Gut bacteria are required for the benefits of black raspberries in ApcMin/+ mice
Abstract
BACKGROUND:
The gut microbiota plays a pivotal role in the development of inflammatory bowel disease and colorectal cancer.
OBJECTIVE:
To determine whether the gut microbiota is required for the chemoprotective effects of black raspberries (BRBs) in ApcMin/+ mice.
METHODS:
ApcMin/+ mice were given (a) a control diet for 8 weeks, or (b) the control diet for 4 weeks and then a 5% BRB diet for additional 4 weeks, or (c) the control diet and antibiotics for 4 weeks followed by the 5% BRB diet and antibiotics for the next 4 weeks. At the end of the study, all the mice were euthanized, and colonic and intestinal polyps were counted. mRNA expression levels of TLR4, NF-κ B1, and COX2 were determined in colon and small intestine of these ApcMin/+ mice by quantitative real-time PCR.
RESULTS:
5% BRBs significantly suppressed intestinal and colonic polyp development in the ApcMin/+ mice, whereas antibiotics significantly abolished BRBs’ chemoprotective effects. BRBs decreased mRNA levels of TLR4, NF-κ B1, and COX2 in colon, whereas significantly enhanced mRNA levels of TLR4 and NF-κ B1 were observed in small intestine of BRB-treated ApcMin/+ mice fed antibiotics.
CONCLUSIONS:
The gut microbiota is required for BRBs’ chemoprotection against polyp development in ApcMin/+ mice.
Abbreviations
Apc | Adenomatous polyposis coli |
AOM | azoxymethane |
BRBs | black raspberries |
CAC | colitis-associated colon cancer |
CI | confidence interval |
COX2 | cyclooxygenase 2 |
CRC | colorectal cancer |
DSS | dextran sodium sulfate |
FAP | familial adenomatous polyposis |
FFAR2 | free fatty acid receptor 2 |
FMT | fecal microbial transplant |
IBD | inflammatory bowel disease |
IKKs | inhibitor of nuclear factor kappa B kinases |
IL-1 | interleukin-1 |
IRAK | IL-1 receptor-associated kinase |
LPS | lipopolysaccharide |
MyD88 | myeloid differentiation factor 88 |
NF-κB | nuclear factor kappa B |
OR | odds ratio |
PAMPs | pathogen-associated molecular patterns |
SCFAs | short-chain fatty acids |
TLRs | Toll-like receptors |
TRAF6 | tumor necrosis factor receptor-associated factor 6 |
UC | ulcerative colitis |
1Introduction
The human gastrointestinal tract harbors trillions of microbes [1, 2], and accumulating evidence suggests a pivotal role for the gut microbiota in the development of inflammatory bowel disease (IBD) [3] and colorectal cancer (CRC) [4–8]. Thus, CRC patients host a distinct microbiota compared with healthy subjects [9]. Also, the gut microbiota gradually changes with disease progression across stages of CRC [10]. In addition, fecal bacteria from CRC patients can promote intestinal carcinogenesis in mice [11]. In light of these findings, alterations in microbial composition have the potential to become noninvasive diagnostic and prognostic tools for CRC in humans [12–14].
Pathobiotic bacteria can infiltrate through the gut epithelial surface barrier, invade adjacent normal tissues, generate toxic products, and further promote colonic inflammation and tumorigenesis [15]. Animal studies have reported that mice treated with antibiotics were less susceptible to colitis-associated colon cancer (CAC) induced by the azoxymethane (AOM)/dextran sodium sulfate (DSS) [16, 17]. In these studies, antibiotics suppressed colon cancer development by inhibiting bacteria-induced colonic inflammation. However, it is important to note that majority of sporadic CRC patients show no history of IBD [6, 8], and large epidemiologic studies suggest a positive association between frequent use of antibiotics and a higher risk of developing CRC [18–21]. For instance, the Nurses’ Health Study in the United States showed that women who had taken antibiotics for more than 2 months between ages 20 and 39 years or between 40 and 59 years had respective multivariable odds ratios (OR) of 1.36 (95% confidence interval (CI): 1.03–1.79) and 1.69 (95% CI: 1.24–2.31) of developing CRC after age 60 [18]. Therefore, maintaining a healthy and balanced gut microbiota is essential for preventing CRC.
Fecal microbial transplantation (FMT) is a dramatic method of replacing bacteria in dysbiosis patients with more beneficial bacteria. Many clinical trials have shown clinical improvements in some recipients with active ulcerative colitis (UC) [22–27] and Crohn’ s disease [28]. However, FMT may not be suitable for preventing or treating CRC, which usually develops gradually over 10 or more years. On the other hand, consuming prebiotics in the diet could be an elegant way to promote a healthy gut microbiota and modulate dysbiosis [29]. Prebiotics—typically dietary fibers and their fermented metabolites, short-chain fatty acids (SCFAs)—are drawing enormous attention due to their overall health benefits [30–32]. Meta-analyses have shown that high intake of dietary fiber, especially fiber from whole grains and cereals, negatively associates with the risk of developing CRC [33].
Our laboratory focuses on dietary constituents, such as black raspberries (BRBs), that have chemoprotective effects against colon cancer. BRBs are enriched in dietary fiber and many chemopreventive compounds such as anthocyanins and ellagitannins. We previously showed that BRBs were beneficial to patients with CRC [34, 35] or familial adenomatous polyposis (FAP) [36]. 5% BRBs in the diet, containing ∼ 3.8μ mol anthocyanins/g and 2.25% fiber, suppressed colorectal carcinogenesis and modulated immune cells in mouse models of CRC [37–39]. BRBs changed the gut microbiota of rats in ways that might contribute to their chemopreventive effects [40]. Importantly, we demonstrated that functional free fatty acid receptor 2 (FFAR2)—the receptor for SCFAs—is required for BRB-mediated effects [39], suggesting that bacterial fermentation of BRBs and their components is a critical mechanism. In addition, toll-like receptors (TLRs), as well as its downstream nuclear factor kappa B (NF-κ B) and cyclooxygenase 2 (COX2), has been shown to associate with human CRC [41–44]. We previously demonstrated that 5% BRBs suppressed the protein levels of NF-κ B and COX2 in DSS-induced UC in mice [45], an inflammatory colonic injury that can dramatically increase the risk of CRC. Therefore, the current study aimed to directly determine whether BRBs need the gut microbiota to exert their protective effects in ApcMin/+ mice, as well as to examine BRBs’ effects on The TLR4/NF-κ B/COX2 pathway.
2Materials and methods
2.1Animals and BRBs
All animal study protocols were approved by the Medical College of Wisconsin Animal Care and Use Committee under the animal protocol AUA00002430 “ Identification of specific berry types and berry components that exhibit anti-inflammatory and anti-cancer activities in different animal cancer models.” Breeding pairs of ApcMin/+ mice were purchased from the Jackson Laboratory (Bar Harbor, ME).
A synthetic diet from the American Institute of Nutrition (AIN-76A; Dyets Inc., Bethlehem, PA) was used as the control diet. BRB powder was purchased from Berri Products LLC (Corvallis, OR) and stored at 4° C. The sugar and starch content of the BRB diet was adjusted to create an isocaloric diet [37–40, 46].
2.2Animal experiments
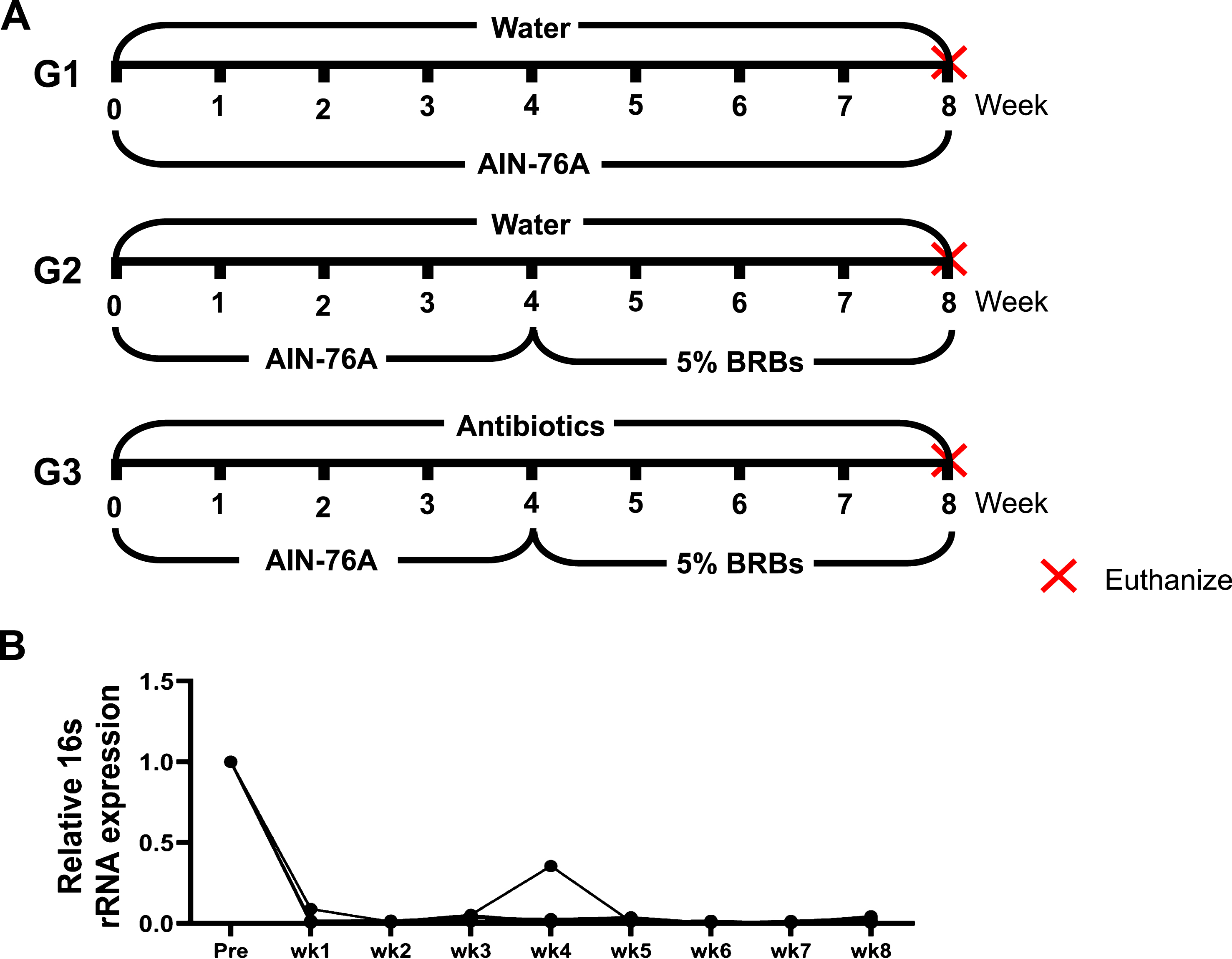
Four- to five-week-old ApcMin/+ mice were randomly assigned to three study groups. The mice in groups 1 and 2 (G1 and G2) were fed regular drinking water and the control AIN-76A diet for 4 weeks. Then the mice in G1 continued on the control diet, while the mice in G2 changed to 5% BRB diet. The mice in group 3 (G3) were first given the control diet and antibiotics in the drinking water (1 g/L ampicillin, 1 g/L neomycin, 1 g/L metronidazole, and 0.5 g/L vancomycin) for 4 weeks. For the next 4 weeks, they were fed 5% BRBs along with the antibiotic treatment (Fig. 1A). At the end of the study, all the mice were euthanized by CO2 asphyxiation, and the number of colonic and intestinal polyps was determined. Whole tissues of the colon and small intestine of all the mice were collected, fixed in formalin, and embedded in paraffin (FFPE). Hematoxylin and eosin (H& E)-stained tissue sections were evaluated histopathologically by our pathologists.
Fig.1
(A) Animal experimental protocol of the current study. ApcMin/+ mice were randomly assigned to three study groups. The mice in G1 were fed regular drinking water and the control diet for 8 weeks. The mice in G2 were fed regular drinking water and the control diet for 4 weeks, and then change to 5% BRBs for additional 4 weeks. The mice in G3 were first given the control diet and antibiotics (1 g/L ampicillin, 1 g/L neomycin, 1 g/L metronidazole, and 0.5 g/L vancomycin) in the drinking water for 4 weeks. For the next 4 weeks, they were fed 5% BRBs along with the antibiotic treatment. (B) Antibiotics in the drinking water substantially decreased gut bacterial populations in ApcMin/+ mice. wk: week.
2.3Quantitative real-time PCR
RNA was isolated from FFPE samples of colon and small intestine according to the manufacturer’ s instructions (Recover All Total Nucleic Acid Isolation Kit for formalin-fixed and paraffin-embedded tissues, Ambion, Grand Island, NY). Quantitative PCR was performed to measure the relative expression levels of TLR4 (Mm.PT.58.41780308.g), NF-κ B1 (Mm.PT.58.30400172), and COX2 (Mm.PT.58.17730756). Respective primers were purchased from Integrated Device Technology (San Jose, CA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene. Δ Ct-method was performed to analyze the relative expression levels of the respective genes.
2.4Measurements of fecal microbial populations by quantitative real-time PCR
Fecal specimens were collected every week for 4 weeks from the antibiotic-treated ApcMin/+ mice. Fecal DNA was isolated according to the manufacturer’ s instructions (PowerSoil® DNA Isolation Kit, MO Bio laboratories, Carlsbad, CA). Universal primers were designed to measure the overall populations of bacteria (Forward: ACTCCTACGGGAGGCAGCAGT; Reverse: ATTACCGCGGCTGCTGGC) as previously described [47–51].
2.5Statistical analysis
One-way ANOVA and post-hoc analysis were performed using SigmaPlot (Systat Software, San Jose, CA) to analyze polyp number and relative gene expression. A p value less than 0.05 was considered statistically significant.
3Results and Discussion
3.1Gut bacteria are required for BRBs’ anti-tumor effects in ApcMin/+ mice
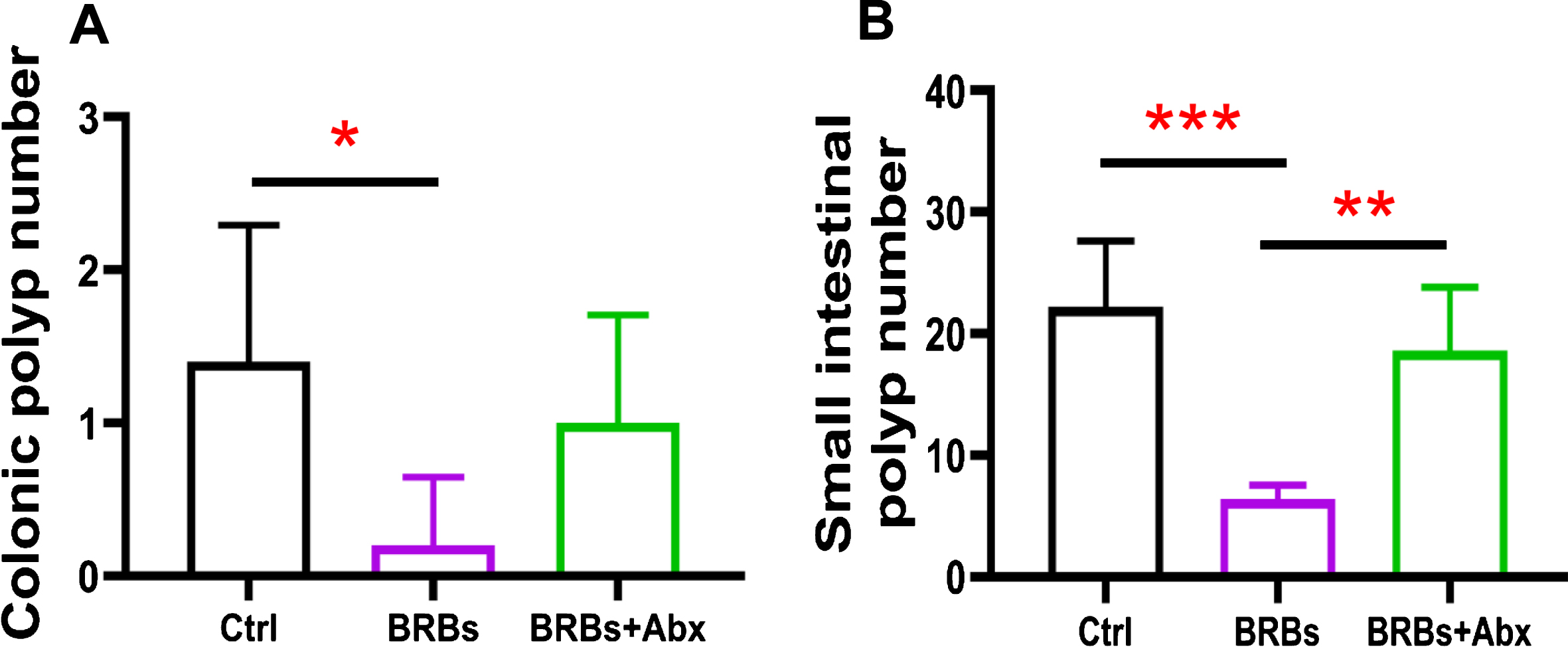
The Adenomatous polyposis coli (Apc) gene, a tumor suppressor, functions to induce β -catenin degradation and suppress the Wnt signaling pathway, and mutations in the Apc gene contribute strongly to CRC [52, 53]. Moreover, multiple intestinal neoplasia mice (ApcMin/+), which carry a truncating mutation at codon 850 of the Apc gene, develop multiple colonic and intestinal polyps, making them a widely used model of human CRC [53]. We fed ApcMin/+ mice with either a control diet (G1) or 5% BRBs from week 4 to week 8 (G2). Another group of ApcMin/+ mice were first given the control diet and antibiotics in the drinking water for 4 weeks. Starting at week 4, the mice received the 5% BRB diet along with antibiotics for 4 weeks (G3) (Fig. 1A). After 1 week of the antibiotic treatment, there was a substantial decrease in the overall fecal bacterial population (Fig. 1B). At the end of study, we examined the number of colonic and intestinal polyps. The results confirmed those of our previous studies [38, 39]: that 5% BRBs in the diet significantly suppress colonic (Fig. 2A) and intestinal (Fig. 2B) tumor development in ApcMin/+ mice. However, antibiotics completely abolished the anti-tumor effects of the BRB diet in small intestine (Fig. 2), suggesting that gut microbiota is required for BRBs’ beneficial effects in ApcMin/+ mice.
Fig.2
Gut bacteria are required for the benefits of BRBs in ApcMin/+ mice. BRBs significantly decreased the number of polyps in colon (A) and small intestine (B) of ApcMin/+ mice, but antibiotics abolished those BRB-mediated chemoprotective effects. Ctrl: control diet; Abx: antibiotics. n = 5 per group; * p < 0.05; ** p < 0.01; *** p < 0.001.
Although large epidemiologic studies have suggested a positive association between frequent use of antibiotics and risk of CRC in humans [18–21], this conclusion is controversial in animal models as it is dependent on the model. In one study, antibiotics promoted intestinal polyp development in ApcMin/+ mice [54], while in another study that used the CAC model, antibiotics suppressed tumor growth induced by AOM/DSS [55]. The underlying mechanism of the latter study suggests that antibiotics might inhibit bacteria-induced inflammation, thereby suppressing tumor development. Other studies have reported similar results: severe colonic inflammation exacerbated polyposis and antibiotics rescued that symptom [56, 57]. Interestingly, one study observed that antibiotics depleted the gut microbiota but induced mild inflammation in the gastrointestinal track of wild-type mice [58], suggesting that colonic inflammation and dysbiosis cannot be cured simply by depleting gut bacteria.
The dynamic interactions between a host and its gut microbiota and between diet and the microbiota play critical roles in the prevention and treatment of CRC [59–61]. A healthy dietary pattern has been shown to associate with a lower risk of CRC [33]. Microbiome studies have identified several “ good” bacterial species, such as Akkermansia muciniphila, and some “ bad” bacterial species, including Escherichia coli and Fusobacterium spp [7]. Therefore, maintaining a balanced and healthy gut microbiota by adopting a healthy lifestyle would help lower the risk of developing CRC [6, 32, 62].
3.2BRBs suppress the TLR4/NF-κ B/COX2 pathway in ApcMin/+ mice
TLRs are important regulators of intestinal epithelial homoeostasis. They belong to the interleukin (IL)-1 superfamily of transmembrane receptors that recognize pathogen-associated molecular patterns (PAMPs). In particular, lipopolysaccharide (LPS) fragments from degraded outer membrane of gram-negative bacteria interact with TLR4, leading to the recruitment of downstream adaptor molecules, such as myeloid differentiation factor 88 (MyD88), IL-1 receptor-associated kinase (IRAK), and tumor necrosis factor receptor-associated factor 6 (TRAF6) [63]. The complex then phosphorylates the inhibitor of NF-κ B kinases (IKKs), activating NF-κ B. NF-κ B signaling plays an essential role in inflammation and cell survival. One of its downstream targets is COX2 [63]. Therefore, we examined the expression of TLR4/NF-κ B/COX2 pathway in BRB-treated ApcMin/+ mice.
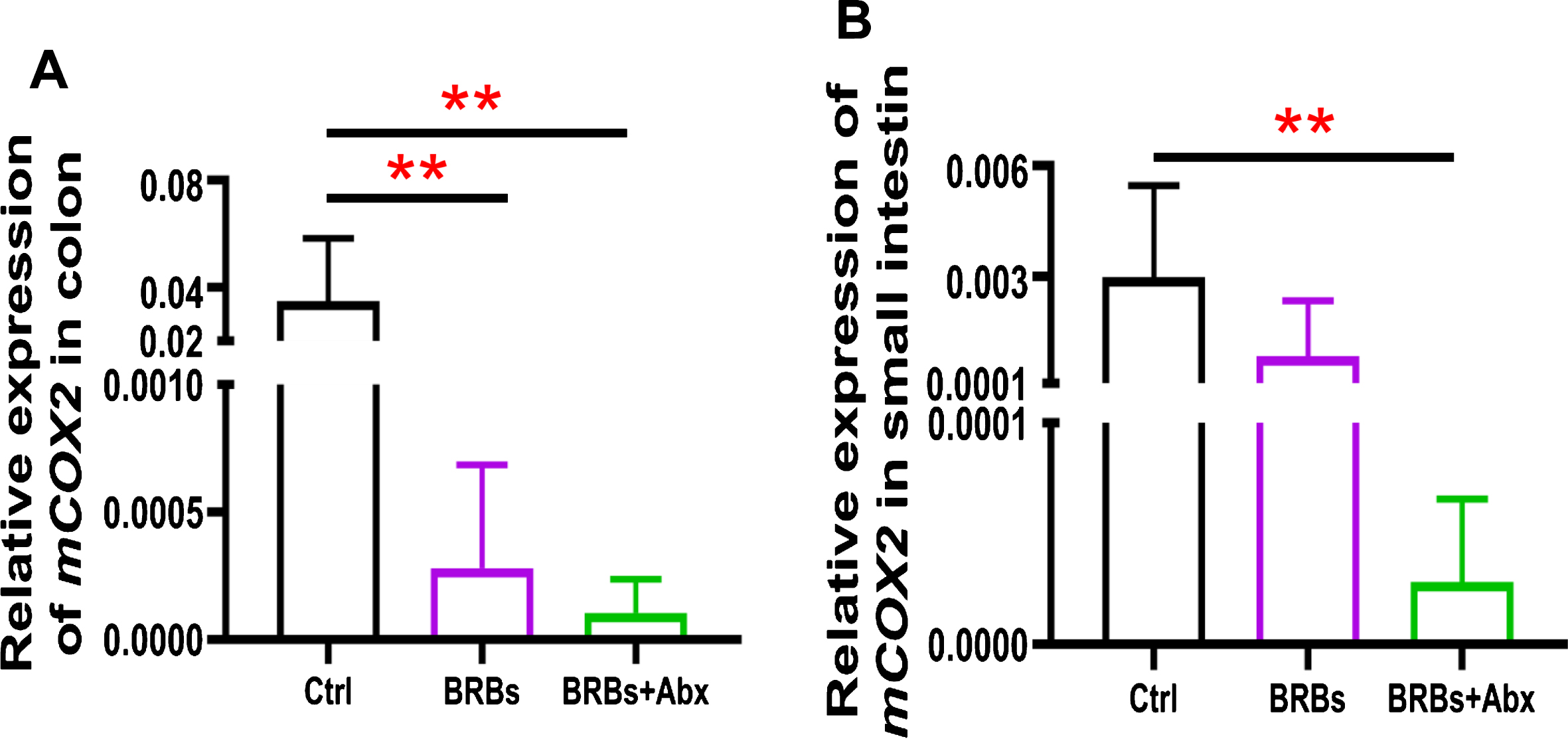
We found that 5% BRBs in the diet significantly decreased mRNA levels of TLR4 (Fig. 3A), NF-κ B (Fig. 4A), and COX2 (Fig. 5A) in colon of the ApcMin/+ mice. However, the TLR4/NF-κ B/COX2 pathway was not significantly changed by BRBs (Figs. 3B, 4B, and 5B) in small intestine, suggesting that other mechanisms contributed to the BRBs’ anti-tumor effects in small intestine. Intriguingly, significantly enhanced mRNA levels of TLR4 (Fig. 3B) and NF-κ B (Fig. 4B) were observed in small intestine of the BRB-treated ApcMin/+ mice fed antibiotics. Our results agree with those of Grasa L et al., who reported that antibiotics induced mild inflammation and TLR4 expression in the gastrointestinal tract of wild-type mice [58]. It is likely that the antibiotic treatment eliminated majority of the gut bacteria, whereas some other (or unknown) bacteria were selected and able to grow and induce a mild inflammatory response. Further investigations need to examine the mechanisms underlying the increased expression of TLR4 and NF-κ B promoted by antibiotics in the small intestine, as well as whether this enhancement contributes to antibiotics’ ability to counter the beneficial effects of BRBs.
Fig.3
mRNA expression levels of TLR4 in colon (A) and small intestine (B) of ApcMin/+ mice. Ctrl: control diet; Abx: antibiotics. ** p < 0.01; *** p < 0.001; **** p < 0.0001.
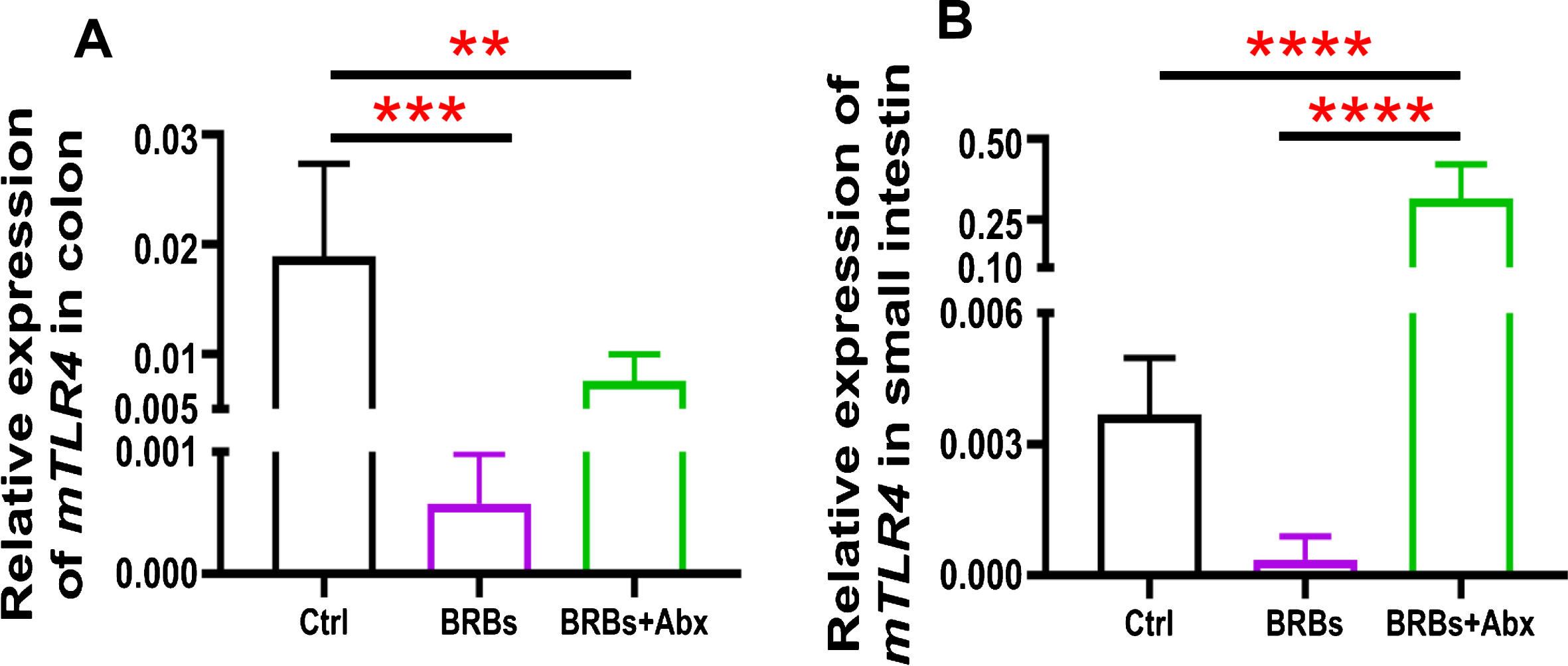
Fig.4
mRNA expression levels of NF-κ B1 in colon (A) and small intestine (B) of ApcMin/+ mice. Ctrl: control diet; Abx: antibiotics. ** p < 0.01; **** p < 0.0001.
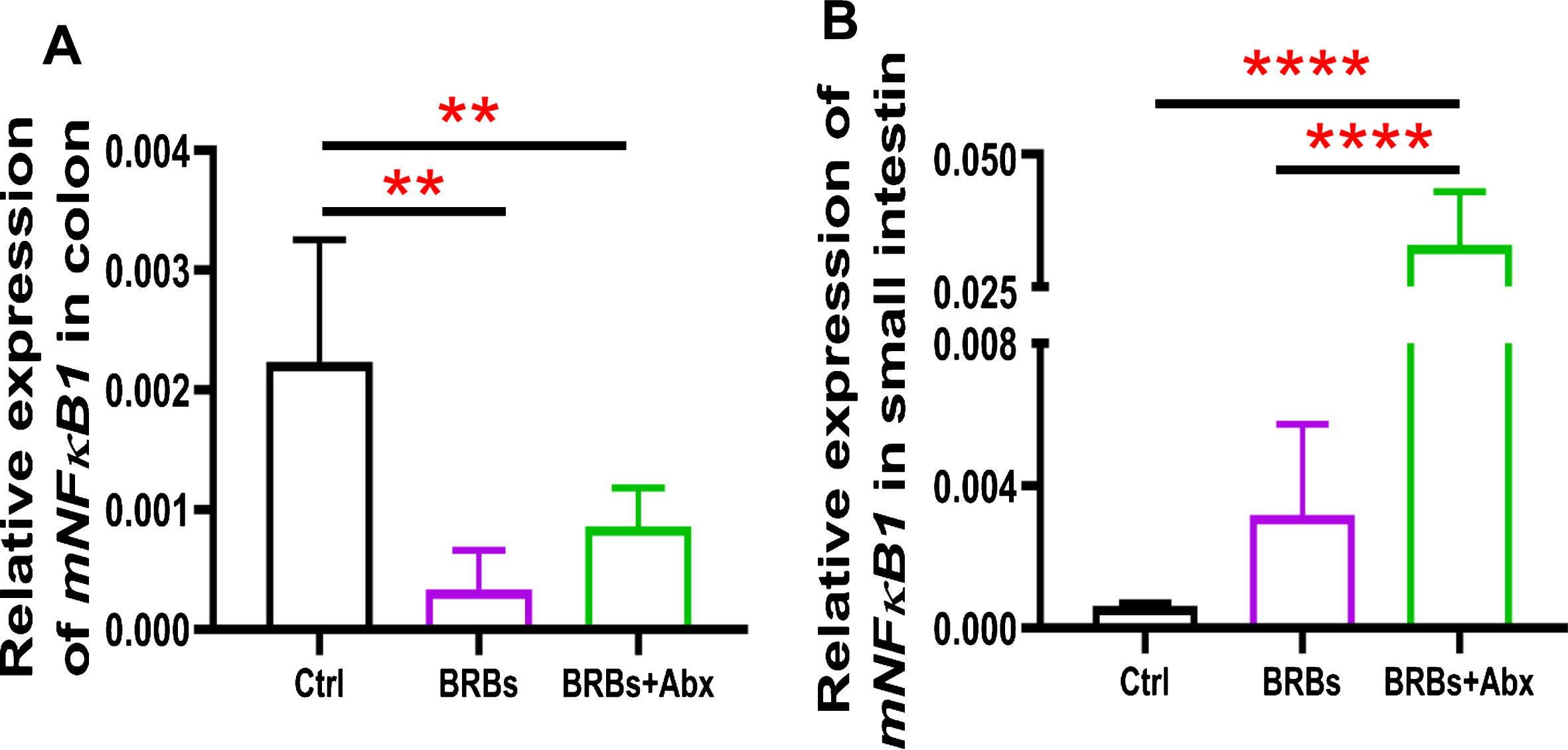
Fig.5
mRNA expression levels of COX2 in colon (A) and small intestine (B) of ApcMin/+ mice. Ctrl: control diet; Abx: antibiotics. ** p < 0.01.
The TLR4/NF-κ B/COX2 pathway has been examined in CRC patients and animal models, but the results have been surprisingly inconsistent. For example, studies showed that human intestinal epithelial cells normally express low levels of TLR4 [63], while specimens from UC [41] or CRC patients [42, 43], as well as from the CRC mouse model [41], showed increased expression of TLR4. In addition, high expression of TLR4 and MyD88 has been shown to associate with liver metastasis and poor prognosis in patients with CRC [44]. In contrast, another study demonstrated that normal colon epithelium and CRC cell lines were positive for TLR4, and specimens from metastatic CRC patients showed either loss of expression or strong downregulation of TLR4 in comparison with normal tissue and non-metastatic tumors [64]. Interestingly, one group found that TLR4 expression by tumor cells associated with a lower rate of tumor recurrence, whereas TLR4 expression by fibroblasts associated with a high rate of tumor recurrence [65]. Therefore, the TLR4/NF-κ B/COX2 pathway might play different roles in different compartments of the tumor microenvironment.
4Conclusions
In the current study, we investigated the relationship between the gut microbiota and BRB-mediated chemoprotective effects against CRC. We demonstrated that antibiotic treatment abolished BRBs’ anti-tumor effects, indicating that the gut microbiota is required for BRBs’ beneficial effects in ApcMin/+ mice. In addition, BRBs suppressed the TLR4/NF-κ B/COX2 pathway in colon of ApcMin/+ mice, whereas antibiotic treatment increased the expression of TLR4 and NF-κ B in small intestine of the BRB-treated ApcMin/+ mice.
Competing interests
No potential competing interest was disclosed.
Funding
We thank the National Institutes of Health (5 R01 CA148818) and American Cancer Society (RSG-13-138-01—CNE to L.-S. Wang) for funding this study.
Authors’ contribution
Study design and acquisition, analysis, and interpretation of data: Pan Pan, Kiyoko Oshima, Yi-Wen Huang, Martha Yearsley, Jianying Zhang, Mark Arnold, Jianhua Yu, and Li-Shu Wang.
Manuscript writing and revising: Pan Pan and Li-Shu Wang.
Final approval: Pan Pan and Li-Shu Wang.
Acknowledgments
We thank Hzin-Tsu Wang and Chieh-Ti Kuo for their assistance in this study.
References
[1] | Ley RE , Peterson DA , Gordon JI . Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) ;124: (4):837–48. doi: 10.1016/j.cell.2006.02.017. |
[2] | Qin J , Li R , Raes J , Arumugam M , Burgdorf KS , Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) ;464: (7285):59–65. doi: 10.1038/nature08821. |
[3] | Shanahan F . The microbiota in inflammatory bowel disease: Friend, bystander, and sometime-villain. Nutr Rev. (2012) ;70: (Suppl 1):S31–7. doi: 10.1111/j.1753-4887.2012.00502.x. |
[4] | Jobin C . Colorectal cancer: Looking for answers in the microbiota. Cancer Discov. (2013) ;3: (4):384–7. doi: 10.1158/2159-8290.CD-13-0042. |
[5] | Shanahan F , O’ Toole PW . Host-microbe interactions and spatial variation of cancer in the gut. Nat Rev Cancer. (2014) ;14: (8):511–2. doi: 10.1038/nrc3765. |
[6] | Chen J , Pitmon E , Wang K . Microbiome, inflammation and colorectal cancer. Semin Immunol. (2017) ;32: :43–53. doi: 10.1016/j.smim.2017.09.006. |
[7] | Sears CL , Garrett WS . Microbes, microbiota, and colon cancer. Cell Host Microbe. (2014) ;15: (3):317–28. doi: 10.1016/j.chom.2014.02.007. |
[8] | Irrazabal T , Belcheva A , Girardin SE , Martin A , Philpott DJ . The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell. (2014) ;54: (2):309–20. doi: 10.1016/j.molcel.2014.03.039. |
[9] | Flemer B , Lynch DB , Brown JM , Jeffery IB , Ryan FJ , Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. (2017) ;66: (4):633–43. doi: 10.1136/gutjnl-2015-309595. |
[10] | Nakatsu G , Li X , Zhou H , Sheng J , Wong SH , Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun (2015) ;6: :8727. doi: 10.1038/ncomms9727. |
[11] | Wong SH , Zhao L , Zhang X , Nakatsu G , Han J , Xu W, et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology. (2017) ;153: (6):1621–33. doi: 10.1053/j.gastro.2017.08.022. |
[12] | Shah MS , DeSantis TZ , Weinmaier T , McMurdie PJ , Cope JL , Altrichter A, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. (2018) ;67: (5):882–91. doi: 10.1136/gutjnl-2016-313189. |
[13] | Liang Q , Chiu J , Chen Y , Huang Y , Higashimori A , Fang J, et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. (2017) ;23: (8):2061–70. doi: 10.1158/1078-0432.CCR-16-1599. |
[14] | Baxter NT , Ruffin MTt , Rogers MA , Schloss PD . Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. (2016) ;8: (1):37. doi: 10.1186/s13073-016-0290-3. |
[15] | Sheflin AM , Whitney AK , Weir TL . Cancer-promoting effects of microbial dysbiosis. Curr Oncol Rep. (2014) ;16: (10):406. doi: 10.1007/s11912-014-0406-0. |
[16] | Tanaka Y , Ito S , Isobe K . Vancomycin-sensitive bacteria trigger development of colitis-associated colon cancer by attracting neutrophils. Sci Rep (2016) ;6: :23920. doi: 10.1038/srep23920. |
[17] | Klimesova K , Kverka M , Zakostelska Z , Hudcovic T , Hrncir T , Stepankova R, et al. Altered gut microbiota promotes colitis-associated cancer in IL-1 receptor-associated kinase M-deficient mice. Inflamm Bowel Dis. (2013) ;19: (6):1266–77. doi: 10.1097/MIB.0b013e318281330a. |
[18] | Cao Y , Wu K , Mehta R , Drew DA , Song M , Lochhead P, et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut. (2018) ;67: (4):672–8. doi: 10.1136/gutjnl-2016-313413. |
[19] | Dik VK , van Oijen MG , Smeets HM , Siersema PD . Frequent Use of Antibiotics Is Associated with Colorectal Cancer Risk: Results of a Nested Case-Control Study. Dig Dis Sci. (2016) ;61: (1):255–64. doi: 10.1007/s10620-015-3828-0. |
[20] | Boursi B , Haynes K , Mamtani R , Yang YX . Impact of antibiotic exposure on the risk of colorectal cancer. Pharmacoepidemiol Drug Saf. (2015) ;24: (5):534–42. doi: 10.1002/pds.3765. |
[21] | Wang JL , Chang CH , Lin JW , Wu LC , Chuang LM , Lai MS . Infection, antibiotic therapy and risk of colorectal cancer: A nationwide nested case-control study in patients with Type 2 diabetes mellitus. Int J Cancer. (2014) ;135: (4):956–67. doi: 10.1002/ijc.28738. |
[22] | Uygun A , Ozturk K , Demirci H , Oger C , Avci IY , Turker T, et al. Fecal microbiota transplantation is a rescue treatment modality for refractory ulcerative colitis. Medicine (Baltimore). (2017) ;96: (16):e6479. doi: 10.1097/MD.0000000000006479. |
[23] | Fuentes S , Rossen NG , van der Spek MJ , Hartman JH , Huuskonen L , Korpela K, et al. Microbial shifts and signatures of long-term remission in ulcerative colitis after faecal microbiota transplantation. ISME J. (2017) ;11: (8):1877–89. doi: 10.1038/ismej.2017.44. |
[24] | Paramsothy S , Kamm MA , Kaakoush NO , Walsh AJ , van den Bogaerde J , Samuel D, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: A randomised placebo-controlled trial. Lancet. (2017) ;389: (10075):1218–28. doi: 10.1016/s0140-6736(17)30182-4. |
[25] | Ishikawa D , Sasaki T , Osada T , Kuwahara-Arai K , Haga K , Shibuya T, et al. Changes in Intestinal Microbiota Following Combination Therapy with Fecal Microbial Transplantation and Antibiotics for Ulcerative Colitis. Inflamm Bowel Dis. (2017) ;23: (1):116–25. doi: 10.1097/MIB.0000000000000975. |
[26] | Nishida A , Imaeda H , Ohno M , Inatomi O , Bamba S , Sugimoto M, et al. Efficacy and safety of single fecal microbiota transplantation for Japanese patients with mild to moderately active ulcerative colitis. J Gastroenterol. (2017) ;52: (4):476–82. doi: 10.1007/s00535-016-1271-4. |
[27] | Moayyedi P , Surette MG , Kim PT , Libertucci J , Wolfe M , Onischi C, et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology. (2015) ;149: (1):102–9 e6. doi: 10.1053/j.gastro.2015.04.001. |
[28] | Suskind DL , Brittnacher MJ , Wahbeh G , Shaffer ML , Hayden HS , Qin X, et al. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn’ s disease. Inflamm Bowel Dis. (2015) ;21: (3):556–63. doi: 10.1097/MIB.0000000000000307. |
[29] | Di Bartolomeo F , Startek JB , Van den Ende W . Prebiotics to fight diseases: Reality or fiction? Phytother Res, (2013) ;27: (10):1457–73. doi: 10.1002/ptr.4901. |
[30] | Meyer D . Health benefits of prebiotic fibers. Adv Food Nutr Res. (2015) ;74: :47–91. doi: 10.1016/bs.afnr.2014.11.002. |
[31] | Verspreet J , Damen B , Broekaert WF , Verbeke K , Delcour JA , Courtin CM . A Critical Look at Prebiotics Within the Dietary Fiber Concept. Annu Rev Food Sci Technol. (2016) ;7: :167–90. doi: 10.1146/annurev-food-081315-032749. |
[32] | Seidel DV , Azcarate-Peril MA , Chapkin RS , Turner ND . Shaping functional gut microbiota using dietary bioactives to reduce colon cancer risk. Semin Cancer Biol. (2017) ;46: :191–204. doi: 10.1016/j.semcancer.2017.06.009. |
[33] | Pan P , Yu J , Wang LS . Colon Cancer: What We Eat. Surg Oncol Clin N Am. (2018) ;27: (2):243–67. doi: 10.1016/j.soc.2017.11.002. |
[34] | Wang LS , Arnold M , Huang YW , Sardo C , Seguin C , Martin E, et al. Modulation of genetic and epigenetic biomarkers of colorectal cancer in humans by black raspberries: A phase I pilot study. Clin Cancer Res. (2011) ;17: (3):598–610. doi: 10.1158/1078-0432.CCR-10-1260. |
[35] | Pan P , Skaer CW , Stirdivant SM , Young MR , Stoner GD , Lechner JF, et al. Beneficial Regulation of Metabolic Profiles by Black Raspberries in Human Colorectal Cancer Patients. Cancer Prev Res (Phila). (2015) ;8: (8):743–50. doi: 10.1158/1940-6207.CAPR-15-0065. |
[36] | Wang LS , Burke CA , Hasson H , Kuo CT , Molmenti CL , Seguin C, et al. A phase Ib study of the effects of black raspberries on rectal polyps in patients with familial adenomatous polyposis. Cancer Prev Res (Phila). (2014) ;7: (7):666–74. doi: 10.1158/1940-6207.CAPR-14-0052. |
[37] | Pan P , Kang S , Wang Y , Liu K , Oshima K , Huang Y-W, et al. Black Raspberries Enhance Natural Killer Cell Infiltration into the Colon and Suppress the Progression of Colorectal Cancer. Frontiers in Immunology (2017) ;8: (997). 10.3389/fimmu.2017.00997. |
[38] | Pan P , Skaer CW , Wang HT , Stirdivant SM , Young MR , Oshima K, et al. Black raspberries suppress colonic adenoma development in ApcMin/+ mice: Relation to metabolite profiles. Carcinogenesis. (2015) ;36: (10):1245–53. doi: 10.1093/carcin/bgv117. |
[39] | Pan P , C WS , Wang HT , Oshima K , Huang YW , Yu J, et al. Loss of free fatty acid receptor 2 enhances colonic adenoma development and reduces the chemopreventive effects of black raspberries in ApcMin/+ mice. Carcinogenesis. (2017) ;38: (1):86–93. doi: 10.1093/carcin/bgw122. |
[40] | Pan P , Lam V , Salzman N , Huang YW , Yu J , Zhang J, et al. Black Raspberries and Their Anthocyanin and Fiber Fractions Alter the Composition and Diversity of Gut Microbiota in F-344 Rats. Nutr Cancer. (2017) ;1–9. doi: 10.1080/01635581.2017.1340491. |
[41] | Fukata M , Chen A , Vamadevan AS , Cohen J , Breglio K , Krishnareddy S, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. (2007) ;133: (6):1869–81. doi: 10.1053/j.gastro.2007.09.008. |
[42] | Cammarota R , Bertolini V , Pennesi G , Bucci EO , Gottardi O , Garlanda C, et al. The tumor microenvironment of colorectal cancer: Stromal TLR-4 expression as a potential prognostic marker. J Transl Med. (2010) ;8: :112. doi: 10.1186/1479-5876-8-112. |
[43] | Santaolalla R , Sussman DA , Ruiz JR , Davies JM , Pastorini C , Espana CL, et al. TLR4 activates the beta-catenin pathway to cause intestinal neoplasia. PLoS One. (2013) ;8: (5):e63298. doi: 10.1371/journal.pone.0063298. |
[44] | Wang EL , Qian ZR , Nakasono M , Tanahashi T , Yoshimoto K , Bando Y, et al. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer. (2010) ;102: (5):908–15. doi: 10.1038/sj.bjc.6605558. |
[45] | Montrose DC , Horelik NA , Madigan JP , Stoner GD , Wang LS , Bruno RS, et al. Anti-inflammatory effects of freeze-dried black raspberry powder in ulcerative colitis. Carcinogenesis. (2011) ;32: (3):343–50. doi: 10.1093/carcin/bgq248. |
[46] | Pan P , Skaer CW , Wang HT , Kreiser MA , Stirdivant SM , Oshima K, et al. Systemic Metabolite Changes in Wild-type C57BL/6 Mice Fed Black Raspberries. Nutr Cancer. (2017) ;69: (2):299–306. doi: 10.1080/01635581.2017.1263748. |
[47] | Kang M , Mischel RA , Bhave S , Komla E , Cho A , Huang C, et al. The effect of gut microbiome on tolerance to morphine mediated antinociception in mice. Scientific Reports. (2017) ;7: :42658. doi: 10.1038/srep42658. |
[48] | Lopez I , Ruiz-Larrea F , Cocolin L , Orr E , Phister T , Marshall M, et al. Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol. (2003) ;69: (11):6801–7. |
[49] | Lazarevic V , Gaia N , Girard M , Schrenzel J . Decontamination of 16S rRNA gene amplicon sequence datasets based on bacterial load assessment by qPCR. BMC Microbiol. (2016) ;16: :73. doi: 10.1186/s12866-016-0689-4. |
[50] | Ahmed S , Macfarlane GT , Fite A , McBain AJ , Gilbert P , Macfarlane S . Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. (2007) ;73: (22):7435–42. doi: 10.1128/AEM.01143-07. |
[51] | Clifford RJ , Milillo M , Prestwood J , Quintero R , Zurawski DV , Kwak YI, et al. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. (2012) ;7: (11):e84855. doi: 10.1371/journal.pone.0048558. |
[52] | Weitz J , Koch M , Debus J , Hohler T , Galle PR , Buchler MW . Colorectal cancer. Lancet. (2005) ;365: (9454):153–65. doi: 10.1016/S0140-6736(05)17706-X. |
[53] | McCart AE , Vickaryous NK , Silver A . Apc mice: models, modifiers and mutants. Pathol Res Pract. (2008) ;204: (7):479–90. doi: 10.1016/j.pr2008.03.004. |
[54] | Kaur K , Saxena A , Debnath I , O’ Brien JL , Ajami NJ , Auchtung TA, et al. Antibiotic-mediated bacteriome depletion in Apc(Min/+) mice is associated with reduction in mucus-producing goblet cells and increased colorectal cancer progression. Cancer Med. (2018) ;7: (5):2003–12. doi: 10.1002/cam4.1460. |
[55] | Zackular JP , Baxter NT , Iverson KD , Sadler WD , Petrosino JF , Chen GY, et al. The gut microbiome modulates colon tumorigenesis. MBio. (2013) ;4: (6):e00692–13. doi: 10.1128/mBio.00692-13. |
[56] | Dennis KL , Wang Y , Blatner NR , Wang S , Saadalla A , Trudeau E, et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res. (2013) ;73: (19):5905–13. doi: 10.1158/0008-5472.CAN-13-1511. |
[57] | Grivennikov SI , Wang K , Mucida D , Stewart CA , Schnabl B , Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. (2012) ;491: (7423):254–8. doi: 10.1038/nature11465. |
[58] | Grasa L , Abecia L , Forcen R , Castro M , de Jalon JA , Latorre E, et al. Antibiotic-Induced Depletion of Murine Microbiota Induces Mild Inflammation and Changes in Toll-Like Receptor Patterns and Intestinal Motility. Microb Ecol. (2015) ;70: (3):835–48. doi: 10.1007/s00248-015-0613-8. |
[59] | Akin H , Tozun N . Diet, microbiota, and colorectal cancer. J Clin Gastroenterol. (2014) ;48: (Suppl 1):S67–9. doi: 10.1097/MCG.0000000000000252. |
[60] | Niederreiter L , Adolph TE , Tilg H . Food, microbiome and colorectal cancer. Dig Liver Dis. (2018) ;50: (7):647–52. doi: 10.1016/j.dld.2018.03.030. |
[61] | Shanahan F . The colonic microbiota in health and disease. Curr Opin Gastroenterol. (2013) ;29: (1):49–54. doi: 10.1097/MOG.0b013e32835a3493. |
[62] | Kang M , Martin A . Microbiome and colorectal cancer: Unraveling host-microbiota interactions in colitis-associated colorectal cancer development. Semin Immunol. (2017) ;32: :3–13. doi: 10.1016/j.smim.2017.04.003. |
[63] | Fukata M , Abreu MT . TLR4 signalling in the intestine in health and disease. Biochem Soc Trans. (2007) ;35: (Pt 6):1473–8. doi: 10.1042/BST0351473. |
[64] | Simiantonaki N , Kurzik-Dumke U , Karyofylli G , Jayasinghe C , Michel-Schmidt R , Kirkpatrick CJ . Reduced expression of TLR4 is associated with the metastatic status of human colorectal cancer. International journal of molecular medicine. (2007) ;20: (1):21–9. |
[65] | Eiro N , Gonzalez L , Gonzalez LO , Fernandez-Garcia B , Andicoechea A , Barbon E, et al. Toll-like receptor-4 expression by stromal fibroblasts is associated with poor prognosis in colorectal cancer. Journal of immunotherapy (Hagerstown, Md: 1997). (2013) ;36: (6):342–9. doi: 10.1097/CJI.0b013e31829d85e6. |




