Validation of the Correlation Between Single Nucleotide Polymorphism rs307826 in VEGFR3 and Outcome in Metastatic Clear-Cell Renal Cell Carcinoma Patients Treated with Sunitinib
Abstract
BACKGROUND:
Previously, we have shown a correlation between single nucleotide polymorphism (SNP) rs307826 in vascular endothelial growth factor receptor-3 (VEGFR3) and outcome in metastatic clear-cell renal cell carcinoma (m-ccRCC) patients treated with sunitinib.
OBJECTIVE:
We aimed to validate this finding in an independent patient series.
METHODS:
m-ccRCC patients receiving sunitinib as first-line targeted therapy were included in a validation cohort. Endpoints were response rate (RR), progression-free survival (PFS) and overall survival (OS). We also updated survival data of our discovery cohort as described previously.
RESULTS:
Eighty-four patients were included in the validation cohort. rs307826 AG/GG-carriers had a shorter PFS (8 versus 12 months, p = 0.04) and a trend towards a shorter OS (18 versus 27 months, p = 0.22) compared to AA-carriers. In the total series of 168 patients (from the discovery cohort, as described previously, and the validation cohort), rs307826 AG/GG-carriers had a poorer RR (29% versus 53%, p = 0.008), PFS (8 versus 15 months, p = 0.0002) and OS (22 versus 31 months, p = 0.004) compared to AA-carriers. rs307826 was independently associated with PFS and OS on multivariate analysis.
CONCLUSION:
VEGFR3 rs307826 seems to be associated with outcome on sunitinib in m-ccRCC. Its impact highlights the role of VEGFR3 in ccRCC pathogenesis and as a target of sunitinib.
INTRODUCTION
Inactivation of the von Hippel–Lindau (VHL) tumor suppressor gene is the most frequent molecular alteration in clear-cell renal cell carcinoma (ccRCC). Inactivated VHL leads to elevated protein levels of hypoxia-induced factor-α which up upregulates the vascular endothelial growth factor (VEGF) dependent pro-angiogenesis pathway. Targeted therapies directed against circulating VEGF or against the VEGF-receptors (VEGFR)-1, -2, and -3 have significantly improved the perspectives of patients with metastatic ccRCC (m-ccRCC). Sunitinib is an orally administered tyrosine kinase receptor inhibitor (TKI) that targets VEGFR1, VEGFR2 and VEGFR3. In a randomized controlled trial sunitinib significantly prolonged progression-free survival (PFS) (11 versus 5 months, p < 0.001) as compared to interferon alpha [1, 2]. Median overall survival (OS) was respectively 26.4 and 21.8 months (p = 0.051). Sunitinib is a standard treatment option in ccRCC, but other VEGFR-TKIs like sorafenib, pazopanib, cabozantinib, tivozanib and axitinib are also used in different stages of the disease. Only recently, sunitinib was superseded by immune checkpoint inhibitors (ICPI) [3, 4] or combinations of axitinib and ICPIs [5, 6] in first-line treatment of m-ccRCC.
Although around 40% of RCC patients receiving sunitinib experience an objective response and 45% achieve disease stabilization, 15% will experience progressive disease (PD) at first evaluation probably due to intrinsic resistance or due to other factors. Moreover, even patients with an initial clinical benefit will finally progress due to acquired resistance or for other reasons. The identification of biomarkers able to predict intrinsic resistance could avoid unnecessary costs and side effects, guiding alternative treatment decisions. On the other hand, the identification of biomarkers for acquired resistance could provide novel directions to develop therapies that block these resistance pathways. Several studies have shown that VEGFR-TKIs are the most efficient in m-ccRCCs with activated VEGF-dependent angiogenesis [7, 8]. Sarcomatoid dedifferentiated ccRCCs, which have less angiogenesis, are resistant to VEGFR-TKIs [9]. We have also proposed single nucleotide polymorphism (SNP) rs9582036 in VEGFR1 as a biomarker predicting outcome in m-ccRCC treated with sunitinib, although this finding still needs further validation [10]. Recently, two independent research groups have shown a correlation between SNP rs307826 in VEGFR3 and outcome on sunitinib in m-ccRCC. rs307826 in VEGFR3 is an adenosine (A) >guanine (G) change that leads to a T494A amino acid substitution in VEGFR3. In Caucasians, the minor allele (G) frequency is around 10% and around 20% of the population carries the AG/GG-genotype. On a series of 88 m-ccRCC patients, we showed a better PFS (20 versus 9 months; p = 0.022) and a better OS (34 versus 22 months; p = 0.0058) in AA-carriers compared to AG/GG-carriers. AG/GG-genotypes were frequent (60%) in patients with early PD as best response and rare (22%) in patients with partial response (PR) (p = 0.02) [11]. Similarly, on a series of 89 m-ccRCC patients treated with sunitinib, a Spanish research group showed that the rs307826 genotype AG/GG was correlated with reduced PFS [12].
The aim of the present study was to validate the association of rs307826 in VEGFR3 to first-line sunitinib outcome in an independent cohort of m-ccRCC patients.
MATERIALS AND METHODS
For this retrospective study, germ-line DNA samples were collected in the CIT-rein kidney tumor bank (frozen normal kidney tissue), in patients treated at the University Hospitals Leuven (peripheral blood samples) and in patients included in the Belgian multicentric METASUN trial (peripheral blood samples, NCT02570789). The French-Belgian multicentric CIT-rein kidney tumor bank contains more than 250 frozen kidney tumor samples collected at 20 academic or teaching hospitals. We selected patients with pathologically confirmed ccRCC treated in first-line with sunitinib. Eligible patients could have received cytokines (Interferon-alpha) as systemic treatment for kidney tumors before starting sunitinib, but they could not have received ICPIs. To make sure that the effect of the first-line VEGFR-TKI was accurately measured, patients had to take sunitinib during at least one complete cycle of 28 days and had to reach at least the first evaluation by CT scan. All the patients were treated in routine clinical practice. Drug schedule, dose-reduction policy and timing of radiological assessments were left to the discretion of the attending doctors in accordance with contemporary local practice guidelines. The vast majority of patients started sunitinib therapy at the standard dose. In some cases, sunitinib was started at reduced dose. In our database, we also selected all the m-ccRCC patients treated with nivolumab in consecutive therapy lines after a previous VEGFR-TKI at the University Hospitals Leuven. Nivolumab was administered intravenously every two weeks at 3 mg/kg or every two weeks at a flat dose of 240 mg or every four weeks at a flat dose of 480 mg. Response evaluation was done by CT scan every twelve weeks. The protocol was approved by the medical ethics review boards of all participating institutions, and signed consent was obtained from all patients. In some cases, we used frozen biologic material from patients who had already died and for whom a general positive advice for the utilization of remaining tissue was given by the institutional board.
DNA was isolated at INSERM UMR1138 in Paris, France, from fresh frozen normal kidney tissue sampled in the nephrectomy specimen using the Qiaquick extraction kit (Qiagen, Valencia, CA, USA) and quantified by fluorometry (Fluoroskan Thermo Lab systems, Cergy-Pontoise, France). DNA was isolated from peripheral blood at the Vesalius Research Center in Leuven with the Qiagen DNA kit (Qiagen, Valencia, CA, USA) and final DNA concentration quantified with Nanodrop (Nanodrop, Wilmington, USA). High-throughput SNP genotyping was performed at the Vesalius Research Center in Leuven, Belgium, using the Sequenom Mass Array platform [13]. Genotyping was performed by investigators blinded for the clinical data.
mRNA was isolated from corresponding Formalin-Fixed Paraffin-Embedded (FFPE) primary kidney tumors at INSERM UMR1138 in Paris or at VIB in Leuven. The tissue block with the highest tumoral content was selected for further processing. After trimming, we cut seven slices of 5μm from every block. The first and last slide were H & E stained and the tumoral section was delignated after microscopic review. We macrodissected blanco slides to include only tumor tissue, using a total surface area of 50–1800 mm² with 5μm thickness. As both primary and metastatic ccRCC tend to grow as enlarging, often encapsulated lesions that push aside surrounding tissue instead of infiltrating it, contamination with surrounding normal tissue was no concern. We extracted RNA with the Maxwell® RSC RNA FFPE kit (Promega) according to the manufacturer’s instructions. We prepared cDNA libraries using the Forward QuantSeq 3’ mRNA-Seq Library Prep Kit for Illumina (Lexogen) according to the manufacturer’s instructions for FFPE tissue. Of note, we used 5μl of RNA as input, incubated the samples for 60 minutes during the “first strand cDNA synthesis” step and performed 16 PCR cycles. cDNA concentrations and fragment length were measured with the QubitTMdsDNA HS assay (Thermofisher) and Bioanalyzer HS DNA electrophoresis (Agilent). We used Illumina cBOT for clonal cluster generation and performed RNAseq using the Illumina HiSeq 4000 kit according to the manufacturer’s instructions. The reads were trimmed to remove adaptor sequences and low-quality regions following Lexogen’s recommendations. Next, the reads were mapped to the human genome (hg19) using HIsat2 (v2.1.0) and quantified using featureCounts (v1.6.4). Raw reads were processed using DESeq2 (v 1.26.0) and normalized using the VarianceStabilizingTransformation (vst function).
When fresh frozen samples were available, the primary kidney tumors were also classified into the molecular ccrcc1–4 classification as described previously [14]. Ccrcc2-tumors have a favorable prognosis and respond better on sunitinib [14] or pazopanib [15]. Ccrcc1- and ccrcc4-tumors are a more aggressive subtype of ccRCC: they have an intermediate and poor prognosis, respectively, and their outcome on sunitinib and pazopanib is usually poor. Copy number loss and gain was also available in fresh frozen samples: we checked copy number loss and gain at chromosome 5q35, which hosts VEGFR3, using the methodology described in our previous publication [14].
Study endpoints were response rate (RR), PFS and OS. We defined PFS as the time between the first day on treatment and the date of radiological progressive disease or death. Patients who had not progressed at database closure were censored at last follow-up. OS was defined as the time between the first day on treatment and the date of death or last date of follow-up. Objective response was assessed by treating doctors by RECIST and classified as complete response (CR), PR, stable disease (SD), or PD. Time between initial diagnosis and development of metachronous metastases as well as median PFS (mPFS) and median OS (mOS) were estimated by the Kaplan-Meier survival analysis. For the multivariate analysis, we collected data on patient characteristics usually associated with mPFS and mOS, such as the 6 risk factors included in the International Metastatic Renal Cell Carcinoma Database Consortium (IMDC) prognostic score and presence or absence of bone metastases [16]. Clinical data were collected at 19 different sites in France and Belgium. Fisher exact, ANOVA or Chi-square tests were used to compare proportions. Results with a two-sided p-value of < 0.05 were considered as significant in the multivariate analysis. Statistical analyses were conducted using GraphPad Prism 5 (GraphPad Software, La Jolla, UCLA) and XLSTAT software (Addinsoft, Paris, France).
RESULTS
We included 168 patients treated with sunitinib in first-line, with a global mPFS of 14 months and a global mOS of 30 months. At database closure (November 2019), 76% of these patients had reached progression and 70% had died. Patient characteristics are reported in Table 1. The majority of patients (>98%) were of Caucasian origin.
Table 1
Patient characteristics at diagnosis and at the start of sunitinib treatment and baseline clinical and biochemical parameters associated with PFS AND OS
| All (n = 168) | AA (n = 126) | AG/GG (n = 42) | p | ||
| At initial diagnosis | |||||
| Gender | Male | 115/168 (68.5%) | 89/126 (70.6%) | 26/42 (61.9%) | 0.34 |
| M1 (synchronous metastases) | 90/164 (54.9%) | 66/123 (53.7%) | 24/41 (58.5%) | 0.72 | |
| Median metastasis-free-survival in patients | 16 months | 16 months | 13 months | 0.36 (*) | |
| with metachronous metastases | |||||
| Fuhrman | Grade 1–3 | 75/163 (46.0%) | 58/122 (47.5%) | 17/41 (41.5%) | 0.59 |
| Grade 4 | 88/163 (54.0%) | 64/122 (52.5%) | 24/41 (58.5%) | ||
| Sarcomatoid dedifferentiation | 0% | 113/151 (74.8%) | 84/112 (75.0%) | 29/39 (74.4%) | 0.35 (*) |
| (compared to tumor volume) | 1–24% | 32/151 (21.2%) | 25/112 (22.3%) | 7/39 (17.9%) | |
| 25% or more | 6/151 (4.0%) | 3/112 (2.7%) | 3/39 (7.7%) | ||
| Molecular ccrcc1–4 classification | ccrcc1 | 30/87 (34%) | 20/62 (32%) | 10/25 (40%) | 0.07(**) |
| ccrcc2 | 41/87 (47%) | 34/62 (55%) | 7/25 (28%) | ||
| ccrcc3 | 3/87 (3%) | 2/62 (3%) | 1/25 (4%) | ||
| ccrcc4 | 13/87 (15%) | 6/62 (10%) | 7/25 (28%) | ||
| At the start of systemic therapy | |||||
| Karnofsky<80 | 28/167 (16.8%) | 18/126 (14.3%) | 10/41 (24.4%) | 0.15 | |
| Neutrophils>7.800/mm3 | 15/165 (9.0%) | 8/123 (6.5%) | 7/42 (16.7%) | 0.06 | |
| Platelets>450.000/mm3 | 21/166 (12.7%) | 15/125 (12.0%) | 6/41 (14.6%) | 0.79 | |
| Hemoglobin low (<12 g/dl (women) | 88/167 (52.7%) | 67/125 (53.6%) | 21/42 (50.0%) | 0.72 | |
| or <14 g/dl (men)) | |||||
| LDH>1.5*ULN | 9/162 (5.6%) | 6/124 (4.8%) | 3/38 (7.9%) | 0.44 | |
| Corrected Calcium >10.2 mg/dl | 12/122 (9.8%) | 9/95 (9.5%) | 3/27 (11.1%) | 0.73 | |
| Time from nephrectomy to systemic | 107/167 (64.1%) | 82/126 (65.1%) | 25/41 (60.1%) | 0.71 | |
| treatment <12 months | |||||
| Cytokines before sunitinib/pazopanib | 26/167 (15.6%) | 17/125 (13.6%) | 9/42 (21.4%) | 0.23 | |
| Site of metastasis | Lung | 122/168 (72.6%) | 90/126 (71.4%) | 32/42 (76.2%) | 0.69 |
| Adenopathies | 79/168 (47.0%) | 63/126 (50.0%) | 16/42 (38.1%) | 0.21 | |
| Liver metastases | 34/168 (20.2%) | 20/126 (15.9%) | 14/42 (33.3%) | 0.03 | |
| Bone metastases | 60/168 (35.7%) | 45/126 (35.7%) | 15/42 (35.7%) | >0.99 | |
| Brain metastases | 13/168 (7.7%) | 10/126 (7.9%) | 3/42 (7.1%) | >0.99 | |
| IMDC | Favorable | 26/162 (16.0%) | 20/123 (16.3%) | 6/39 (15.4%) | 0.35 (**) |
| Intermediate | 98/162 (60.5%) | 75/123 (60.9%) | 23/39 (59.0%) | ||
| Poor | 38/162 (23.5%) | 28/123 (22.8%) | 10/39 (25.6%) | ||
Note: The p-value compares AA and AG/GG-carriers by Fisher exact or (*) Kaplan-Meier estimates, or (**) Chi-square. ULN: upper limit of normal. IMDC: International Metastatic Renal Cell Carcinoma Database Consortium.
Among these 168 patients, 126 (75%) were wild type/wild type (rs307826 AA), 36 (21%) wild type/variant (rs307826 AG) and 6 (4%) variant/variant (rs307826 GG). The observed minor allele frequency was 14.2%, which corresponds to the minor allele frequency for Caucasians in dbSNP (10.2%). The patient characteristics, including IMDC risk groups, were similar between AA- and AG/GG-carriers. Disease evolution between diagnosis and start of systemic therapy was also similar: the proportion of patients presenting with synchronous metastases as well as time between initial diagnosis and development of metastases in patients with metachronous metastases, was similar in AA- and AG/GG-carriers. In 87 patients, fresh frozen samples were available and the primary kidney tumors were classified into the molecular expression-based ccrcc1–4 classification. There was a trend to a higher prevalence of the AA-genotype in the more indolent ccrcc2 subtype and a higher prevalence of the AG/GG-genotype in the more aggressive ccrcc1- and ccrcc4-subtype (p = 0.07).
The patients were divided into a discovery and a validation cohort. The discovery cohort was composed of 84 patients that were included in our previous study. Four of the 88 original patients were excluded, because central pathology review classified them as papillary RCC. Sunitinib was started between November 2005 and July 2011. Using the updated clinical data of our previous discovery cohort, RR was 52% versus 25%, early PD rate 7% versus 30% (p = 0.01), mPFS 18 versus 9 months (p = 0.002) and mOS 34 versus 23 months (p = 0.004) (Table 2) (Fig. 1, panel A + B). Only in 7% of these 84 patients, consecutive therapy with nivolumab was documented.
Table 2
Response rates, median tumor shrinkage, median progression-free survival and median overal survival depending on rs307826 after start of sunitinib
| AA | AG+GG | p | |
| Sunitinib (Discovery) (n = 84) | |||
| PD | 4/58 (7%) | 6/20 (30%) | 0.01 (*) |
| SD | 24/58 (41%) | 9/20 (45%) | |
| PR | 30/58 (52%) | 5/20 (25%) | |
| Median tumor shrinkage | –31% | 0% | 0.003 (**) |
| mPFS | 18 months | 9 months | 0.002 (***) |
| mOS | 34 months | 23 months | 0.004 (***) |
| Sunitinib (Validation) (n = 84) | |||
| PD | 11/62 (18%) | 6/18 (33%) | 0.24 (*) |
| SD | 18/62 (29%) | 6/18 (33%) | |
| PR | 33/62 (53%) | 6/18 (33%) | |
| Median tumor shrinkage | –31% | –4.5% | 0.07 (**) |
| mPFS | 12 months | 8 months | 0.04 (***) |
| mOS | 27 months | 18 months | 0.22 (***) |
| Sunitinib (Total series) (n = 168) | |||
| PD | 15/120 (13%) | 12/38 (32%) | 0.008 (*) |
| SD | 42/120 (35%) | 15/38 (39%) | |
| PR | 63/120 (53%) | 11/38 (29%) | |
| Median tumor shrinkage | –31% | –4% | 0.0005 (**) |
| mPFS | 15 months | 8 months | 0.0002 (**) |
| mOS | 31 months | 22 months | 0.004 (***) |
Note: (*) Chi-square test. (**) Mann-Whitney test. (***) Kaplan-Meier estimates. PD: progressive disease. SD: stable disease. PR: partial response. mPFS: median progression-free survival. mOS: median overall survival.
Fig. 1
Kaplan-meier estimates for progression-free survival and overal survival in patients treated with first-line sunitinib: panel A + B: discovery cohort. panel C + D: validation cohort. panel E + F: total cohort.
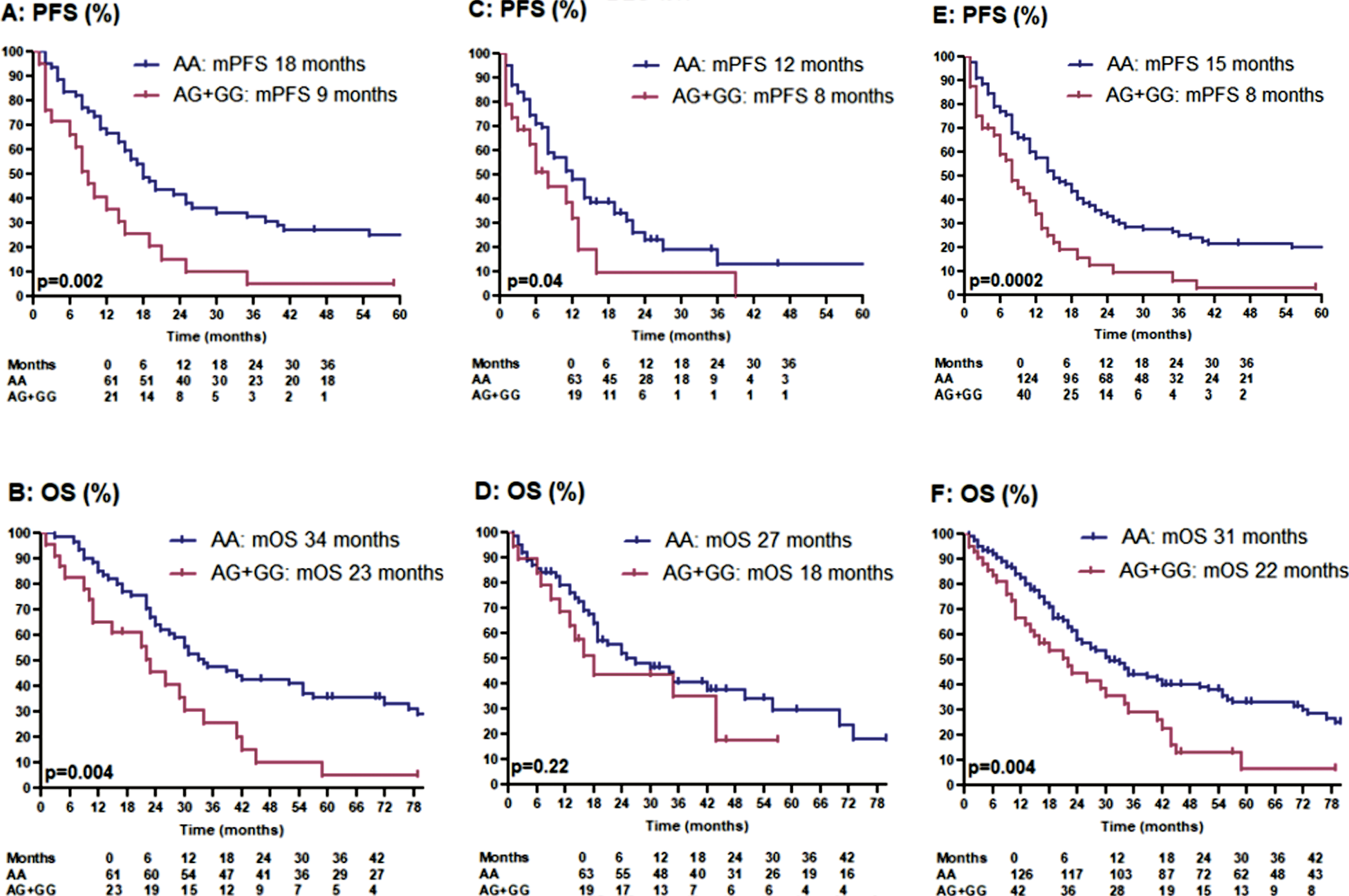
The validation cohort was composed of 84 new patients. In the majority of these patients, sunitinib was started between July 2011 and March 2018. In the validation cohort, RR was 53% versus 33%, early PD rate 18% versus 33% (p = 0.24), mPFS 12 versus 8 months (p = 0.04) and mOS 27 versus 18 months (p = 0.22) (Table 2) (Fig. 1, panel C + D). In 38% of these 84 patients, consecutive therapy with nivolumab (31 patients) or avelumab (1 patient) was documented. Possibly, consecutive ICPIs have positively impacted mOS in AG/GG-carriers.
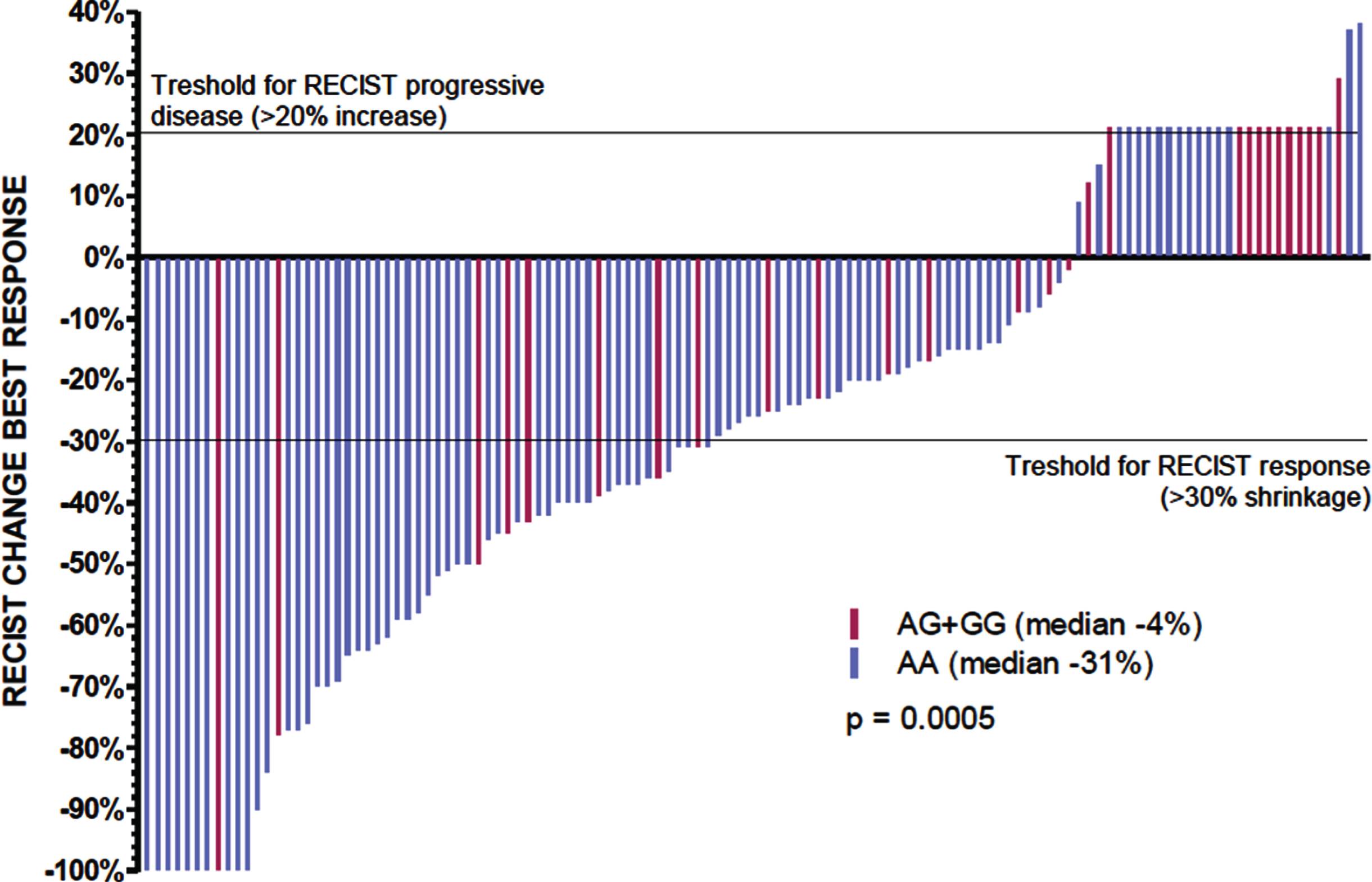
In the total patient series, RR was 53% versus 29%, early PD rate 13% versus 32% (p = 0.008), mPFS 15 versus 8 months (p = 0.0002) and mOS 31 versus 22 months (p = 0.004) in AA- versus AG/GG-carriers (Table 2) (Fig. 1, panel E + F). The precise percentage of tumor shrinkage or increase was documented in 95 AA-carriers and 28 AG/GG-carriers and was significantly better in AA-carriers (median change compared to baseline –31% versus –4% in AG/GG-carriers; p = 0.0005) (Fig. 2).
Fig. 2
Tumor shrinkage depending on rs307826 in sunitinib treated patients.
In a bivariate analysis including the IMDC risk score and rs307826, both the IMDC risk score and rs307826 remained as independently associated with PFS (p = 0.035 and p = 0.002, respectively) and OS (p = 0.008 and p = 0.009, respectively). The impact of rs307826 was similar in IMDC good, intermediate and poor risk patients, both for PFS and for OS, as shown in Supplemental Figure 1, panel A and B. Similarly, the impact of rs307826 was also visible in the more indolent ccrcc2 and the more aggressive ccrcc1 and ccrcc4-tumors (Supplemental Figure 1, panel C and D).
The following criteria were included in the multivariate analysis: rs307826, baseline hemoglobin, baseline platelets, Karnofsky Performance Status, interval between initial diagnosis and start of systemic therapy <12 months, baseline neutrophil count, the presence of bone metastases and baseline calcium levels. In the final multivariable model, the following criteria remained as independently associated with poorer PFS and OS: increased neutrophil count, Karnofsky Performance Status <80, interval between initial diagnosis and start of systemic therapy shorter than one year, the presence of bone metastases and presence of the AG/GG variant in rs307826. For the latter, the hazard ratio (HR) for PFS was 1.81 (95% CI 1.17–2.78; p = 0.007) and for OS 1.59 (95% CI 1.03–2.47; p = 0.037) (Table 3).
Table 3
Results of the multivariate analysis
| Variable | p | Hazard ratio | 95% Confidence interval |
| PFS | |||
| Neutrophils <7.800/mm² | <0.0001 | 3.55 | 1.91–6.59 |
| KPS >70 | 0.031 | 1.73 | 1.05–2.85 |
| Interval primary diagnosis to start systemic therapy >12 months | 0.026 | 1.56 | 1.05–2.32 |
| No bone metastases | 0.031 | 1.53 | 1.04–2.24 |
| rs307826 AA-genotype | 0.007 | 1.81 | 1.17–2.78 |
| OS | |||
| Neutrophils<7.800/mm² | <0.0001 | 5.14 | 2.57–10.30 |
| KPS>70 | 0.049 | 1.77 | 1.00–3.12 |
| Interval primary diagnosis to start systemic therapy >12 months | 0.000 | 2.25 | 1.43–3.54 |
| No bone metastases | 0.007 | 1.73 | 1.16–2.57 |
| rs307826 AA-genotype | 0.037 | 1.59 | 1.03–2.47 |
Note: PFS: progression-free survival. OS: overall survival. KPS: Karnofsky Performance Status.
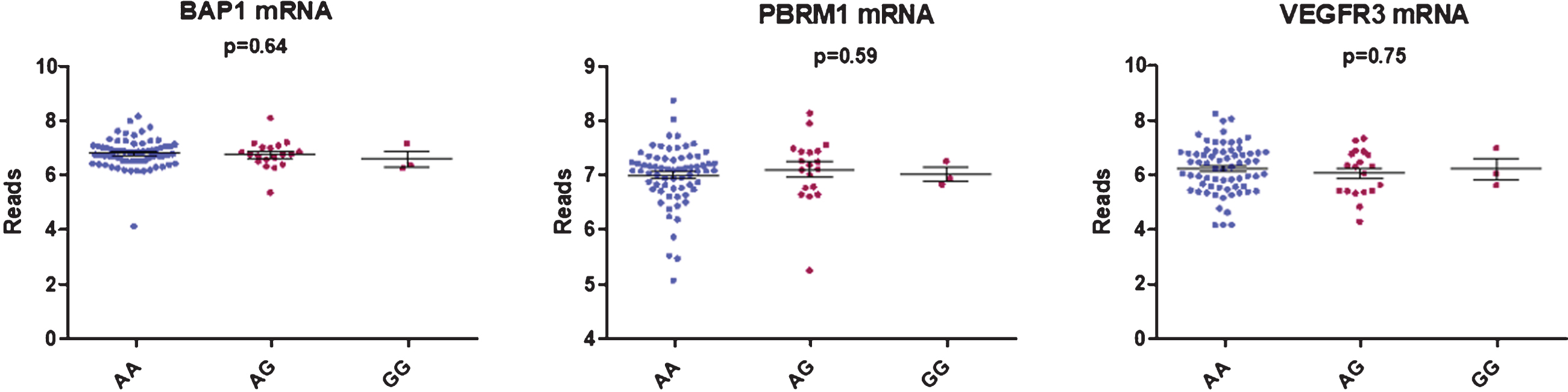
We did not find any correlation between rs307826 and intratumoral VEGFR3, PBRM1 or BAP1 mRNA expression levels as detected in 90 FFPE tumoral kidney samples (AA (n = 68) versus AG (n = 22) versus GG (n = 3)) (Fig. 3). Neither did we detect, in 79 patients with available data, any impact of 5q35 amplifications on VEGFR3 mRNA expression levels.
Fig. 3
Boxplot showing the correlation between BAP1, PBRM1 and VEGFR3 mRNA expression and rs307826 genotypes (ANOVA).
On a series of 89 m-ccRCC patients (patient characteristics in Supplemental Table 1) treated with nivolumab in second or later line, rs307826 did not impact outcome in terms of RR, PFS or OS (Supplemental Table 2 and Supplemental Figure 2, panel A and B).
DISCUSSION
The aim of the present study was to validate the association of SNP rs307826 in VEGFR3 with outcome on first-line sunitinib in an independent validation cohort of m-ccRCC patients.
In this independent validation cohort, we have found a comparable impact on outcome of rs307826 in VEGFR3 as in the discovery series published previously: AG/GG-carriers have a poorer RR and PFS as compared to AA-carriers. We observed a trend towards shorter OS as well, however, this correlation might be weakened due to the increased use of ICPIs in consecutive therapy lines. This effect on RR, PFS and OS was not seen in a series of 89 m-ccRCC patients treated with nivolumab in second or later therapy line.
An overview of the literature on this topic (Table 4) reveals contradictive results, as often seen in SNP-research projects [17]. However, several studies, mostly in RCC, but also in neuro-endocrine tumors (NETs), treated with VEGFR-TKIs point towards better outcome in AA-carriers.
Table 4
Summary of the findings in literature on correlation between rs307826 and outcome
| Study | Tumor type | Therapy | End-point | n | Favorable genotype | Outcome | Hazard (or relative risk) (95% CI) | p |
| In RCC treated with sunitinib | ||||||||
| Beuselinck et al. | RCC | Sunitinib | PFS | 168 | AA better | AA: 15 months | 0.40 (0.24–0.65) | 0.0002 |
| (present study) | AG/GG: 8 months | |||||||
| RR | AA better | AA: PR 53% | Relative risk | 0.015 | ||||
| AG/GG: PR 29% | 0.55 (0.33–0.93) | |||||||
| OS | AA better | AA: 31 months | 0.49 (0.31–1.80) | 0.004 | ||||
| AG/GG: 22 months | ||||||||
| Garcia-Donas et al. [12] | RCC | Sunitinib | PFS | 89 | AA better | AA: 13.7 months | 0.28 (0.14–0.57) | 0.00049 |
| AG: 3.6 months | ||||||||
| RR | AA better | AA: less PD as | 0.11 (0.02–0.52) | 0.0051 | ||||
| best response | ||||||||
| OS | Negative | NR | 0.56 (0.21–1.54) | 0.26 | ||||
| Motzer et al. [18] | RCC | Sunitinib | PFS | 202 | AA better (trend) | AA versus AG | 0.67 (0.39–1.15) | 0.145 |
| RR | Negative | AA versus AG | 1.00 (0.5–2.2) | 1.00 | ||||
| OS | Negative | NR | NR | NR | ||||
| Van der veldt et al. [20] | RCC | Sunitinib | PFS | 129 | Negative | NR | NR | NR |
| OS | Negative | NR | NR | NR | ||||
| Dornbusch et al. [19] | RCC | Sunitinib | PFS | 117 | Negative | AA: 13 months | 1.41 (0.73–2.50) | 0.24 |
| AG/GG: 23 months | ||||||||
| OS | Negative | NR | 0.73 (0.35–1.52) | 0.40 | ||||
| In RCC treated with pazopanib | ||||||||
| Xu et al. [21, 22] | RCC | Pazopanib | PFS | 380 | Negative | NR | NR | 0.61 |
| RR | 368 | AA better (trend) | NR | NR | 0.07 | |||
| OS | 241 | AA better (trend) | NR | 0.74 (0.46–1.18) | 0.2 | |||
| In neuro-endocrine tumors treated with sunitinib or pazopanib | ||||||||
| Grande et al. [24] | NET | Pazopanib | PFS | 44 | AA better (trend) | AA: 12 months | 0.14 (0.02–1.04) | 0.055 |
| AG: 2 months | ||||||||
| Jimenez-Fonseca et al. [23] | NET | Sunitinib | PFS | 43 | Negative | NR | 0.76 (0.37–1.56) | 0.46 |
| RR | Negative | NR | 0.70 (0.06–8.33) | 0.78 | ||||
| OS | AA better | NR | 0.27 (0.10–0.74) | 0.01 | ||||
| In RCC: in peculiar situations | ||||||||
| Garrigos et al. [29] | RCC | Post-nephrectomy | OS | 73 | AA better | AA: 127 months | 0.28 | 0.03 |
| AG/GG: 96 months | ||||||||
| Crona et al. [25] | RCC | Sorafenib | PFS | 295 | GG poorer | NR | 2.92 (1.19–7.19) | 0.019 |
| or placebo | ||||||||
| OS | GG poorer | GG: 194 days | 13.8 (3.0–62.6) | 0.00012 | ||||
| AG/AA: 394 days | ||||||||
NOTE: RCC: renal cell carcinoma. NET: neuro-endocrine tumors. PFS: progression-free survival. OS: overall survival. RR: response rate. NR: not reported.
Four studies are available in literature in metastatic RCC (m-RCC) patients treated with sunitinib. In a series of 89 patients treated with sunitinib, time-to-progression for rs307826 GA-carriers was 3.6 months versus 13.7 months for AA-carriers (p = 0.00049). Early PD was observed more frequently in AG-carriers. No significant association with OS was observed (p = 0.26, but HR 0.56) [12]. In a series of 202 patients, a trend for better PFS (HR 0.67, p = 0.145) was found for AA-carriers versus AG-carriers. No impact on RR was found. However, only 70% of the patients included in the study cohort were genotyped and in the genotyped population, PFS was better than in the non-genotyped population. As a consequence, the authors state that they may have missed a certain number of AG/GG carriers [18]. The two remaining studies are negative studies (for PFS and OS), one of 117 patients [19] and one of 129 patients [20]. For the latter two studies, no precise data on HR were published. One study is available in m-RCC patients treated with pazopanib in the pivotal phase III trial. No correlation with PFS was found, but a trend towards improved RR and OS in AA-carriers [21, 22]. In NETs treated with VEGFR-TKIs, two groups have studied the impact of rs307826 on outcome. In 43 patients treated with sunitinib, Jimenez-Fonseca has shown a better OS (p = 0.01) for AA-carriers compared to AG/GG-carriers. No significant impact on RR or PFS was observed, although the HR was in favor of the AA-carriers (0.70 for RR and 0.76 for PFS, respectively)[23]. In 44 patients with NETs from pancreatic origin, treated with pazopanib, Grande et al. have shown a trend to better PFS (HR 0.14; p = 0.055) for AA-carriers compared to AG-carriers (12 versus 2 months, respectively). Data on RR and OS were not available [24]. Finally, two other studies in m-RCC patients have shown a significant impact of rs307826 on outcome. In a series of 73 patients, OS after nephrectomy was significantly better in AA-carriers compared to AG/GG-carriers (127 versus 96 months; HR 0.28; p = 0.03). Finally, in 295 m-RCC patients, a poorer PFS and OS were observed in GG-carriers compared to AA/AG-carriers. As the authors only published the pooled data of patients treated with sorafenib or placebo, it was not possible to detect the impact of rs307826 in sorafenib-treated patients only. Outcome in AA- and AG-carriers seemed to be similar [25].
Few data have been published on the possible pathophysiological impact of rs307826 in VEGFR3. VEGFR3 signaling is involved in embryonic angiogenesis, adult lymphangiogenesis and tumoral angiogenesis [26, 27]. Crona et al. have shown that VEGFR3 rs307826 variant carriers have increased VEGFR3 phosphorylation (an effect potentiated by VEGF-C stimulation), membrane trafficking and receptor activation. Both in HUVECs as in human embryonic kidney cells, VEGFR3 rs307826 variant carrier cells were more resistant to sorafenib cytotoxicity compared to wild type cells [25].
We did not observe any correlation between rs307826 genotypes and intratumoral VEGFR3 mRNA expression levels. In 63 primary ccRCC tumors, Garcia-Donas et al. have shown thatVEGFR3 expression, as detected by immunohistochemistry, was higher in rs307826 AA-carriers compared to AG-carriers (p = 0.002) [28].
We did not find any difference in PBRM1 and BAP1 mRNA expression and the rs307826 genotype. ccRCCs with BAP1 mutations are known to present a more aggressive behavior, while ccRCCs with PBRM1 mutations are known to present a more indolent disease course. This observation is in coherence with the lack of difference in baseline IMDC risk in patients with different genotypes. Finally, the impact of rs307826 was clearly visible within each IMDC risk group and within the molecular ccrcc1, ccrcc2 and ccrcc4 subgroup.
Our study has several limitations. The global impact of rs307826 is not consistent and not clinically relevant enough to make patient decisions based on this SNP. Furthermore, our study was a retrospective analysis of patients treated in several centers without a central protocol dictating dosing schedule and dose modifications or timing of radiological assessments. Finally, because our patients were mainly Caucasians, the relevance of these polymorphisms needs to be assessed in other ethnic groups, in whom the described polymorphism may be less frequent. It remains to be studied if the impact of rs307826 is predictive or prognostic, although we did not observe any impact of rs307826 in nivolumab treated patients.
However, despite these weaknesses, these results teach us something about the involvement of VEGFR3 in RCC pathogenesis and/or sunitinib pharmacodynamics: VEGFR3 seems to be involved in ccRCC pathogenesis or seems to be a functional target of sunitinib.
CONCLUSION
Our study shows additional evidence that VEGFR3 rs307826 might have an impact on outcome in m-ccRCC patients treated with VEGFR-TKIs. It highlights the involvement of VEGFR3 in RCC pathogenesis and pharmacodynamics.
FUNDING
Benoit Beuselinck is senior clinical investigator of the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium). Diether Lambrechts is supported by the Stichting Tegen Kanker.
AUTHOR CONTRIBUTIONS
Lab work: Van Brussel T, Couchy G
Study development: Beuselinck B, Lambrechts D, Wolter P, Zucman-Rossi J, Elaidi R, Oudard S, Machiels JP
Pathology review: Rioux-Leclercq N, Baldewijns M
Patient inclusion: Elaidi R, Oudard S, Roussel E, Albersen M, Debruyne PR, Machiels JP, Richard V, Verschaeve V, Laguerre B, Beuselinck B
Writing of the text: Beuselinck B, Verbiest A, Vanmechelen M.
CONFLICT OF INTEREST
BB received an unrestricted research grant from Bristol Myers-Squibb and honorarium from Merck, Pfizer, Bristol-Myers-Squibb, Ipsen and Astra Zeneca.
SO received honorarium from Merck, Pfizer, Bristol-Myers-Squibb, bayer, Roche and Sanofi.
The other authors have no conflicts of interest to report.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/KCA-200086.
ACKNOWLEDGMENTS
We are grateful to all the patients who collaborated to this study.
REFERENCES
[1] | Motzer RJ , Hutson TE , Tomczak P , Michaelson MD , Bukowski RM , Rixe O , et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. The New England Journal of Medicine. (2007) ;356: (2):115–24. |
[2] | Motzer RJ , Hutson TE , Tomczak P , Michaelson MD , Bukowski RM , Oudard S , et al. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. (2009) ;27: (22):3584–90. |
[3] | Motzer RJ , Tannir NM , McDermott DF , Aren Frontera O , Melichar B , Choueiri TK , et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. The New England Journal of Medicine. (2018) ;378: (14):1277–90. |
[4] | Motzer RJ , Rini BI , McDermott DF , Aren Frontera O , Hammers HJ , Carducci MA , et al. Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. The Lancet Oncology. (2019) ;20: (10):1370–85. |
[5] | Rini BI , Plimack ER , Stus V , Gafanov R , Hawkins R , Nosov D , et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. The New England Journal of Medicine. (2019) ;380: (12):1116–27. |
[6] | Motzer RJ , Penkov K , Haanen J , Rini B , Albiges L , Campbell MT , et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. The New England Journal of Medicine. (2019) ;380: (12):1103–15. |
[7] | Beuselinck B , Verbiest A , Couchy G , Job S , de Reynies A , Meiller C , et al. Pro-angiogenic gene expression is associated with better outcome on sunitinib in metastatic clear-cell renal cell carcinoma. Acta Oncologica. (2018) ;57: (4):498–508. |
[8] | McDermott DF , Huseni MA , Atkins MB , Motzer RJ , Rini BI , Escudier B , et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nature Medicine. (2018) ;24: (6):749–57. |
[9] | Beuselinck B , Lerut E , Wolter P , Dumez H , Berkers J , Van Poppel H , et al. Sarcomatoid Dedifferentiation in Metastatic Clear Cell Renal Cell Carcinoma and Outcome on Treatment With Anti-Vascular Endothelial Growth Factor Receptor Tyrosine Kinase Inhibitors: A Retrospective Analysis. Clinical Genitourinary Cancer. 2014. |
[10] | Beuselinck B , Jean-Baptiste J , Schoffski P , Couchy G , Meiller C , Rolland F , et al. Validation of VEGFR1 rs9582036 as predictive biomarker in metastatic clear-cell renal cell carcinoma patients treated with sunitinib. BJU International. (2016) ;118: (6):890–901. |
[11] | Beuselinck B , Karadimou A , Lambrechts D , Claes B , Wolter P , Couchy G , et al. Single-nucleotide polymorphisms associated with outcome in metastatic renal cell carcinoma treated with sunitinib. British Journal of Cancer. (2013) ;108: (4):887–900. |
[12] | Garcia-Donas J , Esteban E , Leandro-Garcia LJ , Castellano DE , del Alba AG , Climent MA , et al. Single nucleotide polymorphism associations with response and toxic effects in patients with advanced renal-cell carcinoma treated with first-line sunitinib: a multicentre, observational, prospective study. The Lancet Oncology. (2011) ;12: (12):1143–50. |
[13] | Reumers J , De Rijk P , Zhao H , Liekens A , Smeets D , Cleary J , et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nature Biotechnology. (2012) ;30: (1):61–8. |
[14] | Beuselinck B , Job S , Becht E , Karadimou A , Verkarre V , Couchy G , et al. Molecular subtypes of clear cell renal cell carcinoma are associated with sunitinib response in the metastatic setting. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. (2015) ;21: (6):1329–39. |
[15] | Verbiest A , Couchy G , Job S , Zucman-Rossi J , Caruana L , Lerut E , et al. Molecular Subtypes of Clear Cell Renal Cell Carcinoma Are Associated With Outcome During Pazopanib Therapy in the Metastatic Setting. Clinical Genitourinary Cancer. 2017. |
[16] | Beuselinck B , Oudard S , Rixe O , Wolter P , Blesius A , Ayllon J , et al. Negative impact of bone metastasis on outcome in clear-cell renal cell carcinoma treated with sunitinib. Annals of Oncology: Official Journal of the European Society for Medical Oncology/ESMO. (2011) ;22: (4):794–800. |
[17] | Beuselinck B , Zucman-Rossi J . Kidney cancer: Single nucleotide polymorphisms in mRCC-is their time up? Nature Reviews Urology. (2015) ;12: (8):424–6. |
[18] | Motzer RJ , Hutson TE , Hudes GR , Figlin RA , Martini JF , English PA , et al. Investigation of novel circulating proteins, germ line single-nucleotide polymorphisms, and molecular tumor markers as potential efficacy biomarkers of first-line sunitinib therapy for advanced renal cell carcinoma. Cancer Chemotherapy and Pharmacology. (2014) ;74: (4):739–50. |
[19] | Dornbusch J , Zacharis A , Meinhardt M , Erdmann K , Wolff I , Froehner M , et al. Analyses of potential predictive markers and survival data for a response to sunitinib in patients with metastatic renal cell carcinoma. PloS One. (2013) ;8: (9):e76386. |
[20] | van der Veldt AA , Eechoute K , Gelderblom H , Gietema J , Guchelaar HJ , van Erp NP , et al. Genetic polymorphisms associated with a prolonged progression-free survival in patients with metastatic renal cell cancer treated with sunitinib. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. (2011) ;17: (3):620–9. |
[21] | Xu CF , Bing NX , Ball HA , Rajagopalan D , Sternberg CN , Hutson TE , et al. Pazopanib efficacy in renal cell carcinoma: evidence for predictive genetic markers in angiogenesis-related and exposure-related genes. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. (2011) ;29: (18):2557–64. |
[22] | Xu CF , Johnson T , Garcia-Donas J , Choueiri TK , Sternberg CN , Davis ID , et al. IL8 polymorphisms and overall survival in pazopanib- or sunitinib-treated patients with renal cell carcinoma. British Journal of Cancer. (2015) ;112: Suppl:1190–8. |
[23] | Jimenez-Fonseca P , Martin MN , Carmona-Bayonas A , Calvo A , Fernandez-Mateos J , Redrado M , et al. Biomarkers and polymorphisms in pancreatic neuroendocrine tumors treated with sunitinib. Oncotarget. (2018) ;9: (97):36894–905. |
[24] | Grande E , Capdevila J , Castellano D , Teule A , Duran I , Fuster J , et al. Pazopanib in pretreated advanced neuroendocrine tumors: a phase II, open-label trial of the Spanish Task Force Group for Neuroendocrine Tumors (GETNE). Annals of Oncology: Official Journal of the European Society for Medical Oncology / ESMO. (2015) ;26: (9):1987–93. |
[25] | Crona DJ , Skol AD , Leppanen VM , Glubb DM , Etheridge AS , Hilliard E , et al. Genetic variants of VEGFA and FLT4 are determinants of survival in renal cell carcinoma patients treated with sorafenib. Cancer Research. 2018. |
[26] | Partanen TA , Alitalo K , Miettinen M . Lack of lymphatic vascular specificity of vascular endothelial growth factor receptor 3 in 185 vascular tumors. Cancer. (1999) ;86: (11):2406–12. |
[27] | Valtola R , Salven P , Heikkila P , Taipale J , Joensuu H , Rehn M , et al. VEGFR-3 and its ligand VEGF-C are associated with angiogenesis in breast cancer. Am J Pathol. (1999) ;154: (5):1381–90. |
[28] | Garcia-Donas J , Leandro-Garcia LJ , Gonzalez Del Alba A , Morente M , Alemany I , Esteban E , et al. Prospective study assessing hypoxia-related proteins as markers for the outcome of treatment with sunitinib in advanced clear-cell renal cell carcinoma. Annals of Oncology: Official Journal of the European Society for Medical Oncology / ESMO. (2013) ;24: (9):2409–14. |
[29] | Garrigos C , Espinosa M , Salinas A , Osman I , Medina R , Taron M , et al. Single nucleotide polymorphisms as prognostic and predictive biomarkers in renal cell carcinoma. Oncotarget. (2017) ;8: (63):106551–64. |




