Germplasm resources for verticillium wilt resistance breeding and genetics in strawberry (Fragaria)
Abstract
BACKGROUND:
The fungal disease verticillium wilt has been recognized as an obstacle to strawberry production since its initial description in 1931. The full potential of genetic resistance as a solution to this problem has yet to be determined or realized.
OBJECTIVE:
Our investigations are concerned with defining new sources of resistance to verticillium wilt disease in cultivated and wild strawberry germplasm, and with advancing genetic studies on the basis of resistance/susceptibility.
METHODS:
We screened 23 diploid, 1 decaploid, and 26 octoploid Fragaria (strawberry) germplasm accessions and cultigens for response to root-dip inoculation with Verticillium dahliae isolate V1. Pedigree relationships of 10 studied cultigens were examined. Crosses were performed between resistant and susceptible accessions.
RESULTS:
Variability in inoculation response existed within and between species at diploid and octoploid levels. Very or moderately resistant accessions were found within each of three diploid and three octoploid species. Moderately or very susceptible accessions were documented within F. vesca and each octoploid species. Segregation for resistance/susceptibility was evident in progeny populations.
CONCLUSIONS:
The verticillium wilt resistance ratings reported here and discussed in relation to prior studies adds to the body of publically available knowledge about sources of wilt resistance and susceptibility in Fragaria germplasm.
1Introduction
The fungal disease verticillium wilt has been a major obstacle to U.S. strawberry production since first identified and described by Thomas [1]. In most major U.S. strawberry production areas, methyl bromide and chloropicrin have been the foundation for control of soil-borne disease organisms [2], including Verticillium dahliae Kleb., the causal agent of verticillium wilt. However, the classification of methyl bromide as an ozone depleting chemical under the Montreal Protocol in 1987, and by the U.S. Environmental Protection Agency in 1993, has prompted the development of alternative chemical treatment practices [3]. Non-chemical alternatives such as steam and solarization treatments have shown promise, but have not been demonstrated to be cost effective [2]. Use of non-pathogenic V. dahliae isolates as competitors has been explored, but was limited because of infestation by other pathogens [4]. Verticillium wilt was a serious disease before the advent of soil fumigation [5, 6], and its control is an ongoing concern in affected production areas.
At least partial relief from the problems posed by verticillium wilt could be provided by incorporation of genetic resistance into new commercial strawberry varieties. While enhanced wilt resistance has been obtained in strawberry via the introduction of a chitinase gene [7], the genetic engineering approach would also negate the opportunity to exploit the burgeoning organic production niche. The skyrocketing demand for organic strawberries from $2 million in 1997 to $55 million in 2009 has stimulated intensive interest among breeders in the prospect of developing wilt resistant strawberry varieties [8]. The foregoing realities emphasize the need to identify and exploit naturally occurring sources of genetic resistance and the need to breed for resistance.
Quantitative genetic variation for field resistance to verticillium wilt was documented in cultivated strawberry breeding populations in the mid-twentieth century [9–11] and more recently [5, 12–14], indicating the potential for enhancing resistance through breeding. Shaw et al. [15] have asserted that genetic resistance to wilt is best regarded as one component of an integrated management system that also includes efforts to reduce pest populations at all growth stages. Even so, it is evident that considerable potential exists for improving wilt resistance through germplasm evaluation and breeding, and that the full potential for genetic resistance to verticillium wilt in the cultivated strawberry has yet to be defined or realized.
The strawberry genus Fragaria is remarkably diverse, encompassing more than 23 species and spanning ploidy levels from diploid (2n = 2x = 14) to decaploid (2n = 10x = 70) [16, 17]. The octoploid (2n = 8x = 56) cultivated strawberry, Fragaria×ananassa, is known to have arisen via hybridization between its octoploid ancestors, F. chiloensis and F. virginiana, in the mid-1700s [18]. Phylogenetic and genomic studies have implicated ancestral forms of diploids F. vesca and F. iinumae as likely genome donors to the octoploids [19]; however, the evolutionary pathway from the diploid to the octoploid level is yet to be delineated [20].
Each of the ancestral octoploids has multiple subspecies or formae: F. chiloensis has formae chiloensis, patagonica, and subspecies lucida and pacifica; while F. virginiana has subspecies virginiana, grayana, glauca, and platypetala [21]. An effort to reconstruct Fragaria×ananassa by crossing representatives of F. chiloensis and F. virginiana was initiated [22] and is ongoing [23]. A broad sampling of the diversity present in F. chiloensis and F. virginiana is represented by the 38 carefully chosen accessions that comprise the USDA “supercore” collection [24] maintained by the National Clonal Germplasm Repository (NCGR) in Corvallis Oregon.
Our ongoing investigations are concerned with defining new sources of resistance in cultivated and wild strawberry germplasm, and with advancing genetic studies on the basis of resistance/susceptibility. In the present study, we evaluated representatives of five diploid species, three octoploid species, and –serendipitously –one representative of decaploid F. cascadensis. At the time that this study was initiated, NCGR accession CFRA 110 was identified by the NCGR as octoploid F. virginiana ssp. platypetala. However, it has since been determined to be decaploid [16], and to belong to the newly defined decaploid species, F. cascadensis [25]. The octoploids in our study included 17 members of the NCGR supercore set, and ten cultigens (cultivars and advanced breeding selections) of relevance to our breeding programs. In addition to reporting our resistance/susceptibility ratings for 50 germplasm accessions and cultigens, we place our results within the context of prior knowledge through integrated discussion of previous verticillium wilt studies in strawberry, both wild and cultivated e.g. [9, 10, 12, 26–31].
2Materials and methods
Plant and fungal materials. Information about the studied plant accessions, their geographic origins (wild-collected materials) or breeding program sources, and their identification numbers (local and/or Plant Introduction) is provided for diploids and polyploids, respectively, in Tables 1 and 2. Subspecies designations are not provided for California accessions of F. vesca due to uncertainty in differentiating the subspecies Fragaria vesca ssp. bracteata from F. vesca ssp. californica and their possible hybrids. Among diploid accessions (Table 1); CFRA364.002, CFRA333.001, and CFRA520.001 were obtained from the NCGR, accessions from Hokkaido, Japan were collected by Thomas M. Davis and Kim Hummer [32], and GS1J was collected by Gunter Staudt, while all other UNH-numbered accessions were collected by Davis.
The polyploid accessions of octoploids Fragaria virginiana and F. chiloensis, and decaploid F. cascadensis belonging to the Fragaria supercore collection were obtained from the NCGR. The studied F.×ananassa accessions were of interest to our breeding programs. Plants of ‘Sparkle’ and ‘Tristar’ were purchased from Nourse Farms, Whately MA, while the other cultivars and breeding lines were provided by Andrew R. Jamieson. F.×ananassa cultivars ‘Annapolis’, ‘Cavendish’, ‘Evangeline’, ‘Mira’, ‘Wendy’ (aka ‘AC Wendy’), and ‘Laurel’ (tested as K93-20), and numbered breeding clones K05-9 and M903 were developed at the Agri-Food Canada Kentville Research Station, Nova Scotia, Canada. Cultivar ‘Sparkle’ is a 1942 release from the New Jersey State University breeding program [18], while ‘Tristar’ is a 1981 release from the USDA Beltsville program.
Screening methods. A series of trials was conducted, in which from five to 20 genotypes were screened. In each trial, each genotype was represented by three orfour inoculated and two uninoculated (control) plants. Pots containing inoculated or uninoculated plants were maintained within separate containment trays, and plants were randomly distributed within the trays. Some genotypes were included in multiple trials.
Verticillium dahliae isolate V1 was obtained from Mansun Kong at Driscoll Strawberry Associates, Watsonville, CA, USA, and was originally isolated from an infected strawberry plant (M. Kong, personal communication). Plants were maintained in the UNH MacFarlane Greenhouse facility in Pro-Mix Mycorrhizaetrademark (Premier Tech Horticulture LTD, Canada), and were propagated by rooting stolons that were still attached to mother plants to produce runner plantlets. Runner nodes were pinned onto the surface of Metro-mix 360 (Hummerttrademark International, Missouri) medium in 4” standard round polypropylene pots (Dillen Products Company, Ltd., Middlefield, Ohio) using staples made from plastic-coated wire. Plantlets were allowed to root for two weeks prior to separation from the mother plant and inoculation. Ten plantlets were rooted from each mother plant, and eight were ultimately used in each trial (four inoculated plantlets and four uninoculated controls). Concurrent with plant propagation, fungal inoculum was cultured in an appropriate volume (∼10 ml for each plant to be inoculated) of autoclaved Difcotrademark Czapek-Dox broth (BD Biosciences). The broth, contained in 1L flasks, was inoculated in a laminar flow hood with two to four ∼1 cm2 pieces of fungus-covered Czapek-Dox agar from fungal culture plates. The inoculated broth was then incubated for two weeks at room temperature on an orbital shaker at 200 RPM.
On the day of inoculation, the fungal culture was strained through two layers of cheesecloth and one layer of Miracloth (EMD Millipore, Billerica, Massachusetts), and then centrifuged at 10,000×g for 5 min. The culture medium was decanted and the conidial pellet was resuspended in a volume of sterile distilled water equivalent to that of the decanted culture medium. Conidia were quantified using a hemacytometer (Bright Light Counting Chamber Improved Neubauer, Hausser Scientific, Horsham Pa), and the suspension was diluted, if necessary, with sterile distilled water to ∼2×107 conidia/ml. Immediately prior to root dipping, rooted plantlets were separated from mother plants and trimmed to remove runners. Soil was shaken from roots prior to dipping. Root dipping was performed in 50-ml polyvinyl chloride pipet basins (Fisher Scientific, Pittsburgh, PA). Root systems were immersed, two at a time per basin, for 5 min in either 20 ml fungal spore suspension or 20 ml sterile distilled water (uninoculated control). After dipping, the plants were replanted in new 4” pots of sterile (autoclaved) Metromix medium, then moved to a greenhouse under ambient light and temperature conditions or to a 22C temperature-controlled growth room under broad-spectrum (140μmoles/m2/sec) fluorescent lights, then maintained in containment trays with minimal watering for a period of at least eight weeks.
Plant verticillium wilt disease ratings. At the end of the observational period, each individual plant was rated relative to controls according to the following rating scale, as exemplified by plants shown in Fig. 1. A rating of 1 (healthy) was given to plants closely resembling uninoculated controls. A rating of 1.5 (slightly symptomatic) indicated mild stunting and/or mild outer leaf necrosis and/or browning. A rating of 2.0 (mildly symptomatic) indicated distinct stunting and/or distorted growth, more leaf necrosis and browning, and perhaps one or two dead leaves. A rating of 2.5 (very symptomatic) was given to plants with severe stunting, leaf necrosis and browning, yet still having one to a few green leaves. A 3.0 (dead) rating indicated that all of the leaves were necrotic and the plant was considered to be nearly or completely dead.
A mean disease rating was then calculated for each germplasm accession or cultigen on the basis of the respective total number of rated plants. As guided by previous literature [9, 31, 33] and to facilitate comparison with prior studies, we then assigned a qualitative classification to each accession or cultigen based upon its mean disease rating. For this purpose, the scale of mean disease ratings was partitioned into five ordered categories. The category ranges and corresponding classifications (in parentheses) were: 1.0 to 1.3 (very resistant = VR), 1.4 to 1.7 (moderately resistant = MR), 1.8 to 2.2 (intermediate = I), 2.3 to 2.6 (moderately susceptible = MS), and 2.7 to 3.0 (very susceptible = VS). For example, an accession with a mean disease rating of 1.5 would have been classified as MR.
Pedigree analysis. As an aid to understanding the genetic relationships among studied octoploid cultigens and their known ancestors and descendants, pedigree relationships were determined by extracting relevant information from the RosBREED “Breeding Information Management System” (BIMS) crop reference set for Fragaria (http://www.rosaceae.org/breeders_toolbox), and integrating it with information from published cultivar release announcements to create a project-specific database. Verticillium resistance ratings from the present study (Table 2) and resistance categories from various literature sources (Table 3) were then added to the database. We traced the pedigrees of the studied cultigens to find common ancestors. Finally, the pedigree relationships and verticillium resistance ratings, where available, were depicted in the form of an annotated pedigree map, which was constructed using Pedimaptrademark software (Wageningen UR –Plant Breeding). The BIMS crop reference data set and current Pedimap resource are products of the USDA-NIFA-SCRI RosBREED and EU-FruitBreedomics (www.fruitbreedomics.com) projects.
3Results
The results of our verticillium inoculation response screenings are summarized for the diploid and polyploid genotypes, respectively, in Tables 1 and 2. In the tables, the accessions and cultigens are ordered by species, subspecies (or formae), and mean disease rating. The numbers of plants used to calculate mean disease ratings varied among accessions and cultigens for two reasons. Although each accession or cultigen was initially represented by four inoculated plants per trial, plus controls, in a few cases a plantlet was lost due to causes not related to verticillium wilt, and only three inoculated plants were rated. Also, some accessions that displayed relatively high levels of resistance or susceptibility were chosen for inclusion as comparators in multiple trials. In early trials, diploid Fragaria vesca accession TMD2 was identified as moderately resistant and BC30 was identified as very susceptible, while among octoploid accessions Fragaria virginiana CFRA1699 was identified as very resistant and CFRA1455 and CFRA1408 were identified as moderately to very susceptible. These five accessions were used as resistant and susceptible controls, respectively, in subsequent trials, and therefore are represented by larger numbers of inoculated plants as indicated in the “N” column in Tables 1 and 2. Importantly, these comparator accessions displayed consistent resistant or susceptible responses across trials under both greenhouse and growth room conditions.
Diploid accessions. At the diploid level, the mean disease ratings of the 15 F. vesca accessions ranged from 1.5 to 3.0 (Table 1). The most resistant F. vesca accessions, all with mean ratings of 1.5, were one accession from British Columbia (BC5) and two from Mendocino County, California (U2A and TMD2); while the most susceptible, all with mean ratings of 2.9 to 3.0, were three accessions from British Columbia (BC3, BC30, and GS1J), two from California (DN2A and HP3A) and one from New Hampshire (‘Pawtuckaway’). The three representatives of F. vesca subsp. americana (WC6, WC8, and ‘Pawtuckaway’) all had mean ratings in the moderately-to-very susceptible range (2.6 to 3.0), while both resistance and susceptibility was seen among the F. vesca accessions from California and among those from British Columbia. The other diploid accessions included three species (F. bucharica, F. iinumae, and F. viridis) and one interspecific hybrid (CFRA364) that were each represented by a single accession and had mean ratings in the very-to-moderately resistant range of 1.0 to 1.7 (Table 1). Among the four accessions of F. nipponica, mean ratings ranged from 1.3 (very resistant) to 2.0 (intermediate).
Wild polyploid accessions. The mean ratings of octoploids F. chiloensis and F. virginiana ranged from 1.6 to 3.0 and 1.2 to 3.0, respectively (Table 2). The only wild octoploid accession categorized as “very resistant” was F. virginiana subsp. virginiana CFRA1699, while three and four accessions of F. chiloensis and F. virginiana, respectively, were categorized as “very susceptible”. The single representative of decaploid F. cascadensis (CFRA110) was also categorized as very susceptible (mean rating 3.0).
Octoploid cultigens- ratings and pedigree analysis. Among the ten cultigens of F.×ananassa, mean ratings ranged from 1.0 to 3.0 (Table 2). ‘Tristar’ (1.0) and breeding clone M903 (1.3) were categorized as very resistant, while ‘Mira’, ‘Evangeline’, ‘Laurel’, and breeding clone K05-9 were categorized as very susceptible. Of the remaining four cultigens, three were intermediate and one was moderately susceptible.
We examined the ancestries of rated cultigens with the aid of the constructed pedigree (Fig. 2), with the exception of very susceptible rated breeding clone K05-9 which, as an open-pollinated selection from an unknown Driscoll variety has unknown parentage and so could not be integrated into the pedigree. Previous verticillium resistance ratings were unavailable for both parents of ‘Tristar’, ‘Evangeline’ and ‘M903’, while ratings for only one parent each were available for ‘Wendy’, and ‘Annapolis’. Among the four cultigens for which both parents had available ratings, the very susceptible ‘Mira’ had very susceptible parents ‘Scott’ and ‘Honeoye’; the very susceptible ‘Laurel’ had intermediate parents ‘Allstar’ and ‘Cavendish’; the very susceptible ‘Sparkle’ had as parents intermediate ‘Fairfax’ and very resistant ‘Aberdeen’; and the intermediate ‘Cavendish’ had as parents very susceptible ‘Glooscap’ and intermediate ‘Annapolis’. Similarly, in some instances where all ratings came from literature sources, very resistant ‘Etna’ had very susceptible parents ‘Belrubi’ and ‘Marlate’; moderately resistant ‘Temple’ had as parents very resistant ‘Aberdeen’ and intermediate ‘Fairfax’; and very susceptible ‘Bounty’ had as parents very susceptible ‘Jerseybelle’ and very resistant ‘Senga Sengana’ (Fig. 2).
Genetic analysis. Based upon the foregoing results, we identified promising intra-specific crossing combinations between moderately (MR) or very resistant (VR) and very susceptible (VS) parents, at the diploid (F. vesca) and octoploid (F. virginiana) levels, respectively: diploid TMD2 (MR)×BC30 (VS); octoploid CFRA1408 (VS)×CFRA1699 (VR), and octoploid CFRA1455 (MS)×CFRA1699 (VR). We have performed the indicated crosses and generated F1 hybrid progeny. The results from an initial screening (unpublished data) are indicative of segregation forresistance/susceptibility in the F1 progeny of each cross, suggesting that one or both parents in each cross are heterozygous for genetic determinants of resistance.
4Discussion
Phenotypic assessment provides guidance to the collection and archiving of plant germplasm, and generates knowledge resources of value to breeders and geneticists interested in understanding and manipulating specific traits. The trait of interest to the present study is resistance/susceptibility to the destructive fungal disease, verticillium wilt. This report advances knowledge about the occurrence and inheritance of wilt resistance and susceptibility in wild and cultivated strawberry germplasm in three ways. First, we present new disease resistance/susceptibility data for 50 Fragaria germplasm accessions and cultigens representing three levels of ploidy. Second, we define and informatively display pedigree relationships among a selected group of resistant and susceptible strawberry cultigens, thereby graphically documenting both the extent of, and the gaps in, present knowledge about verticillium susceptibility in a coherent assemblage of important strawberry germplasm. Finally, within the following discussion, we summarize and synthesize prior knowledge from diverse, publicly available sources, For the latter purpose, relevant (and in some cases obscure) information sources were identified using searches of scientific literature and the USDA Germplasm Resources Information Network (GRIN) database (http://www.ars-grin.gov/), and keyword web searches.
Diploid accessions. In our screenings of diploid germplasm, broad variability in disease responses was observed among the sixteen Fragaria vesca accessions, ranging from moderately resistant to very susceptible. The evident biodiversity within F. vesca provides an attractive opportunity for association genetics studies within this species, which among strawberry species is currently unique in having an available reference genome sequence [34]. Among the other diploids tested, all ratings fell within the intermediate to very resistant range. As in our own study, Olbricht et al. [31] detected variation for verticillium wilt resistance/susceptibility both within and between Fragaria species. Among ten diploid species tested by Olbricht et al. [31], susceptibility was reported in at least one representative each of five diploid species, including the only identified F. vesca accession included in their study. Perhaps importantly, consistent “tolerance,” where the pathogen was internally present but the host plant was asymptomatic, was found only in F. iinumae, while in our own study the only tested representative of F. iinumae ranked among the most resistant of our diploid accessions (Table 1). Like F. vesca, F. iinumae is of particular interest because of its status as an ancestral subgenome donor to the octoploid species [19]. Wild polyploid accessions. High levels of susceptibility were found in at least one representative of each of the two octoploid species, F. chiloensis and F. virginiana, and in the only representative of decaploid F. cascadensis. Moderate to elevated levels of resistance also occurred in each of the wild octoploid species; however, of the wild octoploids only F. virginiana subsp. virginiana accession Montreal River 10 (CFRA1699) was rated as very resistant. CFRA1699 has also been determined to possess a number of favorable horticultural traits, including resistance to common foliar diseases, and has been used extensively in the F.×ananassa reconstruction project [23, 35, 36]. Another accession, LH50-4 (CFRA1697), used extensively in the F.×ananassa reconstruction project due to its day-neutrality trait, cold hardiness, fruit color, and resistance to root knot nematode [23, 35, 36] was rated as very susceptible to verticillium wilt in our study.
Several prior studies of verticillium resistance in wild octoploid Fragaria germplasm have appeared, mostly in pre-1970’s literature [33, 37–40], as list in Table 3 and/or summarized below. These early reports documented, with varying degrees of precision, the sites of origin (collection) of the studied materials, and thus point subsequent investigators to potentially valuable sites for future collections. Although the rating and classification schemes employed by previous authors have varied, all were sufficiently categorical and co-sequential as to allow meaningful comparisons among studies, including our own results as presented here.
Of the two ancestral octoploid species, comparatively less attention has been given to the occurrence and transmission of verticillium resistance in F. virginiana. In a multi-inoculation greenhouse trial [38], no resistance was found among two F. virginiana clones and 39 seedlings, but the source or origin of the F. virginiana accessions was not provided. In contrast, Varney et al. [9] found the so called “Sheldon” clone of F. virginiana to be very resistant. In a multi-inoculation greenhouse trial, Newton and van Adrichem [41] found no resistance in 49 evaluated Ontario F. virginiana seedlings, while we rated an Ontario accession, CFRA1699, as very resistant.
Greater attention has been given to F. chiloensis. Newton and van Adrichem [41] found 8 resistant out of 40 evaluated Oregon coast F. chiloensis seedlings. Wilhelm [38] found no resistance in F. chiloensis from Ambato, Peru. However, we found Peruvian F. chiloensis CFRA372, an accession used in the F.×ananassa reconstruction project for its fruit qualities and resistance to root lesion nematodes [23], to be moderately resistant to verticillium wilt. Additional F. chiloensis accessions CFRA24, CFRA1088, CFRA1691, and CFRA34 are also involved in the F.×ananassa reconstruction project [23], and we found these accessions moderately to very susceptible to verticillium wilt. In a field and greenhouse study of 1009 F. chiloensis clones from 14 sites along the California coast from Santa Maria to just north of San Francisco, Bringhurst et al. [33] found that where multiple clones were sampled per site, a range of resistance and susceptibility was detected within sites, suggesting the possibility of segregation for resistance/susceptibility among seedling-derived plants within sites. For instance, from their Bodega Bay site, Bringhurst et al. [33] found one resistant F. chiloensis clone and two susceptible clones, in comparison to 12 resistant clones previously reported by Wilhelm [38].
Bringhurst et al. [33] found the entire range of resistance to susceptibility among six clones from Pigeon Point, the collection site of FRA357, which was categorized as moderately resistant in our study. Similarly, Bringhurst et al. [33] found intermediate to extreme susceptibility at the Scotts Creek site, the collection site for CFRA1692, which was used extensively in horticultural trait evaluations and in the F.×ananassa reconstruction project [23, 36], but for which we do not have verticillium wilt resistance data. We found F. chiloensis accessions CFRA34 (Redwoods Creek Park) and CFRA1691 (Jessie M. Honeyman Memorial State Park, Oregon) both to be very susceptible to verticillium wilt; however as the Bringhurst et al. [33] findings suggest, a more in-depth collection from these sites might recover resistant individuals as well. Future collections including numerous clones for screening would be prudent, as was done by Bringhurst et al. [33], for there is apparent diversity for resistance within individual sites.
Upon finding that one F. chiloensis genotype was highly resistant to verticillium wilt, Maas and Galletta [42] recommended that efforts should be increased to evaluate F. chiloensis clones as a potential source of resistance for use in strawberry breeding. Previously, van Adrichem and Orchard [40] suggested that F. chiloensis might have been the introgressive source of verticillium resistance already employed in some breeding programs. Screenings of progenies derived from cultivar×F. chiloensis crosses [43], including crosses involving susceptible ‘Elsanta’, identified two wild accessions, F. chiloensis ssp. lucida E2/1 (California) and F. chiloensis ssp. pacifica ‘Yaquina’ (Oregon) as potential sources of resistance. In contrast, when ‘Elsanta’ was crossed with the F. chiloensis cultivar ‘Culture’ (Chile), over one third of the progeny plants succumbed to wilt.
Octoploid cultigens. For about half of the cultigens that we assayed in the present study, our literature survey uncovered results of prior wilt resistance testing (Table 3). Although the prior studies varied in relation to environment (field or greenhouse) and to methodological aspects such as inoculation techniques, Verticillium isolates, and inoculum concentrations, our results from specific cultivars were generally consistent with those previously reported (Table 3). For instance, we rated ‘Tristar’ as very resistant, while it has previously been rated as very resistant, tolerant, and moderately resistant (see Table 3 and citations therein). We rated ‘Annapolis’ as intermediate, and it was also rated as such by Maas et al. [29]. We rated ‘Cavendish’ as intermediate, while it has elsewhere been rated as moderately resistant [44]. Thus, we have reason to be confident that our results provide an accurate and useful description of the tested germplasm.
Upon gathering information about ancestors of cultigens employed in the present study, some disagreements among prior studies were evident in the literature. Different ratings were found for ‘Howard17’ (Table 3), which is suspected of having two different clones [10]. Disagreements among studies may also stem from differing experimental conditions. Resistance to different sources of Verticillium dahliae isolates resulted in different resistance ratings [42, 45], and susceptibility to wilt was synergistically increased when coincident with infection by the nematode Pratylenchus penetrans [46, 47].
The pedigree diagram (Fig. 2) depicts information available to us about the ancestries of nine of the ten strawberry cultigens tested, tracing back as many as 12 generations from the most recent ‘Wendy’ release. All but two of these (‘Sparkle’ and ‘Tristar’) are products of the Nova Scotia breeding program. Notably, verticillium wilt resistance ratings are presently available for only 67 out of the 130 cultigens in the pedigree. Of the 63 cultigens with no available ratings, only 12 are named cultivars. The available ratings span a spectrum from very resistant to very susceptible; suggesting that considerable insight into the genetic basis of verticillium wilt resistance/susceptibility could be distilled from the described germplasm, provided that the missing resistance ratings could be filled in.
From the displayed pedigree relationships (Fig. 2), it was evident that resistance can appear in progeny (e.g. ‘Etna’) of a cross between two susceptible parents (e.g. ‘Belrubi’ and ‘Marlate’). Thus, the possible existence of dominant genetic factors for susceptibility must be considered, as must the possibility that resistance could be “introduced” into a susceptible breeding line simply by providing opportunity for resistance to emerge through progeny segregation, even via selfing of susceptible types [40].
‘Tristar’ is described in its release note as resistant to verticillium wilt, leaf blight, powdery mildew, and red stele root rot, as well as tolerant to leaf blight [48]. We confirmed its wilt resistance in the present study. Although, the day-neutrality trait of ‘Tristar’ is attributed to introgression from F. virginiana ssp. glauca via CA65.65-601 [48], the source(s) of wilt resistance is/are unclear, as there were multiple opportunities for either resistance or susceptibility to be transmitted from cultigens in its pedigree, both with and without known verticillium wilt resistance (Fig. 2).
Conclusions. The verticillium wilt resistance ratings reported here for 50 accessions and cultigens adds to the body of publically available knowledge about sources of wilt resistance and susceptibility in Fragaria germplasm. Upon incorporation into the GRIN database, the results obtained for 17 members of the USDA supercore germplasm set will add further value to this collection by expanding the knowledge resources associated with its members. We anticipate that useful insights will be gained through further examination of breeding pedigrees, such as that presented here. However, the power of the pedigree approach is maximized only when phenotype information is available for all pedigree members. Thus, additional resistance phenotyping is warranted to fill existing information gaps. Overall, the high level of progeny segregation and the lack of any consistent pattern of resistance transmission in the pedigree suggest the involvement of multiple genes and the presence of high levels of heterozygosity in these cultigens Thus, opportunities exist to breed for increased resistance.
Acknowledgments
This work was supported in part by a grant from the USDA National Germplasm System (Small Fruits) award #358-2100-038-01S, Screening for Verticillium Wilt Resistance in Wild Diploid and Octoploid Strawberry Germplasm, and by New Hampshire Agricultural Experiment Station Project NH00433, Genomic Tools for Horticultural Crops. This is Scientific Contribution Number 2530 from the New Hampshire Agricultural Experiment Station (NHAES). Funding for “RosBREED: Enabling Marker-Assisted Breeding in Rosaceae” was provided by the Specialty Crop Research Initiative Competitive Grant 2009-51181-05808 of the USDA’s National Institute of Food and Agriculture. We thank Mansun Kong of Driscoll Strawberry Associates, Watsonville, California, for providing the fungal isolate used in this work.
References
1 | Thomas HE (1931) Verticillosis of strawberries Phytopathol 21: 996 |
2 | Samtani JB, Gilbert C, Weber JB, Subbarao KV, Goodhue RE, Fennimore SA (2012) Effect of steam and solarization treatments on pest control, strawberry yield, and economic returns relative to methyl bromide fumigation HortScience 47: 64 70 |
3 | U.S. Environmental Protection Agency.Methyl bromide critical use nomination for preplant soil use for Strawberry Nursery in Open Fields [Internet]. 2013. http://www.epa.gov/ozone/mbr/CUN2013/2013CUNStrawberryNursery.pdf |
4 | Diehl K, Rebensburg P, Lentzsch P (2013) Field Application of Non-Pathogenic Verticillium dahliae; Genotypes for Regulation of Wilt in Strawberry Plants American Journal of Plant Sciences 4: 24 32 |
5 | Shaw DV, Gubler WD, Hansen J (1997) Field resistance of California strawberries to Verticillium dahliae at three conidial inoculum concentrations HortScience 32: 711 713 |
6 | Sjulin TM (2003) The North American small fruit industry -II. Contributions of public and private research in the past 25 years and a view to the future HortScience 38: 960 967 |
7 | Chalavi V, Tabaeizadeh Z, Thibodeau P (2003) Enhanced resistance to Verticillium dahliae in transgenic strawberry plants expressing a Lycopersicon chilense chitinase gene J Amer Soc Hort Sci 128: 747 753 |
8 | Koike ST, Bull CT, Bolda M, Daugovish O (2012) Organic Strawberry Production Manual University of California, Agriculture and Natural Resources |
9 | Varney EH, Moore JN, Scott DH (1959) Field resistance of various strawberry varieties and selections to Verticillium Plant Dis Rptr 43: 567 571 |
10 | Bringhurst RS, Wilhelm S, Voth V (1961) Pathogen variability and breeding verticillium wilt resistant strawberries Phytopathol 51: 786 794 |
11 | Bowen HH, Hough LF, Varney EH (1968) Breeding studies of verticillium wilt resistance in Fragaria×ananassa Duch J Amer Soc Hort Sci 93: 340 351 |
12 | Shaw DV, Gubler WD, Larson KD, Hansen J (1996) Genetic variation for field resistance to Verticillium dahliae evaluated using genotypes and segregating progenies of California strawberries J Amer Soc Hort Sci 121: 625 628 |
13 | Shaw DV, Gordon TR (2003) Genetic response for reaction to verticillium wilt in strawberry with two-stage family and genotypic selection HortScience 38: 432 4 |
14 | Antanaviciute L, Šurbanovski N, Harrison N, McLeary KJ, Simpson DW, Wilson F (2015) Mapping QTL associated with Verticillium dahliae resistance in the cultivated strawberry (Fragaria×ananassa) Horticulture Research 2: 15009 |
15 | Shaw DV, Gordon TR, Larson KD, Kirkpatrick SC (2005) The effect of Verticillium infection in runner plant propagation nurseries on resistant and susceptible strawberry genotypes J Amer Soc Hort Sci 130: 707 710 |
16 | Nathewet P, Hummer KE, Yanagi T, Iwatsubo Y, Sone K (2010) Karyotype analysis in octoploid and decaploid wild strawberries in Fragaria (Rosaceae) Cytologia 75: 277 288 |
17 | Staudt G (2009) Strawberry biogeography, genetics and systematics Acta Hort 71 83 |
18 | Darrow GM (1966) The Strawberry: History, breeding and physiology 1st ed. Holt, Rinehart and Winston |
19 | Folta K, Davis T (2006) Strawberry genes and genomes Critical Rev. Plant Sci 25: 399 415 |
20 | Davis T, Reinhard A, Reavey P, Lin J, Zhang H, Mahoney L (2009) Chloroplast DNA inheritance, ancestry, and sequencing in Fragaria Acta Horticulturae 859: 221 228 |
21 | Staudt G (1999) Systematics and geographic distribution of the American strawberry species, taxonomic studies in the genus Fragaria (Rosaceae:Potentilleae) University of California Press |
22 | Hancock JF, Luby JJ, Dale A (1993) Should we reconstitute the strawberry? Acta Hort 348: 86 93 |
23 | Hancock JF, Finn CE, Luby JJ, Dale A, Callow PW, Serce S (2010) Reconstruction of the strawberry, Fragaria×ananassa, using genotypes of F. virginiana and F. chiloensis HortScience 45: 1006 1013 |
24 | Hancock JF, Hokanson SC, Finn CE, Hummer KE (2000) Introducing a supercore collection of wild octoploid strawberries Acta Hort 567: 77 79 |
25 | Hummer KE (2012) A new species of Fragaria (Rosaceae) from Oregon J Bot Res Inst Texas 6: 9 15 |
26 | Varney EH, Moore JN, Scott DH (1960) Field resistance of 29 additional strawberry varieties and selections to Verticillium Plant Dis. Rptr 44: 370 371 |
27 | vanAdrichem MCJ, Bosher JE (1962) A search for Verticillium resistance in strawberry varieties Can J Plant Sci 42: 365 367 |
28 | Maas JL (1984) Compendium of Strawberry Diseases APS Press St. Paul, Minnesota |
29 | Maas J, Galletta G, Draper A (1989) Resistance in strawberry to races of Phytophthora fragariae and to isolates of Verticillium from North America Acta Hort 265: 521 526 |
30 | Hancock JF, Maas JL, Shanks CH, Breen PJ, Luby JJ (1991) Strawberries (Fragaria) Acta Hort 290: 491 546 |
31 | Olbricht K, Staudt G, Becker P, Szankowski I (2009) Verticillium resistance in Asian Fragaria species: Preliminary results Acta Hort 842: 227 230 |
32 | Hummer KE, Davis T, Iketani H, Imanishi H (2006) American–Japanese expedition to Hokkaido to collect berry crops in 2014 HortScience 41: 993 993 |
33 | Bringhurst RS, Wilhelm S, Voth V (1966) Verticillium wilt resistance in natural populations of Fragaria chiloensis in California Phytopathol 56: 219 222 |
34 | Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL (2011) The genome of woodland strawberry (Fragaria vesca) Nature Genet 43: 109 16 |
35 | Hancock JF, Finn CA, Hokanson SC, Luby JJ, Goulart BL, Demchak K (2001) A multistate comparison of native octoploid strawberries from North and South America J Amer Sci Hort Sci 126: 579 586 |
36 | Stegmeir TL, Finn CE, Warner RM, Hancock JF (2010) Performance of an elite strawberry population derived from wild germplasm of Fragaria chiloensis and F. virginiana HortScience 45: 1140 1145 |
37 | McKeen WE, Bosher JE (1955) Verticillium wilt of strawberries in British Columbia Plant Dis Rptr 39: 371 372 |
38 | Wilhelm S (1955) Verticillium wilt of the strawberry with special reference to resistance Phytopathol 45: 387 391 |
39 | Wilhelm S (1955) Verticillium wilt resistance: Strawberries resistant to verticillium wilt also show resistance to powdery mildew in plant disease studies CA Agri 9: 8 15 |
40 | vanAdrichem MCJ, Orchard WR (1958) Verticillium wilt resistance in the progenies of Fragaria chiloensis from Chile Plant Dis Rptr 42: 1391 1393 |
41 | Newton W, van Adrichem CJ (1958) Resistance to verticillium wilt in F1 generations of self-fertilized species of Fragaria Can J Bot 36: 297 9 |
42 | Maas J, Galletta G (1989) Germplasm evaluation for resistance to fungus-incited diseases Acta Hort 265: 461 72 |
43 | Olbricht K, Ulrich D, Dathe B (2006) Cross breeding with accessions of Fragaria chiloensis resulting in selections with outstanding disease resistance and fruit quality characteristics Acta Hort 708: 507 510 |
44 | Jamieson AR, Sanford KA, Nickerson NL (1991) “Cavendish” Strawberry HortScience 26: 1561 1563 |
45 | Gordon TR, Kirkpatrick SC, Hansen J, Shaw DV (2006) Response of strawberry genotypes to inoculation with isolates of Verticillium dahliae differing in host origin Plant Pathol 55: 766 769 |
46 | Muller J (1973) On the Verticillium-Pratylenchus disease complex in strawberries Gartenbauwissenschaft 38: 43 45 |
47 | Potter JW, Dale A (1991) Root lesion nematode tolerance in wild and cultivated strawberry The strawberry into the 21st century 202 208 Timber Press Portland, Oregon |
48 | Draper AD, Galletta GJ (1981) “Tribute” and “Tristar” everbearing strawberries HortScience 16: 794 5 |
49 | Galletta GJ, Draper AD, Swartz HJ (1981) “Allstar” strawberry HortScience 16: 792 4 |
50 | Galletta GJ, Maas JL, Draper AD (1982) Predicting verticillium wilt reaction of strawberry cultivars and selections Adv in Strawberry Production 1: 21 |
51 | Jamieson AR, Nickerson NL (1989) Recent progress in breeding strawberries for Atlantic Canada Acta Hort 265: 85 90 |
52 | Gaggioli D, Maas JL, Mezzetti B, Rosati P (1989) The use of culture filtrate of the fungus to screen for Verticillium albo-atrum in strawberry Acta Hort 265: 61 68 |
53 | U.S. Department of Agriculture. Germplasm Resources Information Network [Internet]. 2013. Available from: http://www.ars-grin.gov |
54 | Amenduni M, Colella C, D’Amico M, Bubici G, Cirulli M (2004) Strawberry breeding for resistance to verticillium wilt Acta Hort 649: 69 72 |
55 | Scott DH, Lawrence FJ, Draper AD (1979) Strawberry Varieties in the United States U.S. Department of Agriculture |
56 | Daugaard H, Lindhard H (2000) Strawberry cultivars for organic production Gartenbauwissenschaft 65: 213 217 |
57 | Labanowska BH, Meszka B, Bielenin A, Olszak R (2004) A field evaluation of disease and insect resistance of several strawberry cultivars in Poland Acta Hort 649: 255 258 |
58 | Sowik I, Bielenin A, Michalczuk L (2001) In vitro testing of strawberry resistance to Verticillium dahliae and Phytophthora cactorum Scientia Hort 88: 31 40 |
59 | Bordelon B, EllisM, Bessin R.Midwest small fruit and grape spray guide [Internet]. Purdue University; Purdue Extension; 2001. Available from: https://ag.purdue.edu/hla/Hort/Documents/ID-169.pdf |
60 | Galletta GJ, Draper AD, Stiles HD, Swartz HJ (1980) “Scott” strawberry HortScience 15: 541 542 |
61 | Dale A, Vandenberg AA, Wang SL, Ricketson CL (1993) St. Williams, Scotland and Selkirk strawberries Can J Plant Sci 73: 297 300 |
62 | Babadoost M. Verticillium wilt of strawberry [Internet]. University of Illinois at Urbana-Champaign; 2001. Available from: http://web.aces.uiuc.edu/vista/pdf_pubs/707.PDF |
63 | Bringhurst RS, Voth V (1980) Six new strawberry varieties released CA Agri 34: 12 5 |
Figures and Tables
Fig.1
Disease ratings and examples of their respective verticillium wilt phenotypes. Within each panel, a control plant is shown on the left, and an inoculated plant of the same variety is shown on the right along with its disease rating (number in black box).
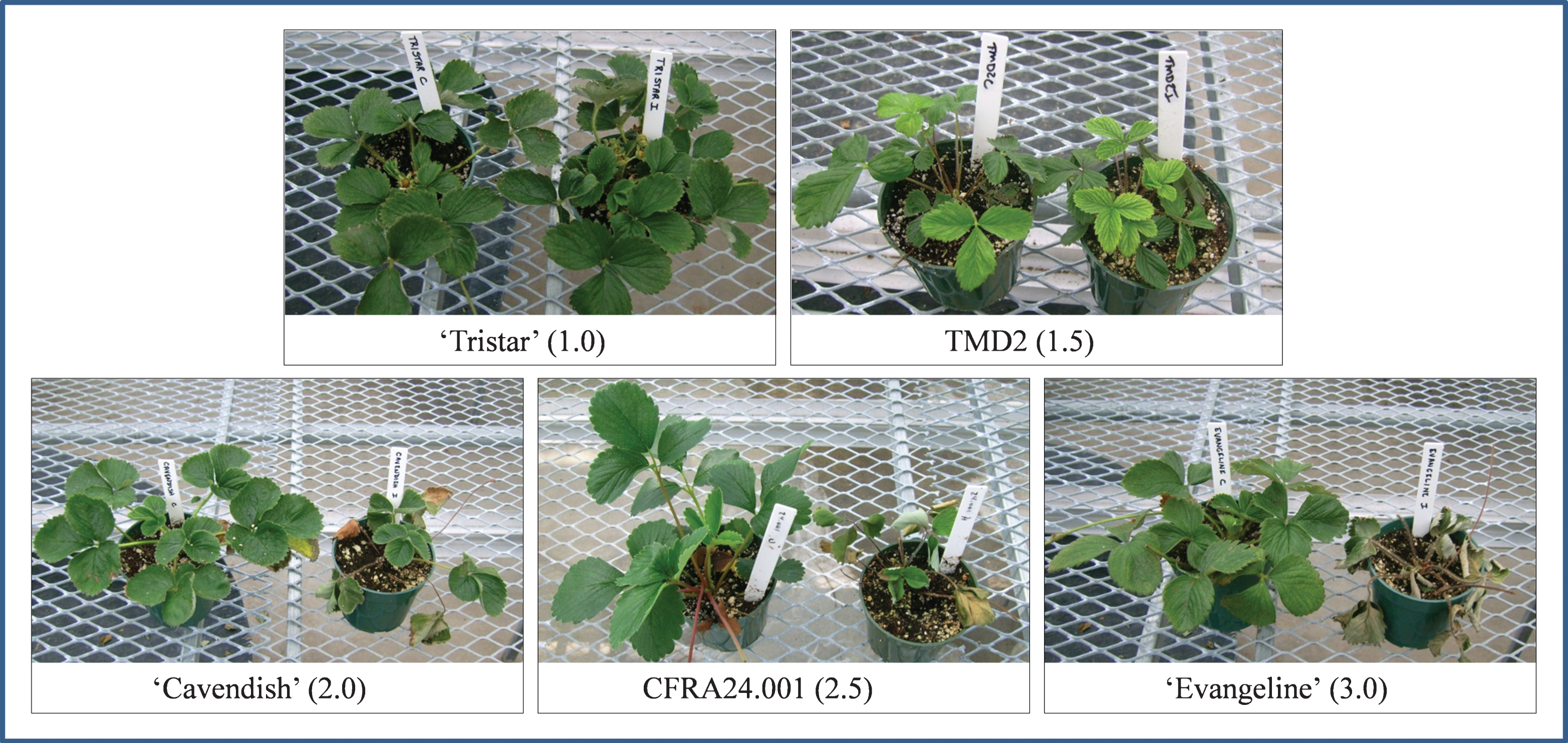
Fig.2
Pedigree showing ancestral relationships and previously reported and newly determined resistance phenotypes for strawberry cultigens. Red and blue lines, respectively, connect individuals to their female and male parents. Box colors indicate degree of resistance (see Key). Phenotypic data are lacking for cultigens in white boxes. Blue framed boxes indicate phenotypic determination by trials at UNH.
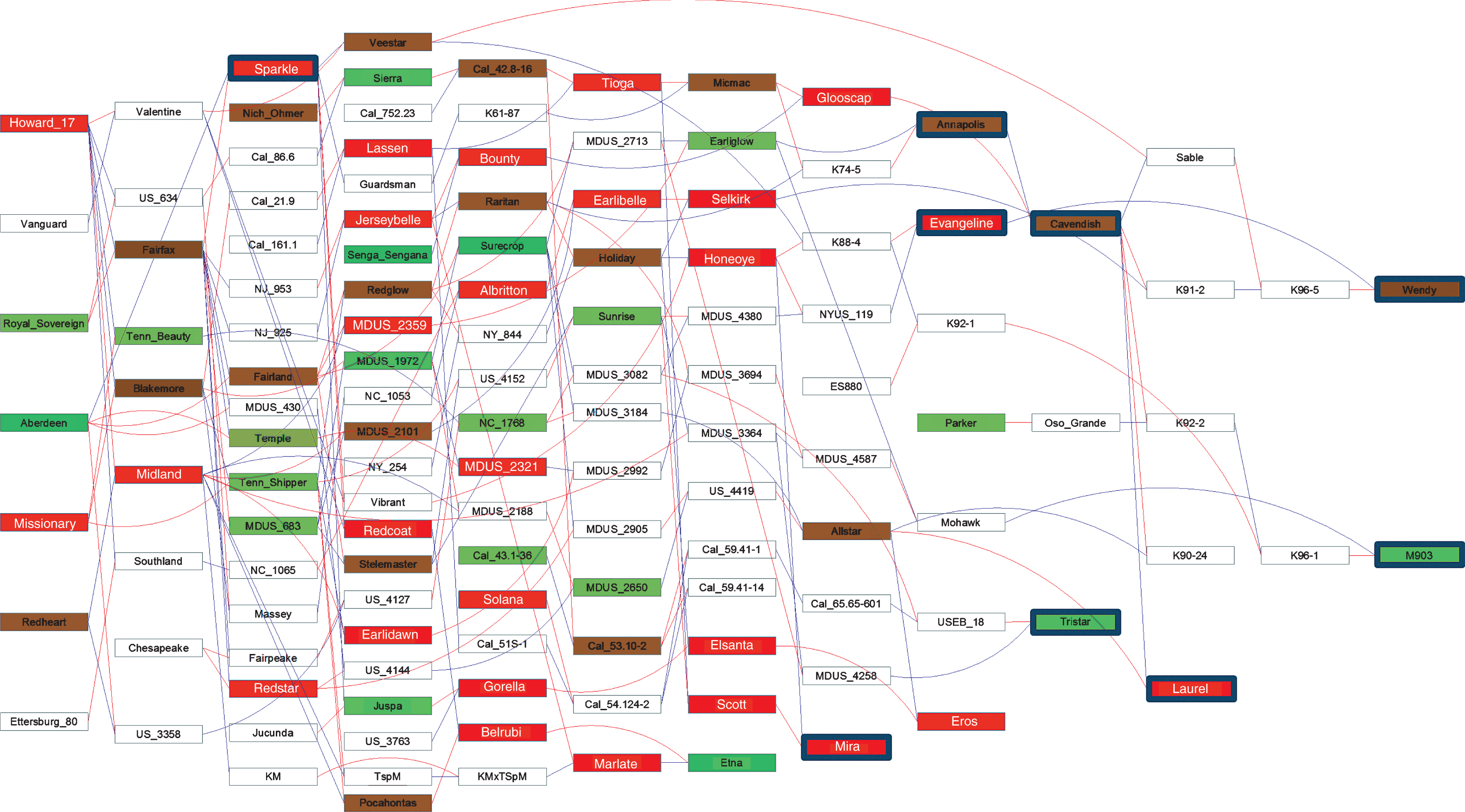
Table 1
The inoculation responses of diploid Fragaria genotypes
| Taxon | Origin | PI numbersz | CFRAz | Local namey | Nx | Meanx | SDx |
| F. vesca ssp.bracteata | BC, Canada | 660763 | 1988.001 | BC5 | 4 | 1.5 | 0.0 |
| F. vesca ssp.w | Mendocino, Co. CA Co., CA,USA | 660765 | 1990.001 | TMD2 | 32 | 1.5 | 0.5 |
| F. vesca ssp.w | Mendocino Co., CA, USA | NA | NA | U2A | 4 | 1.5 | 0.6 |
| F. vesca ssp.w | BC, Canada | NA | NA | BC7 | 4 | 2.0 | 0.0 |
| F. vesca ssp. bracteata | Santa Cruz Co., CA | TBD | 2185 | HP6A | 4 | 2.3 | 0.5 |
| F. vesca ssp. bracteata | Santa Cruz Co., CA | NA | NA | HP7B | 4 | 2.5 | 0.6 |
| F. vesca ssp. americana | Coos Co., NH | TBD | 2186 | WC6 | 4 | 2.5 | 0.6 |
| F. vesca ssp. americana | Coos Co., NH | TBD | 2187 | WC8 | 4 | 2.8 | 0.3 |
| F. vesca ssp.w | Humboldt Co., CA | NA | NA | H1B | 4 | 2.8 | 0.5 |
| F. vesca ssp.w | Del Norte Co., CA | TBD | 2188 | DN2A | 7 | 2.9 | 0.2 |
| F. vesca ssp. bracteata | Santa Cruz Co., CA | TBD | 2189 | HP3A | 4 | 2.9 | 0.3 |
| F. vesca ssp. bracteata | BC, Canada | 660764 | 1989.001 | BC30 | 11 | 2.9 | 0.2 |
| F. vesca ssp. bracteata | BC, Canada | TBD | 2190 | BC3 | 8 | 3.0 | 0.0 |
| F. vesca ssp. bracteata | BC, Canada | TBD | 2191 | GS1J | 4 | 3.0 | 0.0 |
| F. vesca ssp. americana | Rockingham Co.,NH | 657856 | 1948.001 | Pawtuckaway | 4 | 3.0 | 0.0 |
| F. bucharica | Pakistan | 551851 | 520.001 | 880083 Pakistan | 4 | 2.0 | 0.0 |
| F. iinumae | Hokkaido, Japan | 637964 | 1850 | J7 | 4 | 1.3 | 0.5 |
| F. nipponica | Hokkaido, Japan | 637979 | 1866 | J32 | 4 | 1.3 | 0.3 |
| F. nipponica | Hokkaido, Japan | 637980 | 1868 | J34 | 12 | 1.3 | 0.4 |
| F. nipponica | Hokkaido, Japan | 637977 | 1864 | J30A | 3 | 1.7 | 0.6 |
| F. nipponica | Hokkaido, Japan | 637975 | 1862 | J25 | 4 | 2.0 | 0.0 |
| F. vesca×F. viridis | uncertain | 551744 | 364.002 | CA 1450 | 4 | 1.6 | 0.3 |
| F. viridis | Germany | 551741 | 333.001 | CA 72.501-2 | 4 | 1.8 | 0.5 |
zNCGR Fragaria accessions have National Plant Germplasm System (NPGS) Plant Introduction (PI) numbers, and local numbers beginning with the prefix CFRA. TBD = Awaiting assignment of PI number. NA = not applicable, not part of NPGS. yLocal names/numbers used by original collectors. In Table 3, all these are UNH numbers, while various collectors’ local numbers are included in Table 2. xN = number of replicate plants; Mean = the mean disease rating for the indicated N; SD = standard deviation of the mean. wCalifornia accessions of F. vesca subspecies are difficult to differentiate and may be either ssp. bracteata, ssp. californica, or hybrids thereof.
Table 2
Inoculation responses of 8x and 10x (F. cascadensis) Fragaria genotypes. The superscript definitions are as in Table 1
| Taxon | Origin | PI no.z | CFRAz | Local namey | Nx | Meanx | SDx |
| F. chiloensis f. chiloensis | Peru | 551736 | 372.002 | CA1541 | 4 | 1.6 | 0.3 |
| F. chiloensis ssp. lucida | CA, USA | 551728 | 357.002 | CA1367 | 16 | 1.7 | 0.6 |
| F. chiloensis f. patagonica | Chile | 236579 | 24.001 | Darrow 72 | 8 | 2.4 | 0.5 |
| F. chiloensis f. patagonica | Chile | 602570 | 1108.002 | 2 CAR 3B | 4 | 2.5 | 0.6 |
| F. chiloensis f. patagonica | Chile | 612316 | 1088.002 | 2 BRA 1A | 4 | 2.5 | 0.6 |
| F. chiloensis ssp. lucida | OR,USA | 612489 | 1691.001 | HM1 | 12 | 2.9 | 0.4 |
| F. chiloensis f. patagonica | Chile | 552091 | 796.001 | Termas de Chillan-TDC | 4 | 3.0 | 0.0 |
| F. chiloensis ssp. lucida | CA, USA | 551445 | 34.002 | RCP 37 | 6 | 3.0 | 0.0 |
| F. virginiana ssp. virginiana | ON, Canada | 612497 | 1699.001 | Montreal River 10 | 9 | 1.2 | 0.3 |
| F. virginiana ssp. grayana | MS, USA | 612569 | 1414.001 | NC 95-19-1 | 6 | 2.2 | 1.0 |
| F. virginiana ssp. grayana | FL, USA | 612570 | 1435.002 | JP 95-1 | 4 | 2.5 | 1.0 |
| F. virginiana ssp. grayana | GA, USA | 612320 | 1455.001 | JP 95-9-6 | 26 | 2.6 | 0.7 |
| F. virginiana ssp. virginiana | NC,USA | 612325 | 1620.001 | NC96-5-3 | 4 | 2.8 | 0.5 |
| F. virginiana ssp. grayana | MS, USA | 612486 | 1408.001 | NC 95-19-1 | 7 | 2.9 | 0.4 |
| F. virginiana ssp. grayana | FL,USA | 612570 | 1435.001 | JP95-1 | 3 | 3.0 | 0.0 |
| F. virginiana ssp. glauca | MT, USA | 612495 | 1697.001 | LH 50-4 | 4 | 3.0 | 0.0 |
| F. cascadensis (10x) | OR, USA | 551527 | 110.001 | – | 4 | 3.0 | 0.0 |
| F.×ananassa ‘Tristar’ | USDA-MD | 551954 | 663.001 | EB60 | 4 | 1.0 | 0.0 |
| F.×ananassa ‘M903 | NS, Canada | NA | NA | M903 | 3 | 1.3 | 0.6 |
| F.×ananassa ‘Cavendish’ | NS, Canada | 616560 | 1169.000 | K83-4 | 4 | 2.0 | 0.0 |
| F.×ananassa ‘Annapolis’ | NS, Canada | 552257 | 964.001 | K78-4 | 4 | 2.0 | 0.0 |
| F.×ananassa ‘Wendy’ | NS, Canada | NA | NA | K98-6 | 4 | 2.0 | 0.0 |
| F.×ananassa ‘Sparkle’ | NJ, USA | 551559 | 183.001 | ‘Paymaster’ | 3 | 2.5 | 0.6 |
| F.×ananassa ‘Mira’ | NS, Canada | NA | NA | K84-5 | 4 | 2.8 | 0.5 |
| F.×ananassa ‘Evangeline’ | NS, Canada | NA | NA | K93-1 | 4 | 3.0 | 0.0 |
| F.×ananassa K05-9 | NS, Canada | NA | NA | K05-9 | 4 | 3.0 | 0.0 |
| F.×ananassa ‘Laurel’ | NS, Canada | NA | NA | K93-20 | 4 | 3.0 | 0.5 |
Table 3
Information sources for members of the pedigree shown in Fig. 2. The superscripts in the Classification and Information Sources columns relate the classification to the respective source(s) of the information. In the Information Sources column, relevant results from the present study are included and identified as “ UNH result”
| Cultigen | Classificationz | Information sources |
| Aberdeen | VR1, VR/MS2 | 1[10, 29][29], 2Eastern (VR)/California (MS) isolates [42] |
| Allstar | I1, I-T2, MR3 | 1[29], 2[28, 49], 3[50] |
| Annapolis | MR1, I2 | 1[51], 2[29], UNH result |
| Belrubi | VS | [52] |
| Blakemore | MR | [28, 29, 38, 53, 54] |
| Bounty | VS1, I2 | 1[28, 51], 2[29] |
| Cal_39.117-4 | I | [10] |
| Cal_42.8-16 | VR1, MR2 | 1[10], 2[33] |
| Cal_43.1-36 | MR | [10] |
| Cal_53.10-2 | I | [10] |
| Cavendish | I1, MR2 | 1UNH result, 2[44, 51, 53] |
| Earlibelle | MS | [55] |
| Earlidawn | VS1, I2 | 1[9, 28, 53]; 2 [11, 12] |
| Earliglow | T1, MR2, MS3, I4 | 1[50], 2[28, 29], 3[11], 4[12, 53] |
| Elsanta | VS | [31, 53, 56–58] |
| Eros | MS | [53] |
| Etna | R | [52] |
| Evangeline | VS | UNH result |
| Fairfax | I1, MS2 | 1[29], 2[9] |
| Fairland | I | [9] |
| Glooscap | VS | [59] |
| Gorella | VS | [52] |
| Holiday | I | [28] |
| Honeoye | VS | [28, 29, 42, 51–53, 57] |
| Howard_17 | MS1, VR/MR2 | 1[38], 2VR/MR [10] |
| Jerseybelle | MS1, VS2 | 1[28, 29], 2[9, 26, 53] |
| Juspa | MR | [18] |
| Lassen | MS1, VS2 | 1[12, 33], 2[10, 39, 53] |
| Laurel | VS | UNH result |
| Marlate | VS | [28] |
| M903-3 | VR | UNH result |
| MDUS_1972 | VR | [9] |
| MDUS_2321 | MS | [9] |
| MDUS_2359 | MS | [9] |
| MDUS_2650 | VR | [9] |
| MDUS_683 | MR1,VR2, VR/MS3 | 1[26], 2[29], 3[42] Eastern(VR)/California(MS) isolates |
| Micmac | MR1, I2 | 1[29], 2[28] |
| Midland | VS | [9, 28] |
| Mira | VS | UNH result |
| Missionary | MS | [9, 38] |
| NC_1768 | MR | [9] |
| Nich Ohmer | I | [1] |
| Parker | MR | [12] |
| Pocahontas | I | [9, 29] |
| Raritan | MR1, MS2 | 1[29], 2[28, 53] |
| Redcoat | VS | [28, 53] |
| Redglow | I1,MS2 | 1[9, 11], 2[28] |
| Redheart | I | [28] |
| Redstar | VS1, I2 | 1[28] |
| Royal Sovereign | R | [30] |
| Scott | MR1, I to MR2 | 1[29, 53], 2[28, 60] |
| Selkirk | MS | [61] |
| Senga_Sengana | VR/T1, MS2, VS3 | 1[57], T [31], 2combined with nematodes [47], 3[58] |
| Shasta | MR1, MS2, VS3 | 1[39], 2[28, 33, 53], 3[10] |
| Sierra | VR1, MR2 | 1[38, 42], 2[28, 33, 39, 53, 54] |
| Solana | MS1, VS2 | 1[33], 2[10] |
| Sparkle | I1, MS2 | 1[29], 2[9, 28, 53] and UNH result |
| Stelemaster | I | [28] |
| Sunrise | VR1, MR2 | 1[28], 2[29] |
| Surecrop | VR1, MR2 | 1[9, 11, 28], 2[29, 50, 53] |
| Temple | MR | [9] |
| Tenn_Beauty | MR | [62] |
| Tenn_Shipper | MR | [9] |
| Tioga | MS1, VS2 | 1[28], 2[53, 63] |
| Tristar | VR1, T2, MR3 | 1[48], UNH result, 2[53], 3[29, 50] |
| Veestar | MR | [53] |
| Wendy | I | UNH result |
zClassifications (U = Unknown, VR = Very Resistant, MR = Moderately Resistant, I = Intermediate, MS = Moderately Susceptible, VS = Very Susceptible, and T = Tolerant). Note: the term “tolerant (T)“ has been used in differing ways by various authors. The superscript following the qualitative classification corresponds to the superscript preceding the cited literature in the same row.




