Amentoflavone derivatives against SARS-CoV-2 main protease (MPRO): An in silico study
Abstract
Globally, novel coronavirus (nCoV19) outbreak is a great concern to humanity owing to the unavailability of effective medication or vaccine to date. Therefore, the development of drugs having anti-COVID-19 potential is a need of time. In this milieu, in-silico studies have proven to be rapid, inexpensive and effective as compared to other experimental studies. Evidently, natural products have shown significant potential in drug development to curtail different ailments, which have opened a new horizon in the screening of anti-COVID-19 agents. In this study, in-silico analysis were performed on derivatives of amentoflavone (4′, 4′′′-Dimethylamentoflavone, 4′′′, 7-Di-O-Methylamentoflavone, 4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone, 7,4′-Dimethylamentoflavone, 7′-O-Methylamentoflavone, 7-O-methylamentoflavone, Heveaflavone, kayaflavone, and Sciadopitysin) and FDA approved anti-viral drug (camostatmesylate). All the derivatives of amentoflavone and FDA-approved anti-viral drugs were docked against SARS-CoV2 main protease (MPRO). The ten derivatives of amentoflavone showed strong interactions with the MPRO protein. In all cases, derivatives of amentoflavone showed good interaction with the targeted protein and better binding/docking score (–9.0351, –8.8566, –8.8509, –8.7746, –8.6192, –8.2537, –8.0876, –7.9501, –7.6429, and –7.6248 respectively) than FDA approved anti-viral drug. Therefore, derivatives of amentoflavone may be potent leads in drug discovery to combat HCoVs, such as SARS-CoV2. Moreover, to support the outcomes of this study further in-vivo investigations are required.
List of abbreviation
ABL1 | Abelson murine leukemia viral oncogene homolog 1 |
BLAST | Basic Local Alignment Search Tool |
Calcineurin–NFAT | Calcineurin nuclear; factor of activated T-cells |
COVID-19 | Coronavirus disease 2019 |
3CLPRO | 3-chymotrypsin-like protease |
HIV | Human immunodeficiency virus |
hACE2R | Human angiotensin-converting enzyme 2 receptor |
NCBI | National Center for Biotechnology Information |
RdRp | RNA-dependent RNA polymerase |
RSV | Respiratory syncytial virus |
TMPRSS-2 | Transmembrane protease serine 2 |
PLPRO | Papain-like protease |
SAM | S-adenosyl methionine |
SARS-CoV | Severe acute respiratory syndrome coronavirus |
1Introduction
In December 2019 world faced a disaster SARS-CoV-2. The SARS-CoV-2 (also called the novel coronavirus 2019 or nCoV-19) belongs to the Betacorona virus genus similar to SARS HCoV and MERS HCoV [1–3]. TheHCoV-2 s are positive-sense with a length of 30,000 bp and single-stranded RNA viruses. Two groups of proteins characterized in HCoVs are (i) Structural proteins (e.g., Spike (S), Envelope (E), Matrix (M), & Nucleocapsid (N)), and (ii) Non-structural proteins (e.g., RdRp (RNA-dependent RNA polymerase), PLPRO (Papain-like protease), & 3CLPRO (Proteases 3-chymotrypsin-like protease) [3]. The CoVpolyprotein encodes two proteases, which share in its processing and release of the translated non-structural proteins (nsps), the main protease is called 3-chymotrypsin-like protease (e.g., MPROor 3CLPRO) and PLPRO [4]. Viral attachment and entry within the host cell is due to presence of S protein on the outer surface of virion [5]. In a non-randomized clinical trial, synergistic use of hydroxychloroquine and azithromycin was recommended to minimize the effect of COVID-19. Other than this, various other methods including used of antiviral drugs and plasma have been used by clinicians [6, 7].
Evidently, plant-derived natural phytomolecules are thought to be potent source of drug leads of interest and may open a new horizon for drug discovery process [8]. Amentoflavone, a bioflavonoid in nature, is extremely found all over the world [9]. It’s a complex compound where two apigenin structurally are bound with each other by C3′-C8′′ linkage [10]. Literature have shown that amentoflavone possesses various pharmacological properties such as anti-diabetic, antioxidant, neuroprotective, antitumor, antivirus, antifungal, cardio-protective, antiinflammatory, antibacterial, anti-senescence, etc. [11–20]. Results of numerous studies have suggested that amentoflavone possess antiviral potential against dengue, herpes simplex virus 1 (HSV-1), human immunodeficiency virus (HIV), Coxsackievirus B3 (CVB3), respiratory syncytial virus (RSV), and acyclovir (ACV)-resistant strains (e.g., HSV-1/106, HSV-1/153, and HSV-1/Blue) [17, 21–24].
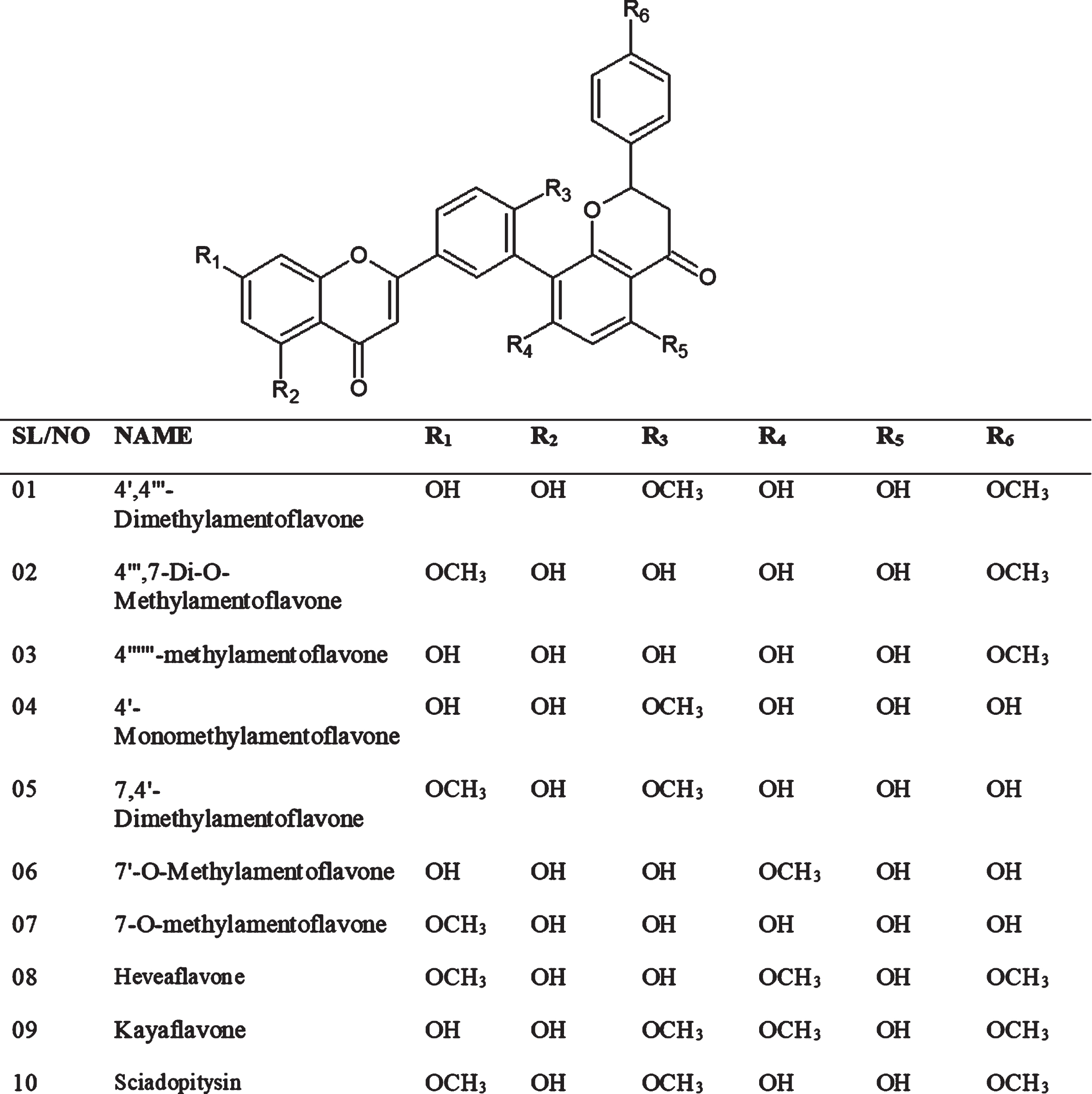
Amentoflavone has several (10) derivatives such as 4′,4′′′-Dimethylamentoflavone, 4′′′,7-Di-O-Methylamentoflavone, 4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone, 7,4′-Dimethylamentoflavone, 7′-O-Methylamentoflavone, 7-O-methylamentoflavone, Heveaflavone, kayaflavone, and Sciadopitysin (Fig. 1) [25]. According to a study, amentoflavonehave shown inhibitory potential (IC50:8.3μM) against SARS-CoV [26], probably due to inhibition of 3CLPRO (3-chymotrypsin-like protease), 3CLPRO has proven to be a valuable target and acquired significant importance in drug design of SARS-CoVs. It is also termed as ‘the Achilles’ or ‘heel of coronaviruses [27, 28].
Fig. 1
The chemical structure of derivatives of amentoflavone.
There are several strategies for the development of new drugs having anti-CoVs potential. Among all, the most focused strategies are: (a) blocking the cellular attachment of the virus with the host cells, and (b) inhibiting the transcription &replication of the virus. Therefore, MPRO (CoV main protease) having vital role in mediation of viral replication & transcription have attained interest of scientist in designing anti-SARS drugs [29–32]. Molecular docking is considered an efficient approach in screening potentially active components against specific target proteins, such as MPRO. Therefore, MPRO is an important target in the design of potential anti-CoV-2 inhibitors. Keeping in view the current situation of SARS-CoV-2 and antiviral properties of amentoflavone and its derivatives, this study shows the in silico analysis of amentoflavone derivatives against SARS-CoV-2 main protease (MPRO).Pharmacokinetic properties of amentoflavone and its derivatives were predicted using pkCSM (https://biosig.unimelb.edu.au/pkcsm/) and Swiss ADME (https://www.swissadme.ch) online software’s.
2Materials and Methods
2.1Proteins/Macromolecule preparation
The three-dimensional structure of SARS-CoV2 main protease (MPRO, 3CLpro) with (PDB accession ID: 6LU7) was retrieved from Protein Data Bank (https://www.rcsb.org/). Worldwide, protein Data bank is a trusted source for the three-dimensional crystal enzyme structure of biological macromolecules [33]. The crystal structures were prepared by removing water molecule and steric clashes. Subsequently the minimized structure was used for further analysis.
2.2Ligand preparation
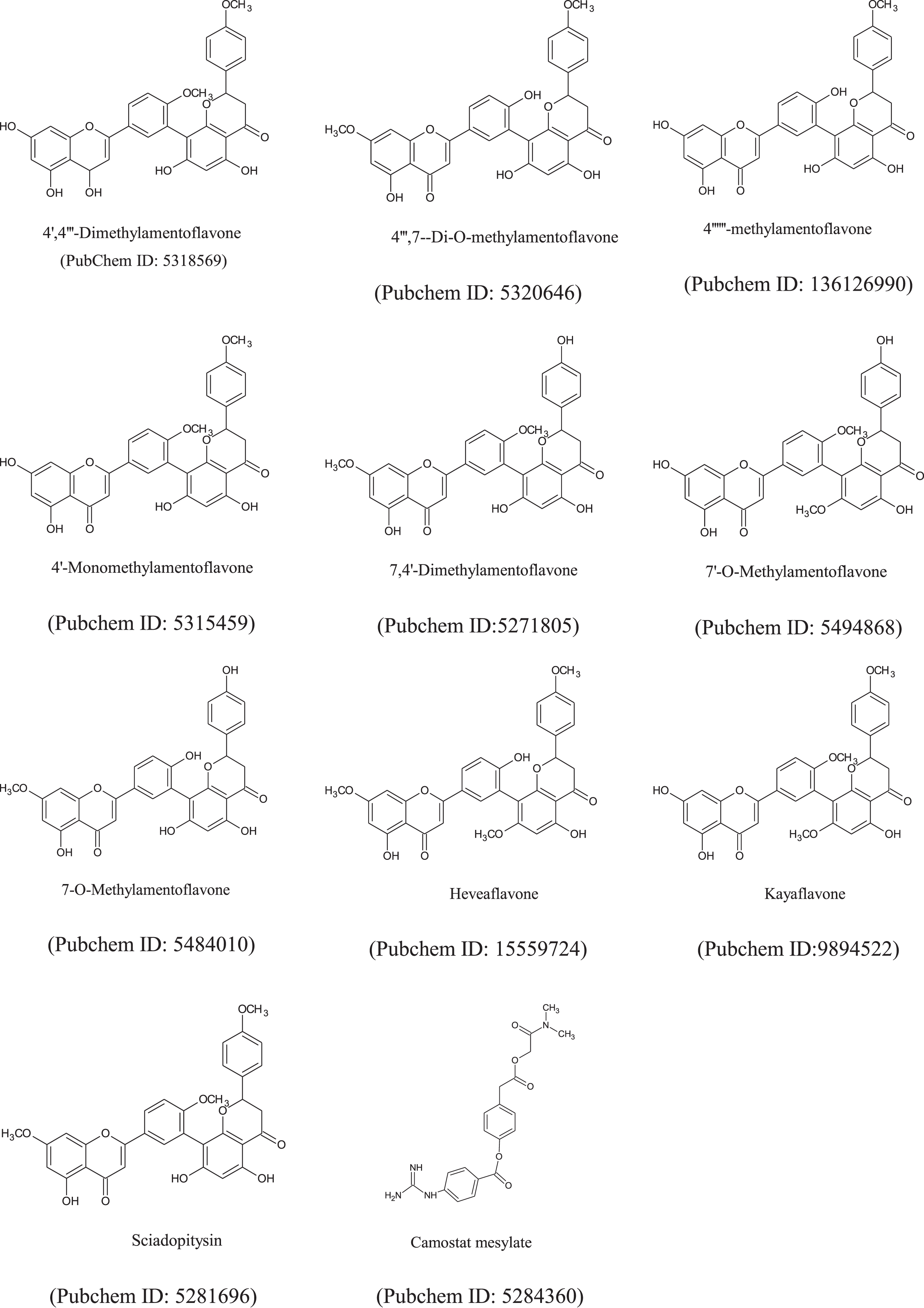
The chemical structure of amentoflavone’s derivatives and antiviral drug (e.g. Camostatmesylate, PubChem ID: 5284360) (Fig. 2), were obtained from the PubChem repository sample) in the ‘sdf’ file format. Chem3D Pro12.0 program packages was used for optimization of all internal energies of the ligands [34]. The Smiles structure of these compounds were also calculated by using ChemDraw (Table S1).
Fig. 2
The chemical structures of amentoflavone’s derivatives and FDA-approved anti-viral drugs.
2.3Docking analysis and binding site
Molecular docking analysis have played a significant role in predicting and verifying the binding modes and interaction poses of ligands within the binding pocket of target proteins. MOE-dock module implemented in the Molecular Operating Environment (MOE) software package [35] was employed for the docking of all the amentoflavone derivatives and FDA approved anti-viral drug against the SARS-CoV-2 main protease (MPRO) protein. The 3D-structure of main protease (MPRO) of SARS-CoV2 (PDB ID: 6LU7) present in Protein Data Bank was used in this study (https://www.rcsb.org/). The protease crystal structure co-crystallized with an engineered peptide inhibitor (N3) having excellent resolution (2.16 Å) comprised of 306 residues length chain [36]. All the solvent molecules were removed prior to molecular docking followed by 3D protonation. In order to get the minimal stable energy conformation, energy was minimized to 0.05 Gradient via MMFF94 s force field implemented in MOE. The 3D structural coordinates for all amentoflavone derivatives and anti-viral drug were built using Molecular Builder Module in MOE. By using the default parameter of MOE, energy of all the compounds were minimized, and all 3D coordinated of the compounds were protonated. Finally, refined target protein (MPRO) structures were used for docking study using the default parameters of MOE. For each of the compounds, total 10 conformations were allowed to be form. Later, the compounds were ranked based on docking score and protein-ligand interaction profile. Further, PyMol was used for molecular interactions of predicted protein–ligand interaction (PLI).
2.4Pharmacokinetic properties analysis
pkCSM and Swiss ADME online software were used for prediction of pharmacokinetic properties of these ten (10) derivatives of amentoflavone [37, 38]. The absorption, distribution, metabolism excretion, and toxicity parameters of these ten derivatives were mainly analyzed. The website was logged, on and the SMILES of the derivatives of amentoflavone data from PubChem were searched and submitted to the website, ADMET mode in pkCSM, [38] and ADME mode in Swiss-ADME was selected [37]. pkCSM is one the latest methodology that is used extensively for predicting and optimizing toxicity and pharmaco-kinetic perspectives relying on distance based graphical signatures [38, 39]. SwissADME is a freely available online tool, which is being employed for prediction of ADME parameters and drug-likeness [37, 40].
3Results
3.1Interaction with the MPRO
Insight into the binding mode of the amentoflavon derivatives, and FDA approved anti-viral drug within the active site of SARS-CoV-2 main protease (MPRO) molecular docking analysis were performed using X-ray crystal structure of SARS-CoV-2 (MPRO) (PDB ID: 6LU7) with an excellent resolution of 2.16 Å. The molecular docking results of the amentoflavon derivatives compounds with the (MPRO) enzyme provide substantial information regarding nature of binding mode, which significantly correlates with the experimental results. It is evident from docking analysis that all the derivatives of amentoflavon showed appropriate orientation towards catalytic residues revealing significant interactions with the catalytic site residues of the target enzymes. The post-molecular docking analysis indicate the importance of the amentoflavon derivatives compounds, that strongly interact with the target enzyme and block the enzymatic activity of SARS-CoV (MPRO). Analysis revealed significant results indicating that selected amentoflavon derivatives have inhibitory effect on SARS-CoV-2 (MPRO). The 10 selected amentoflavon derivatives C1 (4′,4′-Dimethylamentoflavone, 4′), C2 (7-Di-O-Methylamentoflavone), C3 (4′-methylamentoflavone), C4 (4′-Monomethylamentoflavone), C5 (7,4′-Dimethylamentoflavone), C6 (7′-O-Methylamentoflavone), C7 (7-O-methylamentoflavone), C8 (Heveaflavone), C9 (kayaflavone), and C10(Sciadopitysin) were docked with SARS-CoV-2 (MPRO) along with theFDA approved anti-viral drug C11 (camostatmesylate) to compare the results. Table 1 demonstrates the results of molecular docking study.
Table 1
Interaction Details, Docking Score, distance, and Binding Energy, of the Compounds docked in the Active Site of MPRO (SARS-CoV-2 main protease)
| S.No | Interaction details | Docking score | ||||||||
| Ligands | Receptor | Interaction | Distance | E Kcal/mol | ||||||
| 1 | C | 24 | OE1 | GLN | 189 | (A) | H-donor | 3.27 | –0.6 | –8.7746 |
| C | 31 | 5-ring | HIS | 41 | (A) | H-pi | 3.71 | –1.4 | ||
| 6-ring | CB | GLU | 166 | (A) | pi-H | 3.50 | –0.6 | |||
| 2 | O | 19 | NE2 | HIS | 41 | (A) | H-acceptor | 3.23 | –0.8 | –7.6429 |
| 6-ring | CA | ASN | 142 | (A) | pi-H | 4.28 | –0.6 | |||
| 6-ring | N | GLY | 143 | (A) | pi-H | 3.60 | –0.6 | |||
| 3 | C | 28 | OE1 | GLN | 189 | (A) | H-donor | 3.26 | –0.7 | –8.2537 |
| O | 9 | N | GLY | 143 | (A) | H-acceptor | 3.00 | –0.8 | ||
| O | 20 | OG | SER | 46 | (A) | H-acceptor | 2.81 | –0.6 | ||
| C | 35 | 5-ring | HIS | 41 | (A) | H-pi | 3.70 | –1.3 | ||
| 6-ring | CB | GLU | 166 | (A) | pi-H | 3.58 | –0.5 | |||
| 4 | C | 27 | OE1 | GLN | 189 | (A) | H-donor | 3.26 | –0.7 | –8.8509 |
| O | 9 | N | GLY | 143 | (A) | H-acceptor | 3.02 | –0.8 | ||
| O | 19 | OG | SER | 46 | (A) | H-acceptor | 2.80 | –0.5 | ||
| C | 34 | 5-ring | HIS | 41 | (A) | H-pi | 3.71 | –1.3 | ||
| 6-ring | CB | GLU | 166 | (A) | pi-H | 3.61 | –0.6 | |||
| 5 | O | 17 | OE2 | GLU | 166 | (A) | H-donor | 2.79 | –0.8 | –7.6248 |
| C | 40 | SD | MET | 49 | (A) | H-donor | 3.77 | –0.5 | ||
| C | 3 | 5-ring | HIS | 41 | (A) | H-pi | 4.24 | –1.2 | ||
| 6-ring | CA | GLN | 189 | (A) | pi-H | 3.74 | –0.5 | |||
| 6 | O | 18 | OE2 | GLU | 166 | (A) | H-donor | 2.76 | –0.7 | –8.8566 |
| O | 19 | SG | CYS | 145 | (A) | H-donor | 3.74 | –0.8 | ||
| O | 40 | SD | MET | 49 | (A) | H-donor | 3.64 | –0.6 | ||
| O | 19 | NE2 | HIS | 163 | (A) | H-acceptor | 2.89 | –2.3 | ||
| 7 | C | 27 | OE1 | GLN | 189 | (A) | H-donor | 3.28 | –0.7 | –7.9501 |
| O | 9 | N | GLY | 143 | (A) | H-acceptor | 3.01 | –0.8 | ||
| O | 19 | OG | SER | 46 | (A) | H-acceptor | 2.82 | –0.6 | ||
| C | 34 | 5-ring | HIS | 41 | (A) | H-pi | 3.66 | –1.5 | ||
| 6-ring | CB | GLU | 166 | (A) | pi-H | 3.57 | –0.5 | |||
| 8 | C | 27 | OE1 | GLN | 189 | (A) | H-donor | 3.24 | –0.7 | –8.6192 |
| O | 9 | N | GLY | 143 | (A) | H-acceptor | 2.96 | –0.9 | ||
| O | 19 | OG | SER | 46 | (A) | H-acceptor | 2.82 | –0.5 | ||
| C | 33 | 5-ring | HIS | 41 | (A) | H-pi | 3.72 | –1.4 | ||
| 9 | C | 27 | OE1 | GLN | 189 | (A) | H-donor | 3.25 | –0.7 | –9.0351 |
| O | 9 | N | GLY | 143 | (A) | H-acceptor | 2.98 | –0.9 | ||
| O | 19 | OG | SER | 46 | (A) | H-acceptor | 2.81 | –0.5 | ||
| C | 33 | 5-ring | HIS | 41 | (A) | H-pi | 3.73 | –1.3 | ||
| 6-ring | CB | GLU | 166 | (A) | pi-H | 3.65 | –0.5 | |||
| 10 | O | 37 | O | THR | 190 | (A) | H-donor | 3.02 | –0.7 | –8.0876 |
| O | 18 | CA | ASN | 142 | (A) | H-acceptor | 3.32 | –0.6 | ||
| O | 18 | N | GLY | 143 | (A) | H-acceptor | 3.09 | –2.5 | ||
| 11 | N | 9 | SD | MET | 165 | (A) | H-donor | 3.69 | –0.6 | –7.3965 |
| O | 25 | N | GLY | 143 | (A) | H-acceptor | 3.04 | –0.8 | ||
| 6-ring | CG | GLN | 189 | (A) | pi-H | 3.59 | –0.7 | |||
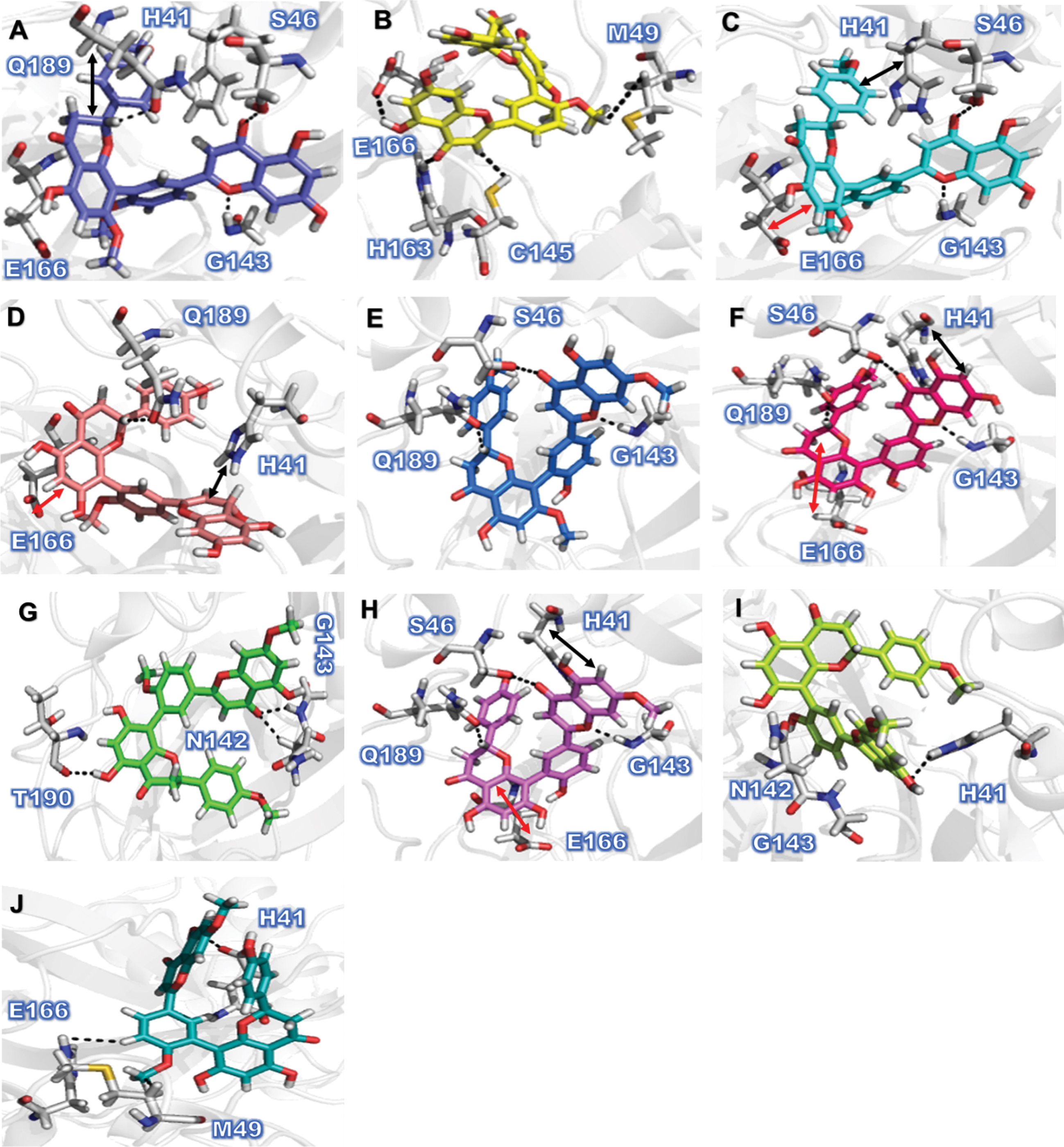
In the present study, inhibition potential of amentoflavon derivatives, ranked by docking score (S) were in the order of Amentoflavone C9 (–9.0351) > C6 (–8.8566), C4 (–8.8509) > C1 (–8.7746) > C8 (–8.6192) > C3 (–8.2537) > C10 (–8.0876) > C7 (–7.9501) > C2 (–7.6429), and C5 (–7.6248). The amentoflavon plant extract, which are potent inhibitor of COVID-19 Mpro, can inhibit multiple steps of the virus replication cycle. All the resulting compounds showed consistent protein-ligand interactions with the amino acid residues of the active site domains of Mpro. Analysis of the predicted binding conformations of our most potent compound 9 revealed that C9 fits straight into the binding cavity of Mpro (Fig. 3A). Visually inspecting the best binding position for C9 showed that it is able to form hydrogen bond interaction with backbone (O) of G143 (2.98 Å), sidechain (O) of S46 (2.81Å), and with (C) of Q189 (3.25Å), while forming H-pi bond with H41 (3.73 Å), and pi-H bond with E166 (3.65Å). Similarly, in case of C6, we noticed different interaction with the target protein, including 3 H-donor interaction with sidechain (O) of E166 (2.76Å), C145 (3.74Å), M49 (3.64Å), and H-acceptor with sidechain (O) of H163 (2.89Å) (Fig. 3B). Comparatively the molecular docking analysis demonstrate, that C4, C1, C8, C3, C7, and C5 amentoflavon derivatives exhibited consistent (PLI) profile with target protein (Mpro) as in case of C9 but not with all resultant residues as shown in (Fig. 3C-G). On the other hand it was also observed that C10 and C2 forming different pattern of PLI, such as C10 form strong H-donor interaction with sidechain (O) of T190 (3.02Å), and 2 H-acceptor with sidechain (O) of N142 (3.32Å), and G143 (3.09Å) (Fig. 3 G, 3I). The overall summary of the molecular docking analysis revealed that amentoflavone derivatives strongly bind with the catalytic residues of Mpro as compared to the FDA approved anti-viral drug. Hence, based on the results of these in-silico studies, further in-vitro and in-vivo studies must be planned to validate the effectiveness of these compounds as potential inhibitors of SARS-CoV–2 Mpro.
Fig. 3
Protein–ligand Interaction (PLI) profile of the amentoflavone derivatives against SARS-CoV–2 Mpro. Figure (1A-1J) Indicates PLI profile for the amentoflavone derivatives, C9 (3A), C6 (3B), C4 (3C), C1 (3D), C8 (3E), C3 (3F), C10 (3G), C7 (3H), C2 (3I), and C5 (3G). Double-sided arrows in black color represent theH-pi interaction, while red color double arrow represent pi-H interaction.
3.2Pharmacokinetic properties analysis
For the analysis and optimization of pharmacokinetic properties, the pkCSM and Swiss ADME approach confer a platform. Here, the molecular weight of The derivatives of amentoflavone such as 4′,4′′′-Dimethylamentoflavone, 4′′′,7-Di-O-Methylamentoflavone, 4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone, 7,4′-Dimethylamentoflavone,7-O-methylamentoflavone, Heveaflavone, kayaflavone, and Sciadopitysin are 566.518, 566.518, 552.491, 552.48, 566.51, 552.48, 552.491, 580.545, 580.545, 580.54 gm/mol, respectively. All the compounds have the same no. of Lipinski rule violation (1), have no AMES toxicity, and same bioavailability score (0.55).
Three amentoflavone derivatives (4′,4′′′-Dimethylamentoflavone, 4′′′,7-Di-O-Methylamentoflavone, and 7,4′-Dimethylamentoflavone) have the same topological polar surface area are 159.80 Å2. The other four derivatives of amentoflavone (4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone, 7-O-methylamentoflavone, and 7-O-methylamentoflavone) also contain the same topological polar surface area are 170.80 Å2. Besides this, Heveaflavone, kayaflavone, and Sciadopitysin have a topological polar surface area such as 148.80 Å2.
In pKCSM, some of the amentoflavone derivatives (4′,4′′′-Dimethylamentoflavone, 4′′′,7-Di-O-Methylamentoflavone, Heveaflavone, kayaflavone, and Sciadopitysin) have been shown hepatotoxicity, but some (7,4′-Dimethylamentoflavone, 4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone,7-O-methylamentoflavone, and 7-O-methylamentoflavone) don’t have any hepatotoxicity. Furthermore, oral rat acute toxicity (LD50) for all the derivatives of amentoflavone ranged between 2.535–3.06 and the maximum tolerated dose (MTD) for human ranged 0.295 to 0.437(log mg/kg/day). Additionally, the total clearance (TC) in the range between 0.571–0.833 (log ml/min/kg), and predicted octanol/water partition coefficient (LogP) were in the range of 5.3886 to 6.043 for all derivatives. Table 2 shows the pharmacokinetic properties of Amentoflavone derivatives predicted by Swiss ADME and pkCSM.
Table 2
Pharmacokinetic properties of Amentoflavone derivatives predicted by Swiss ADME and pkCSM
| Compounds | MW (g/mol) | H-Ac | H-Do | N.rot | TPSA (Å2) | LogP | B.S | LD50 | HT | AT | MTD (log | NLV | TC (log |
| mg/kg/day) | ml/min/kg) | ||||||||||||
| 4′,4′′′-Dimethylamentoflavone | 566.51 | 10 | 4 | 5 | 159.80 | 5.74 | 0.55 | 2.723 | Yes | No | 0.43 | 1 | 0.710 |
| 4′′′,7-Di-O-Methylamentoflavone | 566.51 | 10 | 4 | 5 | 159.80 | 5.74 | 0.55 | 2.854 | Yes | No | 0.423 | 1 | 0.723 |
| 4′′′′′′-methylamentoflavone | 552.49 | 10 | 5 | 4 | 170.80 | 5.3886 | 0.55 | 2.96 | No | No | 0.295 | 1 | 0.619 |
| 4′-Monomethylamentoflavone | 552.48 | 10 | 5 | 4 | 170.80 | 5.437 | 0.55 | 2.56 | No | No | 0.437 | 1 | 0.571 |
| 7,4′-Dimethylamentoflavone | 566.51 | 10 | 4 | 5 | 159.80 | 5.74 | 0.55 | 2.733 | No | No | 0.427 | 1 | 0.646 |
| 7′-O-Methylamentoflavone | 552.48 | 10 | 5 | 4 | 170.80 | 5.437 | 0.55 | 2.548 | No | No | 0.436 | 1 | 0.617 |
| 7-O-methylamentoflavone | 552.49 | 10 | 5 | 4 | 170.80 | 5.437 | 0.55 | 2.535 | No | No | 0.437 | 1 | 0.488 |
| Heveaflavone | 580.54 | 10 | 3 | 6 | 148.80 | 6.043 | 0.55 | 2.997 | Yes | No | 0.412 | 1 | 0.791 |
| Kayaflavone | 580.54 | 10 | 3 | 6 | 148.80 | 6.043 | 0.55 | 2.84 | Yes | No | 0.419 | 1 | 0.794 |
| Sciadopitysin | 580.54 | 10 | 3 | 6 | 148.80 | 6.043 | 0.55 | 3.06 | Yes | No | 0.419 | 1 | 0.833 |
MW: molecular weight; HT: hepatotoxicity; H-Ac: number of hydrogen bond acceptor; AT: AMES toxicity; H-Do: number of hydrogen bond donors; N.rot: numberof rotatable bonds; B.S: Bioavailability Score; LD50: oral rat acute toxicity; MTD: the maximum tolerated dose for human; NLV: number of Lipinski rule violation;LogP: predicted octanol/water partition coefficient; TC, Total clearance;TPSA: topological polar surface area (Å2).
4Discussion
COVID-19 is now a pandemic [41]. Scientists worldwide are working hard to discover and design novel agents having inhibitory potentials. Many anti-viral drugs like Camostat mesylate, Ritonavir, Remdesivir, Lopinavir, and Indinavir, are being studied in clinical trials to validate their anti-Coronavirus effect [42]. Azithromycin and hydroxychloroquine are used to cure affected patients in case of emergency and are published as potential SARS-CoV-2inhibitors [43–45]. All the above stated drugs may be SARS-CoV-2inhibitors but side-effects associated with their use have also been shared. Therefore, it is need of the time to search and discover alternative anti-SARS-CoV-2 agents having no or less side effects and significant inhibitory potential.
Keeping in view the current pandemic situation and unavailability of effective anti-SARS-CoV-2 agents that are safe, natural compounds present in plants and animals and their associated derivatives must be studied in different in-silico, in-vitro and in-vivo studies to find potent lead compound in combating nCoV-19 infections [46]. The derivatives of amentoflavone (biflavonoid) were isolated by Okigawa et al. [9] and have acquired attention of scientists owing to biological activities associated with it.
For preparing an ideal anti-COVID-19 drug, a compound must have four basic criteria: (i) restricting the entry of virus by inhibition of cellular attachment; (ii) inhibiting replication of virus within the host cells; (iii) cytotoxic potential on the prevailing virus; and (iv) protecting the host normal cells against viral originated inflammatory responses and oxidative stress. Evidently, amentoflavone possess antiviral potential via all the above-mentioned pathways. Moreover, it has antioxidant [47, 48], and anti-inflammatory [11, 49, 50], activities. Besides, the derivative of amentoflavone also has antioxidant, and anti-inflammatory properties [51]. The MPRO should have the ability to break host polyproteins and induce the formation of protein for viral replication [52]. Amentoflavone has interaction capability with MPRO [22]. So, it should possible, derivatives of amentoflavone can bind with MPRO. Therefore, derivatives of amentoflavone can inhibit viral infection.
Derivatives of amentoflavone such as 4′,4′′′-Dimethylamentoflavone, 4′′′,7-Di-O-Methylament-oflavone, 4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone, 7,4′-Dimethylamentoflavone, 7′-O-Methylamentoflavone, 7-O-methylamentoflavone, Heveaflavone, kayaflavone, and Sciadopitysin displayed a greater docking score and strong binding interaction with COVID-19 main protease (MPRO) –8.7746, –7.6429, –8.2537, –8.8509, –7.6248, –8.8566, –7.9501, –8.6192, –9.0351, and –8.0876 respectively, as compare to Ritonavir, Lopinavir, and Remdesivir. Furthermore, some flavonoids and polyphenolic plant-derived compounds such as kaempferol, quercetin, demethoxycurcumin, curcumin, catechin, epicatechingallate, and gingerol are investigated to have potent inhibitory properties against main protease of SARS-CoV-2 [8], which claimed that the derivative of amentoflavone has better properties than other proposed inhibitors. Many standard drugs such as Ritonavir, Lopinavir, and Remdesivir displayed a binding affinity with MPRO protein, and derivatives of amentoflavone showed good to moderate binding affinities with MPRO protein through so many same active sites (Table 1).
From the pharmacokinetic properties (predicted) of the selected compounds by Swiss ADME and pkCSM (Table 2), it has been seen that amentoflavone derivatives are safe and may be used as a potent anti-COVID-19 drug as predicted by the website. More research is required concerning the toxicogenic studies of natural metabolites in plant based and/or animal models. Even tough further in-vivo instigations are required authenticate the findings of current study, nevertheless results of our study will provide a baseline information for planning further studies (pre-clinical & clinical) on these compounds. Conclusively, outcomes of this study may inspire researchers in the field of drug design to perform in depth studies on these potent natural compounds.
5Conclusions
4′,4′′′-Dimethylamentoflavone, 4′′′,7-Di-O-Methylamentoflavone, 4′′′′′′-methylamentoflavone, 4′-Monomethylamentoflavone, 7,4′-Dimethylamentoflavone, 7′-O-Methylamentoflavone, 7-O-methyla-mentoflavone, Heveaflavone, kayaflavone, and Sciadopitysin are the derivatives of amentoflavone (biflavonoid) those have antiviral activity in several viruses. Derivatives of amentoflavone does have a good interaction with target protein, according to this molecular docking. The docking socresshown by amentoflavone derivatives were also higher than those found in clinical trial antiviral drugs (Camostatmesylate), which are widely used in several countries to treat COVID-19. Aside from that, pharmacokinetics studies reveal positive results when compared to Camostatmesylate. Amentoflavone derivatives can be envisioned as possible lead compounds against SARS-CoV-2 infection based on the findings of this report. While further in vivo testing is needed to confirm the results presented here, our findings will aid future nonclinical, preclinical, and clinical studies with these compounds, while also inspiring medicinal chemistry scientists to perform appropriate study on this promising natural lead compound and its derivatives.
Data Availability
The data such as source file associated to docking study are available from corresponding author upon request.
Conflicts of interest
Authors declare no potential conflict of interest.
Authors Contributions
Rajib Hossain, Shafi Mahmud, Abul Bashar Ripon Khalipha, Abu Saim Mohammad Saikat, involve in Conceptualization, Dipta Dey, Rasel Ahmed Khan, Abdur Rauf, Abdul Wadood involved in validation, investigation while Humaria Rafique, Sami Bawazeer, Anees Ahmed Khalil, Zainab M. Almarhoon, Yahia N. Mabkhot, Muhammad Torequl Islam and Haroon Khan involve in validation, investigation, data curation and manuscript writing. All authors read and approved the final version of the manuscript.
Supporting information
All data related to this paper is included in the text.
Acknowledgments
We are very much thankful to the International Centre for Empirical Research and Development (ICERD), Bangladesh. The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: 22UQU4350073DSR16. We also thanked Dr. Ajmal, University of Nizwa, Oman for providing technical support for the in-silico study.
Supplementary material
[1] The Supplementary is available in the electronic version of this article: https://dx.doi.org/10.3233/MGC-220077.
References
[1] | Elfiky A.A. , Mahdy S.M. , Elshemey W.M. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses,pp, Journal of Medical Virology 89: (6) ((2017) ), pp. 1040–1047. |
[2] | Chan J.F. , Lau S.K. , To K.K. , Cheng V.C. , Woo P.C. , Yue K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease, Clinical Microbiology Reviews 28: (2) ((2015) ), 465–522. |
[3] | Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 -nCoV, COVID-19) coronavirus, Computers in Biology and Medicine 119: ((2020) ), 1–9.https://doi.org/10.1016/j.compbiomed.2020.103670. |
[4] | Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design, The FEBS Journal 281: (18) ((2014) ), 4085–4096. |
[5] | Ibrahim I.M. , Abdelmalek D.H. , Elfiky A.A. GRP78: A cell’s response to stress, Life Sciences 226: ((2019) ), 156–163. |
[6] | Huang C. , Wang Y. , Li X. et al. Clinical features of patients infected with novel coronavirus in Wuhan, China, The lancet 395: (10223) ((2020) ), 497–506. |
[7] | Gautret P. , Lagier J.C. , Parola P. et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, International Journal of Antimicrobial Agents 56: (1) ((2020) ), 105949. |
[8] | Khaerunnisa S. , Kurniawan H. , Awaluddin R. , Suhartati S. Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study, Preprints, pp. 1–14, 2020, https://doi.org/10.20944/preprints202003.0226.v1. |
[9] | Okigawa M. , Hwa C.W. , Kawano N. , Rahman W. Biflavones in Selaginella species, Phytochemistry 10: (12) ((1971) ), 3286–3287. |
[10] | Yu S. , Yan H. , Zhang L. , Shan M. , Chen P. , Ding A. , Li S.F.Y. A review on the phytochemistry, pharmacology, and pharmacokinetics of amentoflavone, a naturally-occurring biflavonoid, Molecules 22: (2) ((2017) ), pp. 299. |
[11] | Abdallah H.M. , Almowallad F.M. , Esmat A. , Shehata I.A. , Abdel-Sattar E.A. Anti-inflammatory activity of flavonoids from Chrozophora tinctoria, Phytochemistry Letters 13: ((2015) ), 74–80. |
[12] | Ndongo J.T. , Issa M.E. , Messi A.N. , Ngo Mbing, J. , Cuendet, M. , Pegnyemb, D.E. , Bochet, C.G. Cytotoxic flavonoids and other constituents from the stem bark of Ochna schweinfurthiana, Natural Product Research 29: (17) ((2015) ), 1684–1687. |
[13] | Park N.H. , Lee C.W. , Bae J.H. , Na Y.J. Protective effects of amentoflavone on Lamin A-dependent UVB-induced nuclear aberration in normal human fibroblasts, Bioorganic & Medicinal Chemistry Letters 21: (21) ((2011) ), pp. 6482–6484. |
[14] | Shen X. , Niu X. , Li G. , Deng X. , Wang J. Amentoflavone ameliorates Streptococcus suis-induced infection infection in vitro and in vivo, , Applied and Environmental Microbiology 84: (24) ((2018) ), e01804–18. |
[15] | Liu S. , Yang X. , Zhang H. , Zhang J. , Zhou Y. , Wang T. , Hu N. , Deng X. , Bai X. , Wang J. Amentoflavone attenuates Clostridium perfringens gas gangrene by targeting alpha-toxin and perfringolysin O, Frontiers in Pharmacology 11: ((2020) ), pp. 179. |
[16] | Sok Hwang, I. , Lee, J. , Jin, H.G. , Woo, E.R. , Lee, D.G. Amentoflavone stimulates mitochondrial dysfunction and induces apoptotic cell death in Candida albicans, Mycopathologia 173: (4) ((2012) ), 207–218. |
[17] | Coulerie P. , Nour M. , Maciuk A. et al. Structure-activity relationship study of biflavonoids on the Dengue virus polymerase DENV-NS5 RdRp, Planta Medica 79: (14) ((2013) ), pp. 1313–1318. |
[18] | Rong S. , Wan D. , Fan Y. et al. Amentoflavone affects epileptogenesis and exerts neuroprotective effects by inhibiting NLRP3 inflammasome, Frontiers in Pharmacology 10: ((2019) ), pp. 856. |
[19] | Zheng X. , Liu C. , Zhai Y. , Li L. , Wang X. , Feng W. Protection effect of amentoflavone in Selaginella tamariscina against TNF-alpha-induced vascular injury of endothelial cells, Yao xue xue bao = Acta pharmaceutica Sinica 48: (9) ((2013) ), 1503–1509. |
[20] | Su C. , Yang C. , Gong M. , Ke Y. , Yuan P. , Wang X. , Li M. , Zheng X. , Feng W. Antidiabetic activity and potential mechanism of amentoflavone in diabetic mice, Molecules 24: (11) ((2019) ), 2184. |
[21] | Wilsky S. , Sobotta K. , Wiesener N. , Pilas J. , Althof N. , Munder T. , Wutzler P. , Henke A. Inhibition of fatty acid synthase by amentoflavone reduces coxsackievirus B3 replication, Archives of Virology 157: (2) ((2012) ), 259–269. |
[22] | Lin, Y.M. , Anderson, H. , Flavin, M.T. , Pai, Y.H.S. , Mata-Greenwood, E. , Pengsuparp, T. , Pezzuto, J.M. , Schinazi, R.F. , Hughes, S.H. , Chen, F.C. In vitro anti-HIV activity of biflavonoids isolated from Rhus succedanea and Garcinia multiflora, Journal of Natural Products 60: (9) ((1997) ), 884–888. |
[23] | Ma S.C. , But P.P.H. , Ooi V.E.C. , He Y.H. , Lee S.H.S. , Lee S.F. , Lin R.C. Antiviral amentoflavone from Selaginella sinensis, Biological and Pharmaceutical Bulletin 24: (3) ((2001) ), 311–312. |
[24] | Li F. , Song X. , Su G. et al. Amentoflavone inhibits HSV-1 and ACV-resistant strain infection by suppressing viral early infection, Viruses 11: (5) ((2019) ), 466. |
[25] | Ogunwa T. , Ayenitaju F. An insight into the precise molecular interaction and inhibitory potential of amentoflavone and its substituted derivatives on human α-amylase, Archives of Current Research International 10: (1) ((2017) ), 1–14. |
[26] | Ryu Y.B. , Jeong H.J. , Kim J.H. et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition, Bioorganic & Medicinal Chemistry 18: (22) ((2010) ), 7940–7947. |
[27] | Berry M. , Fielding B.C. , Gamieldien J. Potential broad spectrum inhibitors of the coronavirus 3CLpro: A virtual screening and structure-based drug design study, Viruses 7: (12) ((2015) ), 6642–6660. |
[28] | Wu K. , Chen L. , Peng G. , Zhou W. , Pennell C.A. , Mansky L.M. , Geraghty R.J. , Li F. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses, Journal of Virology 85: (11) ((2011) ), 5331–5337. |
[29] | Gagneur A. , Sizun J. , Vallet S. , Legrand M.C. , Picard B. , Talbot P.J. Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study, Journal of Hospital Infection 51: (1) ((2002) ), 59–64. |
[30] | Van Der Hoek, L. , Pyrc, K. , Jebbink, M.F. et al. Identification of a new human coronavirus, Nature Medicine 10: (4) ((2004) ), 368–373. |
[31] | Siddell S.G. , Ziebuhr J. , Snijder E.J. Coranaviruses, Toroviruses and Arteriviruses, In Topley and Wilson’s micro biology and microbial infections (2005), pp. 823–856. Edward Arnold, 2005. |
[32] | Woo P.C. , Lau S.K. , Chu C. et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia, Journal of Virology 79: (2) ((2005) ), 884–895. |
[33] | Burley S.K. , Berman H.M. , Kleywegt G.J. , Markley J.L. , Nakamura H. , Velankar S. Protein Data Bank (PDB): the single global macromolecular structure archive, Protein Crystallography (2017), pp. 627–641. |
[34] | Rahman A. , Ali M.T. , Shawan M.M.A.K. , Sarwar M.G. , Khan M.A.K. , Halim M.A. Halogen-directed drug design for Alzheimer’s disease: A combined density functional and molecular docking study, SpringerPlus 5: (1) ((2016) ), 1–14. |
[35] | Ccgi M. Molecular operating environment (MOE), 2013.08. Chemical Computing Group Inc., Montreal (2016). |
[36] | Jin Z. , Du X. , Xu Y. et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors, Nature 582: (7811) ((2020) ), 289–293. |
[37] | Daina A. , Michielin O. , Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Scientific Reports 7: (1) ((2017) ), 1–13. |
[38] | Pires D.E. , Blundell T.L. , Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures, Journal of Medicinal Chemistry 58: (9) ((2015) ), 4066–4072. |
[39] | Azzam K.A. SwissADME and pkCSM Webservers Predictors: an integrated Online Platform for Accurate and Comprehensive Predictions for In Silico ADME/T Properties of Artemisinin and its Derivatives, Kompleksnoe Ispolzovanie Mineralnogo Syra 325: (2) ((2023) ), pp. 14–21. |
[40] | Bakchi B. , Krishna A.D. , Sreecharan E. , Ganesh V.B.J. , Niharika M. , Maharshi S. , Puttagunta S.B. , Sigalapalli D.K. , Bhandare R.R. , Shaik A.B. An Overview on Applications of SwissADME Web Tool in the Design and Development of Anticancer, Antitubercular and Antimicrobial agents: A Medicinal Chemist’s Perspective, Journal of Molecular Structure 1259: ((2022) ), pp. 132712. |
[41] | Zhou S. , Wang Y. , Zhu T. , Xia L. CT features of coronavirus disease (COVID-19) pneumonia in 62 patients in Wuhan, China, American Journal of Roentgenology 214: (6) ((2020) ), pp. 1287–1294. |
[42] | Babadaei M.M.N. , Hasan A. , Vahdani Y. , Bloukh S.H. , Sharifi M. , Kachooei E. , Haghighat S. , Falahati M. Development of remdesivir repositioning as a nucleotide analog against COVID-19 RNA dependent RNA polymerase, Journal of Biomolecular Structure and Dynamics (2020), pp. 1–9. |
[43] | Wang M. , Cao R. , Zhang L. , Yang X. , Liu J. , Xu M. , Shi Z. , Hu Z. , Zhong W. , Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus -nCoV) in vitro,, Cell Research 30: (3) ((2020) ), 269–271. |
[44] | Vincent M.J. , Bergeron E. , Benjannet S. , Erickson B.R. , Rollin P.E. , Ksiazek T.G. , Seidah N.G. , Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread, Virology Journal 2: (1) ((2005) ), 1–10. |
[45] | Heald-Sargent T. , Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence, Viruses 4: (4) ((2012) ), pp. 557–580. |
[46] | Jassim S.A.A. , Naji M.A. Novel antiviral agents: a medicinal plant perspective, Journal of Applied Microbiology 95: (3) ((2003) ), 412–427. |
[47] | Arwa P.S. , Zeraik M.L. , Farias Ximenes, V. , Da Fonseca, L.M. , Da Silva Bolzani V. , Siqueira Silva D.H. Redox-active biflavonoids from Garcinia brasiliensis as inhibitors of neutrophil oxidative burst and human erythrocyte membrane damage, Journal of Ethnopharmacology 174: ((2015) ), 410–418. |
[48] | Li X. , Wang L. , Han W. , Mai W. , Han L. Amentoflavone protects against hydroxyl radical-induced DNA damage via antioxidant mechanism, Turkish Journal of Biochemistry/Turk Biyokimya Dergisi 39: (1) ((2014) ), 30–36. |
[49] | Ishola I.O. , Chaturvedi J.P. , Rai S. , Rajasekar N. , Adeyemi O.O. , Shukla R. , Narender T. Evaluation of amentoflavone isolated from Cnestis ferruginea Vahl ex DC (Connaraceae) on production of inflammatory mediators in LPS stimulated rat astrocytoma cell line (C6) and THP-1 cells, Journal of Ethnopharmacology 146: (2) ((2013) ), 440–448. |
[50] | Oh J. , Rho H.S. , Yang Y. et al. Extracellular signal-regulated kinase is a direct target of the anti-inflammatory compound amentoflavone derived from Torreya nucifera, Mediators of Inflammation 2013: ((2013) ), pp. 1–11. |
[51] | Buonano M. , Attías D.G. de and G.A. Ávila de, Protiumneglectum, Podocalyx loranthoides, and Brosimum utile source offlavonoids and other phenolic compounds with antioxidant activity, Rev Fac Farm 71: ((2008) ), 8–14. |
[52] | Wu C. , Liu Y. , Yang Y. et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods, Acta Pharmaceutica Sinica B 10: (5) ((2020) ), pp. 766–788. |




