Drosophila as a Model for Microbiota Studies of Neurodegeneration
Abstract
Accumulating evidence show that the gut microbiota is deeply involved not only in host nutrient metabolism but also in immune function, endocrine regulation, and chronic disease. In neurodegenerative conditions such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis, the gut-brain axis, the bidirectional interaction between the brain and the gut, provides new route of pathological spread and potential therapeutic targets. Although studies of gut microbiota have been conducted mainly in mice, mammalian gut microbiota is highly diverse, complex, and sensitive to environmental changes. Drosophila melanogaster, a fruit fly, has many advantages as a laboratory animal: short life cycle, numerous and genetically homogenous offspring, less ethical concerns, availability of many genetic models, and low maintenance costs. Drosophila has a simpler gut microbiota than mammals and can be made to remain sterile or to have standardized gut microbiota by simple established methods. Research on the microbiota of Drosophila has revealed new molecules that regulate the brain-gut axis, and it has been shown that dysbiosis of the fly microbiota worsens lifespan, motor function, and neurodegeneration in AD and PD models. The results shown in fly studies represents a fundamental part of the immune and proteomic process involving gut-microbiota interactions that are highly conserved. Even though the fly’s gut microbiota are not simple mimics of humans, flies are a valuable system to learn the molecular mechanisms of how the gut microbiota affect host health and behavior.
INTRODUCTION
Trillion of microbes such as bacteria, fungi, and viruses live inside our body and on all of our surfaces, with the largest concentration found in the gut [1]. Among such symbiotic microbes, the microorganisms on the surface of the gut are called gut microbiota. The gut microbiota has recently been shown to play an important role in the physiology and disease. Our symbiotic partners provide immune and metabolic functions by employing various molecular and genetic signaling pathways to protect the host from pathogens [2, 3]. Gut microbiota form biofilms, a layer of bacterial extracellular matrix, which protects them from attack [4]. The gut microbiota is affected by the host’s genetic background, age, diet, comorbidities, and antibiotic use [5–7]. Disruption of gut microbiota is referred to dysbiosis and can contribute to various diseases. Recently, the involvement of gut microbiome has been reported in many areas such as aging, obesity, diabetes, fatty liver disease, cancer, and neurodegenerative diseases [5, 8–12]. Furthermore, accumulating evidence indicate that dysbiosis of the gut microbiota can cause low-grade inflammation which exacerbates chronic diseases such as atherosclerosis, asthma, and chronic kidney disease [13–16].
The brain and the gut strongly influence each other. This bidirectional relationship is called the “gut-brain axis” [17, 18]. Although much about the gut-brain axis is still unclear, it is well known that mental stress can cause diarrhea and constipation through changes in the gut microbiota. The relationship between gut microbiota and brain function has recently been analyzed in experiments using mice. Diaz Heijtz et al. compared the behavior of specific pathogen free mice with a normal gut microbiota with germ-free mice and showed that mice with microbiota gradually became accustomed to unfamiliar environments and reduce their amount of movement, whereas germ-free mice continued to move indefinitely in unfamiliar environments and tend to feel less fear. Furthermore, the gene expression of brain-derived neurotrophic factor (BDNF) was lower in germ-free mice than in mice with normal gut microbiota, suggesting that gut microbiota influence the development of BDNF-mediated neural circuits that control anxiety-related behavior [19]. Hsiao et al. reported using a mouse model of autism spectrum disorders that a metabolite called 4-ethylphenylsulfate produced by gut microbiota caused autism spectrum disorder-related behaviors, and that these phenotypes improved when Bacteroides fragilis was taken orally [20]. Furthermore, Sampson et al. reported that Parkinson’s disease (PD) model alpha synuclein (α-syn) overexpressing mice that are germ free are protected from the disease [21]. Thus, evidence is accumulating that the gut microbiota is significantly involved in various neurologic disorders.
Drosophila melanogaster (henceforth Droso-phila), one of many species of fruit fly, has classically been used as an optimal research model in the field of genetic engineering. In recent years it has been also attracting attention as an experimental animal for gut microbiota research in neurodegenerative diseases [22, 23]. Drosophila research has addressed the involvement of the microbiota on physical function, longevity, aging, and obesity. Furthermore, since 2018, a series of articles have been published analyzing gut microbiota in fly models of the neurodegenerative disease [24–27]. Drosophila has many advantages as a laboratory animal: short life cycle, numerous and genetically homogenous offspring, less ethical concerns, availability of many genetic models, and low maintenance costs. Drosophila has orthologs for about 75%of human disease-related genes and the fly’s nervous system shares basic structure and functional features with humans [28]. Various neurodegenerative diseases such as Alzheimer’s disease (AD), PD, amyotrophic lateral sclerosis (ALS), Charcot-Marie-Tooth disease, and spinocerebellar atrophy adopt Drosophila disease model [29–33]. Moreover, Drosophila has a simpler gut microbiome than mice, which can be changed to any bacterial composition by experimental manipulation [34]. This makes Drosophila a suitable model for the study of the gut microbiome.
In this review, we discuss the history, physiology, anatomy of the gastrointestinal tract, and gut microbiota of Drosophila. We will then present research on Drosophila gut microbiota and neurodegenerative disease, and finally discuss how the findings from Drosophila gut microbiota can be adopted for therapeutic development of neurodegenerative diseases.
THE HISTORY, PHYSIOLOGY, AND ANATOMY OF DROSOPHILA
Drosophila has contributed to the Nobel Prize in Physiology or Medicine at least five times [35–37]. The first case was Thomas H. Morgan in 1933, who demonstrated the chromosome theory of inheritance, which states that chromosomes are carriers of genes. By collecting mutations in Drosophila and conducting mating experiments among them, Morgan produced the first chromosome map. He was able to locate the genes on the chromosomes by comparing a chromosome map with the pattern of salivary gland chromosomes of Drosophila and proved that the genes were on the chromosomes. The second case was Hermann J. Muller in 1946, who discovered that X-ray radiation could induce gene mutations in Drosophila. In the 1920 s he conducted an experiment in which male Drosophila was irradiated with X-rays, proving that the effects of radiation exposure can be transmitted to offspring and that radiation can artificially generate gene mutations. The third case was Edward B. Lewis, Christiane Nusslein-Volhard, and Eric F. Weischaus. They identified the genes involved in embryonic development using Drosophila with random mutations. Once embryonic development was delayed, the mutated gene was found to be involved, thus revealing genes necessary for Drosophila embryonic development. Many of identified genes were found in other species, indicating that similar developmental mechanisms in Drosophila apply to organisms other than flies. They were awarded the Nobel Prize in Physiology or Medicine in 1995. The fourth was Bruce A. Beutler, Jules A. Hoffmann, and Ralph M. Steinman. Hoffmann discovered the role of the Drosophila Toll gene in the immune system. Beutler demonstrated that one of the mammalian Toll-like receptors, TLR4, is a transmembrane component of the receptor complex of the cell wall of Gram-negative bacteria. Together with Steinman’s discovery of dendritic cells, the mechanism by which the innate immune system initiates the acquired immune system was identified, and these three researchers were awarded the Nobel Prize in Physiology or Medicine in 2011. The fifth case was Jeffrey C. Hall, Michael Rosbash, and Michael W. Young. They cloned a Drosophila clock gene (named “period”) and found that the mRNA and proteins encoded by period change in response to circadian oscillations. They were awarded the Nobel Prize in Physiology or Medicine in 2017 for their elucidation of the molecular mechanisms of circadian rhythms. Thus, many important biological discoveries have been made with Drosophila, and it continues to demonstrate its value as an attractive model animal not only in genetic engineering but also in other fields of biology.
The life cycle of Drosophila is about 10 days: the first instar larvae hatch in about one day at 25°C after spawning. The first instar larvae molt into the second instar larvae after one day at 25°C, and the third instar larvae after one more day. Third instar larvae live for two days and then pupate. They spend four to four and half days as pupae. Under eutrophic conditions, adult females are ready to lay eggs two to three days after emergence. The entire developmental period of Drosophila is easily affected by temperature, with a twofold delay in developmental time at 18°C. Development almost completely ceases at 12°C, and it dies at temperature below at 5°C. Drosophila is also not tolerant to high temperatures, and will die if reared continuously above 30°C. Drosophila survives for two to three months in captivity, and the maximum number of eggs laid by one female is about 300 to 500 [28].
One of the most useful experimental tools in Drosophila is the GAL4-UAS targeted expression system, in which GAL4 is a yeast-derived transcription factor that binds to UAS sequences and activates the expression of downstream genes. The main feature of this system is that the GAL4 strain which determines “where to express”, and the UAS strain which determines “what to express”, are generated independently. By analyzing the phenotypes obtained from the progeny of crosses between the targeted GAL4 and UAS strains, it is possible to efficiently perform overexpression in various combinations. Many strains that specifically express GAL4 in various tissues and cells and strains that have UAS upstream of many genes have been created and are available from stock centers around the world [38]. Furthermore, in Drosophila, the GAL4-UAS system can be combined with RNA interference (RNAi) to suppress specific gene expression in any tissue or cell. Transgenic lines expressing double-stranded RNAs for target mRNAs are being created and are available for most of the protein coding genes in Drosophila [39].
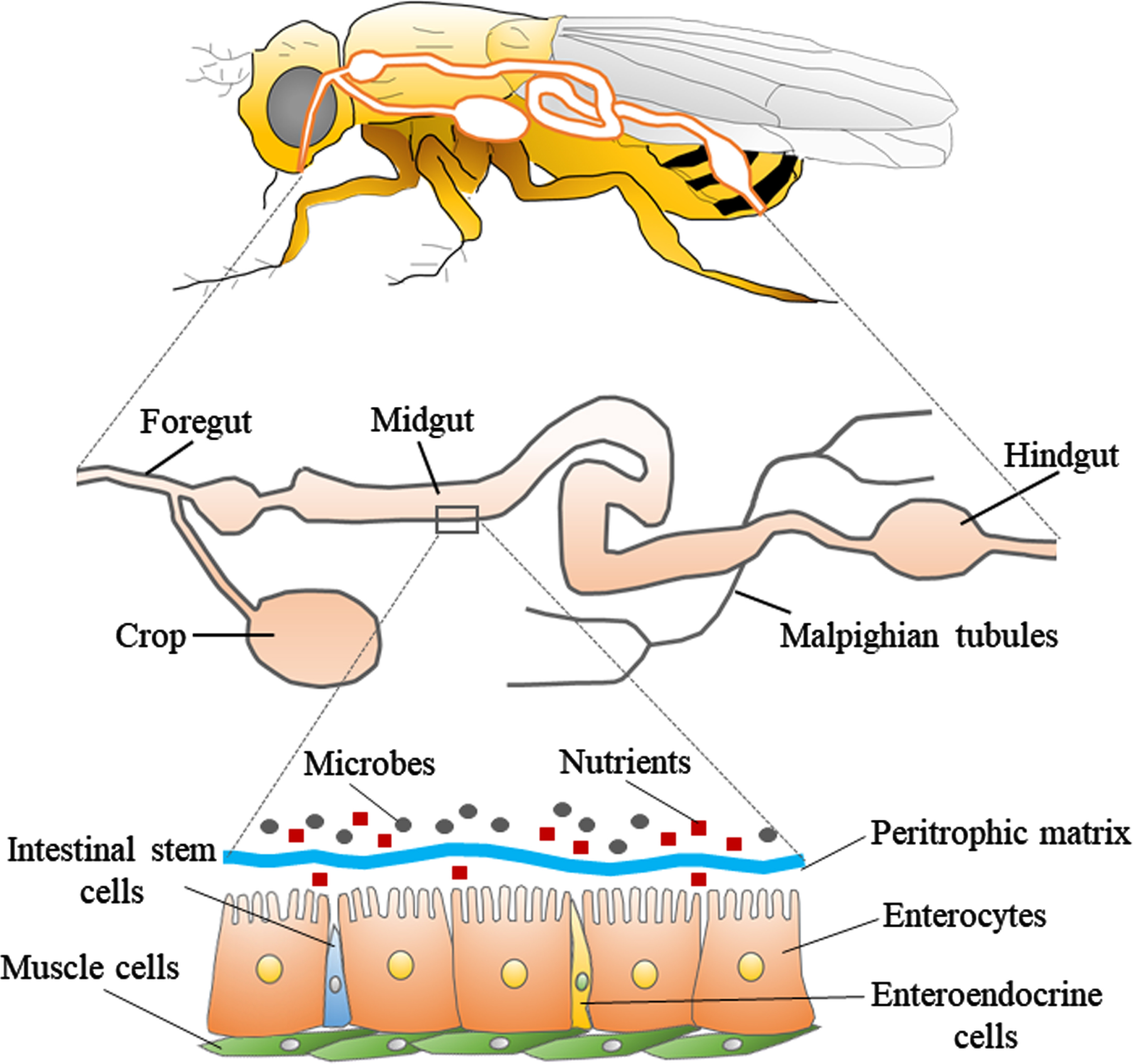
The digestive tract of Drosophila consists of three parts: foregut, midgut, and hindgut (Fig. 1). The foregut contains the oral cavity, esophagus, and crop (present only in adult fly), and is responsible for the physical preparation and salivary digestion of food. The midgut is the center of digestion and absorption, with over 300 digestive enzymes at work. The hindgut absorbs substances such as water and nutrients released from the Malpighian tubule, which is the equivalent of the mammalian kidney. The ingested food first enters a pouch-like structure called the crop. It then passes through the midgut, and is excreted through the hindgut. The midgut of the fly is composed of a single layer of epithelial cells, which contain the enterocytes, enteroendocrine cells, and intestinal stem cells, and is covered with muscle on the outside. On the luminal side of gut, there is a membrane composed of chitin fibrils and chitin-binding proteins called the peritrophic matrix [40, 41]. This membrane is known to allow only products degraded by digestive enzymes present on the membrane to pass through, and does not allow undigested food or bacteria to pass. Therefore, the peritrophic matrix is considered to be a physical barrier that protects the gut epithelium from pathogens. Although the role of the peritrophic matrix and its components in Drosophila is still unclear, the peritrophic matrix of many insects is thought to contain lysozyme and peptidoglycan degrading enzymes with antimicrobial activity, and the membrane may also serve as an essential site for gut immune responses [42]. Flies have a much simpler gut microbiome consisting of only five to 20 microbial species, compared to humans which have about 500–1,000 microbial species in their gut [43]. Furthermore, relatively simple established methods can be used to generate germ-free flies or flies with standardized gut microbiota (such flies with standardized gut microbiota are called “gnotobiotic flies”) [34]. In detail, we can obtain axenic flies by simply soaking the eggs in sodium hypochlorite and rearing them in sterile food. If we want to give them any bacteria, we can simply mix the bacteria into sterile food. Previous research showed that even human-derived Lactobacillus could be colonized in germ-free flies [44]. Compared to specific-pathogen-free mice, which can only be obtained by dissecting pregnant mice, the simplicity of sterile manipulation of Drosophila is one of the greatest advantages of using Drosophila in the study of gut microbiota.
Fig. 1
Structure of the digestive tract in Drosophila. The upper panel shows a schematic diagram of the body and digestive tract of Drosophila. The middle panel shows the general parts of the digestive tract, which is divided into three parts: foregut, midgut, and hindgut. The lower panel shows the fine structure of a portion of the midgut. The layer consisting of enterocytes, enteroendocrine cells, and intestinal stem cells is covered with muscle cells. There is a peritrophic matrix between the enterocytes and the microbes, which physically separates the enterocytes from the microbes.
The simple and easily regulated gut microbiome of Drosophila helps us to understand the impact of gut microbiota on host physiology as follows. Storelli et al. reported that rearing germ-free Drosophila larvae under nutrient deprivation significantly reduce the growth rate of the larvae. When Lactobacillus plantarum was reintroduced in the germ-free larvae, the decrease in growth rate was restored. Furthermore, they showed that L. plantarum genetically affected the TOR (Target of Rapamycin)-dependent nutrient sensing system [45]. Shin et al. found that rearing germ-free Drosophila monoassociated with pyrroloquinoline quinone-dependent alcohol dehydrogenase mutant bacteria delayed larval development and reduced body size. Such effect was restored by reintroduction of Acetobacter pomorum. Furthermore, they showed that this recovery effect was due to the acetic acid produced by A. pomorum modulating the larval insulin signaling pathway [46]. It is becoming clear that gut microbiome can influence a variety of Drosophila phenotypes, including aging and behavior [47–49]. Although there have been fewer studies of the gut microbiome in Drosophila than in mice, physiological and anatomical simplicity in the use of Drosophila will aid the new discoveries, and help to find new pathways that will be applied to mammals.
THE NEURODEGENERATIVE DISEASES AND GUT MICROBIOTA
The gut and brain influence each other through four pathways: the endocrine, immunological, metabolic and neural pathways. In response to change in the gut environment, gut hormones such as 5-hydroxytriptamine (5-HT) and cytokines secreted by enteroendocrine cells and immune cells, respectively, reach the brain through the bloodstream. Bacterial metabolites and neurotransmitters such as fatty-acids, γ-aminobutyric acid (GABA), and precursor of 5-HT also go through the circulatory system and pass through the blood-brain barrier to signal the brain [50]. Afferent and efferent neurons of the vagus nerve connect the gut and brain stem. Recently, studies with rodents indicate that molecules such as GABA and α-syn can be delivered to the brain via the vagus nerve in the trans-synaptic way [51–55]. This work suggests that the gut-brain axis plays a more important role than we had imagined.
There are two keywords for the relationship be-tween gut microbiota and neurodegenerative diseases: inflammation and cross-seeding (Fig. 2) [56–58]. A dysbiotic gut microbiome and damaged enteric epithelia can provoke systemic inflammation via activation of immune cells and release of cytokines [59, 60]. The hallmark of neurodegenerative diseases, such as AD and PD, is the misfolding and aggregated proteins in the brain, such as amyloid-β (Aβ) and α-syn, respectively. Minter et al. reported that Aβ deposition in a mouse model of familial AD was reduced by changes in gut microbial composition due to long-term use of antibiotics, whereas soluble Aβ was increased, and the same time, the expression of circulating cytokines was altered and the number of plaque-localized astrocytes and microglia was reduced [12]. Harach et al. found alteration in the gut microbiota of familial-AD model mice (amyloid-beta precursor protein, APP, transgenic mice) compared to wild-type mice. They also showed that germ-free APP transgenic mice had dramatically reduced Aβ deposition in the brain compared to mice with normal gut microbiota. Interestingly, while recolonization of gut microbiota in germ-free APP transgenic mice increased brain Aβ levels, microbes from standard (not germ-free) APP transgenic mice increased brain Aβ levels more effectively than that from wild-type mice [61]. AD patients and AD model mice showed different gut microbial composition from healthy controls and wild-type mice, and a stress response pathway such as the MAPK signaling pathway was enhanced [62, 63]. Zhan et al. reported that lipopolysaccharide, a bioactive substance produced by Gram-negative bacilli, and Escherichia coli (E. coli) fragments co-localized in Aβ plaques in the postmortem brains of AD patients, and that lipopolysaccharide was also deposited in perivascular areas of the brain, suggesting that microbial-mediated disruption of the blood-brain barrier might be involved in the neurodegeneration of AD [64]. These reports indicate that systemic inflammation and gut leakage play important roles in AD.
Fig. 2
Inflammation and cross-seeding: two hypotheses of abnormal aggregated protein ccumulation involving the gut. The left panel shows the “cross-seeding” hypothesis. Aggregated proteins (especially α-syn) produced in the gut propagate in a prion-like manner through the vagus nerve to the brainstem. Bacterial amyloid (e.g., curli) promote abnormal protein aggregation through cross-seeding. The right panel shows the “inflammation” hypothesis. Dysbiosis of gut microbiota activate the innate immune system and increase the production of proinflammatory cytokines and induce inflammation in the brain via systemic circulation.
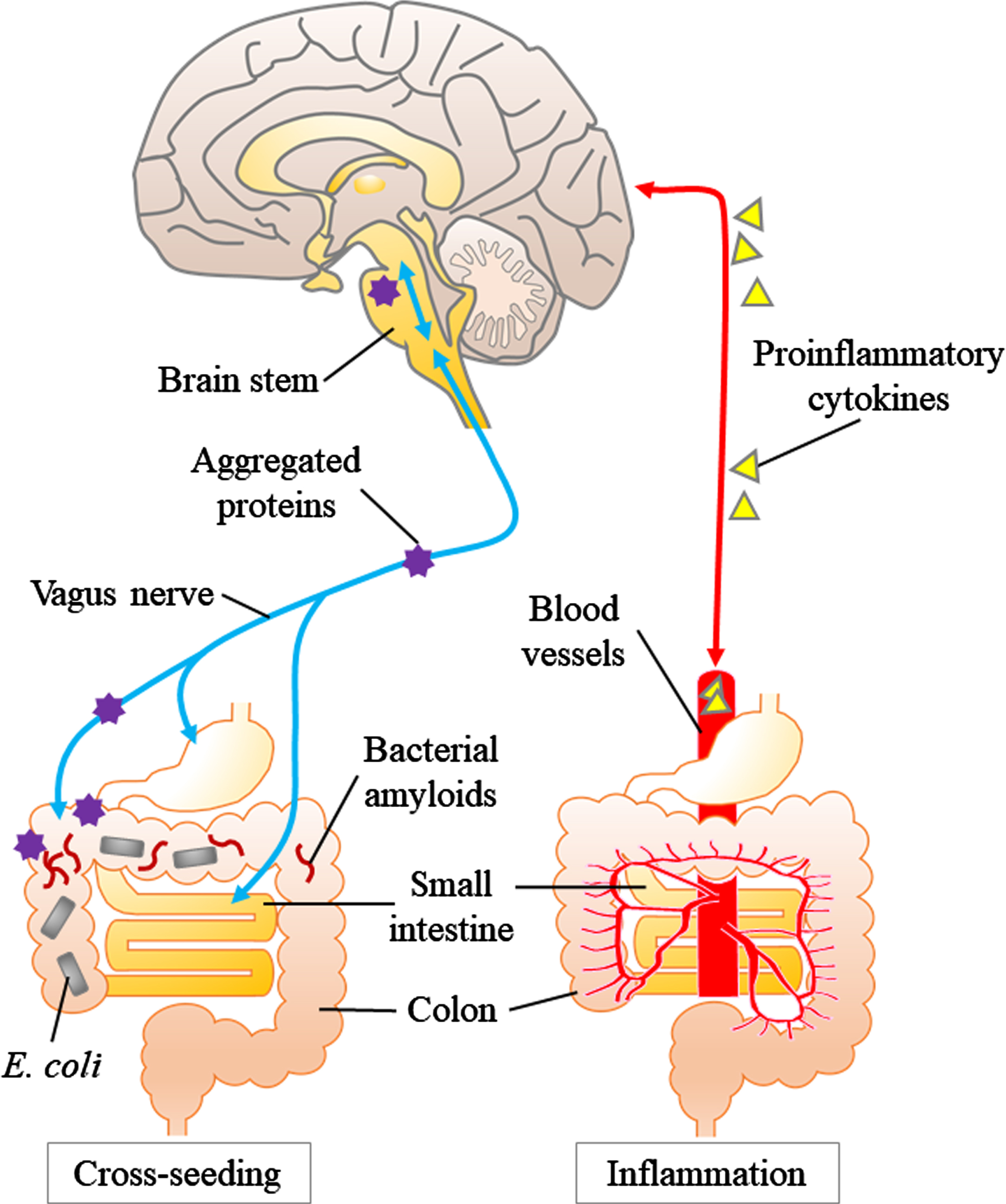
In the experimental setting, the abnormal assemblies of these proteins originate from the disease-specific misfolded proteins and finally adopt an amyloid polymer structure [65]. Many members of the human microbiome including E. coli produce extracellular fibers, which form amyloids. Curli, the first-described bacterial amyloid produced by E. coli, shares structural and pathological properties with human amyloids [66]. Besides, curli fibers induce high level releasing of proinflammatory cytokines such as tumor necrosis factor-alpha, interleukin-6, and interleukin-8 in human cells [67]. Moreover, the bacterial amyloid curli activates Toll-like receptor 2, which also recognizes and targets human amyloids such as Aβ and α-syn [68, 69]. Altogether, dysbiosis of the gut microbiota can cause inflammation and lead to neurodegeneration.
The other keyword, “cross-seeding”, is related to direct abnormal proteinaceous changes in neurodegeneration. In many neurodegenerative diseases, experimental evidences support the concept that disease-specific proteins such as Aβ and α-syn acquire pathogenicity by a prion-like molecular process: the process of amyloid fibril formation proceeds after initiation by the “seed” that serves as the template [70, 71] These pathogenic protein assemblies are also known to propagate from the periphery to the brain in a prion-like manner [65, 72]. A key question is where does the process begin? In PD constipation precedes the onset of motor symptoms many PD persons, showing that PD is closely related to the gastrointestinal tract [73]. As Braak showed in PD, Lewy bodies, which are composed of α-syn, have a pathway from the dorsal nucleus of the vagus nerve upward to the substantia nigra [74]. Cersosimo showed that colonic biopsy in PD patients revealed accumulation of α-syn in the submucosal plexus in 72%of PD patients, while no accumulation was observed in the control group [75, 76]. Svensson et al. showed that the incidence of PD was significantly reduced in people who underwent total vagus nerve resection for the treatment of duodenal ulcer [77]. This report supports the hypothesis that pathological changes in PD begin first in the enteric plexus and propagate through the vagus nerve to the brain. Dysbiosis of the gut microbiota can weaken the barrier function of the gut epithelium and trigger the accumulation of α-syn in the enteric plexus. Thus, bacterial amyloids such as curli may initiate protein aggregation in enteric plexus. A pathway has also been proposed by Braak et al. and others [78] from the olfactory epithelium to the brain. In the roof of the nose the olfactory receptors AR processes of neurons that reside outside the central nervous system that are directly connected to the olfactory bulb as well as the entorhinal cortex. Prion-like protein misfolding may be initiated in olfactory neurons thorough exposure to bacterial products in the roof of the nose [79].
Is there a precedent for the gut factors to cause initiation of a protein folding disorder? This is precisely our current understanding of Creutzfeldt Jakob disease, bovine spongiform encephalopathy as well as kuru. In these conditions exposure to the prion protein in the gut thorough food initiates templated misfolding of the cellular form of the prion protein in the neurons of the gut which is subsequently transmitted to the brain through the autonomic nervous system, as well as the blood. The ability of functional bacterial amyloid proteins such as curli and others to initiate prion-like misfolding of neuronal proteins α-synuclein end Aβ have been reported in in vitro studies [80, 81]. Although the enteric nervous and immune systems are much simpler than those of mammals, an innervated digestive tract and a primitive immune system (including specialized hemocytes involved in phagocytosis) of Drosophila can serve as an appropriate model for studying gut-brain communication [82, 83].
DROSOPHILA FOR MICROBIOTA STUDY OF ALZHEIMER’S DISEASE
Wu et al. used transgenic flies expressing human Aβ42 as an AD model and showed in 2017 that the gut-brain axis is a fundamental factor in neurodegeneration of AD. Gram-negative rod (Ecc15) infection shortened the lifespan of Aβ42-transgenic flies, impaired motor function, and increased cell death in the brain. In these flies, phagocytes were more easily recruited to the brain as a result of enhanced migration of brain phagocytes, and these cells were more likely to produce reactive oxygen species with the elevated JNK pathway. When phagocytes were genetically knocked out, the JNK pathway was suppressed and the flies’ lifespan was extended. This paper clearly showed that in the AD model Drosophila, changes in gut bacteria can activate immune cells to produce inflammatory cytotoxicity of central nervous system [84]. In recent years, several papers have reported on the therapeutic effects of specific bacteria and substrates (called probiotics and prebiotics, respectively) or combination (called symbiotics) that are considered to be good for health promotion on Drosophila models of AD [85–87]. Among these, Tan et al. and Liu et al. used human Aβ42 transgenic flies as an AD model, and Westfall et al. used transgenic flies expressing human APP and β-secretase 1 (BACE1) enzymes as an AD model. All reports used Lactobacillus as probiotics and showed that Lactobacillus administration improved phenotypes such as disrupted compound eye formation, shortened survival period, impaired motor function, and Aβ deposition. Although the number of papers is still limited, the combination of Drosophila’s advantages, such as the ease of regulation of gut microbiome and the homology of neural tissue with humans, will help us to study the role of gut microbiome in AD patients (Table 1).
Table 1
Summary of Drosophila Microbiome Research on AD and PD.
| Disease | Year | Flies | Summary | Ref |
| AD | 2017 | Aβ42-transgenic flies | Gram-negative rod infection shortened the lifespan, impaired motor function, and increased cell death in the brain. | 84 |
| AD | 2019 | Flies expressing Human APP and BACE1 | Lactobacillus administration improved phenotypes | 85 |
| AD | 2020 | Aβ42-transgenic flies | 86 | |
| AD | 2020 | Aβ42-transgenic flies | 87 | |
| PD | 2019 | α-syn transgenic flies | Phenolic acid metabolites were effective in delaying the deterioration of motor function | 90 |
| PD | 2020 | Endogenous PINK1 mutant flies | EGCG improved motor function and prolonged survival. | 91 |
AD, Alzheimer’s disease; PD, Parkinson’s disease; APP, Amyloid-beta precursor protein; BASE1, beta-site amyloid precursor protein cleaving enzyme 1; Aβ42, Amyloid beta 42; α-syn, alpha-synuclein; EGCG, epigallocatechin gallate.
DROSOPHILA FOR MICROBIOTA STUDY OF PARKINSON’S DISEASE AND OTHER NEURODEGENERATIVE DISEASES
It has been shown that the PD model Drosophila exhibits reduced locomotion, shortened lifespan, and neurodegeneration [88, 89]. There have been two reports on the gut microbiota in the Drosophila PD model. Ho et al. showed that phenolic acid metabolites were effective in delaying the deterioration of motor function in human α-syn transgenic flies, and that phenolic acid metabolites inhibited protofibril formation by binding α-syn in vitro. The other report is by Xu et al. [91] (Table 1). When Drosophila mutants with endogenous PINK1 were fed Epigallocatechin-3-gallate (EGCG), a potent polyphenol, improved motor function and prolonged survival were observed. The abundance of gut microbial composition was increased in these ECGC-treated flies, and when such abundance was reduced by antibiotic administration, the treatment effect of EGCG disappeared. By transcriptomic profiling using PINK1 mutant flies, they showed that the expression of genes involved in fly-specific immune hormones was altered in PINK1 mutant flies, and that these gene expression changes were restored by ECGC consumption.
In ALS, another significant neurodegenerative disease, the relationship between the phenotype of ALS and gut microbiota has been investigated using mouse models. Blacher et al. showed that Superoxide dismutase-1 (SOD1) transgenic mice exhibit abnormalities in gut bacteria and changes in the composition of metabolites by bacteria before the onset of ALS, and that the prognosis is worse when these mice are raised in sterile conditions or after treatment with antibiotics. The authors showed that the abundance of Akkermansia muciniphila continued to decrease in SOD1 transgenic mice as the disease progressed. They also showed that colonization of antibiotic-treated SOD1 transgenic mice with A. muciniphila improved motor symptoms, whereas colonization with Ruminococcus torques or Parabacteroides distasonis worsened symptoms. The authors also found that the symptoms worsened when colonized with Ruminococcus torques or Parabacteroides distasonis. The authors further showed that colonization with A. muciniphila increased the concentration of nicotinamide in the cerebrospinal fluid of SOD1 transgenic mice. Similar improvement (vitamin B3) in motor symptoms was observed when nicotinamide was administered [92]. Burberry et al. showed that immune cells such as neutrophils infiltrate the spinal cord and microglia are activated in the C9orf72 mouse model. They also found that the diversity of the gut microbiota of C9orf72 transgenic mice was reduced in pro-inflammatory environments, compared to pro-survival environments. They further showed that reducing the microbial load with broad-spectrum antibiotics or transferring microbes from mice grown in a pro-survival environment can improve the inflammation-associated phenotype and extend the lifespan of C9orf72 transgenic mice raised in a pro-inflammatory environment [93]. The results in mice suggest that the gut microbiota changes with the onset and progression of ALS, that microbial-derived metabolites such as nicotinamide alter the progression of the disease, and that the gut microbiota may alter the motor function and lifespan of ALS model mice through immune responses. However, since gut microbiota is very sensitive to environmental changes, the results of the mouse study should be validated using other models. For now, there are no studies on gut microbiota using Drosophila model of ALS, but it is likely that they will be used in the near future.
APPLICATION AND LIMITATION OF DROSOPHILA GUT-BRAIN AXIS RESEARCH
We have introduced the role of the gut microbiota in neurodegenerative diseases, and the characteristics of Drosophila as a laboratory animal, for the study of gut microbiota in neurodegenerative diseases. A significant paper on the gut-brain axis was published in 2021 using Drosophila. Kim et al. have identified an amino acid sensing mechanism in Drosophila that leads to behavioral changes and is partially driven by the gut microbiota. In flies deficient in essential amino acids derived from the diet or produced by the microbiota, elevated expression of the neuropeptide CNMamide (CNMa) in the anterior midgut gut cells occurs via the Gcn2 and Tor pathways, which in turn modulates feeding behavior via the activity of neurons expressing CNMa receptors. This was then shown to modulate feeding behavior via the activity of neurons expressing CNMa receptors [94]. It is interesting that one of the specific molecules that directly controls the brain-gut axis has been identified.
Drosophila is also a useful laboratory animal to test the effects of probiotics. Westfall et al. showed that new probiotics and symbiotics prolonged survival in Drosophila and improved gene expression related to insulin signaling, fat metabolism, immunity, and oxidative stress. They showed that probiotics and symbiotics prolonged Drosophila survival and improved gene expression related to insulin signaling, fat metabolism, immunity and oxidative stress [95].
The study of gut microbiota by Drosophila has some limitations. Firstly, due to physiological and developmental differences, it is difficult to directly apply the results of Drosophila gut microbiota studies to mammals. Secondly, most invertebrates, including Drosophila, do not have the acquired immune system found in mammals [82]. Therefore, the inflammatory response that can be reproduced in fly disease models is likely to different than the inflammatory response in humans. It is difficult to explain complex changes in gut bacteria in disease with only one experimental model. To decipher the complexity of gut microbiota, we need information from multiple models. The results shown in fly studies may represent a fundamental part of the whole picture of host and gut microbiota interactions that are conserved in multiple species. Even though the fly’s gut microbiota are not simple mimics of humans, flies will be a wonderful system to learn the molecular mechanisms of how the gut microbiota affect the host’s health and behavior.
CONCLUSION
Drosophila is a useful laboratory animal for the study of gut microbiota and neurodegenerative diseases. Studies of Drosophila will allow us to test the hypothesis that the gut microbiota is involved in human neurodegeneration. Disease modeling by Drosophila has just started using microbiota studies and is expected to be applied to a wider field in the future.
ACKNOWLEDGMENTS
This work was supported in part by a Grant-in-Aid (19K16924 to F.K-M.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Japan Society for the Promotion of Science (R.P. Friedland L20552).
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/21-5031r1).
REFERENCES
[1] | Sharon G , Sampson TR , Geschwind DH , Mazmanian SK ((2016) ) The central nervous system and the gut microbiome. Cell 167: , 915–932. |
[2] | Thursby E , Juge N ((2017) ) Introduction to the human gut microbiota. Biochem J 474: , 1823–1836. |
[3] | Cani PD ((2018) ) Human gut microbiome: Hopes, threats and promises. Gut 67: , 1716–1725. |
[4] | Rakoff-Nahoum S , Foster KR , Comstock LE ((2016) ) The evolution of cooperation within the gut microbiota. Nature 533: , 255–259. |
[5] | Wen L , Duffy A ((2017) ) Factors influencing the gut microbiota, inflammation, and type 2 diabetes. J Nutr 147: , 1468S–1475S. |
[6] | Bajic JE , Johnston IN , Howarth GS , Hutchinson MR ((2018) ) From the bottom-up: Chemotherapy and gut-brain axis dysregulation. Front Behav Neurosci 12: , 104. |
[7] | Vallianou N , Dalamaga M , Stratigou T , Karampela I , Tsigalou C ((2021) ) Do antibiotics cause obesity through long-term alterations in the gut microbiome? A review of current evidence. Curr Obes Rep 10: , 244–262. |
[8] | Qin J , Li Y , Cai Z , Li S , Zhu J , Zhang F , Liang S , Zhang W , Guan Y , Shen D , Peng Y , Zhang D , Jie Z , Wu W , Qin Y , Xue W , Li J , Han L , Lu D , Wu P , Dai Y , Sun X , Li Z , Tang A , Zhong S , Li X , Chen W , Xu R , Wang M , Feng Q , Gong M , Yu J , Zhang Y , Zhang M , Hansen T , Sanchez G , Raes J , Falony G , Okuda S , Almeida M , LeChatelier E , Renault P , Pons N , Batto JM , Zhang Z , Chen H , Yang R , Zheng W , Li S , Yang H , Wang J , Ehrlich SD , Nielsen R , Pedersen O , Kristiansen K , Wang J ((2012) ) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490: , 55–60. |
[9] | Hernandez-Ceballos W , Cordova-Gallardo J , Mendez-Sanchez N ((2021) ) Gut microbiota in metabolic-associated fatty liver disease and in other chronic metabolic diseases. J Clin Transl Hepatol 9: , 227–238. |
[10] | Tilg H , Adolph TE , Gerner RR , Moschen AR ((2018) ) The intestinal microbiota in colorectal cancer. Cancer Cell 33: , 954–964. |
[11] | Shanahan F , van Sinderen D , O’Toole PW , Stanton C ((2017) ) Feeding the microbiota: Transducer of nutrient signals for the host. Gut 66: , 1709–1717. |
[12] | Martin-Mateos R , Albillos A ((2021) ) The role of the gut-liver axis in metabolic dysfunction-associated fatty liver disease. Front Immunol 12: , 660179. |
[13] | Tilg H , Zmora N , Adolph TE , Elinav E ((2020) ) The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol 20: , 40–54. |
[14] | Back M , Yurdagul A Jr., Tabas I , Oorni K , Kovanen PT ((2019) ) Inflammation and its resolution in atherosclerosis: Mediators and therapeutic opportunities. Nat Rev Cardiol 16: , 389–406. |
[15] | Fahy JV ((2015) ) Type 2 inflammation in asthma–present in most, absent in many. Nat Rev Immunol 15: , 57–65. |
[16] | Rapa SF , Di Iorio BR , Campiglia P , Heidland A , Marzocco S ((2019) ) Inflammation and oxidative stress in chronic kidney disease-potential therapeutic role of minerals, vitamins and plant-derived metabolites. Int J Mol Sci 21: , 263. |
[17] | Mayer EA ((2011) ) Gut feelings: The emerging biology of gut-brain communication. Nat Rev Neurosci 12: , 453–466. |
[18] | Fung TC , Olson CA , Hsiao EY ((2017) ) Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 20: , 145–155. |
[19] | Diaz Heijtz R , Wang S , Anuar F , Qian Y , Bjorkholm B , Samuelsson A , Hibberd ML , Forssberg H , Pettersson S ((2011) ) Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 108: , 3047–3052. |
[20] | Hsiao EY , McBride SW , Hsien S , Sharon G , Hyde ER , McCue T , Codelli JA , Chow J , Reisman SE , Petrosino JF , Patterson PH , Mazmanian SK ((2013) ) Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155: , 1451–1463. |
[21] | Sampson TR , Debelius JW , Thron T , Janssen S , Shastri GG , Ilhan ZE , Challis C , Schretter CE , Rocha S , Gradinaru V , Chesselet MF , Keshavarzian A , Shannon KM , Krajmalnik-Brown R , Wittung-Stafshede P , Knight R , Mazmanian SK ((2016) ) Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167: , 1469–1480 e1412. |
[22] | Douglas AE ((2018) ) The Drosophila model for microbiome research. Lab Anim (NY) 47: , 157–164. |
[23] | Yamaguchi M , Omori K , Asada S , Yoshida H ((2021) ) Epigenetic regulation of ALS and CMT: A lesson from Drosophila models. Int J Mol Sci 22: , 491. |
[24] | Martino ME , Ma D , Leulier F ((2017) ) Microbial influence on Drosophila biology. Curr Opin Microbiol 38: , 165–170. |
[25] | Xia J , Gravato-Nobre M , Ligoxygakis P ((2019) ) Convergence of longevity and immunity: Lessons from animal models. Biogerontology 20: , 271–278. |
[26] | Kong Y , Jiang B , Luo X ((2018) ) Gut microbiota influences Alzheimer’s disease pathogenesis by regulating acetate in Drosophila model. Future Microbiol 13: , 1117–1128. |
[27] | Batista LL , Malta SM , Guerra Silva HC , Borges LDF , Rocha LO , da Silva JR , Rodrigues TS , Venturini G , Padilha K , da Costa Pereira A , Espindola FS , Ueira-Vieira C ((2021) ) Kefir metabolites in a fly model for Alzheimer’s disease. Sci Rep 11: , 11262. |
[28] | Yamaguchi M , Yoshida H ((2018) ) Drosophila as a model organism. Adv Exp Med Biol 1076: , 1–10. |
[29] | Shulman JM , Chipendo P , Chibnik LB , Aubin C , Tran D , Keenan BT , Kramer PL , Schneider JA , Bennett DA , Feany MB , De Jager PL ((2011) ) Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet 88: , 232–238. |
[30] | Whitworth AJ , Theodore DA , Greene JC , Benes H , Wes PD , Pallanck LJ ((2005) ) Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc Natl Acad Sci U S A 102: , 8024–8029. |
[31] | Neumann M , Sampathu DM , Kwong LK , Truax AC , Micsenyi MC , Chou TT , Bruce J , Schuck T , Grossman M , Clark CM , McCluskey LF , Miller BL , Masliah E , Mackenzie IR , Feldman H , Feiden W , Kretzschmar HA , Trojanowski JQ , Lee VM ((2006) ) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: , 130–133. |
[32] | Warrick JM , Paulson HL , Gray-Board GL , Bui QT , Fischbeck KH , Pittman RN , Bonini NM ((1998) ) Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell 93: , 939–949. |
[33] | Higuchi Y , Okunushi R , Hara T , Hashiguchi A , Yuan J , Yoshimura A , Murayama K , Ohtake A , Ando M , Hiramatsu Y , Ishihara S , Tanabe H , Okamoto Y , Matsuura E , Ueda T , Toda T , Yamashita S , Yamada K , Koide T , Yaguchi H , Mitsui J , Ishiura H , Yoshimura J , Doi K , Morishita S , Sato K , Nakagawa M , Tsuji S , Takashima H ((2018) ) Mutations in COA7 cause spinocerebellar ataxia with axonal neuropathy. Brain 141: , 1622–1636. |
[34] | Koyle ML , Veloz M , Judd AM , Wong AC , Newell PD , Douglas AE , Chaston JM (2016) Rearing the fruit fly Drosophila melanogaster under axenic and gnotobiotic conditions. J Vis Exp, 54219. |
[35] | Rubin GM , Lewis EB ((2000) ) A brief history of Drosophila’s contributions to genome research. Science 287: , 2216–2218. |
[36] | Ramet M ((2012) ) The fruit fly Drosophila melanogaster unfolds the secrets of innate immunity. Acta Paediatr 101: , 900–905. |
[37] | Brown K ((2017) ) An interview with Christiane Nusslein-Volhard. Development 144: , 3851–3854. |
[38] | Brand AH , Perrimon N ((1993) ) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: , 401–415. |
[39] | Takano-Shimizu-Kouno T , Ohsako T ((2018) ) Humanized flies and resources for cross-species study. Adv Exp Med Biol 1076: , 277–288. |
[40] | King DG ((1988) ) Cellular organization and peritrophic membrane formation in the cardia (proventriculus) of Drosophila melanogaster. J Morphol 196: , 253–282. |
[41] | Hegedus D , Erlandson M , Gillott C , Toprak U ((2009) ) New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol 54: , 285–302. |
[42] | Kuraishi T , Binggeli O , Opota O , Buchon N , Lemaitre B ((2011) ) Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc Natl Acad Sci U S A 108: , 15966–15971. |
[43] | Douglas AE ((2019) ) Simple animal models for microbiome research. Nat Rev Microbiol 17: , 764–775. |
[44] | Obadia B , Guvener ZT , Zhang V , Ceja-Navarro JA , Brodie EL , Ja WW , Ludington WB ((2017) ) Probabilistic invasion underlies natural gut microbiome stability. Curr Biol 27: , 1999–2006 e1998. |
[45] | Storelli G , Defaye A , Erkosar B , Hols P , Royet J , Leulier F ((2011) ) Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14: , 403–414. |
[46] | Shin SC , Kim SH , You H , Kim B , Kim AC , Lee KA , Yoon JH , Ryu JH , Lee WJ ((2011) ) Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334: , 670–674. |
[47] | Lee HY , Lee SH , Lee JH , Lee WJ , Min KJ ((2019) ) The role of commensal microbes in the lifespan of Drosophila melanogaster. Aging (Albany NY) 11: , 4611–4640. |
[48] | Douglas AE ((2018) ) Gut microbes alter the walking activity of fruit flies. Nature 563: , 331–332. |
[49] | Schretter CE , Vielmetter J , Bartos I , Marka Z , Marka S , Argade S , Mazmanian SK ((2018) ) A gut microbial factor modulates locomotor behaviour in Drosophila. Nature 563: , 402–406. |
[50] | Collins SM , Surette M , Bercik P ((2012) ) The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 10: , 735–742. |
[51] | Bravo JA , Forsythe P , Chew MV , Escaravage E , Savignac HM , Dinan TG , Bienenstock J , Cryan JF ((2011) ) Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A 108: , 16050–16055. |
[52] | Klingelhoefer L , Reichmann H ((2015) ) Pathogenesis of Parkinson disease–the gut-brain axis and environmental factors. Nat Rev Neurol 11: , 625–636. |
[53] | Lionnet A , Leclair-Visonneau L , Neunlist M , Murayama S , Takao M , Adler CH , Derkinderen P , Beach TG ((2018) ) Does Parkinson’s disease start in the gut? Acta Neuropathol 135: , 1–12. |
[54] | Holmqvist S , Chutna O , Bousset L , Aldrin-Kirk P , Li W , Bjorklund T , Wang ZY , Roybon L , Melki R , Li JY ((2014) ) Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol 128: , 805–820. |
[55] | Van Den Berge N , Ferreira N , Gram H , Mikkelsen TW , Alstrup AKO , Casadei N , Tsung-Pin P , Riess O , Nyengaard JR , Tamguney G , Jensen PH , Borghammer P ((2019) ) Evidence for bidirectional and trans-synaptic parasympathetic and sympathetic propagation of alpha-synuclein in rats. Acta Neuropathol 138: , 535–550. |
[56] | Miller AL , Bessho S , Grando K , Tukel C ((2021) ) Microbiome or infections: Amyloid-containing biofilms as a trigger for complex human diseases. Front Immunol 12: , 638867. |
[57] | Friedland RP ((2015) ) Mechanisms of molecular mimicry involving the microbiota in neurodegeneration. J Alzheimers Dis 45: , 349–362. |
[58] | Friedland RP , McMillan JD , Kurlawala Z ((2020) ) What are the molecular mechanisms by which functional bacterial amyloids influence amyloid beta deposition and neuroinflammation in neurodegenerative disorders? Int J Mol Sci 21: , 1652. |
[59] | Montacute R , Foley K , Forman R , Else KJ , Cruickshank SM , Allan SM ((2017) ) Enhanced susceptibility of triple transgenic Alzheimer’s disease (3xTg-AD) mice to acute infection. J Neuroinflammation 14: , 50. |
[60] | De la Fuente M ((2021) ) The role of the microbiota-gut-brain axis in the health and illness condition: A focus on Alzheimer’s disease. J Alzheimers Dis 81: , 1345–1360. |
[61] | Harach T , Marungruang N , Duthilleul N , Cheatham V , Mc Coy KD , Frisoni G , Neher JJ , Fak F , Jucker M , Lasser T , Bolmont T ((2017) ) Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep 7: , 41802. |
[62] | Li B , He Y , Ma J , Huang P , Du J , Cao L , Wang Y , Xiao Q , Tang H , Chen S ((2019) ) Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement 15: , 1357–1366. |
[63] | Li Z , Zhu H , Guo Y , Du X , Qin C ((2020) ) Gut microbiota regulate cognitive deficits and amyloid deposition in a model of Alzheimer’s disease. J Neurochem 155: , 448–461. |
[64] | Zhan X , Stamova B , Jin LW , DeCarli C , Phinney B , Sharp FR ((2016) ) Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 87: , 2324–2332. |
[65] | Jucker M , Walker LC ((2018) ) Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat Neurosci 21: , 1341–1349. |
[66] | Chapman MR , Robinson LS , Pinkner JS , Roth R , Heuser J , Hammar M , Normark S , Hultgren SJ ((2002) ) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295: , 851–855. |
[67] | Bian Z , Brauner A , Li Y , Normark S ((2000) ) Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis 181: , 602–612. |
[68] | Tukel C , Wilson RP , Nishimori JH , Pezeshki M , Chromy BA , Baumler AJ ((2009) ) Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe 6: , 45–53. |
[69] | Tukel C , Nishimori JH , Wilson RP , Winter MG , Keestra AM , van Putten JP , Baumler AJ ((2010) ) Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12: , 1495–1505. |
[70] | Lundmark K , Westermark GT , Olsen A , Westermark P ((2005) ) Protein fibrils in nature can enhance amyloid protein A amyloidosis in mice: Cross-seeding as a disease mechanism. Proc Natl Acad Sci U S A 102: , 6098–6102. |
[71] | Eisenberg D , Jucker M ((2012) ) The amyloid state of proteins in human diseases. Cell 148: , 1188–1203. |
[72] | Jucker M , Walker LC ((2013) ) Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature 501: , 45–51. |
[73] | Noyce AJ , Bestwick JP , Silveira-Moriyama L , Hawkes CH , Giovannoni G , Lees AJ , Schrag A ((2012) ) Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann Neurol 72: , 893–901. |
[74] | Braak H , Del Tredici K , Bratzke H , Hamm-Clement J , Sandmann-Keil D , Rub U ((2002) ) Staging of the intracerebral inclusion body pathology associated with idiopathic Parkinson’s disease (preclinical and clinical stages). J Neurol 249 Suppl 3: , III/1–5. |
[75] | Lebouvier T , Neunlist M , Bruley des Varannes S , Coron E , Drouard A , N’Guyen JM , Chaumette T , Tasselli M , Paillusson S , Flamand M , Galmiche JP , Damier P , Derkinderen P ((2010) ) Colonic biopsies to assess the neuropathology of Parkinson’s disease and its relationship with symptoms. PLoS One 5: , e12728. |
[76] | Cersosimo MG ((2015) ) Gastrointestinal biopsies for the diagnosis of alpha-synuclein pathology in Parkinson’s disease. Gastroenterol Res Pract 2015: , 476041. |
[77] | Svensson E , Horvath-Puho E , Thomsen RW , Djurhuus JC , Pedersen L , Borghammer P , Sorensen HT ((2015) ) Vagotomy and subsequent risk of Parkinson’s disease. Ann Neurol 78: , 522–529. |
[78] | Friedland RP , Chapman MR ((2017) ) The role of microbial amyloid in neurodegeneration. PLoS Pathog 13: , e1006654. |
[79] | De Luca CMG , Elia AE , Portaleone SM , Cazzaniga FA , Rossi M , Bistaffa E , De Cecco E , Narkiewicz J , Salzano G , Carletta O , Romito L , Devigili G , Soliveri P , Tiraboschi P , Legname G , Tagliavini F , Eleopra R , Giaccone G , Moda F ((2019) ) Efficient RT-QuIC seeding activity for alpha-synuclein in olfactory mucosa samples of patients with Parkinson’s disease and multiple system atrophy. Transl Neurodegener 8: , 24. |
[80] | Javed I , Zhang Z , Adamcik J , Andrikopoulos N , Li Y , Otzen DE , Lin S , Mezzenga R , Davis TP , Ding F , Ke PC ((2020) ) Accelerated amyloid beta pathogenesis by bacterial amyloid FapC. Adv Sci (Weinh) 7: , 2001299. |
[81] | Perov S , Lidor O , Salinas N , Golan N , Tayeb-Fligelman E , Deshmukh M , Willbold D , Landau M ((2019) ) Structural insights into Curli CsgA cross-beta fibril architecture inspire repurposing of anti-amyloid compounds as anti-biofilm agents. PLoS Pathog 15: , e1007978. |
[82] | Lemaitre B , Hoffmann J ((2007) ) The host defense of Drosophila melanogaster. Annu Rev Immunol 25: , 697–743. |
[83] | Lemaitre B , Miguel-Aliaga I ((2013) ) The digestive tract of Drosophila melanogaster. Annu Rev Genet 47: , 377–404. |
[84] | Wu SC , Cao ZS , Chang KM , Juang JL ((2017) ) Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat Commun 8: , 24. |
[85] | Westfall S , Lomis N , Prakash S ((2019) ) A novel synbiotic delays Alzheimer’s disease onset via combinatorial gut-brain-axis signaling in Drosophila melanogaster. PLoS One 14: , e0214985. |
[86] | Tan FHP , Liu G , Lau SA , Jaafar MH , Park YH , Azzam G , Li Y , Liong MT ((2020) ) Lactobacillus probiotics improved the gut microbiota profile of a Drosophila melanogaster Alzheimer’s disease model and alleviated neurodegeneration in the eye. Benef Microbes 11: , 79–89. |
[87] | Liu G , Tan FH , Lau SA , Jaafar MH , Chung FY , Azzam G , Liong MT , Li Y (2020) Lactic acid bacteria feeding reversed the malformed eye structures and ameliorated gut microbiota profiles of Drosophila melanogaster Alzheimer’s disease model. J Appl Microbiol, doi:10.1111/jam.14773. |
[88] | Feany MB , Bender WW ((2000) ) A Drosophila model of Parkinson’s disease. Nature 404: , 394–398. |
[89] | Aggarwal A , Reichert H , VijayRaghavan K ((2019) ) A locomotor assay reveals deficits in heterozygous Parkinson’s disease model and proprioceptive mutants in adult Drosophila. Proc Natl Acad Sci U S A 116: , 24830–24839. |
[90] | Ho L , Zhao D , Ono K , Ruan K , Mogno I , Tsuji M , Carry E , Brathwaite J , Sims S , Frolinger T , Westfall S , Mazzola P , Wu Q , Hao K , Lloyd TE , Simon JE , Faith J , Pasinetti GM ((2019) ) Heterogeneity in gut microbiota drive polyphenol metabolism that influences alpha-synuclein misfolding and toxicity. J Nutr Biochem 64: , 170–181. |
[91] | Xu Y , Xie M , Xue J , Xiang L , Li Y , Xiao J , Xiao G , Wang HL ((2020) ) EGCG ameliorates neuronal and behavioral defects by remodeling gut microbiota and TotM expression in Drosophila models of Parkinson’s disease. FASEB J 34: , 5931–5950. |
[92] | Blacher E , Bashiardes S , Shapiro H , Rothschild D , Mor U , Dori-Bachash M , Kleimeyer C , Moresi C , Harnik Y , Zur M , Zabari M , Brik RB , Kviatcovsky D , Zmora N , Cohen Y , Bar N , Levi I , Amar N , Mehlman T , Brandis A , Biton I , Kuperman Y , Tsoory M , Alfahel L , Harmelin A , Schwartz M , Israelson A , Arike L , Johansson MEV , Hansson GC , Gotkine M , Segal E , Elinav E ((2019) ) Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572: , 474–480. |
[93] | Burberry A , Wells MF , Limone F , Couto A , Smith KS , Keaney J , Gillet G , van Gastel N , Wang JY , Pietilainen O , Qian M , Eggan P , Cantrell C , Mok J , Kadiu I , Scadden DT , Eggan K ((2020) ) C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature 582: , 89–94. |
[94] | Kim B , Kanai MI , Oh Y , Kyung M , Kim EK , Jang IH , Lee JH , Kim SG , Suh GSB , Lee WJ ((2021) ) Response of the microbiome-gut-brain axis in Drosophila to amino acid deficit. Nature 593: , 570–574. |
[95] | Westfall S , Lomis N , Prakash S ((2018) ) Longevity extension in Drosophila through gut-brain communication. Sci Rep 8: , 8362. |



