Molecular characterization of berry skin color reversion on grape somatic variants
Abstract
BACKGROUND:
During grapevine domestication somatic variation has been used as a source of diversity for clonal selection.
OBJECTIVE:
This work provides additional information on the molecular mechanisms responsible for berry skin color reversion on a subset of somatic variants for berry skin color never investigated before.
METHODS:
The berry color locus and its surrounding genomic regionwere genetically characterized through a layer-specific approach, which has already been proven to be a successful method to decipher the molecular mechanisms responsible for color reversions on somatic variants.
RESULTS:
Deletions of different extent and positions were detected among less-pigmented/unpigmented variants derived from a pigmented wild-type. These deletions affected only the inner cell layer in the less pigmented variants and both cell layers in the unpigmented variants. Regarding the pigmented variants derived from an unpigmented wild-type, only one group was distinguished by the Gret1 retrotransposon partial excision from the VvMybA1 promoter. Moreover, within this latter group, VvMybA2 showed an important role regarding the phenotypic variation, through the recovery of the functional G allele.
CONCLUSIONS:
This investigation focused on the berry color locus using somatic variants for berry skin color, promotes a better understanding of the evolutionary events behind their origin and variability, opening an opportunity for their use in the genetic improvement of varieties.
1Introduction
Domestication of the wild Vitis vinifera ssp. sylvestris to Vitis vinifera ssp. sativa species was a complex, long-term, and ongoing evolutionary process that involved multiple loci [1]. This process dramatically changed the grape biology in order to ensure higher sugar content, greater yield, regular production and adaptation to different environments [2].
Today, most of the cultivated grapevine varieties are the result of continuous selection and vegetative propagation over centuries, spontaneously generated by sexual reproduction and via somatic mutations. These events led to grape varieties which have enough similar vegetative and reproductive features to allow them to freely interbreed [3]. Clones are usually selected for a particular character or combination of characters which, when propagated by appropriate means, retain those characters and are genetically identical, i.e.,true-to-type [4].
Although vegetative propagation has been used as a conservative strategy to maintain and propagate clones, somatic mutations may occur naturally when a spontaneous morpho-physiological mutation appears on a shoot, leading to clonal polymorphism [3, 5]. When a mutation appears in a single cell of a shoot apical meristem layer, the following mitotic divisions lead to the emergence of a mutated section [6], giving rise to a periclinal chimera. These structures corresponds to specific types of genetic mosaicism in which one or two entire meristematic cell layers are genetically different from the others, and are developmentally independent from the adjacent layers [7]. Usually they are stable and do not threaten plant fitness, and are maintained by vegetative propagation. However, occasionally these cellular rearrangements in the periclinal chimera lead to homogenization of the genotype to the whole plant [5] in a two-step process. First, a somatic mutation emerges in one cell of the shoot apical meristem and then is propagated by cell division to an entire cell layer, generating a stable chimeric structure. Subsequently, the replacement of one cell layer by another can lead to a homogenization of the genotype of the whole plant, representing the second step of the process [8]. Therefore, divergent genotypes and, to some extent divergent phenotypes, can appear due to these molecular and cellular mechanisms, representing a valuable source of heritable variation that has been widely exploited by winemakers [5, 9]. There are many examples of spontaneous variant traits in grapevines, including berry skin and flesh color or flavor, size and compactness of bunches, canopy growth, date of ripening and productivity [3, 5].
At the genetic level, the most well-documented polymorphisms leading to qualitative variation within varieties are those that affect berry skin color. Currently, several pigmented varieties have certified clones with different skin color shades, such as Aramon, Grenache, Pinot or Terret. Moreover, unpigmented skin berry varieties such as Savagnin, Chardonnay and Chasselas also comprise pigmented clones [5].
The first comparative study of molecular polymorphisms arising during vegetative propagation on the whole genome scale was done on spontaneously generated grape clones. Despite the small number of SNP (single nucleotide polymorphism) and indel events observed, mobile elements were identified as being involved in most of the polymorphisms detected [10].
Overall, the grapevine somatic variants for berry skin color can be divided in two main groups: A) unpigmented and less-pigmented variants from a pigmented ancestor, namely white and grey/pink-skinned variants derived from a black-skinned ancestor; B) pigmented variants from an unpigmented wild-type, namely red/pink-skinned variants derived from a white-skinned wild-type [11–13].
Research on grape berry color shows that the color locus is a cluster of four Myb and Myb-like genes, spread along a 200 kb-region located on chromosome 2, which comprises VvMybA1 and VvMybA2, two adjacent transcription factors that regulate the transcription of VvUFGT (UDP-glucose:flavonoid 3-O-glucosyltransferase), a key point in the anthocyanin pathway. In white-skinned cultivars, the absence of anthocyanins has been related with the insertion of the Gret1, a 10,422 bp long retrotransposon, in the promoter region of VvMybA1 gene combined with two non-conservative mutations in the coding sequence of VvMybA2, a non-synonymous point mutation that leads to an amino acid substitution (change of arginine residue at position 44 in the red allele [G] altered to leucine in the white allele [T]) and a 2-bp deletion (CA) altering the reading frame, that results in a smaller protein [14–17]. An additional point mutation of VvMybA2 gene (C22) related to berry pigmentation was described by Carrasco et al. [18] in wild grapevine accessions with low anthocyanin content. These mutations in both genes lead to the loss of transcription factors expression, which consequently prevents anthocyanin biosynthesis.
Researchers have recently focused on the somatic variation affecting berry skin color in different varieties, using a layer-specific approach to identify the molecular mechanisms responsible for the polymorphisms occurring at berry color locus and to understand the evolutionary events behind their origin [5, 13, 19]. Vezzulli et al. [19] observed a homozygous-like region in Pinot Gris and Pinot Blanc clones, suggesting that the mutation impairing the color locus was directly related with the presence of deletions, extending for at least 4.2 Mb for the L2 cell layer of Pinot Gris and ranging from 100 to 179 kb for both cell layers of Pinot Blanc. Based on these results, they concluded that Pinot Noir gave rise to Pinot Blanc and Pinot Gris independently, suggesting a parallel evolutionary model. Considering the same family of somatic variants, Pelsy et al. [5] investigated a collection of Pinot Noir, Pinot Gris and Pinot Blanc clones and proposed a model integrating both mutation and cell layer rearrangements to explain the mechanism of clone diversification. On the other hand, Migliaro et al. [13] identified polymorphisms that enable the distinction of less pigmented/unpigmented and pigmented mutants from the corresponding wild-type genotype, concluding that phenotypic variation is due to deletion events that can result in either gain- or loss-of-gene function. Deletions of different extent and position were detected among less pigmented/unpigmented variants derived from a pigmented wild-type. Regarding the pigmented variants derived from an unpigmented wild-type, the same authors observed that the main mechanism for color gain was the partial Gret1 retrotransposon excision from the VvMybA1 promoter, leaving only the solo-3’LTR region.
The present study was focused on the genetic characterization of the berry skin color locus and its surrounding genomic region, aiming to uncover the molecular mechanisms responsible for the skin color phenotype reversion on a set of somatic variants for berry skin color, analyzed here for the first time. The results obtained represent an important addition to the current knowledge about the structural dynamics along the distal arm of chromosome 2 and the evolutionary events behind the origin of somatic variants for berry skin color.
2Materials and methods
2.1Plant material
The molecular mechanisms of color variation in grapevine somatic variants for berry skin color were analyzed in 26 accessions belonging to 10 V. vinifera spp. sativa varieties, including the Pinot family (Pinot Noir, Pinot Gris and Pinot Blanc) which was used as a reference (Table 1). The different accessions were provided by the Institut National de la Recherche Agronomique – INRA, Centre of Grapevine Genetic Resources (Montpellier, France); Universidade de Trás-os-Montes e Alto Douro – UTAD (Vila Real, Portugal) and the Instituto Nacional de Investigação Agrária e Veterinária – INIAV, Coleção Ampelográfica Nacional (Dois Portos, Portugal). All accessions were divided in two groups (A and B) according to their botanical origin (Table 1).
Table 1
List of the studied somatic variants for berry skin color divided in two groups (A and B), respective berry skin color, repository and accession number
| Cultivar | Berry skin color1 | Repository2 | Accession number |
| Group A | |||
| Aramon | B | INRA/CGGR | 22Mtp1 |
| Aramon Gris | G | INRA/CGGR | 23Mtp2 |
| Aramon Blanc | W | INRA/CGGR | 24Mtp1 |
| Aspiran Noir | B | INRA/CGGR | 1Mtp3 |
| Aspiran Gris | G | INRA/CGGR | 3Mtp2 |
| Aspiran Blanc | W | INRA/CGGR | 2Mtp1 |
| Grolleau | B | INRA/CGGR | 297Mtp11 |
| Grolleau Gris | G | INRA/CGGR | 305Mtp2 |
| Grolleau Blanc | W | INRA/CGGR | 356Mtp2 |
| Pinot Noir | B | UTAD | F2.2.3 |
| Pinot Gris | G | UTAD | F2.13.2 |
| Pinot Blanc | W | UTAD | F2.13.1 |
| Pique-poul Noir | B | INRA/CGGR | 11Mtp8 |
| Pique-poul Gris | G | INRA/CGGR | 12Mtp5 |
| Pique-poul Blanc | W | INRA/CGGR | 13Mtp7 |
| Terret Noir | B | INRA/CGGR | 14Mtp3 |
| Terret Gris | G | INRA/CGGR | 15Mtp2 |
| Terret Blanc | W | INRA/CGGR | 16Mtp3 |
| Group B | |||
| Folgasao Roxo | R | INIAV/CAN | 52709 |
| Folgasão | W | INIAV/CAN | 52708 |
| Malvasia Cândida Roxo | R | INIAV/CAN | 50911 |
| Malvasia Cândida | W | INIAV/CAN | 50810 |
| Malvasia Fina Roxo | R | UTAD | F2.3.1 |
| Malvasia Fina | W | UTAD | F2.13.5 |
| Mourisco | B | INIAV/CAN | 52002 |
| Mourisco Roxo | R | INIAV/CAN | 52001 |
1B – Black; G – Grey; R – Red; W – White. 2INRA – Institut National de la Recherche Agronomique, CGGR – Centre of Grapevine Genetic Resources (Montpellier, France); UTAD - Universidade de Trás-os-Montes e Alto Douro (Vila Real, Portugal); INIAV - Instituto Nacional de Investigação Agrária e Veterinária, CAN – Coleção Ampelográfica Nacional (Dois Portos, Portugal).
2.2Genomic DNA extraction
To implement the layer-specific approach, two genomic DNA samples for each accession were isolated: one from 100 to 200 mg of young leaf (L1+L2 derived tissue), corresponding to stage E (2 to 3 leaves unfolded) of the Baggiolini scale, and the other from 200 to 300 mg of woody shoot pith (L2 derived tissue) at stage A (winter bud) of the same scale, for a total of 52 tissue-specific DNA samples. The leaf and woody shoot pith materials were ground in a TissueLyser II (Qiagen, Hilden, Germany) and DNA extraction was performed using the DNeasy 96 Plant Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. In each analysis, the genetic difference between the L1+L2 (leaf) and L2 (woody shoot pith) corresponds to the make-up of L1 cell layer. DNA concentration was determined by UV spectrometer (Nanodrop® ND-1000, ThermoFisher Scientific, USA). DNA quality was also checked with a 1% (w/v) agarose gel electrophoresis using 1x TBE buffer followed by ethidium bromide staining. Necessary dilutions were performed (approximately 10 ng/μL) and kept at 4º C for further use. Subsequent molecular analyses were performed independently on both DNA samples from each accession.
2.3Single sequence repeat (SSR) analyses
To determine whether the berry color variants used in the study were true-to-type, i.e. if all berry skin color variants were actually bud sports of the original variety (wild type) genotype, a set of 10 markers (VVS2, VVMD5, VVMD7, VVMD27, VrZAG62, VrZAG79, VMC4f3, VVMD28, VVMD32 and VVIv67), which included the Organisation Internationale de la Vigne et du Vin (OIV) core set established by the European Project GENRES#81 for grapevine cultivars identification, was used. Two multiplex PCRs were carried out with the OIV SSR core set as reported by Castro et al. [20], the first one involving VVS2, VVMD5 and VVMD7 (set A), and the second VVMD27, ssrVrZAG62 and ssrVrZAG79 (set B). Individual reactions were performed with the remaining four primer pairs (VMC4f3, VVMD28, VVMD32 and VVIv67) according to the same authors. The SSR allelic profile of each accession was compared with its published reference profile (Table 2).
Table 2
Primer information of the microssatellite markers used to all the studied loci
| SSR Locus | LG | Use | 5’-3’ Forward primer sequence | 5’-3’ Reverse primer sequence | Reference |
| VVS21,2 | 11 | True-to-type confirmation | CAGCCCGTAAATGTATCCATC | AAATTCAAAATTCTAATTCAACTGG | Thomas and Scott (1993); This et al. (2004) |
| VVMD51,2 | 16 | True-to-type confirmation | CTAGAGCTACGCCAATCCAA | TATACCAAAAATCATATTCCTAAA | Bowers et al. (1996, 1999); This et al. (2004) |
| VVMD71,2 | 7 | True-to-type confirmation | AGAGTTGCGGAGAACAGGAT | CGAACCTTCACACGCTTGAT | Bowers et al. (1996, 1999); This et al. (2004) |
| VVMD271,2 | 5 | True-to-type confirmation | GTACCAGATCTGAATACATCCGTAAGT | ACGGGTATAGAGCAAACGGTGT | Bowers et al. (1999); This et al. (2004) |
| VrZAG621,2 | 7 | True-to-type confirmation | GGTGAAATGGGCACCGAACACACGC | CCATGTCTCTCCTCAGCTTCTCAGC | Sefc (1999); This et al. (2004) |
| VrZag791,2 | 5 | True-to-type confirmation | AGATTGTGGAGGAGGGAACAAACCG | TGCCCCCATTTTCAAACTCCCTTCC | Sefc (1999); This et al. (2004) |
| VMC4f3 | 12 | True-to-type confirmation | AAAGCACTATGGTGGGTGTAAA | TAACCAATACATGCATCAAGGA | Di Gaspero et al. (2000) |
| VVMD282 | 3 | True-to-type confirmation | AACAATTCAATGAAAAGAGAGAGAGAGA | TCATCAATTTCGTATCTCTATTTGCTG | Bowers et al. (1999) |
| VVMD322 | 4 | True-to-type confirmation | TATGATTTTTTAGGGGGGTGAGG | GGAAAGATGGGATGACTCGC | Bowers et al. (1999) |
| VVIv67 | 15 | True-to-type confirmation | TATAACTTCTCATAGGGTTTCC | TTGGAGTCCATCAAATTCATCT | Merdinoglu et al. (2005) |
| SC8_0146_010 | 2 | Delition delimitation | – | – | Adam-Blondon A.F., personal communication |
| SC8_0146_026 | 2 | Delition delimitation | – | – | Adam-Blondon A.F., personal communication |
| VVNTM1 | 2 | Delition delimitation | CCACGCCACTATTGCTAAAC | TGCACCGTATCAAGATCATGTC | Fournier-Level et al. (2009) |
| VVNTM2 | 2 | Delition delimitation | TACCTGCTAACAATGCATTATG | TATTTGGTTTTTTCTAAATAGA | Fournier-Level et al. (2009) |
| VVNTM3 | 2 | Delition delimitation | TGCTGACCTGAATCATTTTCTACT | GATGTTCTGGAGAGATGCTTATC | Fournier-Level et al. (2009) |
| VVNTM4 | 2 | Delition delimitation | TTTGCATGACTGCTTGGTGTAT | CCCATTGCTAAACCCTACTCCT | Fournier-Level et al. (2009) |
| VVNTM5 | 2 | Delition delimitation | AGGAGGAATCCACATCAAAAGA | TGATTCAAAGGAATAAATAACCATCA | Fournier-Level et al. (2009) |
| VVNTM6 | 2 | Delition delimitation | CCTTCTTGGACACCCATACAAA | TTCCCTATCAACAAACTTGAGG | Fournier-Level et al. (2009) |
| VVIU20 | 2 | Delition delimitation | ACAACCTTAATGCTTCTACCAA | TCACCATGGAGATTTTCTGTAG | Merdinoglu et al. (2005) |
| VMC7G3 | 2 | Delition delimitation | TTACTAGTGCTGTCCTGCTCCA | TGCTTCCTCTCTTTCAACTTTCA | Pellerone et al. (2001) |
1OIV core set, 2VIVC database.
Another set of 10 markers (SC8_0146_010, SC8_0146_026, VVNTM1, VVNTM2, VVNTM3, VVNTM4, VVNMT5, VVNTM6, VVIU20, VMC7G3) surrounding the berry color locus and distributed along the distal arm of chromosome 2 was used to investigate polymorphisms in this region (Table 2). PCR reactions were performed as described by Vezzulli et al. [19]. Capillary electrophoresis was carried out in an ABI 3130xl Genetic Analyzer (Life Technologies, Foster City, CA, USA) and the fragments were sized with Peak Scanner V1.0 software (PE Applied Biosystems, CA, USA) using the GeneScan 500 LIZ size standard (Life Technologies) as an internal ladder.
2.4VvMybA1 and VvMybA2 gene structure
The VvMybA1 and VvMybA2 gene polymorphisms (the detection of functional and non-functional alleles) were also investigated.
Regarding characterization of the VvMybA1 locus, the presence or absence of Gret1 retroelement in the VvMybA1 promoter region was detected using the primers a (5’-AAAAAGGGGGGCAATGTAGGGACCC-3’) and d3 (5’-CCTGCAGCTTTTTCG GCATCT-3’) as described by Lijavetzky et al. [14]. PCR amplifications and detection of amplified fragments were performed as reported in Lijavetzky et al. [14]. In order to detect putative functional alleles, the primers used were F2 (5’-GGACGTTAAAAAATGGTTGCACGTG-3’) described by Azuma et al. [21] and R1 (5’-GAACCTCCTTTTTGAAGTGGTGACT-3’) by Lijavetzky et al. [14]. PCR amplifications and detection were performed as reported in Azuma et al. [21] and Carrasco et al. [18], respectively.
For the VvMybA2 gene, the single nucleotide polymorphism (SNP) related to berry color VvMybA2R44 [11] was investigated by a SNaPshot assay. VvMybA2 amplification, SNP genotyping by ABI PRISM SNaPshot Multiplex kit (Life Technologies Corporation, Carlsbad, California) and SNP detection on ABI PRISM 310 Genetic Analyzer Sequencer were performed, as reported in Carrasco et al. [18].
3Results
3.1Varietal identification
Ten SSR loci were used to ascertain the genetic identity of the somatic variants and their relative ancestor to confirm their trueness-to-type. This fingerprinting system revealed that the pigmented and unpigmented variants of each variety are closely related, showing the same genetic profile, which confirms that all the berry skin color variants analyzed were bud sports of their respective wild-type genotype. Cultivar identification was achieved by comparing each SSR profile with the European Vitis database (http://www.eu-vitis.de/) or public profile(Table S1).
3.2VvMybA1 and VvMybA2 allelic composition
To characterize the structural dynamics of the berry color locus two Myb-related genes, VvMybA1 and VvMybA2, were analyzed, since they are known to be functionally involved in berry pigmentation.
Regarding these two Myb-related genes, their allelic composition allowed inferring some of the color differences observed among the studied variants. For all the wild-type pigmented ancestors of the group A, the VvMybA1 promoter was heterozygous, containing both the Gret1 (non-functional VvMybA1ITA) and non-Gret1 (functional VvMybA1AFL) alleles in the two cell layers, as well as in the L1+L2 cell layer of the red/grey-skinned derivate variants (Table 3). In turn, only the Gret1 (non-functional VvMybA1ITA) allele, present in homozygosity, was detected in the L2 of red/grey-skinned variants and in both cell layers of unpigmented variants (Table 3).
Table 3
Genetic profile of berry color locus and its surrounding genomic region on black-to-grey and/ or white revertant somatic variants and its ancestors.
| Cultivar | Layer | Berry skin | Molecular marker and genomic coordinate on chromosome 2 (in Mb) | |||||||||||
| color1 | SC8_010 | SC8_026 | VVNTM1 | VVNTM2 | VvMybA2R44 | VvMybA1 | VVNTM3 | VVNTM5 | VVNTM6 | VVNTM4 | VVIU20 | VMC7G3 | ||
| 12674 | 12970 | 14149 | 14151 | 14181 | 14248 | 14288 | 14325 | 14330 | 14384 | 16539 | 18270 | |||
| Aramon | L1+L2 | B | ho | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | he | he | he |
| L2 | ho | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | he | he | he | ||
| Aramon Gris | L1+L2 | G | ho | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | he | he | he |
| L2 | ho | he | he | ho | T | Gret1 | ho | he | he | he | he | he | ||
| Aramon Blanc | L1+L2 | W | he | he | he | ho | T | Gret1 | ho | ho | ho | he | he | he |
| L2 | he | he | he | ho | T | Gret1 | ho | ho | ho | he | he | he | ||
| Aspiran Noir | L1+L2 | B | ho | he | he | ho | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | ho | he | he | ho | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he | ||
| Aspiran Gris | L1+L2 | G | ho | he | he | ho | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | ho | he | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Aspiran Blanc | L1+L2 | W | ho | he | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he |
| L2 | ho | he | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Grolleau | L1+L2 | B | ho | ho | ho | ho | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | ho | ho | ho | ho | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he | ||
| Grolleau Gris | L1+L2 | G | ho | ho | ho | ho | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Grolleau Blanc | L1+L2 | W | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he |
| L2 | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Pinot Noir | L1+L2 | B | he | ho | he | he | T/G | Gret1/Non-Gret1 | he | he | ho | ho | he | he |
| L2 | he | ho | he | he | T/G | Gret1/Non-Gret1 | he | he | ho | ho | he | he | ||
| Pinot Gris | L1+L2 | G | he | ho | he | he | T/G | Gret1/Non-Gret1 | he | he | ho | ho | he | he |
| L2 | he | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | ho | ho | ||
| Pinot Blanc | L1+L2 | W | he | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he |
| L2 | he | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Pique -poul Noir | L1+L2 | B | he | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | he | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he | ||
| Pique-poul Gris | L1+L2 | G | he | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | he | he | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Pique-poul Blanc | L1+L2 | W | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he |
| L2 | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
| Terret Noir | L1+L2 | B | he | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | he | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he | ||
| Terret Gris | L1+L2 | G | he | he | he | he | T/G | Gret1/Non-Gret1 | ho | he | he | ho | he | he |
| L2 | he | he | ho | ho | T | Gret1 | ho | ho | he | ho | he | he | ||
| Terret Blanc | L1+L2 | W | he | he | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he |
| L2 | he | he | ho | ho | T | Gret1 | ho | ho | ho | ho | he | he | ||
1B – Black, G – Grey, W – White. The grey background indicates the deleted region. he – heterozygous; ho – homozygous; Gret1 – non-functional allele; Non-Gret1 – functional allele.
The VvMybA1 gene did not allow discrimination of most variants of group B where the wild-type ancestor corresponds to a white-skinned variant, since they share the same allelic composition for this gene (Table 4). The partial excision of the Gret1 retrotransposon was only observed in the Mourisco variety of group B (Table 4). The presence of the functional G allele at VvMybA2R44 position was detected in some less-pigmented light red variants (Malvasia Fina Roxo, Folgasão Roxo and Malvasia Cândida Roxo) derived from a white-skinned ancestor (Table 4).
Table 4
Genetic profile of berry color locus and its surrounding genomic region on white-to-red revertant somatic variants and its ancestors.
| Cultivar | Layer | Berry skin | Molecular marker and genomic coordinate on chromosome 2 (in Mb) | |||||||||||
| color1 | SC8_010 | SC8_026 | VVNTM1 | VVNTM2 | VvMybA2R44 | VvMybA1 | VVNTM3 | VVNTM5 | VVNTM6 | VVNTM4 | VVIU20 | VMC7G3 | ||
| 12674 | 12970 | 14149 | 14151 | 14181 | 14248 | 14288 | 14325 | 14330 | 14384 | 16539 | 18270 | |||
| Folgasão Roxo | L1+L2 | R | he | he | ho | ho | T/G | Gret1/? | ho | ho | ho | ho | ho | ho |
| L2 | he | he | ho | ho | T/G | Gret1/? | ho | ho | ho | ho | ho | ho | ||
| Folgasão | L1+L2 | W | he | he | ho | ho | T | Gret1 | ho | ho | ho | ho | ho | ho |
| L2 | he | he | ho | ho | T | Gret1 | ho | ho | ho | ho | ho | ho | ||
| Malvasia Cândida | L1+L2 | R | he | he | ho | ho | T/G | Gret1/? | ho | he | ho | ho | ho | ho |
| Roxo | L2 | he | he | ho | ho | T/G | Gret1/? | ho | he | ho | ho | ho | ho | |
| Malvasia Cândida | L1+L2 | W | he | he | ho | ho | T | Gret1 | ho | he | ho | ho | ho | ho |
| L2 | he | he | ho | ho | T | Gret1 | ho | he | ho | ho | ho | ho | ||
| Malvasia Fina | L1+L2 | R | ho | ho | ho | ho | T/G | Gret1/? | ho | ho | ho | ho | ho | ho |
| Roxo | L2 | ho | ho | ho | ho | T/G | Gret1/? | ho | ho | ho | ho | ho | ho | |
| Malvasia Fina | L1+L2 | W | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | ho | ho |
| L2 | ho | ho | ho | ho | T | Gret1 | ho | ho | ho | ho | ho | ho | ||
| Mourisco Tinto | L1+L2 | B | he | he | ho | ho | T/G | Non-Gret1 | he | he | ho | ho | he | he |
| L2 | he | he | ho | ho | T/G | Non-Gret1 | ho | he | ho | ho | he | he | ||
| Mourisco Roxo | L1+L2 | R | he | he | ho | ho | T/G | Non-Gret1 | he | he | ho | ho | he | he |
| L2 | he | he | ho | ho | T/G | Non-Gret1 | ho | he | ho | ho | he | he | ||
1R – Red; B – Black; W – White. The grey background indicates the putatively homozygous regions. he – heterozygous; ho – homozygous; Gret1 – non-functional allele; ? – undetected functional allele; Non-Gret1 – functional allele.
3.3Genetics behind black-to-grey and/or white skin color reversions
As previously mentioned, all the pigmented ancestor accessions of group A were heterozygous, containing the Gret1 (VvMybA1ITA) and non-Gret1 (VvMybA1AFL) alleles of VvMybA1 in both cell layers (Table 3). Moreover, the clonal differences between the pigmented and the derived unpigmented variants was shown to be caused by deletions of different extents, resulting in the removal of the functional colored allele of both VvMybA genes, leading to the white phenotype, as was also observed by Migliaro et al. [13]. In cultivars derived from asexual reproduction, such as the ones analyzed here, the emergence of the homozygous-like pattern compared with the ancestor (usually heterozygous) can only be assigned to a deletion (grey-shaded region in Table 3). The homozygous-like term corresponds to the genetic hemizygous term (which refers to a null allele, called VvMybA1PNB), which will be used henceforth.
The black-skinned Pinot Noir was homozygous for two microsatellite markers (VVNTM6 and VVNTM4) and heterozygous for eight, as well as for the Gret1 retrotransposon insertion and VvMybA2 SNP, in either L1+L2- or L2- cell layers. Pinot Gris differed from Pinot Noir in having only one allele in the region between the VVNTM1 and VMC7G3 SSR markers, as well as only the Gret1 allele in L2-derived tissues. In turn, Pinot Blanc was hemizygous in a smaller region, between SC08_010 and VVNTM5, both on L1+L2-, and L2-derived tissues (Table 3). A similar situation was observed for Pique-poul and Aramon varieties, where both less-pigmented and unpigmented variants probably derived from the wild-type pigmented ancestor in an independent way due to the different deletion extent and position. Pique-poul Blanc differ from Pique-poul Noir since it has only one allele in the region between the SC8_010 and VVNTM6 SSR markers, as well as only the Gret1 allele (VvMybA1ITA) in both L1+L2- and L2-derived tissues. On the other hand, the hemizygous profile of Aramon Blanc was between the VVNTM2 and VVNTM6 SSR markers. Regarding the less-pigmented variants of both varieties, they only showed a difference from their wild-type pigmented ancestor in the L2 cell layer. Pique-poul Gris revealed the presence of only one allele in the region encompassed by the VVNTM1 and VVNTM6 markers, also affecting the non-Gret1 allele. Aramon Gris, similar to the less pigmented variants already described, can only be distinguished from its wild-type cultivar in L2-derived tissues, between the VVNTM2 and VVNTM3 SSR markers (Table 3).
For Aspiran Gris and Aspiran Blanc, one allele was observed from VVNTM1 to VVNTM6 SSR markers in the L2 cell layer of Aspiran Gris and in the L1+L2-derived tissues and the L2 cell layer of Aspiran Blanc. The same deletion pattern was also detected for Terret Gris and Terret Blanc (Table 3). Finally, in Grolleau Gris, as in the two previous cases (Aspiran and Terret), the homozygous status was restricted to the L2 cell layer, this being the main difference when compared with its wild-type. This deletion extended, at least, from the VvMybA2 marker region to VVNTM6 SSR marker. Due to the homozygosity of the black-skinned ancestor Grolleau upstream of the VvMybA2 marker it was not possible to have an accurate resolution of the deletion on the 5’ border. The same deletion pattern observed for Grolleau Gris was found in Grolleau Blanc, but in this color variant it was also present in L1+L2-derived tissues. Besides that, the homozygosity of the black-skinned ancestor for Grolleau and Terret in VVNTM4 SSR marker also prevents a more precise delimitation of the deletion on the 3’ border (Table 3).
3.4Genetics behind white-to-red skin color reversion
The molecular characterization of group B (pigmented variants derived from an unpigmented wild-type) followed the same layer-specific approach described above for group A. No genetic differences between layers were found within each variety, comparing the wild-type ancestral and its corresponding variants based on the markers used in this study (Table 4).
Although showing the same genetic pattern in L1+L2- and L2- derived tissues, the red and black-skinned Mourisco Roxo and Mourisco Tinto cultivars were both homozygous for a putative functional allele, described as a double insertion of 111- and 44-bp within the promoter region of VvMybA1, which should correspond to VvMybA1SUB allele. The remaining variants of group B were putatively homozygous for the Gret1 insertion (VvMybA1ITA allele) in the entire shoot apical meristem.
Regarding the results obtained for VvMybA2, all cultivars showed to be homozygous for the non-functional T allele, except Folgasão Roxo, Malvasia Cândida Roxo and Malvasia Fina Roxo, which carried the functional G allele (Table 4).
With the exception of Mourisco, in the remaining varieties of group B the markers used were homozygous and monomorphic along an extensive genomic region on the distal arm of chromosome 2. While in Malvasia Fina this putatively homozygous and monomorphic region is present along the entire distal arm of chromosome 2, in Folgasão and Malvasia Cândida this homozygosity is of a smaller length, restricted to the region between VVNTM1 and VMC4f3 and between VVNTM1 and VVNTM3 SSR markers, respectively (Table 4).
4Discussion
4.1True-to-type confirmation
One of the most challenging and unresolved problems for viticulture and germplasm management worldwide concerns the fact that grapevine is a species with alternative nomenclatures, with several cases of synonymies and homonymies. For this reason, it is particularly relevant to confirm that somatic variants correspond to true-to-type of the wild-type ancestor variety at the molecular level. Based on the results obtained by SSR genotyping, possible homonymy cases, as previously observed by Ferreira et al. [22] for cultivars of the varieties Alvarelhão, Bastardo, Carrega and Touriga, were discarded.
4.2Berry skin color reversions associated to somatic variation
The genetic analysis of group A varieties plainly showed that the deletion length encompassing the non-Gret1 allele (VvMybA1AFL) of VvMybA1 is highly variable among different varieties and also within the same variety, resulting in different color phenotypes. Thus, this mutational event on the distal arm of chromosome 2 is likely to happen in a repeated and independent way. All white-skinned variants of group A can be differentiated from the respective wild-type counterpart. In the white-skinned variants, both layers are affected by the deletion and, consequently, the hemizygous state is detectable in the entire shoot apical meristem. For the less-pigmented variants from group A, a layer-specific molecular approach is required to discriminate them from the pigmented ancestor, because the same hemizygous state described for the white-skinned variants was only detected in the L2 cell layer. Thus, all less pigmented variants showed a chimerical status, while in all unpigmented ones the deletion was conserved, involving the entire shoot apical meristem.
Although no differences in the deletion extension of grey- and white-skinned variants could be observed in Aramon, Aspiran, Groulleau and Terret varieties with the layer-specific molecular approach adopted, these data do not exclude the hypothesis that the white-skinned variant evolved from the grey-skinned one, or that these variants evolved independently from their pigmented wild-type ancestor. Thus, it could be helpful to use an integrative analysis considering the available historical information. Migliaro et al. [13] concluded that Tempranillo Gris probably evolved from Tempranillo due to an independent mutational event, considering the historical documentation about the direct selection of Tempranillo Blanco from Tempranillo. The authors inferred that the loss of the functional allele associated with the berry skin color was followed by L1 colonization and displacement to the L2 cell layer by the mutant cell, avoiding the grey berry chimerical status. Therefore, the same process could have occurred for Aramon, Aspiran, Groulleau and Terret varieties where no differences in the deletion extension of grey- and white-skinned mutants could be detected. However, the sequential evolutionary model, described by Walker et al. [15] for Cabernet Sauvignon cannot be excluded since the grey-skinned berries could represent a periclinal chimera of the black-skinned wild-type ancestor. In turn, the white-skinned berries could be the bud sport of the grey-skinned through a process that includes a deletion in the berry color locus in the L2 cells, while the white berry phenotype results from a cellular rearrangement (displacement) in the grey-skinned berries where the L2 cell layer (unpigmented) replaces the L1 cells (pigmented). Furthermore, although considered a rare event due to the stability of the anticlinal cell divisions the opposite phenomenon, L1 cell invasion of the inner layer (a process called replacement), can also beconsidered [9].
The main mechanism found for color gain in cultivars that belong to group B is the partial excision of Gret1, called VvMybA1bRUO allele, which leaves the last part of the retrotransposon (solo-3’LTR region) at VvMybA1 promotor as described by Migliaro et al. [13]. However, a similar mechanism was only observed in the Mourisco variety. Considering the results obtained for the Mourisco variety, both cultivars (Mourisco Roxo and Mourisco Tinto) should have derived from an unpigmented wild-type ancestor, homozygous for the Gret1 insertion (VvMybA1ITA). At least the L2 layer must have been affected by the subsequent partial excision of the Gret1, although it is not possible to verify if the mutation resulting in the partial excision of Gret1 affected the L1 layer as well, since this layer-specific approach was not applied on tissues containing only L1 cells. Thus, the skin color differences observed among these two somatic variants are probably due to specific interferences on the anthocyanin biosynthetic pathway regulated by different regulatory transcription factors, since they both showed the same allelic composition for VvMybA1 and VvMybA2 genes.
The same wild-type genotype could be affected by different mutations resulting in color gain, as previously reported for two variants of Italia. The recovery of VvMybA1 expression on the less-pigmented Ruby Okuyama cultivar was caused by intra-LTR recombination within Gret1 retrotransposon, while for the more intense pigmented Benitaka cultivar, color gain was caused by homologous recombination between VvMybA1 and VvMybA3, called VvMybA1BEN allele, which was identified as a novel functional allele that restored VvMybA1transcripts [16].
Although the Gret1 insertion at the promoter region of VvMybA1 has been considered as the main factor determining grape skin color variation and responsible for the recent and extremely rapid diffusion of the white phenotype regarding the grapevine domestication history, the VvMybA2 polymorphisms also seem to play a relevant role in the VvMybA diversification process [23], as observed for some less-pigmented variants of group B, namely Folgasão Roxo, Malvasia Cândida Roxo and Malvasia Fina Roxo.
As previously mentioned, except for the Mourisco variety, in the remaining varieties of group B, the analyzed molecular markers were homozygous and monomorphic along an extensive genomic region of chromosome 2. These results are in accordance with the homozygosity found by Migliaro et al. [13]. Thus, all these findings agree with the history of grape evolution and domestication. Although domestication and breeding are usually associated with a decrease of grape diversity, its influence, on a genome-wide scale, seems to be weak due to the different changes in morphology observed since grape domestication, including a wide range of berry colors. Myles et al. [23] identified a 5-Mb region on chromosome 2 encompassing the MYB transcription factor genes having observed a positive selection for white grapes around this locus, which is in agreement with the intense breeding for lighter berry color and the rapid dissemination of the MYB mutations responsible for reduced pigmentation previously referred to by Fournier-Level et al. [17].
In summary, this layer-specific approach was informative regarding the discrimination of skin color somatic variants with a pigmented ancestor, particularly the VVNTM1 and VVNTM5 SSR markers. Moreover, VVNTM3 and VVNTM4 SSR markers were the less informative markers since they were shown to be homozygous for almost all the somatic variants for berry skin color analyzed. Additionally, for somatic variants derived from an unpigmented ancestor this approach was shown to be not very informative regarding the discrimination of the variants, due to the extensive homozygosity observed either between variants or between layers of eachvariant.
4.3Evolutionary mechanisms behind the origin of skin color somatic variants
Based on layer localization and the differences in the size of the deletion on the distal arm of chromosome 2, it is suggested that the less-pigmented and unpigmented somatic variants analyzed in this study can be ascribed from a pigmented ancestor (group A) in accordance with two theoretical models: 1) the Sequential model, which is the first evolutionary model proposed to explain the somatic color mutants development within a variety, also reported as ‘Cabernet Sauvignon-model’ by Walker et al. [11]; according to this model, the black-skinned wild-type ancestor gives rise to a less-pigmented variant which in turn gives rise to the unpigmented variant (Fig. 1A); and 2) the Parallel model, first proposed for Pinot family by Vezzulli et al. [19], where the black-skinned berry ancestor gives rise to the less-pigmented (Pinot Gris) and the unpigmented (Pinot Blanc) variants independently (Fig. 1B). Based on the results obtained, three varieties seem to follow the Sequential model, namely; Aspiran, Groulleau and Terret. The remaining varieties (Aramon, Pinot and Pique-poul) seem to be in agreement with the Parallel model.
Fig.1
Schematic representation of the evolutionary models describing the structural dynamics at the berry color locus in the L1 and L2 cell layers. Variants of the Group A can originate either through the Sequential model (A) or the Parallel model (B). The origin of variants from the Group B can be explained by the Revertant model (C), from one out of two different mechanisms, the partial excision of the Gret1 retrotransposon from the VvMybA1 promotor or the recovery of the G functional allele of VvMybA2. On the right side are represented the structures of the different alleles and genotypes found in the somatic variants for berry skin color studied.
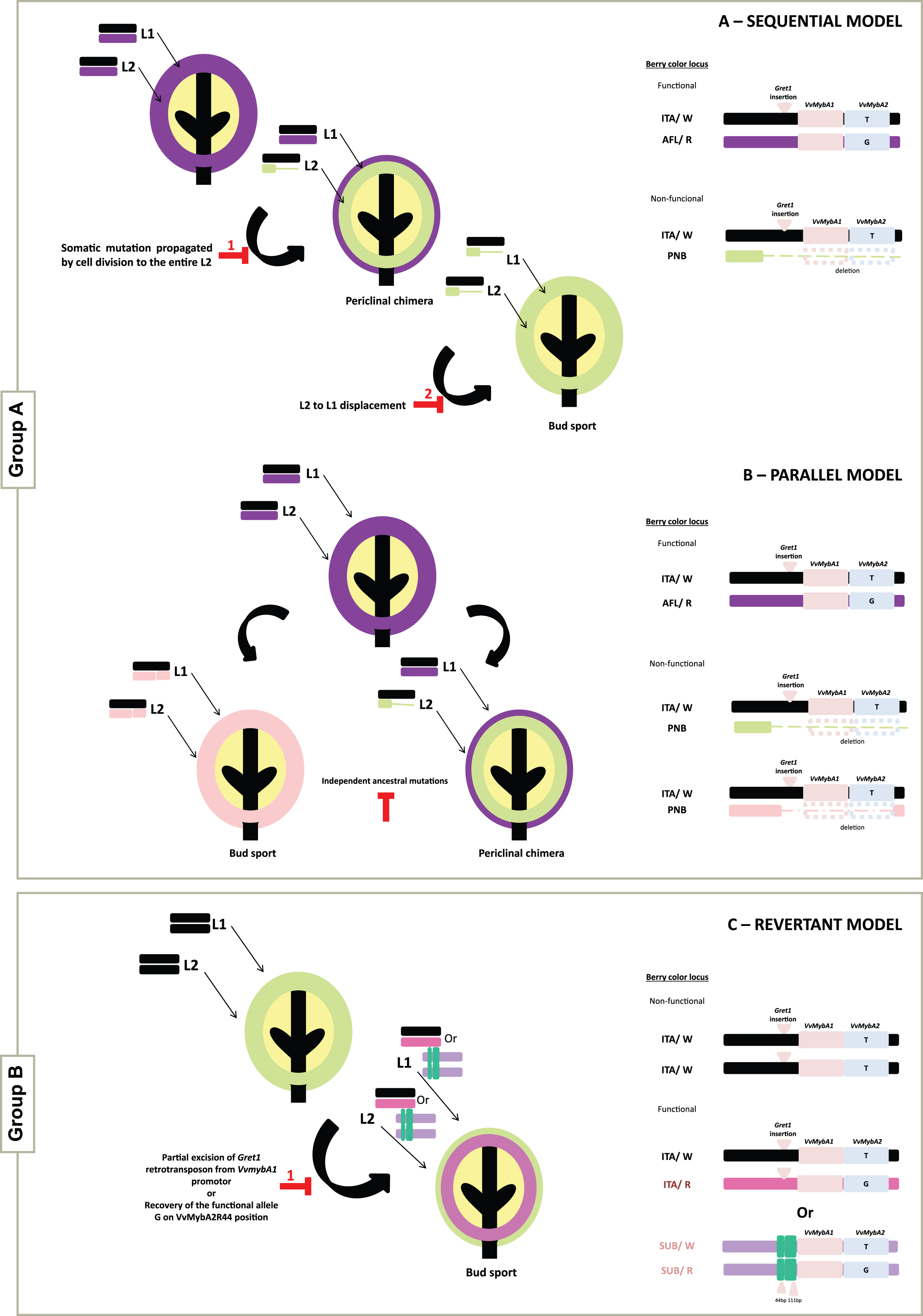
Regarding the experimental data obtained for group B, where the wild-type ancestor is a white-skinned cultivar giving rise to a less-pigmented variant, a Revertant model (Fig. 1C) is proposed. As mentioned above, the main mechanism described as being responsible for color gain involves the partial excision of Gret1 retrotransposon from the VvMybA1 promoter. However, this mechanism can only be applied to the Mourisco variety, in both pigmented variants. Our data suggests a novel mechanism for the genetic make-up of less-pigmented variants evolving from an unpigmented ancestor. For the cases of the less-pigmented variants (Folgasão Roxo, Malvasia Cândida Roxo and Malvasia Fina Roxo) color gain seems to result from the recovery of the functional G allele on VvMybA2R44 position. This event might be playing a fundamental role, probably by restoring VvMybA2 transcripts (Fig. 1C).
5Conclusions
These findings represent a significant breakthrough regarding the mechanisms behind the formation of somatic variants for berry skin color, namely unpigmented or pigmented-related grape cultivars with different color shades, since such molecular information has not been previously available for somatic variants derived from the pigmented ancestors Aramon, Aspiran, Grolleau, Pique-poul and Terret, nor for the unpigmented ancestors, Folgasão, Malvasia Cândida, Malvasia Fina and Mourisco.
These data support the conclusion that, besides VvMybA1 and VvMybA2 playing an important role regarding the phenotypic variation observed among the berry skin color somatic variants due to black-to-grey and/or white reversions, the molecular mechanism leading to skin color variation results from different deletion patterns encompassing the berry color locus. Moreover, the experimental data regarding the white-to-red reversion suggest that VvMybA2 gene might play a fundamental role for color gain through the recovery of functional G allele on VvMybA2R44 position.
This approach was also shown to be particularly informative regarding the discrimination of somatic variants for berry skin color with a pigmented ancestor, therefore it could be used to solve problems of cultivar identification and newly appearing somatic variants for berry skin color.
Conflict of Interest
The authors have no conflict of interest to report.
Acknowledgments
This work was supported by European Investment Funds through FEDER/COMPETE/POCI – Operational Competitiveness and Internationalization Programme, under Project POCI-01-0145-FEDER-006958, and National Funds by FCT - Portuguese Foundation for Science and Technology, under the project UID/AGR/04033/2013. V. Ferreira is also indebted to FCT for her grant (SFRH/BD/96400/2013). Work in R. Arroyo-García lab is funded by the Project Grants RTA2014-00029-C03-01 and RTA2014-00016-C02-01. The authors would like to thank INIAV (Instituto Nacional de Investigação Agrária e Veterinária), INRA (Institut National de la Recherche Agronomique) and UTAD (University of Trás-os-Montes and Alto Douro) for kindly providing the plant material.
Supplementary material
[1] The supplementary material is available in the electronic version of this article: 10.3233/JBR-170289.
References
[1] | Terral J-F , Tabard E , Bouby L , Ivorra S , Pastor T , Figueiral I , et al. Evolution and history of grapevine (Vitis vinifera) under domestication: New morphometric perspectives to understand seed domestication syndrome and reveal origins of ancient European cultivars. Ann Bot. (2010) ;105: :443–55. doi: 10.1093/aob/mcp298 |
[2] | This P , Lacombe T , Thomas MR . Historical origins and genetic diversity of wine grapes. Trends Genet. (2006) ;22: :511–9. doi: 10.1016/j.tig.2006.07.008 |
[3] | Keller M . The Science of Grapevines: Anatomy and Physiology. 2nd ed. San Diego, CA: Academic Press; (2010) . doi: 10.1016/B978-0-12-374881-2.00012-X |
[4] | Brickell CD , Alexander C , David JC , Hetterscheid WL , et al. International Code of Nomenclature for Cultivated Plants. vol. 10: . 8th ed. Leuven, Belgium: Scripta Horticulturae; (2009) . |
[5] | Pelsy F , Dumas V , Bévilacqua L , Hocquigny S , Merdinoglu D . Chromosome replacement and deletion lead to clonal polymorphism of berry color in grapevine. PLoS Genet. (2015) ;11: :e1005081–doi: 10.1371/journal.pgen.1005081 |
[6] | D’Amato F . Role of somatic mutations in the evolution of higher plants. Caryologia. (1997) ;50: :1–15. doi: 10.1080/00087114.1997.10797380 |
[7] | Dermen H . Nature of plant sports. Am Hortic Mag. (1960) ;39: :123–73. |
[8] | Hocquigny S , Pelsy F , Dumas V , Kindt S , Heloir M-C , Merdinoglu D . Diversification within grapevine cultivars goes through chimeric states. Genome. (2004) ;47: :579–89. doi: 10.1139/g04-006 |
[9] | Pelsy F . Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity (Edinb). (2010) ;104: :331–40. doi: 10.1038/hdy.2009.161 |
[10] | Carrier G , Le Cunff L , Dereeper A , Legrand D , Sabot F , Bouchez O , et al. Transposable elements are a major cause of somatic polymorphism in Vitis vinifera L. PLoS One. (2012) ;7: :e32973–doi: 10.1371/journal.pone.0032973 |
[11] | Walker AR , Lee E , Bogs J , McDavid DAJ , Thomas MR , Robinson SP . White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. (2007) ;49: :772–85. doi: 10.1111/j.1365-313X.2006.02997.x |
[12] | Giannetto S , Velasco R , Troggio M , Malacarne G , Storchi P , Cancellier S , et al. A PCR-based diagnostic tool for distinguishing grape skin color mutants. Plant Sci. (2008) ;175: :402–9. doi: 10.1016/j.plantsci.2008.05.010 |
[13] | Migliaro D , Crespan M , Muñoz-Organero G , Velasco R , Moser C , Vezzulli S . Structural dynamics at the berry colour locus in Vitis vinifera L. somatic variants. Aust J Grape Wine Res. (2014) ;20: :485–95. doi: 10.1111/ajgw.12103 |
[14] | Lijavetzky D , Ruiz-García L , Cabezas JA , De Andrés MT , Bravo G , Ibáñez A , et al. Molecular genetics of berry colour variation in table grape. Mol Genet Genomics. (2006) ;276: :427–35. doi: 10.1007/s00438-006-0149-1 |
[15] | Walker AR , Lee E , Robinson SP . Two new grape cultivars, bud sports of Cabernet Sauvignon bearing pale-coloured berries, are the result of deletion of two regulatory genes of the berry colour locus. Plant Mol Biol. (2006) ;62: :623–35. doi: 10.1007/s11103-006-9043-9 |
[16] | Azuma A , Kobayashi S , Goto-Yamamoto N , Shiraishi M , Mitani N , Yakushiji H , et al. Color recovery in berries of grape (Vitis vinifera L.) “Benitaka”, a bud sport of “Italia”, is caused by a novel allele at the VvmybA1 locus. Plant Sci. (2009) ;176: :470–8. doi: 10.1016/j.plantsci.2008.12.015 |
[17] | Fournier-Level A , Lacombe T , Le Cunff L , Boursiquot J-MM , This P . Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.). Heredity (Edinb). (2010) ;104: :351–62. doi: 10.1038/hdy.2009.148 |
[18] | Carrasco D , De Lorenzis G , Maghradze D , Revilla E , Bellido A , Failla O , et al. Allelic variation in the VvMYBA1 and VvMYBA2 domestication genes in natural grapevine populations (Vitis vinifera subssylvestris). Plant Syst Evol. (2015) ;301: :1613–24. doi: 10.1007/s00606-014-1181-y |
[19] | Vezzulli S , Leonardelli L , Malossini U , Stefanini M , Velasco R , Moser C . Pinot blanc and Pinot gris arose as independent somatic mutations of Pinot noir. J Exp Bot. (2012) ;63: :6359–69. doi: 10.1093/jxb/ers290 |
[20] | Castro I , Martín JP , Ortiz JM , Pinto-Carnide O . Varietal discrimination and genetic relationships of Vitis vinifera L. cultivars from two major Controlled Appellation (DOC) regions in Portugal. Sci Hortic (Amsterdam). (2011) ;127: :507–14. doi: 10.1016/j.scienta.2010.11.018 |
[21] | Azuma A , Kobayashi S , Mitani N , Shiraishi M , Yamada M , Ueno T , et al. Genomic and genetic analysis of Myb -related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor Appl Genet. (2008) ;117: :1009–19. doi: 10.1007/s00122-008-0840-1 |
[22] | Ferreira V , Fernandes F , Pinto-Carnide O , Valentão P , Falco V , Martín JP , et al. Identification of Vitis vinifera L. grape berry skin color mutants and polyphenolic profile. Food Chem. (2016) ;194: :117–27. doi: 10.1016/j.foodchem.2015.07.142 |
[23] | Myles S , Boyko AR , Owens CL , Brown PJ , Grassi F , Aradhya MK , et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci U S A. (2011) ;108: :3530–5. doi: 10.1073/pnas.1009363108 |




