Emerging Role of Environmental Epitranscriptomics and RNA Modifications in Parkinson’s Disease
Abstract
Environmental risk factors and gene-environment interactions play a critical role in Parkinson’s disease (PD). However, the relatively large contribution of environmental risk factors in the overwhelming majority of PD cases has been widely neglected in the field. A “PD prevention agenda” proposed in this journal laid out a set of research priorities focused on preventing PD through modification of environmental risk factors. This agenda includes a call for preclinical studies to employ new high-throughput methods for analyzing transcriptomics and epigenomics to provide a deeper understanding of the effects of exposures linked to PD. Here, we focus on epitranscriptomics as a novel area of research with the potential to add to our understanding of the interplay between genes and environmental exposures in PD. Both epigenetics and epitranscriptomics have been recognized as potential mediators of the complex relationship between genes, environment, and disease. Multiple studies have identified epigenetic alterations, such as DNA methylation, associated with PD and PD-related exposures in human studies and preclinical models. In addition, recent technological advancements have made it possible to study epitranscriptomic RNA modifications, such as RNA N6-methyladenosine (m6A), and a handful of recent studies have begun to explore epitranscriptomics in PD-relevant exposure models. Continued exploration of epitranscriptomic mechanisms in environmentally relevant PD models offers the opportunity to identify biomarkers, pre-degenerative changes that precede symptom onset, and potential mitigation strategies for disease prevention and treatment.
INTRODUCTION
Parkinson’s disease (PD) is the fastest growing neurological disease worldwide and recent estimates of incidence in the US are 1.5 times higher than previous estimates [1–4]. Changing demographics of an aging population and declines in cigarette smoking account for some of this increase; however, the rate of increase outpaces those predicted by these factors alone [1]. In addition, the rate of increase is highest in newly industrialized regions and PD diagnoses within the US cluster in a “PD belt” - regions of the Midwest and Northeast with a history of industrialization [2, 3, 5]. Together, this suggests that environmental factors related to industrialization play a role in PD etiology. Identifying specific factors and elucidating mechanisms by which they increase the risk of PD is critical to reducing the global burden of PD.
While there is debate about the magnitude of the relative contributions of genetic and environmental risk factors in the etiology of sporadic PD (sPD), it is well documented that environmental risk factors and gene-environment interactions play a critical role in disease pathogenesis in most PD cases. Despite this evidence, the role of the environment has been widely neglected in the PD research field [6]. An overwhelming majority of PD research has focused on mutations involved in heritable forms of PD, which account for only 5–10% of PD cases, and genes commonly mutated in the remainder of sPD cases that are not familial or monogenically inherited. Estimates suggest that only about a third of phenotypic variance of sPD can be explained by genetics, further supporting that the environment is a major contributor to PD etiology [7]. These sPD cases are caused by a combination of, and interaction between, genetic and environmental risk factors. Although the focus on disease-linked genes has provided insights into underlying mechanisms of PD pathogenesis, non-genetic factors are understudied and often ignored, hindering the development of comprehensive therapeutic strategies and preventative measures.
In a recent publication in this journal, De Miranda and colleagues proposed an interdisciplinary “PD prevention agenda”—a set of research priorities for both preclinical and clinical research that focuses on preventing PD though modification of environmental factors [6]. As part of this agenda, preclinical studies were encouraged to employ new high-throughput methods for analyzing transcriptomics and epigenomics to provide a deeper understanding of the toxic effects of environmental contaminants linked to PD. Utilizing these methods in environmentally relevant PD models offers the opportunity to identify biomarkers, pre-degenerative changes that precede symptom onset, and potential mitigation strategies for disease prevention and treatment.
Epigenetics, and more recently, the emerging field of epitranscriptomics, have been recognized as potential mediators of the complex relationship between genes, environment and disease because they are sensitive to the environment and regulate gene expression throughout the lifespan (Fig. 1) [8–19]. Thus, exploring both epigenetic and epitranscriptomic changes in PD and models of PD-related exposures is a critical and underexplored avenue of research. By utilizing these high-throughput technologies to explore epigenetic and epitranscriptomic mechanisms in preclinical studies, we can contribute significantly to our understanding of non-genetic factors in PD and to unraveling the complexities of PD etiology.
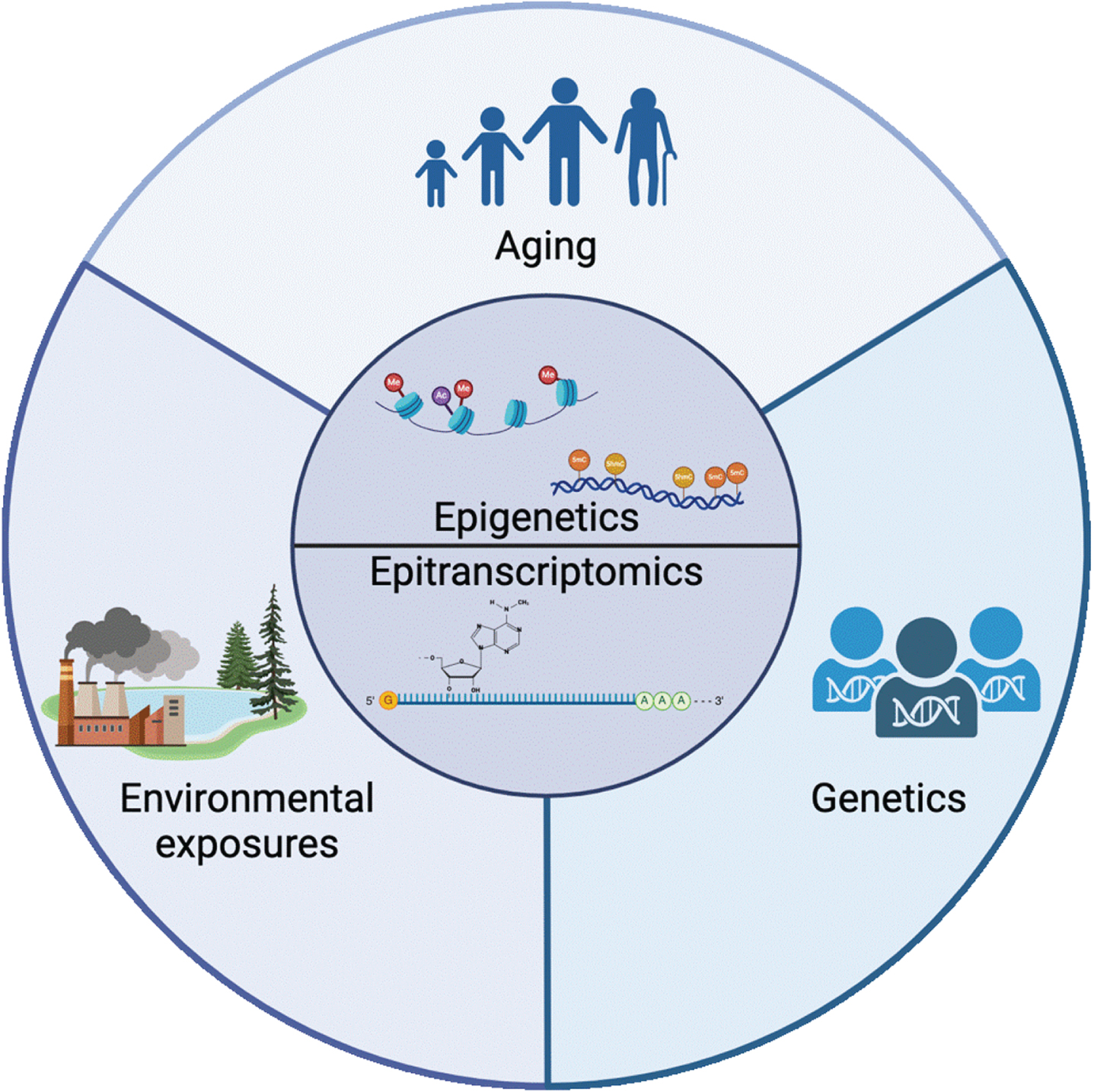
Fig. 1
Epigenetics and epitranscriptomics sit at the intersection of the 3 major classes of risk factors for PD: aging, environmental exposure, and genetics. The mechanisms involved in regulating the epigenome and epitranscriptome are mediators of the complex relationship between aging, the environment, and disease. Created with BioRender.com.
EPIGENETICS
Epigenetics is defined as a set of mechanisms that regulate gene expression without modifying the DNA sequence itself and are meiotically and mitotically heritable in dividing cells [20, 21]. It generally includes posttranslational modification of histones and the regulation of chromatin structure, cytosine modifications, and non-coding RNA-mediated mechanisms (Fig. 2) [22–25]. Because epigenetic marks are sensitive to the environment and play a critical role in regulation of gene expression, they are considered a potential mediator of the relationship between genes, the environment, and disease [8–10].
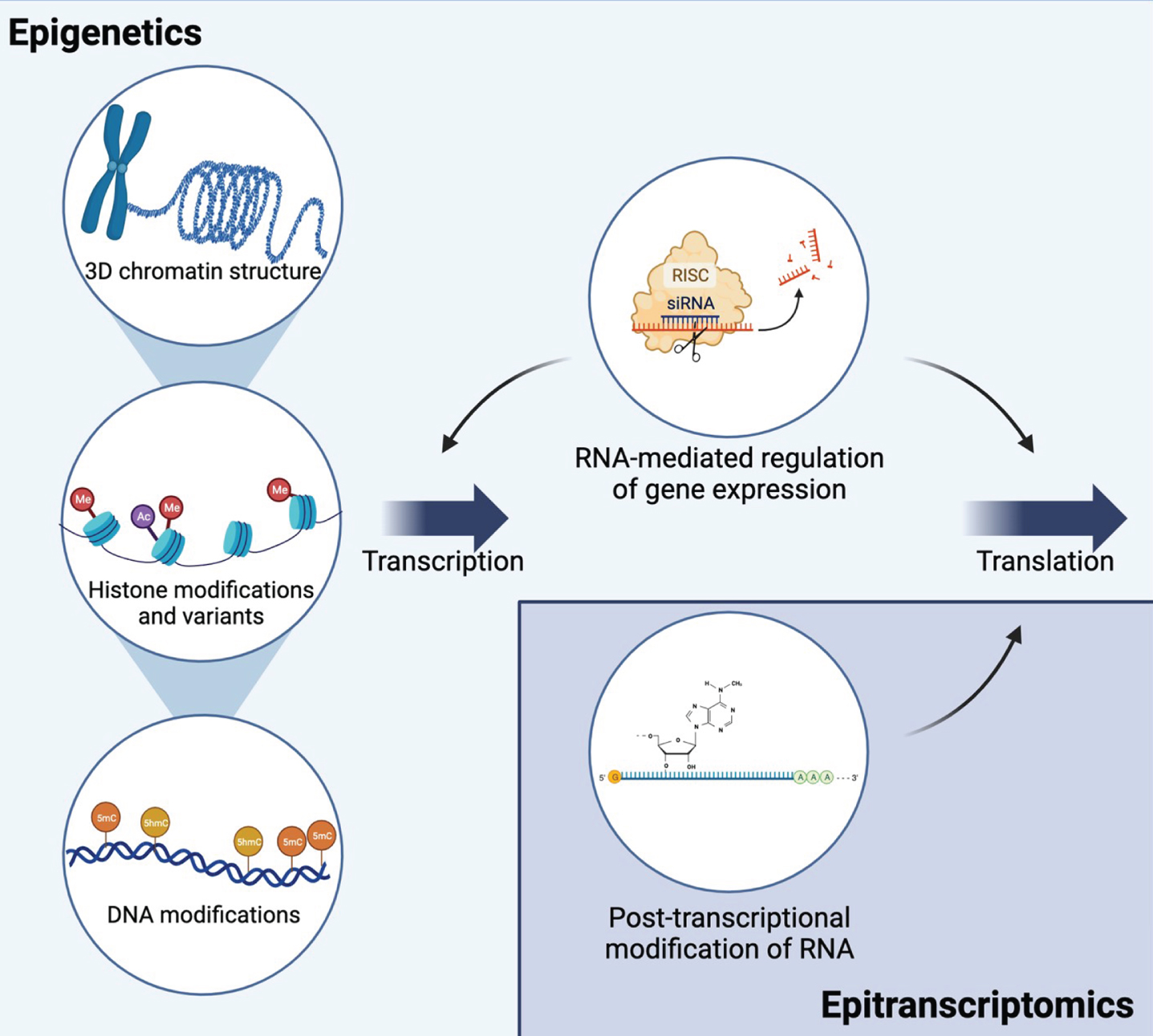
Fig. 2
Epigenetic and epitranscriptomic mechanisms interact to regulate gene expression and translation. Epigenetic mechanisms include mechanisms at the DNA level that regulate transcription: 3D chromatin structure, histone modifications and variants, and DNA modifications. Multiple types of non-coding RNAs regulate both transcription and translation. RNA interference is shown as an example of RNA-mediated gene silencing. Epitranscriptomic mechanisms include multiple types of RNA modifications on all types of RNA. The most common mRNA modification, N6-methyladenosine (m6A), is shown as an example. Together, these mechanisms allow for dynamic regulation of transcription and translation and are influenced by environmental exposures. Created with BioRender.com.
One of the most well studied epigenetic marks is the covalent modification of the fifth position of cytosine in DNA (5-methylcytosine, 5 mC) [25]. More recently, further oxidation of 5 mC to 5-hydroxymethylcytosine (5 hmC) has been recognized as a critical epigenetic mark [26–28]. Each of these marks has a distinct set of “writers” and “readers” that catalyze their generation and recognize these marks [26]. Together, these marks play a critical role in the regulation of gene expression. 5 hmC is thought to be particularly important in the central nervous system, where 5 hmC is highly enriched, and the response to neurotoxicants [24, 26, 29].
Epigenetics in PD
Emerging research has shown distinct DNA methylation patterns in individuals with PD compared to controls [23, 26, 30–34]. Specifically, targeted studies have reported differential DNA methylation at multiple PD-related genes including MAPT, CYP2E1, STX1B and the α-syn gene (SNCA) [35–44]. In addition, epigenome-wide analyses of DNA methylation from postmortem PD brain tissue have identified a number of gene regions that show differential DNA methylation in PD brains including genes previously implicated in PD including PARK7, SLC17A6, and NR4A2 and other genes involved in neurodevelopment, neurotransmitter packaging and release, and axon and neuron projection guidance [45–49]. In addition, studies identified an association between a polymorphism in PD risk and the 5 mC “writer”, DNA methyltransferase 3B (DNMT3B), and the 5 hmC “reader”, TET1 [50, 51]. While much less research has been done on histone modifications in PD, a recent study identified hyperacetylation of histone H3K27 in multiple regions of PD brain tissue and altered H3K27 acetylation was found within known PD genes including SNCA, PARK7, PRKN and MAPT [52]. Together, this data suggests that multiple types of epigenetic regulation may play an important role in PD.
Epigenetics in PD-related exposures
The current state of research exploring the relationship between epigenetic mechanisms and environmental risk factors in PD was recently reviewed in a comprehensive systematic review [53]. Overall, studies of PD-related environmental risk factors in animal and cell models support a role for the epigenome, but human studies to date remain limited, largely due to limited access to brain tissue and lack of exposure data for brain bank subjects [11, 54]. Additional studies in this area are critical for continued elucidation of mechanisms of sPD pathogenesis and identification of novel therapeutic strategies. Targeting of genes and proteins that are epigenetically regulated or aberrant epigenetics patterns themselves with precision treatments, such as epigenome-modifying drugs, represent potential new targets and strategies for disease-modifying therapeutics [55]. However, further research is needed to decipher the complexities of these epigenetic changes and their functional implications.
EPITRANSCRIPTOMICS
Epitranscriptomics focuses on post-transcriptional chemical modifications to RNA molecules and their functional impact on translation, as these modifications play key roles in control of multiple levels of RNA metabolism, including stability, splicing, translation, and localization (Fig. 2) [56–58]. RNA modifications have been identified across all forms of life, and in many RNA species, including mRNAs, rRNAs, tRNAs, and small nuclear RNAs [59]. About 170 RNA modifications exist, and the most well-studied include N6-methyladenosine (m6A), inosine (I), pseudouridine (ψ), 5-methylcytosine (m5C), and 5-hydroxymethylcytosine (hm5C) (Fig. 3) [58, 60, 61]. m6A is the most abundant chemical modification of mRNA, and can impact various aspects of RNA function, including RNA splicing, stability, and translation efficiency [58, 62].
Fig. 3
The most well characterized RNA modifications are m6A, inosine, pseudouridine, and cytosine methylation/hydroxymethylation. Chemical structures of modified and unmodified bases are show, along with enzymes that mediate these modifications and the types of RNA molecules where each modification is typically found. Created with BioRender.com.
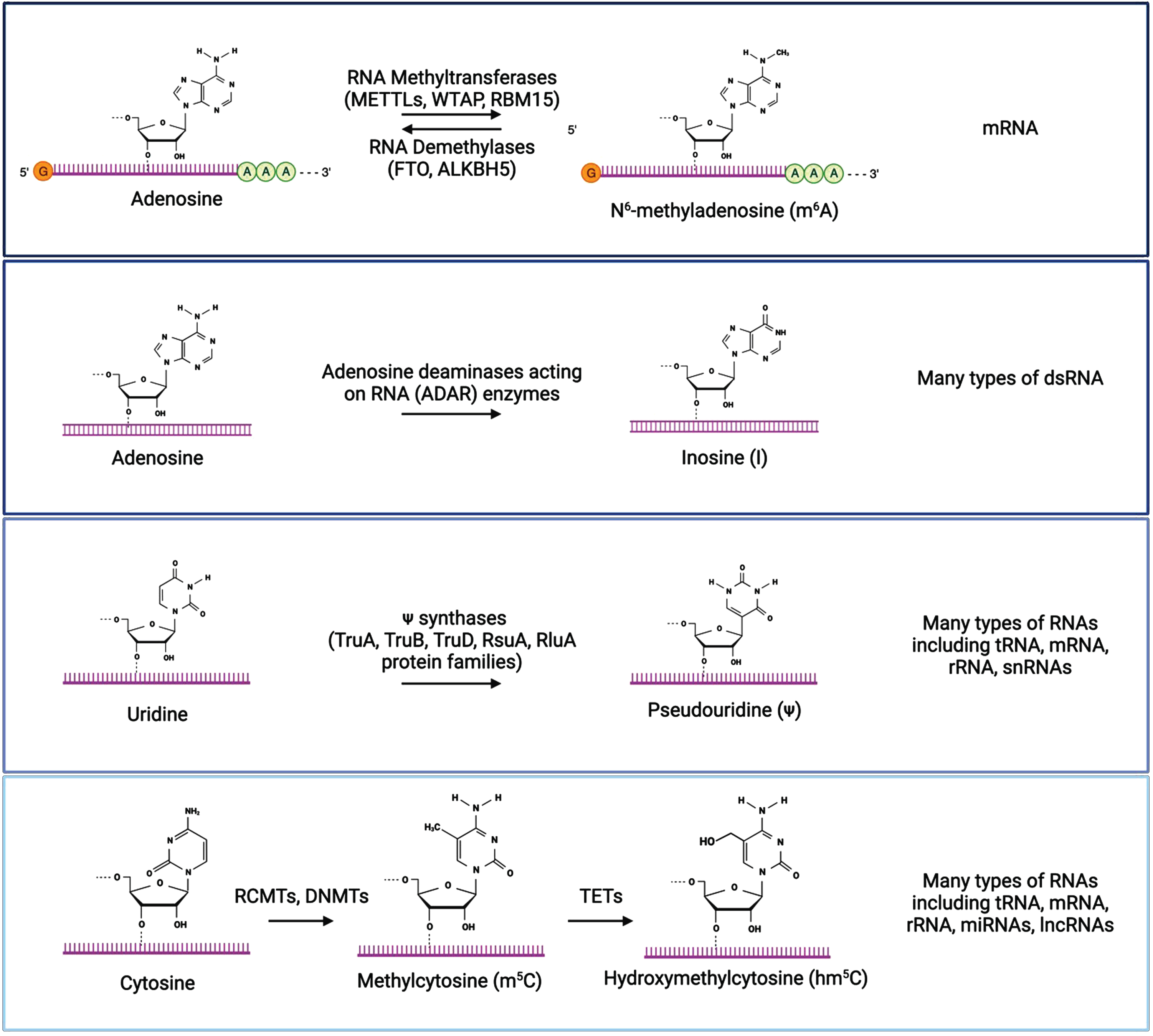
Inosine is produced by the modification of adenosine to inosine on double-stranded RNA (dsRNA) through the deamination of adenosine by adenosine deaminases acting on RNA (ADAR) enzymes [63–65]. This A-to-I modification is recognized as guanosine by the translational machinery, which impacts translation by altering codons, introducing or removing splice sites, or affecting base pairing of the modified RNA either with itself or other RNAs [66, 67]. Inosine plays in important role in the regulation of RNA editing, alternative splicing, and RNA degradation [63]. Pseudouridine is a modification of uridine installed by pseudouridine synthase enzymes [68–73]. It is the most abundant modified nucleotide on RNAs, and plays a role in RNA structure, splicing, and translation efficiency [69, 70]. Methylcytosine (m5C) is a modification of cytosine in RNA that is converted by RNA (C5-cytosine) methyltransferases (RCMTs) and the same DNMTs that add methyl groups to DNA, [74–77]. Similar to the DNA modification, hydroxymethylcytosine (hm5C) is a further modification of methylcytosine generated by the oxidation of m5C by the TET family of enzymes [78, 79]. These modifications are differentially recognized by reader proteins, which can impact mRNA stability and translation, and these modifications play important roles in the regulation of embryonic stem cell differentiation and neurogenesis [74, 80–83]. Dysregulation of these RNA modifications has been implicated in the development and progression of various diseases, including cancer, neurological diseases, and metabolic disorders [59, 84].
N6-methyladenosine
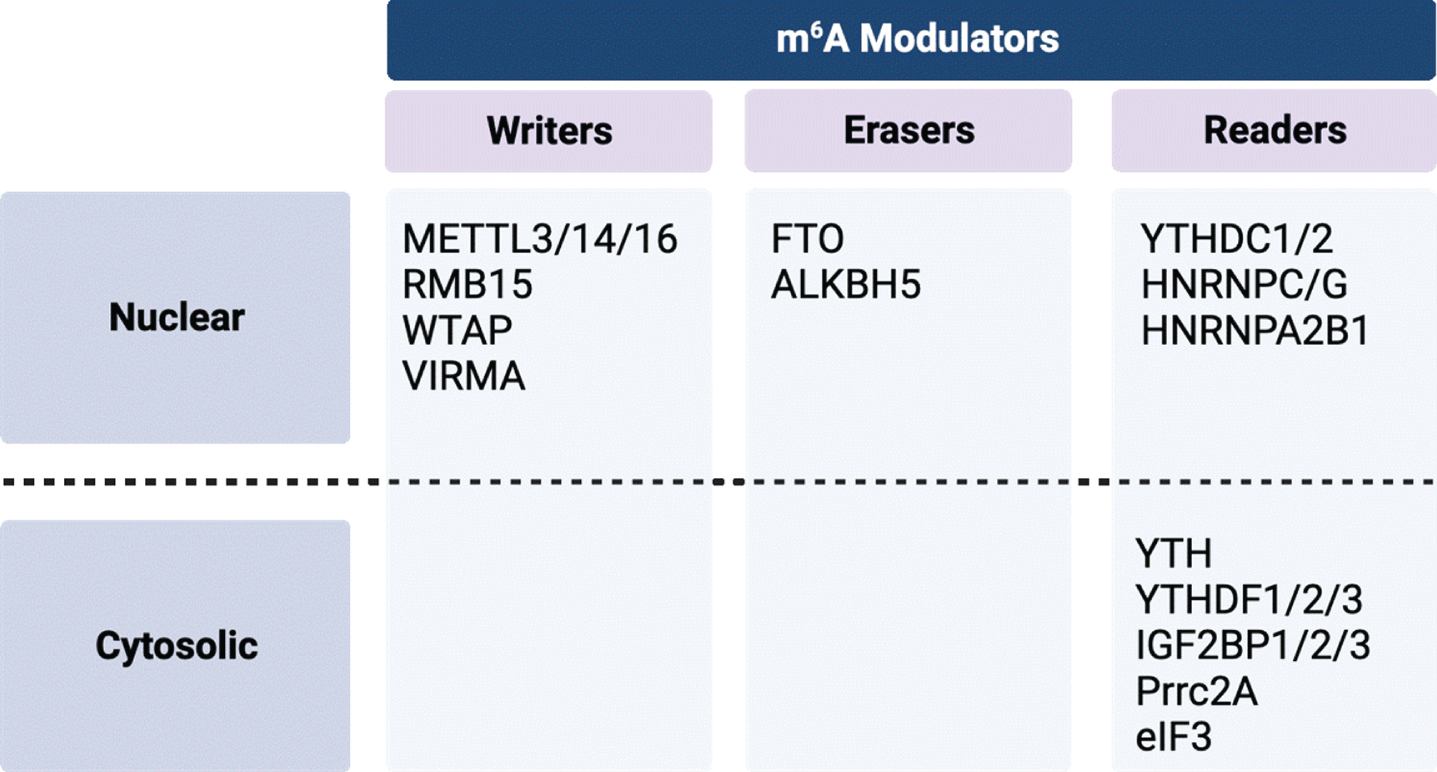
m6A is the most abundant mRNA modification and plays an important role in RNA stability, mRNA translation, alternative splicing, and subcellular RNA localization [85, 86]. m6A modifications are regulated by complexes of proteins termed “writers”, “erasers”, and “readers.” which have been recently reviewed (Fig. 4) [85, 87]. Writers are methyltransferases that convert adenosine to of m6A using the S-adenosylmethionine (SAM) as the methyl donor to catalyze the formation of a methyl group at the sixth N element of adenine in RNA [88]. The most well-characterized writers include the methyltransferase-like (METTL) family of proteins (METTL3, METTL14, and METTL16), WT1 associated protein (WTAP), RNA binding motif protein 15 (RBM15), and vir-like m6A methyltransferase associated (VIRMA). Erasers are demethylases that assist in m6A removal. There two known proteins in this family –the fat mass and obesity associated (FTO) protein and the AlkB homolog 5 (ALKBH5) protein. Readers are cytosolic or nuclear-localized binding proteins that bind m6A modified RNA and regulate gene expression by altering mRNA stability, splicing, and nuclear export. Readers include the YTH domain-containing proteins (YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3) and heterogeneous nuclear ribonucleoproteins (hnRNPs). These protein complexes carry out dynamic and reversible biological processes that can impact the translatability of RNA and potentially contribute to the pathogenesis of disease states [87, 89–102].
Fig. 4
Summary of m6A modulators by class and cellular localization. Created with BioRender.com.
m6A modifications in PD
The importance of RNA modifications, especially m6A, in the development and normal function of the nervous system is well-established and dysregulation of these modifications has been implicated in multiple neurological diseases and aging [85, 86, 103–107]. Studies of m6A in PD models and the human brain have only recently begun to be performed, and more research is needed to develop a comprehensive understanding of m6A activity in relation to PD-associated neurodegeneration.
Existing genetic studies of m6A-SNPs support a role of these modifications in PD. These are SNPs that alter motifs required for m6A modulators to perform their writing, erasing, or reading function or SNPs within the genes encoding these modulators. Two studies that utilized existing GWAS studies, the m6Avar database of m6A-SNPs, existing gene expression data, and eQTL analysis found PD-associated m6A-SNPs within many genes, including the m6A modulator, ALKBH5, which encodes an m6A demethylase; and within motifs required for m6A modulators to function within GAK, which has previously been identified to have two potential PD-linked SNPs in Chinese populations; and ATG16L1, which is important in autophagosome function; and multiple HLA genes [108–111].
A recent analysis of postmortem human PD brain tested for m6A-modified RNAs within neuronal populations in the cerebellum, hippocampus, frontal cortex, and cingulate gyrus [112]. Using a machine learning image analysis method, they concluded m6A-modified RNAs were significantly reduced or mislocalized in cingulate gyrus and hippocampus of subjects with PD, suggesting the possibility of dysregulated epitranscriptomic processes impacting translational control in PD. While this a small preliminary study, this initial finding suggests that further studies of m6A in PD are warranted to confirm and extend these findings.
m6A modifications in PD models
Studies in rodent PD models also support that dysregulation of m6A plays a role in PD (Table 1). A comprehensive study of m6A regulators in the striatum and substantia nigra (SN) of 1-methyl-4-phenyl-1,2,3,6-tetrahydrophyridine (MPTP)-injected C57BL/6 mice (8–12 weeks of age; acute intra-peritoneal injections, 20 mg/kg spread over 8 hours) identified protein expression dysregulation of three m6A writers in the striatum (METTL3, CBLL1, and RBM15), the ALKBH5 eraser in the SN, three readers in the striatum (HNRNPC, IGF2BP3, and FMR1), and four readers in the SN (YTHDF1, HNRNPC, IGF2BP2, and FMR1) by western blot analysis, suggesting dysregulation of the m6A regulatory system in the MPTP model [113]. An additional MPTP study in C57BL/6 mice (4 month old, intra-peritoneal injections, 30 mg/kg/day for 5 days) found increased expression of FTO and decreased expression of METTL14 in the striatum and that exosome-delivery of FTO-targeted siRNAs mitigated the effects of MPTP in the nigrostriatal system [114]. In rats, unilateral SN injection of 6-hydroxydopamine (6-OHDA) (8μg) led to a reduction of m6A and a corresponding elevation of the demethylases FTO in the midbrain and ALKBH5 in the striatum [115]. These findings were recapitulated in PC12 cells where FTO overexpression increased dopaminergic neuronal apoptosis [115]. Collectively, these studies highlight the potential significance of m6A dysregulation in PD pathogenesis and underscore the need to explore these mechanisms in environmentally relevant PD models.
Table 1
Summary of studies of m6a modulators and modifications in PD-relevant models
| Model | Brain Region | Effects on m6A modulators | Effects on m6A modifications | Citations |
| m6A modifications in PD models | ||||
| MPTP, C57BL/6 mice | STR | ↑ CBLL1 (writer) | 113 | |
| ↑ FMR1 (reader) | ||||
| ↓ METTL3 and RBM15 (writers) | ||||
| ↓ IFG2BP3 and HNRNPC (readers) | ||||
| MPTP, C57BL/6 mice | SN | ↑ ALKBH5 (eraser) | 113 | |
| ↑ IGF2BP2 (reader) | ||||
| ↓ YTHDF1, HNRNPC, and FMR1 (readers) | ||||
| MPTP, C57BL/6 mice | STR | ↑ FTO (eraser) | 114 | |
| ↓ METTL14 (writer) | ||||
| 6-OHDA, rat | STR, MB | ↑ FTO and ALKBH5 (erasers) | ↓ m6A | 115 |
| m6A modifications and PD-related environmental exposures | ||||
| PQ, N2A cells | differential m6A modification in circular RNAs and long non-coding RNAs | 131, 132 | ||
| Mn, C57BL/6 mice | STR | ↓ FTO (eraser) | 135 | |
Dopaminergic dysfunction in knockout models of m6A modulators
Knockout models of specific regulators of m6A also indicate a role for these proteins within the dopaminergic system. Knockout of the m6A demethylase, FTO, in C57BL/6 mice increases m6A modification within mRNAs important for synaptic transmission and cell-cell signaling in the striatum and midbrain, as assessed by methylated RNA immunoprecipitation coupled with next-generation sequencing (meRIP-seq) and gene ontology (GO) enrichment analysis [116–118]. In both conventional and conditional FTO knockouts, electrophysiological recordings show an impairment of dopamine receptor type 2 (D2R) and type 3 (D3R)-dependent control of neuronal activity and behavioral responses in midbrain dopaminergic neurons [119]. In addition, conditional deletion of the m6A methyltransferase, METTL14, in the SN of C57BL/6 mice (injection of Cre-recombinase lentivirus into SN floxed METTL14 mice at 8–12 weeks) led to a loss of the dopaminergic marker tyrosine hydroxylase in the SN and impaired motor function and locomotor activity assessed by rotarod, pole test, open-field test and elevated plus maze [120]. These knockout models of specific m6A regulators indicate that m6A plays an important role in dopaminergic neuron function and support a need for more focused research in this area. Taken together, a growing body of evidence from human and rodent studies supports an important regulatory role of m6A modifications in dopaminergic neurons.
m6A modifications and environmental exposures
Emerging data from in vitro, rodent, and human exposure studies suggest that, like epigenetic marks, epitranscriptomic marks are also responsive to environmental exposures [12, 15–19]. This rapidly growing body of evidence indicates that a wide range of exposures, including but not limited to polyaromatic hydrocarbons, endocrine disruptors, dioxins, persistent organic pollutants, certain types of pesticides, heavy metals, air pollution, cigarette smoking, and nanoparticles, have been shown to affect RNA modifications and expression of RNA modulators in many types of cells, animal models and human studies. In addition, a small number of studies have explored the effect of environmental exposures on RNA modifications and expression of RNA in brain or in brain-related cell lines. With these new studies, evidence is growing to support that exposure to many types of neurotoxicants (including dioxin, arsenic, lead, atrazine, cobalt, paraquat) affects both expression of RNA modulators and the patterns of RNA modifications in brain and brain-derived cell lines [18, 121–125].
m6A modifications and PD-related environmental exposures
Research in epitranscriptomics in PD-related environmental exposures is in its infancy with only a few studies reported in the literature to date exploring the effects of the pesticide paraquat (PQ) and manganese (Mn) on epitranscriptomic mechanisms (Table 1). Epidemiological studies report an association between exposure to PQ and increased risk of PD, and mechanistic studies in rodents and cell lines support a role for PQ exposure in dopaminergic dysfunction [126–130]. Two recent studies in N2a neuroblastoma cells found that paraquat exposure led to differential m6A modification in circular RNAs and long non-coding RNAs and that these changes in RNA modifications were mediated by oxidative stress [131, 132]. Mn exposure can lead to manganism, an acquired disorder that shares some clinical features with PD, including the motor symptoms of parkinsonism and dopamine dysfunction [133, 134]. One study explored a potential role for RNA modifications in a mouse model of manganese exposure, reporting decreased levels of FTO in striatum after Mn exposure and that overexpression of FTO in the striatum can rescue Mn-induced motor deficits [135]. Despite this small number of studies, evidence is growing to support that RNA modifications are an important mediator of the effects of environmental exposures in many tissues, including brain, underscoring the need for additional research in this area.
CRITICAL CONSIDERATIONS FOR INCORPORATING EPITRANSCRIPTOMICS INTO PRECLINICAL STUDIES OF ENVIRONMENTAL MODELS OF PD
Historically, comprehensive assessments of epigenomic and epitranscriptomic changes faced constraints posed by technology and cost. Development of methods for cost-effect analysis of the epitranscriptome has lagged behind those for the epigenome. However, recent advances allow research to explore these epitranscriptomic mechanisms with heightened precision and cost-effectiveness [49, 136, 137]. The technology is now available to enable the delineation of specific RNA modifications in environmentally relevant PD models. With these new and evolving methods, both genome-wide and targeted analysis of PD-associated genes are possible and warranted to identify pre-degenerative changes and potential mitigation strategies for disease prevention and treatment.
While these new methods provide new opportunities, successful incorporation of epitranscriptomics into PD-relevant exposure models faces challenges, many of which are similar to those faced by the epigenomics and other omics field. In a recently published chapter, entitled “Best practices for epigenome-wide DNA modification data collection and analysis,” we outlined important considerations for rigor and reproducibility in epigenomic studies and recommendations to address them [138]. Many of these challenges and strategies to address them apply to epitranscriptomic studies as well and are summarized in Fig. 5. Most of these practices are not unique to epitranscriptomics and are covered in detail in our previously published chapter [138]. However, the question of the stability of RNA modifications is unique and important to consider the implications of this. In contrast to DNA, many RNA molecules are short lived so modification of individual RNA molecules is also likely short lived and may be difficult to measure consistently. However, as papers discussed above regarding m6A-SNPs demonstrate, if the mechanisms that lay down and regulate these marks are disrupted by toxicant exposure, aggregate changes in RNA modifications may be more persistent that any individual RNA molecule [108–125, 139].
Fig. 5
Critical considerations for incorporating epitranscriptomics into preclinical studies of environmental models of Parkinson’s disease.
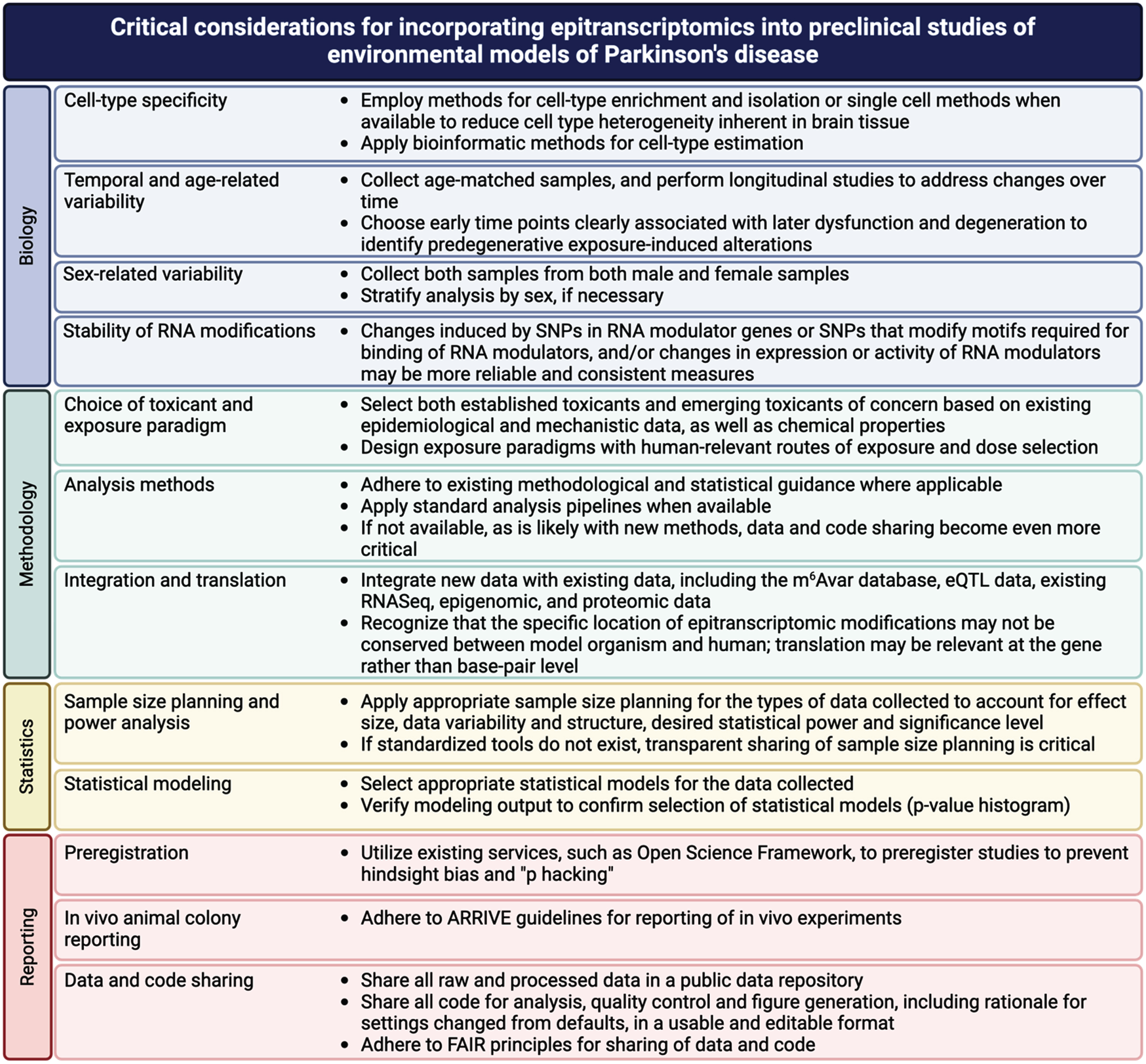
In addition, like epigenetic marks, the functional implications of epitranscriptomic marks associated with exposure can be difficult to assess. Implementation of the strategies highlighted here regarding integration and translation can help to link these marks with functional outcomes. Much as papers discussed above integrated databases GWAS data and eQTL data with data on RNA modifications (m6Avar), integration of new data will be critical to making functional and translational connections [108–111, 140]. Such integration is only possible with open and FAIR sharing of methods, data and code [141–144]. By building on lessons learned in more established fields, epitranscriptomics can avoid the same pitfalls and facilitate discovery of epitranscriptomic mechanisms in environmentally induced increases in PD risk.
CONCLUSIONS
This review focuses the importance of investigating two interrelated areas of study—epigenetics and epitranscriptomics—in the context of sPD, with a focus on epitranscriptomics as an emerging field of research. While there have been substantial strides in unraveling the basic biology of RNA modifications, the application of these findings to PD remains in its infancy [14, 66, 145]. Given the significant role of environmental factors in sPD and the growing evidence that RNA modifications are sensitive to the environment, further exploration of the role of epitranscriptomics in experimental models of sPD offers a valuable avenue for gaining crucial insights into the molecular underpinnings of sPD.
Combined with the ability to develop targeted therapies that modify epigenetic or epitranscriptomic marks holds substantial promise for mitigation exposure-related risk of PD. The strategic design of small molecules or interventions capable of selectively modulating specific modifications or regulators may provide a nuanced approach to altering gene expression and cellular processes, paving the way for innovative disease-modifying treatments. While much research is still needed to generate a comprehensive understanding of RNA modifications in PD-related exposures and to determine the potential therapeutic and preventative relevance, there is great potential of incorporating new high-throughput methods for analyzing epitranscriptomics and epigenomics to provide a deeper understanding how environmental exposures contribute to sPD and insights into mitigation strategies for disease prevention and treatment. The authors have no acknowledgments to report.
ACKNOWLEDGMENTS
The authors have no acknowledgments to report.
FUNDING
This work is supported by NIH R01ES031237.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
REFERENCES
[1] | Dorsey ER , Sherer T , Okun MS , Bloem BR ((2018) ) The emerging evidence of the Parkinson pandemic. J Parkinsons Dis 8: , S3–S8. |
[2] | Willis AW , Roberts E , Beck JC , Fiske B , Ross W , Savica R , Van Den Eeden SK , Tanner CM , Marras C , Parkinson’s Foundation P4 Group ((2022) ) Incidence of Parkinson disease in North America. NPJ Parkinsons Dis 8: , 170. |
[3] | Dorsey ER , Constantinescu R , Thompson JP , Biglan KM , Holloway RG , Kieburtz K , Marshall FJ , Ravina BM , Schifitto G , Siderowf A , Tanner CM ((2007) ) Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 68: , 384–386. |
[4] | Yang W , Hamilton JL , Kopil C , Beck JC , Tanner CM , Albin RL , Ray Dorsey E , Dahodwala N , Cintina I , Hogan P , Thompson T ((2020) ) Current and projected future economic burden of Parkinson’s disease in the U.S. NPJ Parkinsons Dis 6: , 15. |
[5] | Wright Willis A , Evanoff BA , Lian M , Criswell SR , Racette BA ((2010) ) Geographic and ethnic variation in Parkinson disease: A population-based study of US Medicare beneficiaries. Neuroepidemiology 34: , 143–151. |
[6] | De Miranda BR , Goldman SM , Miller GW , Greenamyre JT , Dorsey ER ((2022) ) Preventing Parkinson’s disease: An environmental agenda. J Parkinsons Dis 12: , 45–68. |
[7] | Keller MF , Saad M , Bras J , Bettella F , Nicolaou N , Simón-Sánchez J , Mittag F , Büchel F , Sharma M , Gibbs JR , Schulte C , Moskvina V , Durr A , Holmans P , Kilarski LL , Guerreiro R , Hernandez DG , Brice A , Ylikotila P , Stefánsson H , Majamaa K , Morris HR , Williams N , Gasser T , Heutink P , Wood NW , Hardy J , Martinez M , Singleton AB , Nalls MA; International Parkinson’s Disease Genomics Consortium (IPDGC); Wellcome Trust Case Control Consortium 2 (WTCCC2) ((2012) ) Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum Mol Genet 21: , 4996–5009. |
[8] | Faulk C , Dolinoy DC ((2011) ) Timing is everything: The when and how of environmentally induced changes in the epigenome of animals. Epigenetics 6: , 791–797. |
[9] | Allis CD , Jenuwein T ((2016) ) The molecular hallmarks of epigenetic control. Nat Rev Genet 17: , 487–500. |
[10] | Cavalli G , Heard E ((2019) ) Advances in epigenetics link genetics to the environment and disease. Nature 571: , 489–499. |
[11] | Kochmanski J , VanOeveren SE , Patterson JR , Bernstein AI ((2019) ) Developmental dieldrin exposure alters DNA methylation at genes related to dopaminergic neuron development and Parkinson’s disease in mouse midbrain. Toxicol Sci 169: , 593–607. |
[12] | Cayir A , Byun HM , Barrow TM ((2020) ) Environmental epitranscriptomics. Environ Res 189: , 109885. |
[13] | Liu C , Yang S , Zhang Y , Wang C , Du D , Wang X , Liu T , Liang G ((2021) ) Emerging roles of N6-methyladenosine demethylases and its interaction with environmental toxicants in digestive system cancers. Cancer Manag Res 13: , 7101–7114. |
[14] | Saletore Y , Meyer K , Korlach J , Vilfan ID , Jaffrey S , Mason CE ((2012) ) The birth of the Epitranscriptome: Deciphering the function of RNA modifications. Genome Biol 13: , 175. |
[15] | Cayir A ((2021) ) RNA A-to-I editing, environmental exposure, and human diseases. Crit Rev Toxicol 51: , 456–466. |
[16] | Wu H , Eckhardt CM , Baccarelli AA ((2023) ) Molecular mechanisms of environmental exposures and human disease. Nat Rev Genet 24: , 332–344. |
[17] | Feng Y , Liu T , Xu S , Ren Y , Ge Y , Yin L , Pu Y , Liang G ((2022) ) The role of N6-methyladenosine methylation in environmental exposure-induced health damage. Environ Sci Pollut Res Int 29: , 69153–69175. |
[18] | Zhu X , Fu H , Sun J , Xu Q ((2023) ) Interaction between N6-methyladenosine (m6A) modification and environmental chemical-induced diseases in various organ systems. Chem Biol Interact 373: , 110376. |
[19] | Malovic E , Ealy A , Kanthasamy A , Kanthasamy AG ((2021) ) Emerging roles of N6-methyladenosine (m6A) epitranscriptomics in toxicology. Toxicol Sci 181: , 13–22. |
[20] | Berger SL , Kouzarides T , Shiekhattar R , Shilatifard A ((2009) ) An operational definition of epigenetics. Genes Dev 23: , 781–783. |
[21] | Dupont C , Armant DR , Brenner CA ((2009) ) Epigenetics: Definition, mechanisms and clinical perspective. Semin Reprod Med 27: , 351–357. |
[22] | Goldberg AD , Allis CD , Bernstein E ((2007) ) Epigenetics: A landscape takes shape. Cell 128: , 635–638. |
[23] | Labbe C , Lorenzo-Betancor O , Ross OA ((2016) ) Epigenetic regulation in Parkinson’s disease. Acta Neuropathol 132: , 515–530. |
[24] | Kochmanski J , Bernstein AI ((2020) ) The impact of environmental factors on 5-hydroxymethylcytosine in the brain. Curr Environ Health Rep 7: , 109–120. |
[25] | Smith ZD , Meissner A ((2013) ) DNA methylation: Roles in mammalian development. Nat Rev Genet 14: , 204–220. |
[26] | Cheng Y , Bernstein A , Chen D , Jin P ((2015) ) 5-hydroxymethylcytosine: A new player in brain disorders? . Exp Neurol 268: , 3–9. |
[27] | Kriaucionis S , Heintz N ((2009) ) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: , 929–930. |
[28] | Tahiliani M , Koh KP , Shen Y , Pastor WA , Bandukwala H , Brudno Y , Agarwal S , Iyer LM , Liu DR , Aravind L , Rao A ((2009) ) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: , 930–935. |
[29] | Globisch D , Munzel M , Muller M , Michalakis S , Wagner M , Koch S , Bruckl T , Biel M , Carell T ((2010) ) Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5: , e15367. |
[30] | Jakovcevski M , Akbarian S ((2012) ) Epigenetic mechanisms in neurological disease. Nat Med 18: , 1194–1204. |
[31] | Marques S , Outeiro TF ((2013) ) Epigenetics in Parkinson’s and Alzheimer’s diseases. Subcell Biochem 61: , 507–525. |
[32] | Miranda-Morales E , Meier K , Sandoval-Carrillo A , Salas-Pacheco J , Vazquez-Cardenas P , Arias-Carrion O ((2017) ) Implications of DNA methylation in Parkinson’s disease. Front Mol Neurosci 10: , 225. |
[33] | Lardenoije R , Pishva E , Lunnon K , van den Hove DL ((2018) ) Neuroepigenetics of aging and age-related neurodegenerative disorders. Prog Mol Biol Transl Sci 158: , 49–82. |
[34] | Wullner U , Kaut O , deBoni L , Piston D , Schmitt I ((2016) ) DNA methylation in Parkinson’s disease. J Neurochem 139 Suppl 1: , 108–120. |
[35] | Kaut O , Schmitt I , Wullner U ((2012) ) Genome-scale methylation analysis of Parkinson’s disease patients’ brains reveals DNA hypomethylation and increased mRNA expression of cytochrome P450 2E1. Neurogenetics 13: , 87–91. |
[36] | Coupland KG , Mellick GD , Silburn PA , Mather K , Armstrong NJ , Sachdev PS , Brodaty H , Huang Y , Halliday GM , Hallupp M , Kim WS , Dobson-Stone C , Kwok JB ((2014) ) DNA methylation of the MAPT gene in Parkinson’s disease cohorts and modulation by vitamin E in vitro. Mov Disord 29: , 1606–1614. |
[37] | Jowaed A , Schmitt I , Kaut O , Wullner U ((2010) ) Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci 30: , 6355–6359. |
[38] | Matsumoto L , Takuma H , Tamaoka A , Kurisaki H , Date H , Tsuji S , Iwata A ((2010) ) CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson’s disease. PLoS One 5: , e15522. |
[39] | Desplats P , Spencer B , Coffee E , Patel P , Michael S , Patrick C , Adame A , Rockenstein E , Masliah E ((2011) ) Alpha-synuclein sequesters Dnmt1 from the nucleus: A novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem 286: , 9031–9037. |
[40] | Ai SX , Xu Q , Hu YC , Song CY , Guo JF , Shen L , Wang CR , Yu RL , Yan XX , Tang BS ((2014) ) Hypomethylation of SNCA in blood of patients with sporadic Parkinson’s disease. J Neurol Sci 337: , 123–128. |
[41] | Tan YY , Wu L , Zhao ZB , Wang Y , Xiao Q , Liu J , Wang G , Ma JF , Chen SD ((2014) ) Methylation of alpha-synuclein and leucine-rich repeat kinase 2 in leukocyte DNA of Parkinson’s disease patients. Parkinsonism Relat Disord 20: , 308–313. |
[42] | Song Y , Ding H , Yang J , Lin Q , Xue J , Zhang Y , Chan P , Cai Y ((2014) ) Pyrosequencing analysis of SNCA methylation levels in leukocytes from Parkinson’s disease patients. Neurosci Lett 569: , 85–88. |
[43] | Pihlstrom L , Berge V , Rengmark A , Toft M ((2015) ) Parkinson’s disease correlates with promoter methylation in the alpha-synuclein gene. Mov Disord 30: , 577–580. |
[44] | Schmitt I , Kaut O , Khazneh H , deBoni L , Ahmad A , Berg D , Klein C , Frohlich H , Wullner U ((2015) ) L-dopa increases alpha-synuclein DNA methylation in Parkinson’s disease patients in vivo and in vitro. Mov Disord 30: , 1794–1801. |
[45] | Masliah E , Dumaop W , Galasko D , Desplats P ((2013) ) Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 8: , 1030–1038. |
[46] | Moore K , McKnight AJ , Craig D , O’Neill F ((2014) ) Epigenome-wide association study for Parkinson’s disease. Neuromolecular Med 16: , 845–855. |
[47] | Young JI , Sivasankaran SK , Wang L , Ali A , Mehta A , Davis DA , Dykxhoorn DM , Petito CK , Beecham GW , Martin ER , Mash DC , Pericak-Vance M , Scott WK , Montine TJ , Vance JM ((2019) ) Genome-wide brain DNA methylation analysis suggests epigenetic reprogramming in Parkinson disease. Neurol Genet 5: , e342. |
[48] | Marshall LL , Killinger BA , Ensink E , Li P , Li KX , Cui W , Lubben N , Weiland M , Wang X , Gordevicius J , Coetzee GA , Ma J , Jovinge S , Labrie V ((2020) ) Epigenomic analysis of Parkinson’s disease neurons identifies Tet2 loss as neuroprotective. Nat Neurosci 23: , 1203–1214. |
[49] | Kochmanski J , Kuhn NC , Bernstein AI ((2022) ) Parkinson’s disease-associated, sex-specific changes in DNA methylation at PARK7 (DJ-1), SLC17A6 (VGLUT2), PTPRN2 (IA-2beta), and NR4A2 (NURR1) in cortical neurons. NPJ Parkinsons Dis 8: , 120. |
[50] | Shu L , Qin L , Min S , Pan H , Zhong J , Guo J , Sun Q , Yan X , Chen C , Tang B , Xu Q ((2019) ) Genetic analysis of DNA methylation and hydroxymethylation genes in Parkinson’s disease. Neurobiol Aging 84: , 242e213-242 e216. |
[51] | Pezzi JC , de Bem CM , da Rocha TJ , Schumacher-Schuh AF , Chaves ML , Rieder CR , Hutz MH , Fiegenbaum M , Camozzato AL ((2017) ) Association between DNA methyltransferase gene polymorphism and Parkinson’s disease. Neurosci Lett 639: , 146–150. |
[52] | Toker L , Tran GT , Sundaresan J , Tysnes OB , Alves G , Haugarvoll K , Nido GS , Dolle C , Tzoulis C ((2021) ) Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson’s disease brain. Mol Neurodegener 16: , 31. |
[53] | Tsalenchuk M , Gentleman SM , Marzi SJ ((2023) ) Linking environmental risk factors with epigenetic mechanisms in Parkinson’s disease. NPJ Parkinsons Dis 9: , 123. |
[54] | Song C , Kanthasamy A , Anantharam V , Sun F , Kanthasamy AG ((2010) ) Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: Relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol 77: , 621–632. |
[55] | Kantor B , Tagliafierro L , Gu J , Zamora ME , Ilich E , Grenier C , Huang ZY , Murphy S , Chiba-Falek O ((2018) ) Downregulation of SNCA expression by targeted editing of DNA methylation: A potential strategy for precision therapy in PD. Mol Ther 26: , 2638–2649. |
[56] | Zhou Y , Kong Y , Fan W , Tao T , Xiao Q , Li N , Zhu X ((2020) ) Principles of RNA methylation and their implications for biology and medicine. Biomed Pharmacother 131: , 110731. |
[57] | Hao L , Zhang J , Liu Z , Lin X , Guo J ((2023) ) Epitranscriptomics in the development, functions, and disorders of cancer stem cells. Front Oncol 13: , 1145766. |
[58] | Helm M , Motorin Y ((2017) ) Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat Rev Genet 18: , 275–291. |
[59] | Jonkhout N , Tran J , Smith MA , Schonrock N , Mattick JS , Novoa EM ((2017) ) The RNA modification landscape in human disease. RNA 23: , 1754–1769. |
[60] | Nachtergaele S , He C ((2018) ) Chemical modifications in the life of an mRNA transcript. Annu Rev Genet 52: , 349–372. |
[61] | Boccaletto P , Machnicka MA , Purta E , Piatkowski P , Baginski B , Wirecki TK , de Crecy-Lagard V , Ross R , Limbach PA , Kotter A , Helm M , Bujnicki JM ((2018) ) MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: , D303–D307. |
[62] | Hong J , Xu K , Lee JH ((2022) ) Biological roles of the RNA m(6)A modification and its implications in cancer. Exp Mol Med 54: , 1822–1832. |
[63] | Slotkin W , Nishikura K ((2013) ) Adenosine-to-inosine RNA editing and human disease. Genome Med 5: , 105. |
[64] | George CX , Gan Z , Liu Y , Samuel CE ((2011) ) Adenosine deaminases acting on RNA, RNA editing, and interferon action. J Interferon Cytokine Res 31: , 99–117. |
[65] | Savva YA , Rieder LE , Reenan RA ((2012) ) The ADAR protein family. Genome Biol 13: , 252. |
[66] | Jung Y , Goldman D ((2018) ) Role of RNA modifications in brain and behavior. Genes Brain Behav 17: , e12444. |
[67] | Nishikura K ((2006) ) Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol 7: , 919–931. |
[68] | Carlile TM , Rojas-Duran MF , Zinshteyn B , Shin H , Bartoli KM , Gilbert WV ((2014) ) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: , 143–146. |
[69] | Schwartz S , Bernstein DA , Mumbach MR , Jovanovic M , Herbst RH , Leon-Ricardo BX , Engreitz JM , Guttman M , Satija R , Lander ES , Fink G , Regev A ((2014) ) Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: , 148–162. |
[70] | Lovejoy AF , Riordan DP , Brown PO ((2014) ) Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9: , e110799. |
[71] | Li X , Zhu P , Ma S , Song J , Bai J , Sun F , Yi C ((2015) ) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: , 592–597. |
[72] | Safra M , Nir R , Farouq D , Vainberg Slutskin I , Schwartz S ((2017) ) TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res 27: , 393–406. |
[73] | Hamma T , Ferre-D’Amare AR ((2006) ) Pseudouridine synthases. Chem Biol 13: , 1125–1135. |
[74] | Gao Y , Fang J ((2021) ) RNA 5-methylcytosine modification and its emerging role as an epitranscriptomic mark. RNA Biol 18: , 117–127. |
[75] | Schapira M ((2016) ) Structural chemistry of human RNA methyltransferases. ACS Chem Biol 11: , 575–582. |
[76] | Cheng JX , Chen L , Li Y , Cloe A , Yue M , Wei J , Watanabe KA , Shammo JM , Anastasi J , Shen QJ , Larson RA , He C , Le Beau MM , Vardiman JW ((2018) ) RNA cytosine methylation and methyltransferases mediate chromatin organization and 5-azacytidine response and resistance in leukaemia. Nat Commun 9: , 1163. |
[77] | Li M , Tao Z , Zhao Y , Li L , Zheng J , Li Z , Chen X ((2022) ) 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J Transl Med 20: , 214. |
[78] | Fu L , Guerrero CR , Zhong N , Amato NJ , Liu Y , Liu S , Cai Q , Ji D , Jin SG , Niedernhofer LJ , Pfeifer GP , Xu GL , Wang Y ((2014) ) Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc 136: , 11582–11585. |
[79] | Delatte B , Wang F , Ngoc LV , Collignon E , Bonvin E , Deplus R , Calonne E , Hassabi B , Putmans P , Awe S , Wetzel C , Kreher J , Soin R , Creppe C , Limbach PA , Gueydan C , Kruys V , Brehm A , Minakhina S , Defrance M , Steward R , Fuks F ((2016) ) RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351: , 282–285. |
[80] | Blanco S , Dietmann S , Flores JV , Hussain S , Kutter C , Humphreys P , Lukk M , Lombard P , Treps L , Popis M , Kellner S , Holter SM , Garrett L , Wurst W , Becker L , Klopstock T , Fuchs H , Gailus-Durner V , Hrabe de Angelis M , Karadottir RT , Helm M , Ule J , Gleeson JG , Odom DT , Frye M ((2014) ) Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33: , 2020–2039. |
[81] | Lyko F ((2018) ) The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat Rev Genet 19: , 81–92. |
[82] | Ahmed S , Hossain Z , Uddin M , Taherzadeh G , Sharma A , Shatabda S , Dehzangi A ((2020) ) Accurate prediction of RNA 5-hydroxymethylcytosine modification by utilizing novel position-specific gapped k-mer descriptors. Comput Struct Biotechnol J 18: , 3528–3538. |
[83] | MacArthur IC , Dawlaty MM ((2021) ) TET enzymes and 5-hydroxymethylcytosine in neural progenitor cell biology and neurodevelopment. Front Cell Dev Biol 9: , 645335. |
[84] | Barbieri I , Kouzarides T ((2020) ) Role of RNA modifications in cancer. Nat Rev Cancer 20: , 303–322. |
[85] | Jiang L , Li X , Wang S , Yuan Z , Cheng J ((2022) ) The role and regulatory mechanism of m6A methylation in the nervous system. Front Genet 13: , 962774. |
[86] | Deng J CX , Chen A , Zheng X ((2022) ) m6A RNA methylation in brain injury and neurodegenerative disease. Front Neurol 13: , 995747. |
[87] | Jiang X , Liu B , Nie Z , Duan L , Xiong Q , Jin Z , Yang C , Chen Y ((2021) ) The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6: , 74. |
[88] | Lei C , Wang Q ((2022) ) The progression of N6-methyladenosine study and its role in neuropsychiatric disorders. Int J Mol Sci 23: , 5922. |
[89] | Bokar JA , Shambaugh ME , Polayes D , Matera AG , Rottman FM ((1997) ) Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3: , 1233–1247. |
[90] | Schwartz S , Mumbach MR , Jovanovic M , Wang T , Maciag K , Bushkin GG , Mertins P , Ter-Ovanesyan D , Habib N , Cacchiarelli D , Sanjana NE , Freinkman E , Pacold ME , Satija R , Mikkelsen TS , Hacohen N , Zhang F , Carr SA , Lander ES , Regev A ((2014) ) Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep 8: , 284–296. |
[91] | Pendleton KE , Chen B , Liu K , Hunter OV , Xie Y , Tu BP , Conrad NK ((2017) ) The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169: , 824–835 e814. |
[92] | Agarwala SD , Blitzblau HG , Hochwagen A , Fink GR ((2012) ) RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet 8: , e1002732. |
[93] | Patil DP , Chen CK , Pickering BF , Chow A , Jackson C , Guttman M , Jaffrey SR ((2016) ) m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: , 369–373. |
[94] | Wen J , Lv R , Ma H , Shen H , He C , Wang J , Jiao F , Liu H , Yang P , Tan L , Lan F , Shi YG , He C , Shi Y , Diao J ((2018) ) Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell 69: , 1028–1038.e6. |
[95] | Jia G , Fu Y , Zhao X , Dai Q , Zheng G , Yang Y , Yi C , Lindahl T , Pan T , Yang YG , He C ((2011) ) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: , 885–887. |
[96] | Zheng G , Dahl JA , Niu Y , Fedorcsak P , Huang CM , Li CJ , Vagbo CB , Shi Y , Wang WL , Song SH , Lu Z , Bosmans RP , Dai Q , Hao YJ , Yang X , Zhao WM , Tong WM , Wang XJ , Bogdan F , Furu K , Fu Y , Jia G , Zhao X , Liu J , Krokan HE , Klungland A , Yang YG , He C ((2013) ) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: , 18–29. |
[97] | Xiao W , Adhikari S , Dahal U , Chen YS , Hao YJ , Sun BF , Sun HY , Li A , Ping XL , Lai WY , Wang X , Ma HL , Huang CM , Yang Y , Huang N , Jiang GB , Wang HL , Zhou Q , Wang XJ , Zhao YL , Yang YG ((2016) ) Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 61: , 507–519. |
[98] | Wu B , Su S , Patil DP , Liu H , Gan J , Jaffrey SR , Ma J ((2018) ) Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun 9: , 420. |
[99] | Wang X , Zhao BS , Roundtree IA , Lu Z , Han D , Ma H , Weng X , Chen K , Shi H , He C ((2015) ) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161: , 1388–1399. |
[100] | Shi H , Wang X , Lu Z , Zhao BS , Ma H , Hsu PJ , Liu C , He C ((2017) ) YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res 27: , 315–328. |
[101] | Hsu PJ , Zhu Y , Ma H , Guo Y , Shi X , Liu Y , Qi M , Lu Z , Shi H , Wang J , Cheng Y , Luo G , Dai Q , Liu M , Guo X , Sha J , Shen B , He C ((2017) ) Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27: , 1115–1127. |
[102] | Huang H , Weng H , Sun W , Qin X , Shi H , Wu H , Zhao BS , Mesquita A , Liu C , Yuan CL , Hu YC , Huttelmaier S , Skibbe JR , Su R , Deng X , Dong L , Sun M , Li C , Nachtergaele S , Wang Y , Hu C , Ferchen K , Greis KD , Jiang X , Wei M , Qu L , Guan JL , He C , Yang J , Chen J ((2018) ) Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20: , 285–295. |
[103] | Livneh I , Moshitch-Moshkovitz S , Amariglio N , Rechavi G , Dominissini D ((2020) ) The m(6)A epitranscriptome: Transcriptome plasticity in brain development and function. Nat Rev Neurosci 21: , 36–51. |
[104] | Gao P , Yao F , Pang J , Yin K , Zhu X ((2023) ) m (6)A methylation in cellular senescence of age-associated diseases. Acta Biochim Biophys Sin (Shanghai) 55: , 1168–1183. |
[105] | Mathoux J , Henshall DC , Brennan GP ((2021) ) Regulatory mechanisms of the RNA modification m(6)A and significance in brain function in health and disease. Front Cell Neurosci 15: , 671932. |
[106] | Zhang F , Ignatova VV , Ming GL , Song H (2023) Advances in brain epitranscriptomics research and translational opportunities. Mol Psychiatry, doi: 10.1038/s41380-023-02339-x |
[107] | Dermentzaki G , Lotti F ((2020) ) New insights on the role of N (6)-methyladenosine RNA methylation in the physiology and pathology of the nervous system. Front Mol Biosci 7: , 555372. |
[108] | Qiu X , He H , Huang Y , Wang J , Xiao Y ((2020) ) Genome-wide identification of m(6)A-associated single-nucleotide polymorphisms in Parkinson’s disease. Neurosci Lett 737: , 135315. |
[109] | Chen YP , Song W , Huang R , Chen K , Zhao B , Li J , Yang Y , Shang HF ((2013) ) GAK rs1564282 and DGKQ rs11248060 increase the risk for Parkinson’s disease in a Chinese population. J Clin Neurosci 20: , 880–883. |
[110] | Zhou LL , Zhang X , Bao QQ , Liu RP , Gong MY , Mao GY , Zou M , Zhu JH ((2014) ) Association analysis of PARK16-18 variants and Parkinson’s disease in a Chinese population. J Clin Neurosci 21: , 1029–1032. |
[111] | Guo F , Kang J , Xu J , Wei S , Tao J , Dong Y , Ma Y , Tian H , Guo X , Bi S , Zhang C , Lv H , Shang Z , Jiang Y , Zhang M ((2023) ) Genome-wide identification of m(6)A-associated single nucleotide polymorphisms in complex diseases of nervous system. Neurosci Lett 817: , 137513. |
[112] | Martinez De La Cruz B , Gell C , Markus R , Macdonald I , Fray R , Knight HM ((2023) ) m(6) A mRNA methylation in human brain is disrupted in Lewy body disorders. Neuropathol Appl Neurobiol 49: , e12885. |
[113] | Yu Z , Huang L , Xia Y , Cheng S , Yang C , Chen C , Zou Z , Wang X , Tian X , Jiang X , Zhou L ((2022) ) Analysis of m6A modification regulators in the substantia nigra and striatum of MPTP-induced Parkinson’s disease mice. Neurosci Lett 791: , 136907. |
[114] | Geng Y , Long X , Zhang Y , Wang Y , You G , Guo W , Zhuang G , Zhang Y , Cheng X , Yuan Z , Zan J ((2023) ) FTO-targeted siRNA delivery by MSC-derived exosomes synergistically alleviates dopaminergic neuronal death in Parkinson’s disease via m6A-dependent regulation of ATM mRNA. J Transl Med 21: , 652. |
[115] | Chen X , Yu C , Guo M , Zheng X , Ali S , Huang H , Zhang L , Wang S , Huang Y , Qie S , Wang J ((2019) ) Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem Neurosci 10: , 2355–2363. |
[116] | Dominissini D , Moshitch-Moshkovitz S , Salmon-Divon M , Amariglio N , Rechavi G ((2013) ) Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8: , 176–189. |
[117] | Dominissini D , Moshitch-Moshkovitz S , Amariglio N , Rechavi G ((2015) ) Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-Seq. Methods Enzymol 560: , 131–147. |
[118] | Dominissini D , Moshitch-Moshkovitz S , Schwartz S , Salmon-Divon M , Ungar L , Osenberg S , Cesarkas K , Jacob-Hirsch J , Amariglio N , Kupiec M , Sorek R , Rechavi G ((2012) ) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: , 201–206. |
[119] | Hess ME , Hess S , Meyer KD , Verhagen LA , Koch L , Bronneke HS , Dietrich MO , Jordan SD , Saletore Y , Elemento O , Belgardt BF , Franz T , Horvath TL , Ruther U , Jaffrey SR , Kloppenburg P , Bruning JC ((2013) ) The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci 16: , 1042–1048. |
[120] | Teng Y , Liu Z , Chen X , Liu Y , Geng F , Le W , Jiang H , Yang L ((2021) ) Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J Cell Mol Med 25: , 8567–8572. |
[121] | Rasinger JD , Carroll TS , Lundebye AK , Hogstrand C ((2014) ) Cross-omics gene and protein expression profiling in juvenile female mice highlights disruption of calcium and zinc signalling in the brain following dietary exposure to CB-153, BDE-47, HBCD or TCDD. Toxicology 321: , 1–12. |
[122] | Gohlke JM , Stockton PS , Sieber S , Foley J , Portier CJ ((2009) ) AhR-mediated gene expression in the developing mouse telencephalon. Reprod Toxicol 28: , 321–328. |
[123] | Bai L , Tang Q , Zou Z , Meng P , Tu B , Xia Y , Cheng S , Zhang L , Yang K , Mu S , Wang X , Qin X , Lv B , Cao X , Qin Q , Jiang X , Chen C ((2018) ) m6A demethylase FTO regulates dopaminergic neurotransmission deficits caused by arsenite. Toxicol Sci 165: , 431–446. |
[124] | Li N , Zhang D , Cao S , Qiao M , Zhang P , Zhao Q , Shen Y , Huang X , Song L ((2021) ) The effects of folic acid on RNA m6A methylation in hippocampus as well as learning and memory ability of rats with acute lead exposure. J Funct Foods 76: , 104276. |
[125] | Tang J , Zheng C , Zheng F , Li Y , Wang YL , Aschner M , Guo Z , Yu G , Wu S , Li H ((2020) ) Global N6-methyladenosine profiling of cobalt-exposed cortex and human neuroblastoma H4 cells presents epitranscriptomics alterations in neurodegenerative disease-associated genes. Environ Pollut 266: , 115326. |
[126] | Huang M , Bargues-Carot A , Riaz Z , Wickham H , Zenitsky G , Jin H , Anantharam V , Kanthasamy A , Kanthasamy AG ((2022) ) Impact of environmental risk factors on mitochondrial dysfunction, neuroinflammation, protein misfolding, and oxidative stress in the etiopathogenesis of Parkinson’s disease. Int J Mol Sci 23: , 10808. |
[127] | Weed DL ((2021) ) Does paraquat cause Parkinson’s disease? A review of reviews. Neurotoxicology 86: , 180–184. |
[128] | El-Gamal M , Salama M , Collins-Praino LE , Baetu I , Fathalla AM , Soliman AM , Mohamed W , Moustafa AA ((2021) ) Neurotoxin-induced rodent models of Parkinson’s disease: Benefits and drawbacks. Neurotox Res 39: , 897–923. |
[129] | Boyd WA , Blain RB , Skuce CR , Thayer KA , Rooney AA (2020) NTP Research Report on the Scoping Review of Paraquat Dichloride Exposure and Parkinson’s Disease: Research Report 16. National Toxicology Program, Research Triangle Park (NC). |
[130] | Vaccari C , El Dib R , Gomaa H , Lopes LC , de Camargo JL ((2019) ) Paraquat and Parkinson’s disease: A systematic review and meta-analysis of observational studies. J Toxicol Environ Health B Crit Rev 22: , 172–202. |
[131] | Chen N , Tang J , Su Q , Chou WC , Zheng F , Guo Z , Yu G , Shao W , Li H , Wu S ((2021) ) Paraquat-induced oxidative stress regulates N6-methyladenosine (m(6)A) modification of circular RNAs. Environ Pollut 290: , 117816. |
[132] | Su Q , Chen N , Tang J , Wang J , Chou WC , Zheng F , Shao W , Yu G , Cai P , Guo Z , He M , Li H , Wu S ((2022) ) Paraquat-induced oxidative stress regulates N6-methyladenosine (m(6)A) modification of long noncoding RNAs in Neuro-2a cells. Ecotoxicol Environ Saf 237: , 113503. |
[133] | Budinger D , Barral S , Soo AKS , Kurian MA ((2021) ) The role of manganese dysregulation in neurological disease: Emerging evidence. Lancet Neurol 20: , 956–968. |
[134] | Guilarte TR , Gonzales KK ((2015) ) Manganese-induced parkinsonism is not idiopathic Parkinson’s disease: Environmental and genetic evidence. Toxicol Sci 146: , 204–212. |
[135] | Qi Z , Wang S , Li J , Wen Y , Cui R , Zhang K , Liu Y , Yang X , Zhang L , Xu B , Liu W , Xu Z , Deng Y ((2022) ) Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism.. J Hazard Mater 426: , 128099. |
[136] | Begik O , Mattick JS , Novoa EM ((2022) ) Exploring the epitranscriptome by native RNA sequencing. RNA 28: , 1430–1439. |
[137] | Zhang Y , Lu L , Li X ((2022) ) Detection technologies for RNA modifications. Exp Mol Med 54: , 1601–1616. |
[138] | Kochmanski J , Bernstein AI (2024) Chapter 12 - Best practices for epigenome-wide DNA modification data collection and analysis. In Rigor and Reproducibility in Genetics and Genomics, Dluzen DF, Schmidt MHM, eds. Academic Press, pp. 261-284. |
[139] | Koo HJ , Piao Y , Pak YK ((2012) ) Endoplasmic reticulum stress impairs insulin signaling through mitochondrial damage in SH-SY5Y cells. Neurosignals 20: , 265–280. |
[140] | Zheng Y , Nie P , Peng D , He Z , Liu M , Xie Y , Miao Y , Zuo Z , Ren J ((2018) ) m6AVar: A database of functional variants involved in m6A modification. Nucleic Acids Res 46: , D139–D145. |
[141] | Wilkinson MD , Dumontier M , Aalbersberg IJ , Appleton G , Axton M , Baak A , Blomberg N , Boiten JW , da Silva Santos LB , Bourne PE , Bouwman J , Brookes AJ , Clark T , Crosas M , Dillo I , Dumon O , Edmunds S , Evelo CT , Finkers R , Gonzalez-Beltran A , Gray AJ , Groth P , Goble C , Grethe JS , Heringa J , t Hoen PA , Hooft R , Kuhn T , Kok R , Kok J , Lusher SJ , Martone ME , Mons A , Packer AL , Persson B , Rocca-Serra P , Roos M , van Schaik R , Sansone SA , Schultes E , Sengstag T , Slater T , Strawn G , Swertz MA , Thompson M , van der Lei J , van Mulligen E , Velterop J , Waagmeester A , Wittenburg P , Wolstencroft K , Zhao J , Mons B ((2016) ) The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3: , 160018. |
[142] | Nosek BA , Ebersole CR , DeHaven AC , Mellor DT ((2018) ) The preregistration revolution. Proc Natl Acad Sci U S A 115: , 2600–2606. |
[143] | Foster ED , Deardorff A ((2017) ) Open Science Framework (OSF). J Med Libr Assoc 105: . doi: https://doi.org/10.5195/jmla.2017.88 |
[144] | Percie du Sert N , Hurst V , Ahluwalia A , Alam S , Avey MT , Baker M , Browne WJ , Clark A , Cuthill IC , Dirnagl U , Emerson M , Garner P , Holgate ST , Howells DW , Karp NA , Lazic SE , Lidster K , MacCallum CJ , Macleod M , Pearl EJ , Petersen OH , Rawle F , Reynolds P , Rooney K , Sena ES , Silberberg SD , Steckler T , Wurbel H ((2020) ) The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. BMJ Open Sci 4: , e100115. |
[145] | Shafik AM , Allen EG , Jin P ((2022) ) Epitranscriptomic dynamics in brain development and disease. Mol Psychiatry 27: , 3633–3646. |




