis | 18±1.05 | 22±0.17 | 20±1.06 | 0.9±0.67 |
| Lb. Lactis | 20±1.53 | 18±1.4 | 23±0.38 | 3.5±0.65 |
| Mixed cultures | ||||
| Bf. bifidum/ Lc. Lactis | 31±1.12 | 37±1.54 | 27±0.16 | 2.5±0.34 |
| Bf. bifidum/ Pc. acidilactici | 28±0.71 | 28±0.48 | 25±0.69 | 4±0.12 |
| Bf. bifidum/Sc. boris | 29±1.41 | 29.5±0.15 | 22±1.50 | 1.7±0.39 |
| Bf. bifidum/ Ec. Faecalis | 35±0.83 | 32±1.67 | 34.5±0.45 | 2.5±0.78 |
| Bf. bifidum/Lb. Lactis | 27±0.21 | 33±0.31 | 32±1.18 | 3.7±1.3 |
The strains examined include strains that survived exposure to pH 2 and to 0.3% oxgall for at least 1 h as well as. Values are means±S.E. of triplicate analyses.
Staphyloccus aureus ATCC 6538 was the most sensitive to the presence of Bf. bifidum in mixed culture with Lc. lactis (37±1.54 mm) following by E. coli ATCC 25922 and Bacillus cereus ATCC 10876 to the presence of Bf. bifidum in mixed trial with Ec. Faecalis (35±0.83 mm; 34.5±0.45, respectively). But the inhibitory effect of Bf. bifidum strain in single trial on pathogenic bacteria was somewhat low. However, a loss of inhibitory activity was observed in Lb. Lactis; Pc. acidilactici in single trial and Bf. bifidum in mixed trial with the same strains (Table 6). This finding confirms the results of previous studies [96–98]. Therefore, the mixed cultures of Bf. bifidum strains with LAB strains were more active than the pure one. This result was also been found by Kheadr [99] who noted a remarkable antibacterial activity of B. longum DSM 20097 against both Staphylococcus aureus and Bacillus subtilis. When grown as co-cultures with Lb. paracasei, Lb. acidophilus or Lb. plantarum, B. longum appeared to be more active at inhibiting such indicator organism.
Antimicrobial activity of bifidobacteria was first noticed by Tissier [100], who described various types of antagonistic effect of B. bifidum against E. coli. Other reports described numerous antagonistic activity or specific antimicrobial activity in bifidobacteria linked to the production of lactic and acetic acids, or to bacteriocins [101–103]. Yildirim and Johnson [102] described a bacteriocin (bifidocin B) produced by B. bifidum NCFB 1454, which was found to be active against certain species of Listeria, Bacillus, Enterococcus, Pediococcus and Leuconostoc and is believed to be encoded by a 8.4 kb plasmid.
4Conclusion
In conclusion, the result of screening procedure of LAB and Bifidobacterium strains isolated from different niches as probiotic bacteria showed that BHI 07 Bf. bifidum strain was considered candidates to be used as putative probiotic starter cultures in fermented foods. The efficiency of the antimicrobial activity of the isolated strains needs to be confirmed in vivo and in the food matrix. Further study is needed to characterize the antimicrobial activity of the isolates reported in the present study and evaluate their effectiveness at inhibiting the pathogenic bacteria and their use as an alternative inexpensive and natural remedy to restore and maintain health.
In this context, the result is very important for the selection of probiotic bacteria and may be could have a broad application as probiotic strains. They may play an important role in the food industry as starter cultures, co-cultures or bioprotective cultures, to improve food quality and safety or as probiotic therapeutics appropriate for clinical practice. In addition, sensitivity or intrinsic resistance of the majority of the strains to a recommended set of antibiotics make them safe for use in different products for animal consumption or human for treatment of infection diseases of microbial origin.
Acknowledgments
We are grateful to Mme. MAHMOUDI S. and Mme. BENAMIROUCHE K. for providing some of the pathogenic strains. Also, we thank Mr. Director of Algerian Center of quality control and packaging for having provided laboratory facilities and particular thanks to Mr. BOUKHEIT M. for his technical assistance and full support when needed.
References
[1] | Leroy F , de Vuyst L . Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. (2004) ;15: :67–78. |
[2] | Reddy KB , Raghavendra P , Kumar BG , Misra MC , Prapulla SG . Screening of probiotic properties of lactic acid bacteria isolated from Kanjika, an ayruvedic lactic acid fermented product: An in-vitro evaluation. Journal of General and Applied Microbiology. (2007) ;53: :207–13. |
[3] | Stiles ME , Holzapfel WH . Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. (1997) ;36: :1–29. |
[4] | Coda R , Nionelli L , Rizzello CG , De Angelis M , Tossut P , Gobbetti M . Spelt and emmer flours: Characterization of the lactic acid bacteria microbiota and selection of mixed autochthonous starters for bread making. Journal of Applied Microbiology. (2010) ;108: :925–35. |
[5] | Vandenbergh RA . Lactic acid bacteria, their metabolic products and interference with microbial growth. FEMS Microbiology Reviews. (1993) ;12: :221–38. |
[6] | Kizerwetter-Swida M , Binek M . Selection of potentially probiotic Lactobacillus strains towards their inhibitory activity against poultry enteropathogenic bacteria. Pol J Microbiol. (2005) ;54: :287–94. |
[7] | Coda R , Rizzello CG , Di Cagno R , Trani A , Cardinali G , Gobbetti M . Antifungal activity of Meyerozyma guilliermondii: Identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food Microbiology. (2013) ;33: :243–51. |
[8] | Ammor MS , Mayo B . Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: An update. Meat Science. (2007) ;76: :138–46. |
[9] | Martín B , Garriga M , Hugas M , Aymerich T . Genetic diversity and safety aspects of enterococci from slightly fermented sausages. Journal of Applied Microbiology. (2005) ;98: :1177–90. |
[10] | Rönkä E , Malinen E , Saarela M , Rinta-Koski M , Aarnikunnas J , Palva A . Probiotic and milk technological properties of Lactobacillus brevis. International Journal of Food Microbiology. (2003) ;83: :63–74. |
[11] | Fuller R . Probiotics in man and animals. J Appl Bacteriol. (1989) ;66: :365–78. |
[12] | Gatesoupe FJ . The use of probiotics in aquaculture. Aquaculture. (1999) ;180: :147–65. |
[13] | Lim Sung-Mee1 , Dong-Soon Im . Screening and Characterization of Probiotic Lactic Acid Bacteria Isolated from Korean Fermented Foods. J Microbiol Biotechnol. (2009) ;19: (2):178–86. |
[14] | Metchnikoff E . Lactic acid as inhibiting putrefaction. In: The Prolongation of Life: Optimistic Studies. (1908) . pp. 161–183, Heinemann, London. |
[15] | Tripathi MK , Giri SK . Probiotic functional foods: Survival of probiotics during processing and storage. Journal of Functional Foods. (2014) ;9: (1):225–41. |
[16] | Palacios MC , Haros M , Sanz Y , Rosell MC . Selection of lactic acid bacteria with high phytate degrading activity for application in whole wheat bread making. LWT Food Science and Technology. (2008) ;41: (1):82–92. |
[17] | Bogovič Matijašić B , Stojkovic S , Salobir J , Malovrh S , Rogelj I . Evaluation of the Lactobacillus gasseri K7 and LF221 strains in weaned piglets for their possible probiotic use and their detection in the faeces. Anim Res. (2004) ;53: :35–44. |
[18] | Wang Jingrui , Xing Zhuqing , Tang Wei , Zheng Yongna , Wang Yanping . Isolation, Identification, and Potential Probiotic Characterization of One Lactococcus from Kefir Grain. Food Sci Biotechnol. (2015) ;24: (5):1775–80. |
[19] | Raghavendra Ponnala , Halami Prakash M . Screening, selection and characterization of phytic acid degrading lactic acid bacteria from chicken intestine. International Journal of Food Microbiology. (2009) ;133: :129–34. |
[20] | Famularo G , De Simone C , Pandey V , Sahu AR , Minisola G . Probiotic lactobacilli: An innovative tool to correct the malabsorption syndrome of vegetarians. Medical Hypotheses. (2005) ;65: :1132–5. |
[21] | Uraipan Supansa , Hongpattarakere Tipparat . Antagonistic characteristics against food-borne pathogenic bacteria of lactic acid bacteria and Bifidobacteria isolated from feces of healthy thai infants. Undishapur Microbiol. (2015) ;8: (6):e18264. |
[22] | Borriello SP , Hammes WP , Holzapfel W , Marteau P , Schrezenmeir J , Vaara M , . Safety of probiotics that contain lactobacilli or bifidobacteria. Clin Infect Dis. (2003) ;36: (6):775–80. |
[23] | Abd-El-Malek Y . Traditional Egyptian dairy fermentations. In Stantan WR , Da Silva EJ . (Eds), Global impact of food Microbiology (GIAM)- State of the Art: GIAM and Relevalence to Development in Developing Contries, University of Malaya Press, Kuala Lumpur. (1987) ; pp. 198–208. |
[24] | El Gendy SM . Some traditional fermented dairy products in Egypt. Proceeding of the Eighth Egyptian conference for Dairy Science and Technology, Cairo, Egypt, November 3-5, Egypt, J Dairy Sci. (2001) ;465–79. |
[25] | Ayad EHE , Nashat S , El-Sadek N , Metwaly H , El- Soda M . Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. (2004) ;21: :715–25. |
[26] | Kacem M , Zadi-Karam H , Karam N . Identification of lactic acid bacteria isolated from milk and fermented olive oil in western Algeria. Actes Inst Agron Vet. (2003) ;23: :135–41. |
[27] | Kacem M , Zadi-Karam H , Karam N . Isolation of lactic acid bacteria for its possible use in the fermentation of green algerian olives, Grasas y Aceites. (2004) ;55: :385–93. |
[28] | Herreros MA , Fresno JM , Gonz!alez-Prieto MJ , Tornadijo ME . Technological characterization of lactic acid bacteria isolated from Armada cheese (a Spanish goats’ milk cheese), International Dairy Journal. (2003) ;13: :469–79. |
[29] | Ruiz-Moyano S , Martín A , Benito MJ , Pérez Nevado F , de Guía Córdoba M . Screening of lactic acid bacteria and bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Science. (2008) ;80: :715–21. |
[30] | Kim Pyoung Il , Jung Min Young , Chang Young-Hyo , Kim Saehun , Kim Seong-Jae , Park Yong-Ha . Probiotic properties of Lactobacillus and Bifidobacterium strains isolated from porcine gastrointestinal tract, Appl Microbiol Biotechnol. (2007) ;74: :1103–11. |
[31] | Garcia JL , Garcia E , Arraras A , Garcia P , Ronda C and Lopez R . Cloning, purification and biochemical characterization of the pneumococcal bacteriophage Cp-I lysin. J Virol. (1987) ;61: :2573–80. |
[32] | Terzaghi BE , Sandine WE . Improved Medium for Lactic Streptococci and Their Bacteriophages, Applied Microbiology. (1975) ; pp. 807–813. |
[33] | Beerens H . An elective and selective isolation medium for Bifidobacterium spp. Letters in Applied Microbiology. (1990) ;11: :155–57. |
[34] | Frank JF , Holcomb JE , McGregor JU . Viability of Lactobacillus acidophilus and Bifidobacterium bifidum in soft-serve frozen yogurt. Cult Dairy Prod J. (1991) ;26: :4. |
[35] | Mahmoudi F , Hadadji M , Guessas B , Kihal M . Evaluation of in vitro antagonism and protection against enteropathogenic experimental challenge of different strains of Bifidobacterium. African Journal of Microbiology Research. (2013) ;7: (29):3816–23. |
[36] | Schillinger U , Friedrich-Karl L . Antibacterial Activity of Lactobacillus sake Isolated from Meat. Applied and Environmental Microbiology. (1989) ;1901–06. |
[37] | Chung HS , Kim YB , Chun SL , Ji GE . Screening and selection of acid and bile resistant bifidobacteria. Int J Food Microbiol. (1999) ;47: :25–32. |
[38] | Klingberg TD , Axelsson L , Naterstad K , Elsser D , Budde BB . Identification of potential probiotic starter cultures for Scandinavian-type fermented sausages. International Journal of Food Microbiology. (2005) ;105: :419–31. |
[39] | Vinderola CG , Reinheimer JA . Lactic acid starter and probiotic bacteria, a comparative in vitro study of probiotic characteristics and biological barrier resistance. Food Research International. (2003) ;36: :895–904. |
[40] | Accolas JP , Bloquel R , Didienne R , Régnier J . Propriétés acidifiantes des bactéries lactiques thermophiles en relation avec la fabrication du yoghourt, Lait. (1977) ;57: :1–23. |
[41] | Phillips I , Andrews JM , Bridson E , Cooke EM , Spencer RC , Holt HA , Wise R , Bint AJ , Brown DFJ , Greenwood D , King A , Williams RJ . A Guide to Sensitivity Testing. Report of the Working Party on Antibiotic Sensitivity Testing of the British Society for Antimicrobial Chemotherapy. (1991) . |
[42] | Zhou J , Dong Y , Zhao X , Lee S , Amin A , Ramaswamy S , Domagala J , Musser JM , Drlica K . Selection of Antibiotic-Resistant Bacterial Mutants: Allelic Diversity among Fluoroquinolone-Resistant Mutations. Journal of Infectious Diseases. (2000) ;182: :517–25. |
[43] | Treagan L , Pulliam L . Medical microbiology laboratory procedures. W.B. Saunders Company, Philadelphia. (1982) ; pp. 233–243. |
[44] | Tagg JR , Dajani AS , Wannamaker LW . Bacteriocins of Gram-negative bacteria. Bacteriogical Review. (1976) ;40: (3):722–56. |
[45] | Aslam S , Hamill RJ , Musher DM . Treatment of Clostridium difficile-associated disease: Old therapies and new strategies. Lancet Infect Dis. (2005) ;5: :549–57. |
[46] | Desmazeaud MJ et De Roissart H . Metabolisme général des bactéries lactiques. Bacteries lactiques, aspect fondamentaux et technologiques. Lorica-Uriage. (1994) ;1: :196–207. |
[47] | Guiraud JP . Microbiologie Alimentaire Edition: Dunod, Paris, France. (2003) ;652. |
[48] | Scardovi V. Genus Bifidobacterium Orla-Jensen. In Bergey’s Manual of Systematic Bacteriology, 2: :1418–1434, Edited by Sneath PH A , Mair N.S , Sharpe ME & Holt . Baltimore JG: Williams & Wilkins. (1986) . |
[49] | Baratte-euloge P . Action comparée sur la flore intestinale de trois laits fermentés au Bifidobacterium. Evaluation de propriétés probiotiques et du comportement de la souche BB 536 de Bifidobacterium longum chez l’homme. thèse de Doctorat, Université de nancy I. (1992) )267. |
[50] | Larpent JP . Microbiologie alimentaire. Tec & doc, Lavoisier. Paris. (1997) ;10–72. |
[51] | FAO/WHO. FAO/WHO working group report on drafting guidelines for the evaluation of probiotics in food. Geneva, Swiztherland: World Health Organization (2002) . |
[52] | Huang Y , Adams MC . In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int J Food Microbiol. (2004) ;91: :253–60. |
[53] | Mishra V , Prasad DN . Application of in vitro methods for selection of Lactobacillus casei strains as potential probiotics, Int J Food Microbiol ((2005) ;103: :109–15. |
[54] | Naylin N , Taing O , Hashinaga F , Toshima Y . Antioxidant activity of sugar-tolerant yeast Zygosaccharomyces rouxii, Food Biotechnol. (2005) ;19: :107–20. |
[55] | Dunne C , O’Mahony L , Murphy L , Thornton G , Morrissey D , O’Halloran S , et al. In vitro selection criteria for probiotic bacteria of human origin: Correlation with in vivo findings, Am J Clin Nutr. (2001) ;73: 386S–392S. |
[56] | Kim WS , Ren J , Dunn NW . Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol Lett. (1999) ;171: (1):57–65. |
[57] | Xanthopoulos V , Litopoulou-Tzanetaki E , Tzanetakis N . Characterization of Lactobacillus isolates from infant faeces as dietary adjuncts. Food Microbiol. (2000) ;17: :205–15. |
[58] | Al-Saleh AA , Metwalli AAM , Abu-Tarboush HM . Bile salts and acid tolerance and cholesterol removal from media by some lactic acid bacteria and bifidobacteria. J Saudi Soc for Food and Nutrition. (2006) ;1: :1–17. |
[59] | Holzapfel WH , Haberer P , Snel J , Schillinger U , Huis in’t Veld JHJ . Overview of gut flora and probiotics. Intl J Food Microbiol. (1998) ;41: :85–101. |
[60] | Gilliland SE , Staley TE , Bush LJ . Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J Dairy Sci. (1984) ;67: :3045–51. |
[61] | Goldin BR , Gorbach SL , Saxelin M , Barakat S , Gualtieri L , Salminen S . Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci. (1992) ;37: :121–8. |
[62] | Kim Pyoung Il , Jung Min Young , Chang Young-Hyo , Kim Saehun , Kim Seong-Jae , Park Yong-Ha . Probiotic properties of Lactobacillus and Bifidobacterium strains isolated from porcine gastrointestinal tract. Appl Microbiol Biotechnol. (2007) ;74: :1103–11. |
[63] | CHEN Li-Shui , MA Ying , MAUBOIS Jean-Louis , HE Sheng-Hua , CHEN Li-Jun , LI Hai-Mei . Screening for the potential probiotic yeast strains from raw milk to assimilate cholesterol. Dairy Sci Technol. (2010) ;90: :537–48. |
[64] | Kim K , Kim KH , Storey MK , Voelker DR , Carman GM . Isolation and Characterization of the Saccharomyces cerevisiae EKI1 Gene Encoding Ethanolamine Kinase. Journal of Biological Chemistry. (1999) ;274: (21):14857–66. |
[65] | EFSA, European Commission. Opinion of the Scientific Panel on Additives and Products or Substances used in Animal Feed on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. The EFSA J. (2005) ;223: :1–12. |
[66] | Moubareck C , Gavini F , Vaugien L , Butel MJ , Doucet-Populaire F . Antimicrobial susceptibility of bifidobacteria. J Antimicrob Chemother 66. (2005) ;55: :38–44. |
[67] | Sutter VL , Finegold SM . Susceptibility of Anaerobic Bacteria to 23 Antimicrobial Agents, Antimicrobial Agents and Chemotherapy. (1976) , pp. 736–752. |
[68] | Matteuzzi D , Crociani F , Brigidi P . Antimicrobial susceptibility of Bifidobacterium. Annals of Microbiology. (1983) ; 134 A: :339–49. |
[69] | Lim KS , Huh CS , Baek YJ . Antimicrobial susceptibility of bifidobacteria. Journal of Dairy Science. (1993) ;76: :2168–74. |
[70] | Temmerman R , Pot B , Huys G , Swings J . Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Intemational Journal of Food Microbiology. (2002) ;81: :1–10. |
[71] | Moubareck C , Gavini F , Vaugien L , Butel MJ , Doucet-Populaire F . Antimicrobial susceptibility of bifidobacteria. Journal of Antimicrobial Chemothcrapy. (2005) ;55: :38–44. |
[72] | Charteris WP , Kelly PM , Morelli L , Collins JK . Antibiotic susceptibility of potentially probiotic Lactobacillus species. Journal of Food Protection. (1998) ;61: :1636–43. |
[73] | Charteris WP , Kelly PM , Morelli L , Collins JK . Antibiotic susceptibility of potencially probiotic Bifidobacterium isolates from human and gastrointestioal tract. Letters in Applied Microbiology. (1998) ;26: :333–37. |
[74] | Delgado S , O’sullivan E , Fitzgerald G , Mayo B . Subtractive screening for probiotic properties of Lactobacillus species from the human gastrointestinal tract in the search for new probiotics. Journal of Food Science. (2007) ;72: :M310–M315. |
[75] | Danie1sen M , Wind A . Susceptibility of Lactobacillus spp. To antimicrobial agents. Intemational Journal of Food Microbiology. (2003) ;82: :1–11. |
[76] | Lin MY , Harlander S , Savaiano D . Construction of an integrative food-grade cloning vector for Lactobacillus acidophilus. Appl Microbiol Biotechnol. (1996) ;45: :484–9. |
[77] | Ahn DU , Wolfe FH , Sim JS , Kim DH . Packaging cooked turkey meat patties while hot reduces lipid oxidation. Journal of Food Science. (1992) ;57: :1075–7. |
[78] | Fons M , Hege T , Ladire M , Raibaud P , Ducluzeau R , Maguin E . Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid. (1997) ;37: :199–203. |
[79] | Cataloluk O , Gogebakan B . Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol Lett. (2004) ;236: :7–12. |
[80] | Huys G , D’Haene K , Swings J . Genetic basis of tetracycline and minocycline resistance in potentially probiotic Lactobacillus plantarum strain CCUG 8. Antimicrob Agents Chemother. (2006) ;50: :1550–1. |
[81] | Florez AB , Delgado S and Mayo B . Antimicrobial susceptibility of lactic acid bacteria isolated from a cheese environment. Can J Microbiol. (2005) ;51: :51–8. |
[82] | Raha AR , Ross E , Yusoff K , Manap MY , Ideris A . Characterisation and molecular cloning of an erythromycin resistance plasmid of Lactococcus lactis isolated from chicken cecum. Journal of Biochemistry, Molecular Biology and Biophysics. (2002) ;6: :7–11. |
[83] | Temmerman R , Pot B , Huys G , Swings J . Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int J Food Microbiol. (2003) ;81: :1–10. |
[84] | Temmerman R , Huys G , Swings J . Identification of lactic acid bacteria: Culture-dependent and culture-independent methods. Trends Food Sci. Tech. (2004) ;15: :348–59. |
[85] | Perreten V , Schwarz F , Cresta L , Boeglin M , Dasen G , Teuber M . Antibiotic resistance spread in food. Nature. (1997) ;389: :801–2. |
[86] | Chopra I , Roberts M . Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiology and Molecular Biology Reviews. (2001) ;65: :232–60. |
[87] | Marounek M , Rada V . Susceptibility of poultry Lactobacilli to ionophore antibiotics. J Vet Med. (1995) ;42: :193–6. |
[88] | Finegold SM , Harada NE , Miller LG . Antibiotic susceptibility patterns as aids in classification and characterization of gram-negative anaerobic bacilli. Journal of Bacteriology. (1967) ;94: :1443–50. |
[89] | Ammor MS , Florez AB , van Hoek AH , de Los Reyes-Gavilan CG , Aarts HJ , Margolles A , Mayo B . Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J Mol Microbiol Biotechnol. (2008) ;14: :6–15. |
[90] | Teuber M , Meile L , Schwarz F . Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek. (1999) ;76: :115–37. |
[91] | Salyers AA , Gupta A , Wang Y . Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. (2004) ;12: :412–16. |
[92] | Gasser. Safety of lactic acid bacteria and their occurrence in human clinical infections. Bulletin de l’Institut Pasteur. (1994) ;92: (1):45–67. |
[93] | Ouwehand AC , Salminen S , Isolauri E . Probiotics: An overview of beneficial effects. Antonie Van Leeuwenhoek. (2002) ;82: :279–89. |
[94] | Cross. Microbes versus microbes: Immune signals generated by probiotic Lactobacilli and their role in protection against microbial pathogens. FEMS Immunol Med Microbiol. (2002) ;34: :245–53. |
[95] | Fleming HR , Etchell GL , Costilow RN . Microbial inhibition by isolate of pediococcus from cucumber brine. Appl and Microbiology. (1975) ;30: :104–1042. |
[96] | Juven BJ , Schved F , Lindner P . Antagonistic compounds produced by a chicken intestinal strain of Lactobacillus acidophilus. Journal of Food Protection. (1992) ;55: :157–61. |
[97] | Jin LZ , Ho WY , Abdullah MA , Ali MA , Jalaludin S . Antagonistic effects of intestinal lactobacillus isolates on pathogens of chicken. Lett Appl Microbiol. (1996) ;23: :67–71. |
[98] | Khedkar JN , Dave JM , Sannabhadti SS . Antibacterial activity associated with Bifidobacterium adolescentis. Journal Food Science Technology. (1998) ;35: :527–9. |
[99] | Karimi Torshizi MA , Rahimi SH , Mojgani N , Esmaeilkhanian S , Grime JL . Screening of Indigenous Strains of Lactic Acid Bacteria for Development of a Probiotic for Poultry. Asian-Aust J Anim Sci. (2008) ;21: (10):1495–500. |
[100] | Tissier. The Genera of Lactic Acid Bacteria. (1900) ; pp. 297. |
[101] | Yildirim Z , Johnson MG . Characterization and antimicrobial spectrum of bifidocin B, a bacteriocin produced by Bifidobacterium bifidum NCFB 1454. J Food Prot. (1998) ;61: (1):47–51. |
[102] | Yildirim Z , Johnson MG , Winters DK . Purification and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J Appl Microbiol. (1999) ;86: :45–54. |
[103] | Kheadr EE , Bernoussi N , Lacroix C , Fliss I . Comparison of the sensitivity of commercial strains and infant isolates of bifidobacteria to antibiotics and bacteriocins. International Dairy J. (2004) ;14: :1041–53. |




