A molecular protocol for Early Sex Discrimination (ESD) in Actinidia spp
Abstract
Dioecism and an extended juvenile phase of 3–7 years in kiwifruit hinder the progress in breeding new cultivars. The identification of fruit-bearing females at an early stage of growth is crucial for breeders. Consequently, molecular markers have become a key tool for identifying female and male plants at an early stage of development. Several efforts were made to identify PCR-based sex linked markers in Actinidia; however, those markers are characterized by a highly polymorphic nature affecting the result of the screening reliability, suggesting the need of more suitable, stable markers, characterized by a consistent transferability among genotypes and species. The main goal of this work was to develop a method for the ultimate discrimination of females from male plants at an early stage of growth using sex-linked markers. We developed an Early Sex Discrimination molecular Test (ESD Test) that allows the discrimination of male and female plants using a simple PCR amplification test. We demonstrate that the test could unequivocally identify the gender of an unknown sample both in the most commercially important species A. chinensis and in further 13 Actinidia species tested with the exception of Actinidia latifolia, where markers fail in gender discrimination. Male genotypes could be easily identified and discarded reducing the cost of a breeding program.
1Introduction
The genus Actinidia Lindl. includes about 54 species of climbing plants, all dioecious, originating mainly in central and southern China [1]. Kiwifruit is one of the most recently domesticated fruit crops. The development of new cultivars showing interesting traits for the consumer, e.g., taste, appearance, and health components, has driven the commercial success of this fruit and kiwifruit has become an important horticultural crop over the years, firstly in New Zealand and subsequently in many other countries such as Chile, China and Italy [1, 2].
The kiwifruit is dioecious, with male and female flowers carried on different plants. Male individuals, bearing staminate flowers with numerous stamens and a rudimentary ovary lacking ovules, stand out from female individuals that carry well-developed ovaries but do not produce viable pollen. Gender is monofactorial and it is apparently controlled by a single Mendelian determinant, with the female sex being homogametic (XX) and the male sex heterogametic (XY) [3–5]. Any cross between female and male genotypes usually produces female and male offspring in the ratio of 1 : 1 whatever the ploidy level [3]. Male plants are a waste of land and resources because breeders are usually focused on selecting fruit-bearing individuals. The situation is exacerbated by the long generation cycle which is spread at least between three seasons, interrupted by winter dormancy [6]. Development and introduction of improved cultivars may require many breeding cycles and decades [7]. The management cost of relatively large cross populations over several years is a limiting factor in any woody species-breeding program, particularly in fruit crops like kiwifruit, where expensive support structures are required. Since kiwifruit breeders are seldom interested in selecting males in breeding programs, a great effort has gone into the search for sex markers. The ability to discard male seedlings at a very early stage through the analysis of associated markers would save labor and space in the orchard.
The sex determinant was mapped in the sub-telomeric region of chromosome 17 [8], which is the pseudomolecule 25 in the more recent genome assembly of Actinidia chinensis var. chinensis (genotype Red5) [9]. The suppression of recombination in this region is the key feature of sex chromosomes, allowing the two chromosomes of the pair to evolve separately and the multiple genes involved to be inherited as a single genetic determinant, so that only male and female progeny are produced [10–12]. Therefore, the finding of markers very close to the sex determinants is challenging and it is also difficult to establish the order and the distances that separate markers and sex-genes, considering also the relatively small size of the populations studied. The first attempt to find markers linked to gender was based on the combination of RAPD markers and bulk segregant analysis (BSA). The screening of 500 random primers allowed the discovery of two sex markers, which were then transformed into three SCAR (Sequence-Characterized Amplified Regions) markers, by cloning and sequencing the original RAPD and designing allele-specific primers [13]. These markers were soon deployed routinely in marker-assisted kiwifruit breeding, to eliminate male plants at the seedling stage [14]. The closest gender markers reported up to now are the markers SmX, UDK096, SmY1 and Ke225 spanning 4 cM in the region where the sex phenotype maps. The last one is now preferred in breeding programs in New Zealand (http://www.plantandfood.co.nz), but the primers have not yet been published. On the other hand, markers such as SmX and SmY1 were used in the past but they are not always transferable from one cross population to another and from one species to another and if they are, they sometimes lose their polymorphism [4, 13]. More recently, SNP-based genetic maps have been published that allowed the anchoring of sex-surrounding scaffolds onto the genome assembly and the isolation of new SSR markers from those scaffolds. These new SSR markers could discriminate between male and female progeny in most crosses analyzed [15, 16] whatever the ploidy level of the cross parents. Other markers named A001, A002 and A003 were also successfully amplified in A. rufa and interspecific crosses with A. chinensis var. chinensis [16].
However, because of the highly polymorphic nature of SSR markers, the sex-linked alleles were not always of the same length in the different crosses assayed. Therefore, the identification of the alleles carried by the cross parents is necessary for the analysis of the allele associated with the gender in the progeny. These markers are therefore not suitable for screening germplasm collections where the parentage of accessions is not known in advance [15, 17]. Consequently, new markers associated with the sex determinants that are more appropriate, stable, and characterized by greater transportability among genotypes and species are necessary.
Recently, the two Y-encoded sex determinants in kiwifruit were described by Akagi and co-workers. The Shy Girl (SyGI) gene, which is a Y-encoded cytokinin response regulator that acts as the suppressor of female development [18] is expressed in developing flowers, specifically at the surface of the rudimentary carpels of male flowers. The Friendly Boy (FrBy), a fasciclin-like protein which would maintain male fertility, exhibits strong expression in tapetal cells [19]. Moreover, FrBy acts for the maintenance of male (M) functions, independently of SyGI. These two sex determinants are located at an estimated distance of 500 kb in the specific region of the Y chromosome while they are absent in the corresponding chromosomal female region.
In the present study, we set up an early sex discrimination molecular test (ESD test) using primers designed on the SyGI and FrBy genes that can discriminate between male and female plants with a simple signal of presence/absence both among the species and on genotypes within different ploidies. We also include the housekeeping Ankyrin repeat domain-containing protein (Ank) marker that acts as internal control to test successful PCR amplification.
We developed a simple experimental protocol, which requires only a crude leaf extract, thus reducing the time needed for DNA extraction. The amplicons can be separated on both agarose gel and capillary sequencer. The experimental protocol can be used for rapid selection of female genotypes, reducing the management costs of breeding programs.
2Materials and methods
2.1Plant materials
Young leaves from 43 different male and female Actinidia genotypes were collected from the kiwifruit repository of the experimental farm “A. Servadei” of the University of Udine, Italy. We selected 13 unrelated genotypes from A. chinensis var. chinensis and A. chinensis var. deliciosa and 30 genotypes from 14 other Actinidia species. The genotypes collected are listed in Table 1. Young leaves were collected as duplicate samples during the growing season. In addition, leaves from 31 individuals were collected from two different cross-populations named Ac565 (cross A. chinensis var. chinensis ‘Jintao’ X Ac442 male pollen mix) and Ac567 (cross Ac0171.76 ‘Soreli’ (A. chinensis var. chinensis) × A0074 male ‘Cornell’ (A. arguta)).
Table 1
List of genotypes used in this work
| Genotype | Gender | Species |
| A0181 | M | A. chinensis var. chinensis |
| A0202.32 | F | A. chinensis var. chinensis |
| A0182 | M | A. chinensis var. chinensis |
| Ac453.004 | F | A. chinensis var. chinensis |
| Ac178.15 | M | A. chinensis var. chinensis |
| Ac175.98 | I | A. chinensis var. chinensis |
| A0185.1 Ecor | F | A. chinensis var. deliciosa |
| A0185.2 Ecor | M | A. chinensis var. deliciosa |
| A0102 Lea | I | A. chinensis var. deliciosa |
| A0186 Hayward clone 8 | F | A. chinensis var. deliciosa |
| A0188 Summer 3373 | F | A. chinensis var. deliciosa |
| A0041 Matua | M | A. chinensis var. deliciosa |
| A0042 Tomuri | M | A. chinensis var. deliciosa |
| A0105.2 | F | A. eriantha |
| A0114.4 | F | A. eriantha |
| A0114.2 | M | A. eriantha |
| A0104.4 | F | A. chrysantha |
| A0104.5 | F | A. chrysantha |
| A0104.2 | M | A. chrysantha |
| A0106.6 | F | A. latifolia |
| A0162.5 | M | A. latifolia |
| A0151.1 | F | A. macrosperma |
| A0176 | M | A. macrosperma |
| A0063 | F | A. kolomikta |
| A0064 | F | A. kolomikta |
| A0065 | M | A. kolomikta |
| A0152 | F | A. arisanensis |
| A0175 | F | A. valvata |
| A0132.1 | M | A. valvata |
| A083.3 | M | A. lanceolata |
| A0074 Cornell | M | A. arguta |
| A0091 Issai | F | A. arguta |
| A0068 | M | A. arguta |
| Miss green | F | A. arguta |
| A0103 | M | A. callosa |
| A0124.1 | M | A. rufa |
| A0124.2 | F | A. rufa |
| A0121 | M | A. melanandra |
| A0184 | F | A. melanandra |
| A0180 | F | A. arguta var. purpurea |
| A0069 | F | A. polygama |
| A0070 | M | A. polygama |
| A0050 | F | A. hemsleyana |
Column 1 = accession number or variety name; Column 2 = M=male, F = female, I = inconstant male; Column 3 = species and botanical variety.
22 DNA extraction methods
Genomic DNA of the first replicate of samples was extracted using NucleoSpin® Plant II kit (Macherey-Nagel) starting from lyophilized material ground using the Tissue Lyser (30 Hz×2 min×2v). Briefly, plant samples after homogenization were extracted with Lysis Buffer PL1, which is based on the CTAB (cetyltrimethylammonium bromide) extraction method. DNA quantification was performed using the Nanodrop ND-100 spectrophotometer.
The second replicate of samples was used to test a quick protocol of extraction (Tyrone Possamai 2021, pers comm) which produces a crude extract suitable for DNA analysis in less than one hour. Briefly, lyophilized leaf samples were ground using the Tissue Lyser (30 Hz×2 min×2v). Then, 450μl of lysis buffer were added and the samples were thoroughly mixed. The samples were then incubated for 15–20 min at 65°C. Next 130μl of precipitation buffer were added to the samples, they were mixed for 15 sec, put on ice for 10 min and then centrifuged for 20 min at 15 000×g. Finally, the clear supernatant was collected for analysis.
Lysis Buffer is the AP1 lysis buffer (Qiagen) based on SDS extraction method. This buffer can be made in the laboratory by mixing 0.5%W/V SDS (sodium dodecyl sulfate), 8%W/V PVP-10 (optional), 250 mM sodium chloride, 25 mM NaEDTA and 200 mM Tris –HCl, pH 7.5. Precipitation buffer is the P3 precipitation buffer (Qiagen) containing 5M potassium acetate.
2.3Targets amplification using different methods
Primers were designed within the coding region of the Ankirin repeat domain-containing protein (Ank) in the LG1 of Red5 reference genome (18,539,780–18,539,855, 18,540,915–18,541,078 bp) and Friendly Boy (FrBy) genes using Primer 3 [20]. Primers specific for Shy Girl (SyGI) gene were published by Akagi and co-workers [18]. Primer sequences are reported in Supplementary Table S1. The primer pairs were selected aiming for the amplification of three size-distinguishable PCR products. PCR reactions for the amplification of the three targets were performed using the 5 PRIME HotMaster® Taq DNA Polymerase (QuantaBio) on the samples extracted with NucleoSpin® Plant II kit (Macherey-Nagel). The PCR reaction recipe is given in Tables 2a and 2b. The PCR thermal conditions were 94°C for 2 min, followed by 30 cycles at 94°C for 20 sec, 50°C for 20 sec and 65°C for 30 sec, and a final extension at 65°C for 5 min. The amplification success was verified running PCR products on a 1.5%agarose gel.
Table 2a
PCR reaction set up using FrBy-UD-F/ FrBy-UD-R primers pair
| Solution | Final concentration | Sample volume (μl) |
| Water | Variable | |
| Buffer 10X | 1X | 1 |
| FrBy-UD-F | 0.2μM | 1 |
| FrBy-UD-R | 0.2μM | 1 |
| dNTPs | 0.2 mM | 0.8 |
| 5’ prime Taq Polymerase | 2.5 U | 0.1 |
| Sample DNA | 20 ng | |
| Total volume | 10 |
Table 2b
PCR reaction set up using SyGI-selectF/ SyGI-selectR and Ank-UD-F/ Ank-UD-R primers pairs
| Solution | Final concentration | Sample volume (μl) |
| Water | Variable | |
| Buffer 10X | 1X | 1 |
| SyGI-selectF | 0.2μM | 1 |
| SyGI-selectR | 0.2μM | 1 |
| Ank-UD-F | 0.2μM | 1 |
| Ank-UD-R | 0.2μM | 1 |
| dNTPs | 0.2 mM | 0.8 |
| 5’ prime Taq Polymerase | 2.5 U | 0.1 |
| Sample DNA | 20 ng | |
| Total volume | 10 |
A dilution 2 : 38 was prepared from the crude extracts obtained from the quick extraction protocol. In this case, PCR reactions were performed using MyTaqTM Plant-PCR kit (Bioline). The PCR reaction set up is reported in Tables 3a and 3b. Touch-down PCR cycling program was 95°C for 3 min, followed by 10 cycles at 95°C for 15 sec, 55°C (- 0.5°C at each cycle) for 15 sec and 72°C for 45 sec, and 20 cycles at 95°C for 15 sec, 50°C for 15 sec and 72°C for 45 sec, and a final extension at 72°C for 7 min.
Table 3a
PCR reaction set up using FrBy-UD-F/ FrBy-UD-R primers pair
| Solution | Final concentration | Sample volume (μl) |
| Water | 1 | |
| FrBy-UD-F | 0.2μM | 1 |
| FrBy-UD-R | 0.2μM | 1 |
| MyTaq Plant-Polymerase | 1X | 5 |
| DNA dilution | 2 | |
| Total volume | 10 |
Table 3b
PCR reaction set up using SyGI-selectF/ SyGI-selectR and Ank-UD-F/ Ank-UD-R primers pairs
| Solution | Final concentration | Sample volume (μl) |
| Water | Variable | |
| SyGI-selectF | 0.2μM | 1 |
| SyGI-selectR | 0.2μM | 1 |
| Ank-UD-F | 0.2μM | 1 |
| Ank-UD-R | 0.2μM | 1 |
| MyTaq Plant-Polymerase | 1X | 5 |
| DNA dilution | 2 | |
| Total volume | 10 |
The three gene targets were amplified with FAM or HEX dye-labelled forward primers. The PCR products were diluted 2 to 18 in case of FrBy amplification and 1 to 100 in case of SyGI and Ank amplification, mixed and run on a SeqStudio Genetic Analyzer (Thermo Fisher). The fragments were called and sized using the Microsatellite Analysis Software (MSA), which is a microsatellite genotyping module available on the Thermo Fisher Cloud.
3Results and discussion
3.1Method Set up
The amplification of SyGI gene, Ank gene and FrBy gene allowed the discrimination between male and female plants with a simple signal of presence/absence, both in the Actinidia species and in kiwifruit genotypes with different ploidy [21]. Specifically, male plants amplify three PCR products in contrast to female plants that amplify only the housekeeping gene (Fig. 1). The predicted sizes in A. chinensis genome were 90, 155, and 290 bp for SyGI, Ank, and FrBy markers, respectively.
Fig. 1
Schematic representation of the ESD protocol.
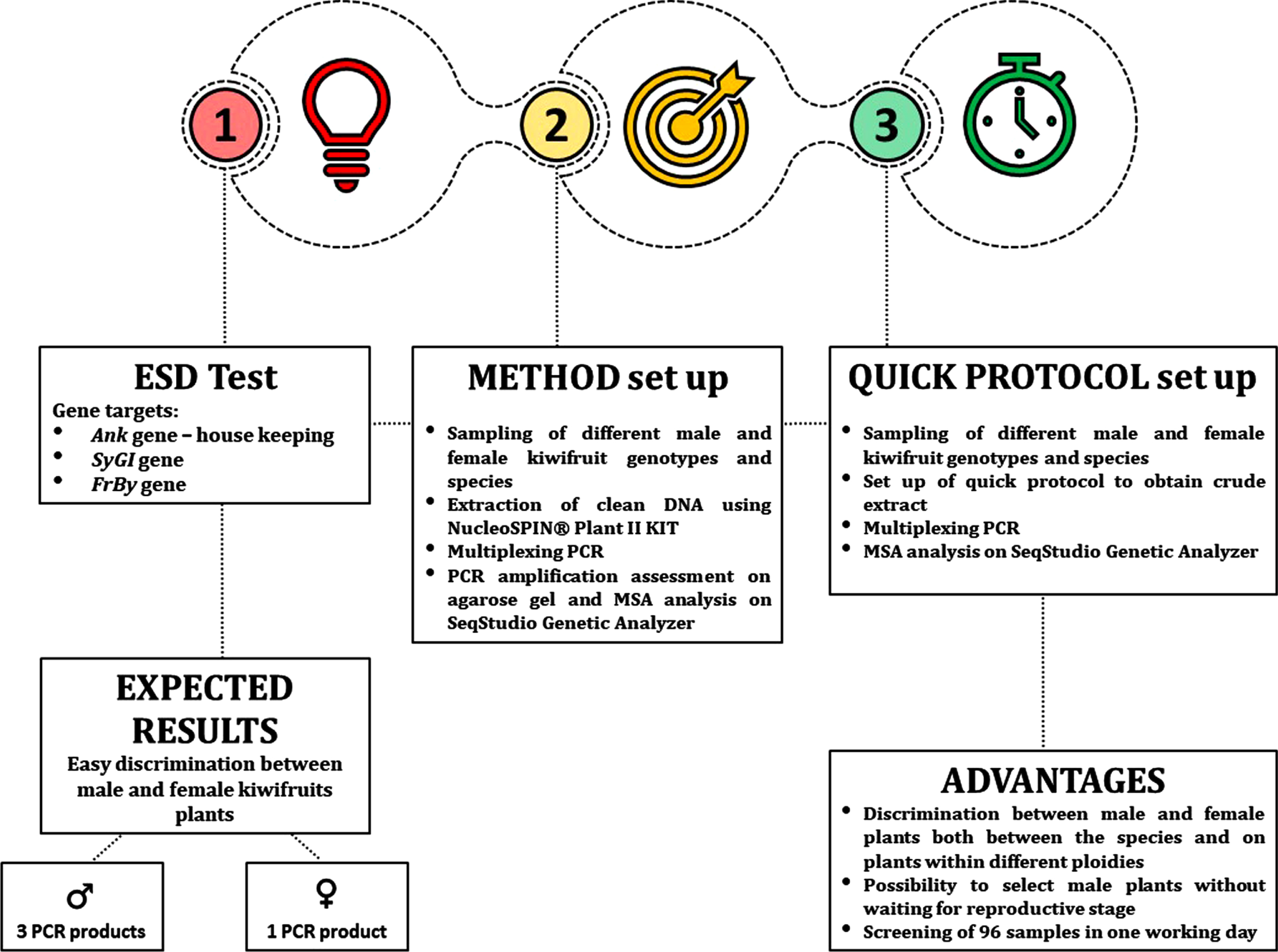
The SyGI primer pair was published by Akagi and co-workers [18] and provides male-specific amplification in most of Actinidia species studied. FrBy and Ank primer pairs were designed specifically for this work. Ank marker acts as internal control to test successful PCR amplification, while our FrBy primers pair also face male-specific amplification and was used as double check of the discriminating test. PCR amplifications were initially set up using genomic DNA of different species of Actinidia, extracted using NucleoSpin® Plant II kit and visualized on agarose gel (Fig. 2). Later, we perfected a quick protocol that allowed the process to be speeded up consistently, allowing up to 192 samples to be screened in a working day.
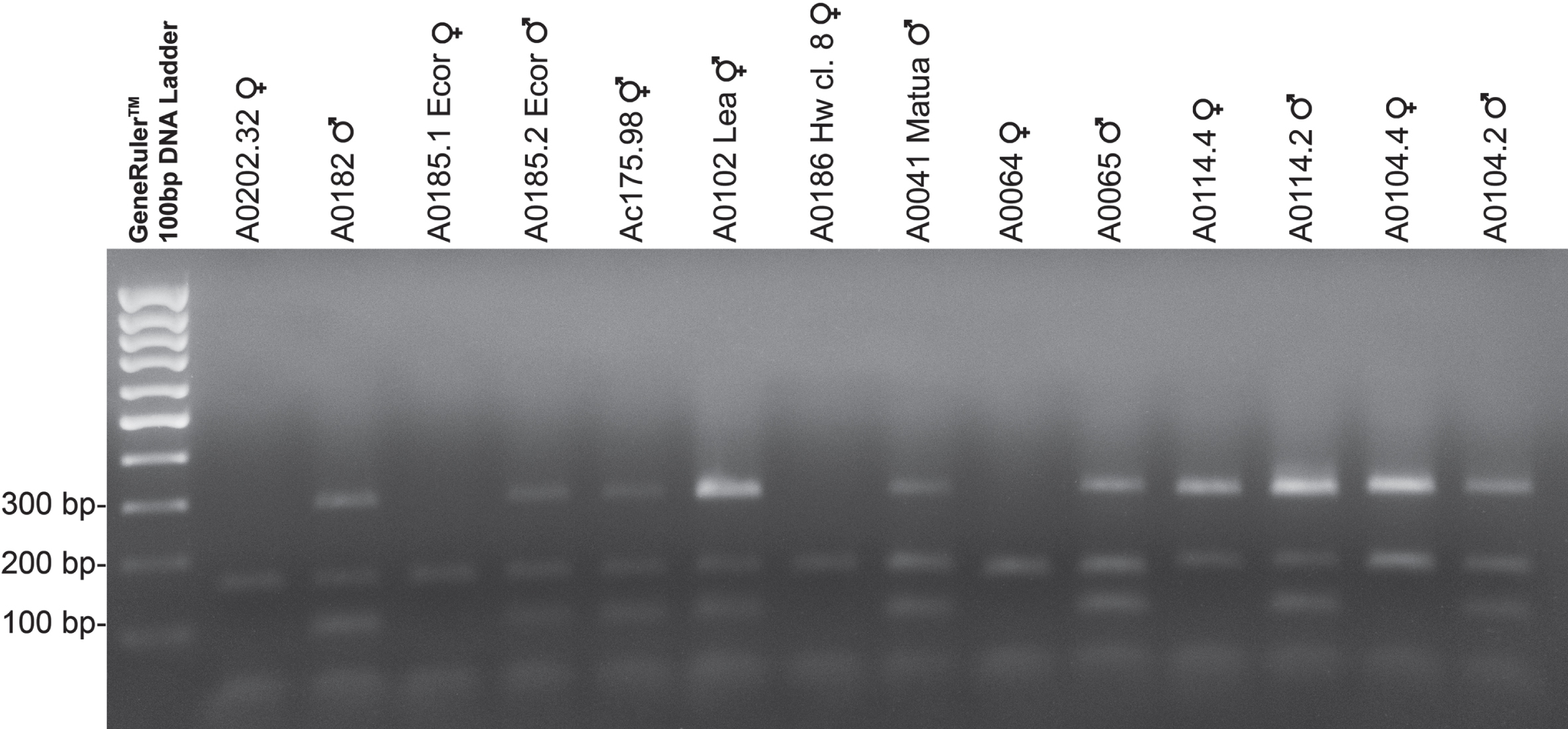
Fig. 2
Panel of amplifications for different genotypes and species of kiwifruit. The panel shows markers amplification in a sample of genotypes and species of kiwifruit. Genotypes were choose to show different case studies. More specifically the panel shows two diploid genotypes of A. chinensis var. chinensis (A0202.32 and A0182), two tetraploid genotypes of A. chinensis var. deliciosa (A0185.1 Ecor and A0185.2 Ecor), two inconstant male genotypes Ac175.98 (A. chinensis var. chinensis) and Ac102 Lea (A. chinensis var. deliciosa), two exaploid genotypes of A. chinensis var. deliciosa (A0186 Hw cl. 8 and A0041 Matua), two genotypes of A. kolomikta (A0064 and A0065), two genotypes of A. eriantha (A0114.4 and A0114.2) and two genotypes of A. chrysantha (A0104.4 and A0104.2). Male and inconstant male plants show three size-distinguishable PCR products: 90 bp, 154 bp and 290 bp respectively for SyGI marker, Ank marker and FrBy marker. The PCR products of the female plants were loaded into the wells 2, 4, 8, 10, 12, 14. In lanes 9 and 10, A. kolomikta female and male genotypes were loaded which showed a peak corresponding to a fragment 144 bp in size in the case of Ank marker (see text for details). GeneRulerTM 100 bp DNA Ladder (Thermo Fisher) was used for sizing the PCR products in the range of 100 to 1,000 bp on agarose gel.
3.2Marker Assisted Selection (MSA) analysis for A. chinensis var. chinensis and A. chinensis var. deliciosa
We tested six different genotypes of A. chinensis var. chinensis and seven different genotypes of A. chinensis var. deliciosa: three male genotypes, two female genotypes and one inconstant male in the case of A. chinensis var. chinensis, and three male genotypes, three female genotypes and one inconstant –male in the case of A. chinensis var. deliciosa. Inconstant males were identified from our kiwifruit repository. These genotypes, also referred to as fruiting males, have bisexual flowers characterized by a small ovary with fewer carpels than a typical female vine, fewer ovules per carpel and shortened, thinner styles with small stigmata and they bear small fruits (20–40 g) with a few dozens of seeds. The ESD test has been effective in the discriminating between male and female for all genotypes.
Pherograms of female genotypes showed only one peak, as expected, corresponding to the fragment of 154 bp from the Ank gene (housekeeping gene) in both A. chinensis var. chinensis and A. chinensis var. deliciosa (Fig. 3). Pherograms of male and inconstant male genotypes showed three peaks, corresponding to the fragments 86 bp, 154 bp and 289 bp, resulting from the amplification of SyGI, Ank and FrBy genes, respectively. Peak size was the same for all genotypes tested with the exception of the genotype Ac178.15. This male of A. chinensis shows a peak of 292 bp in the case of the FrBy gene. Results for all genotypes are summarized in Table 4. Our test could not distinguish between males and inconstant males.
Fig. 3
Representative pherograms of eight genotypes analyzed. Pherograms of two individuals of A. chinensis var. chinensis and two individuals of A. chinensis var. deliciosa, female and male respectively on the left panel. Pherograms of two individuals of the cross-population Ac565 (a controlled cross A. chinensis var. chinensis ‘Jintao’ X Ac442 male pollen mix) and two individuals the cross-population Ac567 (a controlled cross A. chinensis var. chinensis ‘Soreli’ x A. arguta ‘Cornell’ male), female and male respectively on the right panel. Pherograms show the fragments size in base pairs (bp) on the x-axis and the intensity of the peaks on y-axis. The marker Ank (green peak) carries the fragment of 154 bp. The markers SyGI and FrBy (blue peaks) carry the fragment of 86 bp and 289 bp respectively in the case of A. chinensis var. chinensis and A. chinensis var. deliciosa genotypes. Fragment size for these two marker varies in the case of the cross populations. Male individuals of the cross-population Ac565 showed peaks corresponding to the fragments of 86 and 292 bp from the SyGI and FrBy genes respectively and fragments of 88 and 289 bp in the case of the cross population Ac567. In yellow the size-standard peaks.
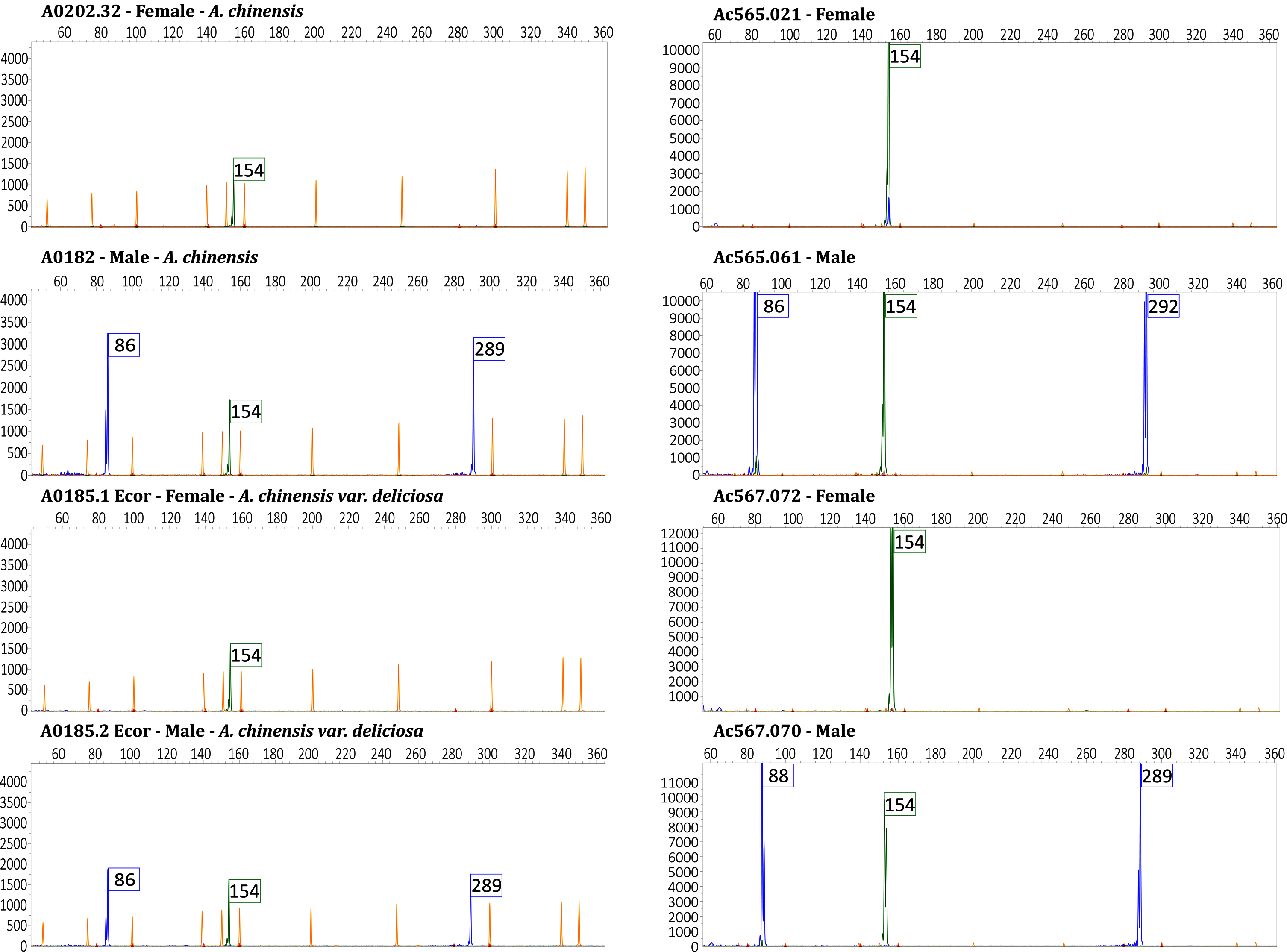
Table 4
Results of MSA analysis for A. chinensis var. chinensis and A. chinensis var. deliciosa
| Genotype | Gender | Species | Marker Name | Gel electrophoresis | Dye Color | Size |
| A0181 | M | A. chinensis var. chinensis | Ank | yes | Green | NA |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| A0202.32 | F | A. chinensis var. chinensis | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0182 | M | A. chinensis var. chinensis | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| Ac0453.004 | F | A. chinensis var. chinensis | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| Ac0178.15 | M | A. chinensis var. chinensis | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 292 | |||
| SyGI | yes | Blue | 86 | |||
| Ac0275.98 | I | A. chinensis var. chinensis | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| A0185.1 Ecor | F | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0185.2 Ecor | M | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| A0102 Lea | I | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| A0186 Hayward clone 8 | F | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0188 Summer 3373 | F | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0041 Matua | M | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| A0042 Tomuri | M | A. chinensis var. deliciosa | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 |
Column 1 = accession number or variety name; Column 2 = M = male, F = female, I = inconstant male; Column 3 = species and botanical variety; Column 4 = marker name as specified in the text; Column 5 = amplicons separated on gel electrophoresis (yes = band detected, no = band not detected); Column 6 = dye colors of the peaks after capillary electrophoresis analysis; Column 7 = Size of the peaks in base pairs (NA = PCR amplification failed; – = PCR amplification not expected).
3.3MSA analysis of different Actinidia species
We tested 30 genotypes from the following species of Actinidia: A. eriantha, A. chrysantha, A. latifolia, A. macrosperma, A. kolomikta, A. arisanensis (the classification of this species is still uncertain), A. valvata, A. lanceolata, A. arguta, A. callosa, A. rufa, A. melanandra, A. polygama and A. hemsleyana (Table 1). We selected one female and one male genotype from each species when available. The housekeeping gene was amplified in both male and female genotypes and pherograms showed a peak corresponding to the fragment 154 bp in size for all Actinidia species with the exception of A. kolomikta which showed a peak corresponding to a fragment of 144 bp in size in both the two female genotypes and in the male one. This deletion can be detected also by gel electrophoresis (Fig. 2). Pherograms of all males in different species showed three peaks resulting from the SyGI, Ank and FrBy markers amplification respectively. SyGI amplification failed only in A. latifolia, presumably because of a mutation in the annealing site of the primers. SyGI amplicon size for the different species ranges from 86 bp of A. rufa, to 91 bp of A. polygama. The amplification of FrBy resulted in the amplification of a 289 bp peak in all species except A. macrosperma, A. valvata and A. polygama, where the fragment size was of 292 bp and A. eriantha where the fragment was of 282 bp. Specific SyGI, FrBy and Ank genes amplicon sizes of all species are summarized in Table 5.
Table 5
Results of MSA analysis of different Actinidia species
| Genotype | Gender | Species | Marker Name | Gel electrophoresis | Dye Color | Size |
| A0105.2 | F | A. eriantha | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 282* | |||
| SyGI | no | Blue | – | |||
| A0114.4 | F | A. eriantha | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 282* | |||
| SyGI | no | Blue | – | |||
| A0114.2 | M | A. eriantha | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 282 | |||
| SyGI | yes | Blue | 88 | |||
| A0104.4 | F | A. chrysantha | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289* | |||
| SyGI | no | Blue | – | |||
| A0104.5 | F | A. chrysantha | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289* | |||
| SyGI | no | Blue | – | |||
| A0104.2 | M | A. chrysantha | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 89 | |||
| A0152 | F | A. arisanensis | Ank | yes | Green | 154 |
| FrBy | no | Blue | 289* | |||
| SyGI | no | Blue | – | |||
| A0106.6 | F | A. latifolia | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289* | |||
| SyGI | no | Blue | – | |||
| A0162.5 | M | A. latifolia | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | no | Blue | NA | |||
| A0151.1 | F | A. macrosperma | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0183 | M | A. macrosperma | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 292 | |||
| SyGI | yes | Blue | 89 | |||
| A0063 | F | A. kolomikta | Ank | yes | Green | 144 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0064 | F | A. kolomikta | Ank | yes | Green | 144 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0065 | M | A. kolomikta | Ank | yes | Green | 144 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 88 | |||
| A0175 | F | A. valvata | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0132.1 | M | A. valvata | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 292 | |||
| SyGI | yes | Blue | 89 | |||
| A0083.3 | M | A. lanceolata | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 89 | |||
| A0074 Cornell | M | A. arguta | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 88 | |||
| A0091 Issai | F | A. arguta | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0068 | M | A. arguta | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 88 | |||
| Miss green | F | A. arguta | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0103 | M | A. callosa | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | no | Blue | 88 | |||
| A0124.1 | M | A. rufa | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 86 | |||
| A0124.2 | F | A. rufa | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0121 | M | A. melanandra | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 289 | |||
| SyGI | yes | Blue | 88 | |||
| A0184 | F | A. melanandra | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0180 | F | A. arguta var. purpurea | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0069 | F | A. polygama | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – | |||
| A0070 | M | A. polygama | Ank | yes | Green | 154 |
| FrBy | yes | Blue | 292 | |||
| SyGI | yes | Blue | 91 | |||
| A0050 | F | A. hemsleyana | Ank | yes | Green | 154 |
| FrBy | no | Blue | – | |||
| SyGI | no | Blue | – |
Column 1 = accession number or variety name; Column 2 = M=male, F = female; Column 3 = species and botanical variety; Column 4 = marker name as specified in the text; Column 5 = amplicons separated on gel electrophoresis (yes = band detected, no = band not detected); Column 6 = dye colors of the peaks after capillary electrophoresis analysis; Column 7 = Size of the peaks in base pairs (NA = PCR amplification failed; – = PCR amplification not expected; *size of the peaks in base pairs reported in italics = unexpected amplification of the FrBy marker in female genotypes).
In four specific cases of female genotypes of A. eriantha, A. chrysantha, A. latifolia and A. arisanensis we observed the unexpected amplification of the FrBy marker. Female genotypes of A. eriantha amplified a fragment of 282 bp in size, while the remaining three female genotypes amplified a fragment of 289 bp in size. At flowering, flowers of A. eriantha were bagged to avoid external pollen contamination. In these conditions, we tested pollen fertility controlled by FrBy gene and if gene function was not impaired, we expected to observe evidence of self-pollination. We did not observe self-pollination and fruit set (Fig. 4). Moreover, we amplified the entire FrBy gene using primers pair described by Akagi and co-workers [19] producing an amplicon of 723 bp, but in the case of A. eriantha female the amplification did not occur. These observations supported the speculation that at least four Actinidia species traces of an interrupted or degenerate FrBy gene sequence are still present, but further investigations are needed.
Fig. 4
Pollen fertility controlled by FrBy gene. A comparison between A. eriantha selfed and open pollinated: fruit set failed after self-pollination of bag-isolated flowers on the left, while open-pollinated fruits are developing normally on the right. See text for details.

3.4MSA analysis of cross-populations
We tested 20 individuals collected from the cross-population named Ac565 (a controlled cross A. chinensis var. chinensis ‘Jintao’ X Ac442 male pollen mix) and 11 individuals from Ac567 (a controlled cross A. chinensis var. chinensis ‘Soreli’ x A. arguta ‘Cornell’ male). Phenotypic evaluation identified 15 male and five female plants in the Ac565 cross population and ten male and one female plants in the Ac567 cross population. The ESD test was effective in discriminating male and female individuals. Pherograms of female genotypes showed only one peak corresponding to the 154 bp fragment from the Ank gene (housekeeping gene) for both cross populations. Pherograms of male individuals showed three peaks corresponding to the fragments of 86, 154, and 292 bp resulting from the SyGI, Ank and FrBy genes amplification respectively in the case of the cross population Ac565 and fragments of 88, 154, and 289 bp in the case of the cross population Ac567 (Fig. 3). Results for all the genotypes are summarized in Table 6.
Table 6
Results of MSA analysis for the cross populations Ac565 and Ac567
| Genotype | Gender | Marker Name | Dye Color | Size |
| Ac565.008 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.020 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.021 | F | Ank | Green | 154 |
| FrBy | Blue | – | ||
| SyGI | Blue | – | ||
| Ac565.024 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.025 | F | Ank | Green | 154 |
| FrBy | Blue | – | ||
| SyGI | Blue | – | ||
| Ac565.033 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.035 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.042 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.044 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.057 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.058 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.059 | F | Ank | Green | 154 |
| FrBy | Blue | – | ||
| SyGI | Blue | – | ||
| Ac565.061 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.066 | F | Ank | Green | 154 |
| FrBy | Blue | – | ||
| SyGI | Blue | – | ||
| Ac565.068 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.071 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.074 | F | Ank | Green | 154 |
| FrBy | Blue | – | ||
| SyGI | Blue | – | ||
| Ac565.077 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.084 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac565.089 | M | Ank | Green | 154 |
| FrBy | Blue | 292 | ||
| SyGI | Blue | 86 | ||
| Ac567.030 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.042 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.048 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.059 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.061 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.066 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.070 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.072 | F | Ank | Green | 154 |
| FrBy | Blue | – | ||
| SyGI | Blue | – | ||
| Ac567.078 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.096 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 | ||
| Ac567.127 | M | Ank | Green | 154 |
| FrBy | Blue | 289 | ||
| SyGI | Blue | 88 |
Column 1 = accession number; Column 2 = M=male, F = female; Column 3 = marker name as specified in the text; Column 4 = dye colors of the peaks after capillary electrophoresis analysis; Column 5 = Size of the peaks in base pairs (– = PCR amplification not expected).
4Conclusion
Methods for determining sex are valuable in dioecious fruit crops because plants cannot be predicted morphologically at an early seedling stage. Prediction of sex at the seedling stage would save a huge amount of resources, such as space required for planting, and the labor of plant management.
DNA based markers for sex-determinants have proved to be a very successful tool for inferring the gender in dioecious plants. Sex-linked molecular markers have been already developed for different woody plants such as papaya (Carica papaya) [22], ginkgo (Ginkgo biloba) [23], date-plum (Diospyros lotus) [24], mulberry (Morus alba L.) [25], Garcinia subelliptica Merr. [26], pistachio (Pistacia vera) [27]. Recently, Chłosta and co-workers [28] exploited the SyGI marker to exclude male and identify female lines in endosperm-derived kiwifruit callus and its regenerants.
In our work, lack of amplification of the SyGI and FrBy markers allows the selection of fruit-bearing females in almost all the genotypes tested. Few exceptions were identified: in female genotypes of A. eriantha, A. chrysantha, A. latifolia and A. arisanensis we observed an unexpected amplification of the FrBy gene, but we ruled out the potential presence of an intact and functional copy of the gene in one of these species. However, with the exception of A. latifolia that did not amplify the SyGI marker probably because a specific mutation in annealing site, for the others 14 species tested the SyGI marker allows the discrimination of male plants from female ones. We did not develop a specific test for A. latifolia with a dedicated primer for gender selection. In conclusion, we demonstrate that the ESD test is effective and reliable in sex discrimination for kiwifruit and several other Actinidia species. The test is an important tool because could help in breeding programs. Moreover, the possibility of exploiting a quick protocol of extraction would greatly reduce the time needed for the screening of a large number of plants. We could screen up to 192 samples in a working day using a low processivity platform such as the SeqStudio Genetic Analyzer (Thermo Fisher).
Author contributions
GC conceived and planned the research; GDM did the experimental work in the laboratory and analysis; GDM and GC wrote the first draft of the text; RT revised the first draft; all authors contributed to the final version of the manuscript.
Acknowledgments
The authors thank Dr. Ross Ferguson for helpful comments and language revision and Dr. Rachele Falchi for helpful suggestion to improve the text manuscript.
Funding
The authors report no funding.
Conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JBR-211530.
References
[1] | Ferguson AR , Huang H . Genetic resources of kiwifruit: domestication and breeding. Horticultural Reviews. (2007) ;33: :1–121. |
[2] | Nishiyama I . Fruits of the Actinidia genus. Vol. 52, Advances in Food and Nutrition Research. Academic Press; (2007) . p. 293–324. |
[3] | Testolin R , Cipriani G , Costa G . Sex segregation ratio and gender expression in the genus Actinidia . Sex Plant Reprod. (1995) ;8: (3):129–32. |
[4] | Harvey CF , Gill GP , Fraser LG , McNeilage MA . Sex determination in Actinidia . 1. Sex-linked markers and progeny sex ratio in diploid A. chinensis. Sex Plant Reprod. (1997) ;10: (3):149–54. |
[5] | Leite Montalvão AP , Kersten B , Fladung M , Müller NA . The diversity and dynamics of sex determination in dioecious plants. Front Plant Sci. (2021) ;11: :2280. Available from: https://doi.org/10.3389/fpls.2020.580488. |
[6] | De Mori G , Zaina G , Franco-Orozco B , Testolin R , De Paoli E , Cipriani G . Targeted mutagenesis of the female-suppressor sygi genein tetraploid kiwifruit by CRISPR/CAS9. Plants. (2020) ;10: (1):62. Available from: https://www.mdpi.com/2223-7747/10/1/62 |
[7] | Nocker SV , Gardiner SE . Breeding better cultivars, faster: applications of new technologies for the rapid deployment of superior horticultural tree crops. Horticulture Research. 2014. 1, 14022. Available from: https://doi.org/10.1038/hortres.2014.22. |
[8] | Huang S , Ding J , Deng D , Tang W , Sun H , Liu D , et al. Draft genome of the kiwifruit Actinidia chinensis . Nat Commun. (2013) ;4: :2640; Available from: https://doi.org/10.1038/ncomms3640. |
[9] | Pilkington SM , Crowhurst R , Hilario E , Nardozza S , Fraser L , Peng Y , et al. A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics. (2018) ;19: (1):1–19. Available from: https://doi.org/10.1186/s12864-018-4656-3. |
[10] | Pilkington SM , Tahir J , Hilario E , Gardiner SE , Chagnä D , Catanach A , et al. Genetic and cytological analyses reveal the recombination landscape of a partially differentiated plant sex chromosome in kiwifruit. BMC Plant Biol. (2019) ;19: (1):172. Available from: https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-019-1766-2. |
[11] | Fraser LG , Tsang GK , Datson PM , De Silva HN , Harvey CF , Gill GP , et al. A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes. BMC Genomics. (2009) ;10: (1):102. Available from: http://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-10-102. |
[12] | McNeilage MA , Fraser LG , Tsang GK , Datson PM , De Silva HN , Crowhurst RN , et al. Molecular genetics and genomics and kiwifruit breeding. In: Acta Horticulturae. International Society for Horticultural Science. (2011) ;913: :63–70. |
[13] | Gill GP , Harvey CF , Gardner RC , Fraser LG . Development of sex-linked PCR markers for gender identification in Actinidia. Theor Appl Genet. (1998) ;97: (3):439–45. doi: 10.1007/s001220050914. |
[14] | Wang T , Gleave AP . Applications of biotechnology in kiwifruit (Actinidia). Innov Biotechnol. 2012; ISBN: 978-953-51-0096-6.Available from: https://www.intechopen.com/books/innovations-in-biotechnology/applications-of-biotechnology-in-kiwifruit-actinidia-. |
[15] | Scaglione D , Fornasiero A , Pinto C , Cattonaro F , Spadotto A , Infante R , et al . A RAD-based linkage map of kiwifruit (Actinidia chinensis Pl.) as a tool to improve the genome assembly and to scan the genomic region of the gender determinant for the marker-assisted breeding. Tree Genet Genomes. (2015) ;11: (6):1–10. Available from https://link.springer.com/article/10.1007/s11295-015-0941-3. |
[16] | Zhang Q , Liu C , Liu Y , Vanburen R , Yao X , Zhong C , et al. High-density interspecific genetic maps of kiwifruit and the identification of sex-specific markers. DNA Research. (2015) ;22: (5):367–375. doi: 10.1093/dnares/dsv019. |
[17] | Testolin R , Cipriani G . Markers, maps, and marker-assisted selection, In The kiwifruit genome; Springer, Cham. (2016) ; p. 85–99. Available from: https://link.springer.com/chapter/10.1007/978-3-319-32274-2_7. |
[18] | Akagi T , Henry IM , Ohtani H , Morimoto T , Beppu K , Kataoka I , et al. A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell. (2018) ;30: (4):780–95. doi: 10.1105/tpc.17.00787. |
[19] | Akagi T , Pilkington SM , Varkonyi-Gasic E , Henry IM , Sugano SS , Sonoda M , et al. Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat Plants. (2019) ;5: (8):801–9. Available from: http://dx.doi.org/10.1101/615666. |
[20] | Untergasser A , Cutcutache I , Koressaar T , Ye J , Faircloth BC , Remm M , et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. (2012) ;40: (15):e115. doi: 10.1093/nar/gks596. |
[21] | Cotrut RC , Stanica F , Cotruţ t R , Stãnicã F , Scapigliati G . Identification of ploidy level on varieties and hybrids of kiwifruit (Actinidia sp.). Scientific Papers. Series B. Horticulture.,Vol. LVII, ISSN 2285-5653. 2013; p. 189-192.Available from: https://www.researchgate.net/publication/322641388 |
[22] | Vashistha P , Yadav A , Dwivedi UN , Yadav K . Genetics of Sex Chromosomes and Sex-linked Molecular Markers in Papaya (Carica papaya L.). Mol Plant Breed. (2016) ;7: (28):1–18. doi: 10.5376/mpb.2016.07.0028). |
[23] | Liao L , Liu J , Dai Y , Li Q , Xie M , Chen Q , et al. Development and application of SCAR markers for sex identification in the dioecious species Ginkgo biloba L. Euphytica. (2009) ;169: (1):49–55.Available from: https://link.springer.com/article/10.1007/s10681-009-9913-8. |
[24] | Akagi T , Kajita K , Kibe T , Morimura H , Tsujimoto T , Nishiyama S , et al. Development of Molecular Markers Associated with Sexuality in Diospyros lotus L. and Their Application in D. kaki Thunb. J. Jap. Soc. Hort. Sci. (2014) ;83: (3):214–221. doi: 10.2503/jjshs1.CH-109. |
[25] | Atsumi R , Nishihara R , Tarora K , Urasaki N , Matsumura H . Identification of dominant genetic markers relevant to male sex determination in mulberry (Morus alba L). Euphytica. (2019) ;215: (11):1–13. Available from: https://link.springer.com/article/10.1007/s10681-019-2511-5. |
[26] | Irei A , Miryeganeh M , Tamashiro M , Saze H , Urasaki N , Tarora K . Development of a male specific genetic marker for Garcinia subelliptica Merr. Tree. Journal of Forest Research. (2021) ;26: (3):222–229. Available from: https://doi.org/10.1080/13416979.2021.1897060. |
[27] | Sahin ZN , Sahin EC , Aydin Y , Uncuoglu AA . Molecular sexual determinants in Pistacia genus by KASP assay. Molecular Biology Reports. 2021. Available from: https://www.researchsquare.com/article/rs-499676/v1 (under review). |
[28] | Chłosta I , Kwolek D , Sliwinska E , Góralski G , Popielarska-Konieczna M . Sex-Linked molecular markers identifyfemale lines in endosperm-derived kiwifruit callus and inregenerants. Plants. (2021) ;10: (3):526. Available from: https://www.mdpi.com/2223-7747/10/3/526/htm |



