ANMerge: A Comprehensive and Accessible Alzheimer’s Disease Patient-Level Dataset
Abstract
Background:
Accessible datasets are of fundamental importance to the advancement of Alzheimer’s disease (AD) research. The AddNeuroMed consortium conducted a longitudinal observational cohort study with the aim to discover AD biomarkers. During this study, a broad selection of data modalities was measured including clinical assessments, magnetic resonance imaging, genotyping, transcriptomic profiling, and blood plasma proteomics. Some of the collected data were shared with third-party researchers. However, this data was incomplete, erroneous, and lacking in interoperability.
Objective:
To provide the research community with an accessible, multimodal, patient-level AD cohort dataset.
Methods:
We systematically addressed several limitations of the originally shared resources and provided additional unreleased data to enhance the dataset.
Results:
In this work, we publish and describe ANMerge, a new version of the AddNeuroMed dataset. ANMerge includes multimodal data from 1,702 study participants and is accessible to the research community via a centralized portal.
Conclusion:
ANMerge is an information rich patient-level data resource that can serve as a discovery and validation cohort for data-driven AD research, such as, for example, machine learning and artificial intelligence approaches.
INTRODUCTION
Alzheimer’s disease (AD) is a progressive disease whose pathology develops years before cognitive symptoms arise and a diagnosis is made by a clinician [1]. Early intervention in non-cognitively impaired, pre-symptomatic disease stages is instrumental to any future disease modifying therapy. Enabling such an early intervention poses the problem of diagnosing a patient with AD before cognitive symptoms indicate disease presence. One approach to establish whether a specific individual is in the pre-symptomatic stages of the disease is a diagnosis based on informative disease biomarkers. The critical prerequisite for discovery and validation of such biomarkers are resourceful patient-level datasets [2]. However, findable AD cohort datasets which are accessible to the research community are scarce.
Open science is a paradigm aimed at increasing societal benefit of research through dissemination and sharing of scientific data. This enables usage and analysis of collected data by the whole research community which subsequently will increase the achieved knowledge gain. Currently, the prime example of following the open science paradigm in the AD field is the Alzheimer’s Disease Neuroimaging Initiative (ADNI) [3]. ADNI is an information rich, comprehe-nsive clinical AD cohort dataset that enables secure, yet easy access to its patient level data for researchers with reasonable study interest. In only a few days, raw data as well as a preprocessed version of ADNI (ADNIMERGE) are accessible via the Laboratory of Neuro Imaging (LONI) service (https://loni.usc.edu/). With regard to clinical data, initial preprocessing, arranging, and cleaning of data is often the most time-consuming step in data analysis. Due to that, a major cumulative time save is possible by sharing an already preprocessed, easy-to-analyze dataset instead of a raw data collection. Here, researchers can simply use the provided ADNIMERGE and thereby avoid investing additional time into data preprocessing and cleaning.
While ADNI is a tremendously important resource, as every cohort dataset, it comes with its own lim-itations and biases [4]. To ensure reliability of observations made in one cohort, validation in data from independent cohorts is necessary [5]. Still, apart from ADNI there are not many AD cohort studies which 1) share their data in a similarly comprehensive version and 2) keep the bureaucracy during an access application as straightforward as ADNI does. From our experience, access applications are often time consuming and if access is granted, shared data is sometimes lacking important information. Therefore, other easily accessible and information rich alternatives besides ADNI are crucial.
In 2005, Lovestone et al. started AddNeuroMed, a project funded by InnoMed, a precursor of the Innovative Medicine Initiative (IMI) [6]. It aimed at collecting longitudinal patient data at multiple sites across Europe to identify urgently needed progre-ssion biomarkers for AD. For this purpose, a broad spectrum of variables was measured including demographics, neuropsychological assessments, genetic variations and transcriptomics, blood plasma prote-omics, and structural magnetic resonance imaging (MRI) of the brain. In 2015, a subset of the colle-cted data was uploaded on Synapse (https://www.synapse.org/). Next to the original AddNeuroMed data, some data from participants of the Maudsley BRC Dementia Case Registry at King’s Health Partners cohort (DCR) and the Alzheimer’s Research Trust UK cohort (ART) was included [7]. Although the shared AddNeuroMed collection is a large dataset, involving more than 1,700 participants, it has only been cited about 65 times. In contrast, ADNI, which involves roughly 2,400 individuals, was cited more than 1,300 times. Compared to the impact ADNI has had on recent research activities, it seems AddNeuroMed has not reached its full potential. One probable reason for the comparably lower data usage might be the findability and the state of the data published on Synapse. The dataset 1) has never been officially published, 2) is not easy to work with due to missing organization, and 3) is not complete with several entries being erroneous or lacking information. To enable the research community to leverage the full potential of this dataset, a lot of data preprocessing efforts are needed and it is vital to point the community toward this unsalvaged resource.
In this work, we present and publish a new, im-proved, and updated version of AddNeuroMed called ANMerge. ANMerge is a comprehensive, preprocessed AD cohort dataset which is again accessible via Synapse (https://doi.org/10.7303/syn22252881). It is fully interoperable in between its modalities, and rigorous data curation was performed to ensure higher information density and usability. Furthermore, we present a detailed overview on which and how much data is available in the dataset. Finally, we highlight the increased preprocessing efforts involved in creating such a dataset. By making ANMerge accessible, we aim to provide the AD research community with an information rich alternative to previously published cohort datasets, and thereby support the discovery and robust validation of scientific insights.
METHODS
Data collection
AddNeuroMed data collection was performed at six different centers across Europe: University of Kuopio, Finland; Aristotle University of Thessal-oniki, Greece; King’s College London, United Kingdom; University of Lodz, Poland; University of Perugia, Italy; and University of Toulouse, France [6]. The participation of those centers highlights AddNeuroMed as a major cross-European effort in AD related data collection. At each site, all protocols and procedures were approved by Institutional Review Boards and informed consent was obtained for all patients according to the Declaration of Helsinki (1991) [8]. In cases where dementia compromised capacity assent from the patient and consent from a relative, according to local law, was obtained.
Exclusion criteria included other neurological or psychiatric diseases, significant unstable systemic ill-ness or organ failure, and alcohol or substance mi-suse. AD diagnosis followed the Diagnostic and Statistical Manual for Mental Diagnosis, fourth edition and National Institute of Neurological and Com-municative Disorders and Stroke–Alzheimer’s Disease and Related Disorders Association criteria [9]. AD patients were included if they exhibited a Mini-Mental State Examination (MMSE) score in the range of 12–28, a Clinical Dementia Rating (CDR) scale score of above 0.5, and were aged 65 years or above. Individuals were considered as mild cognitive impairment (MCI) according to the Petersen criteria [10]. For inclusion, MCI patients aged 65 or above, the MMSE score ranged between 24 and 30, and they scored 0.5 on the CDR. Participants were considered to be cognitively healthy if they showed normal performance on cognitive tests (within 1.5 SD of average for age, gender and education) and scored 0 on the CDR [11].
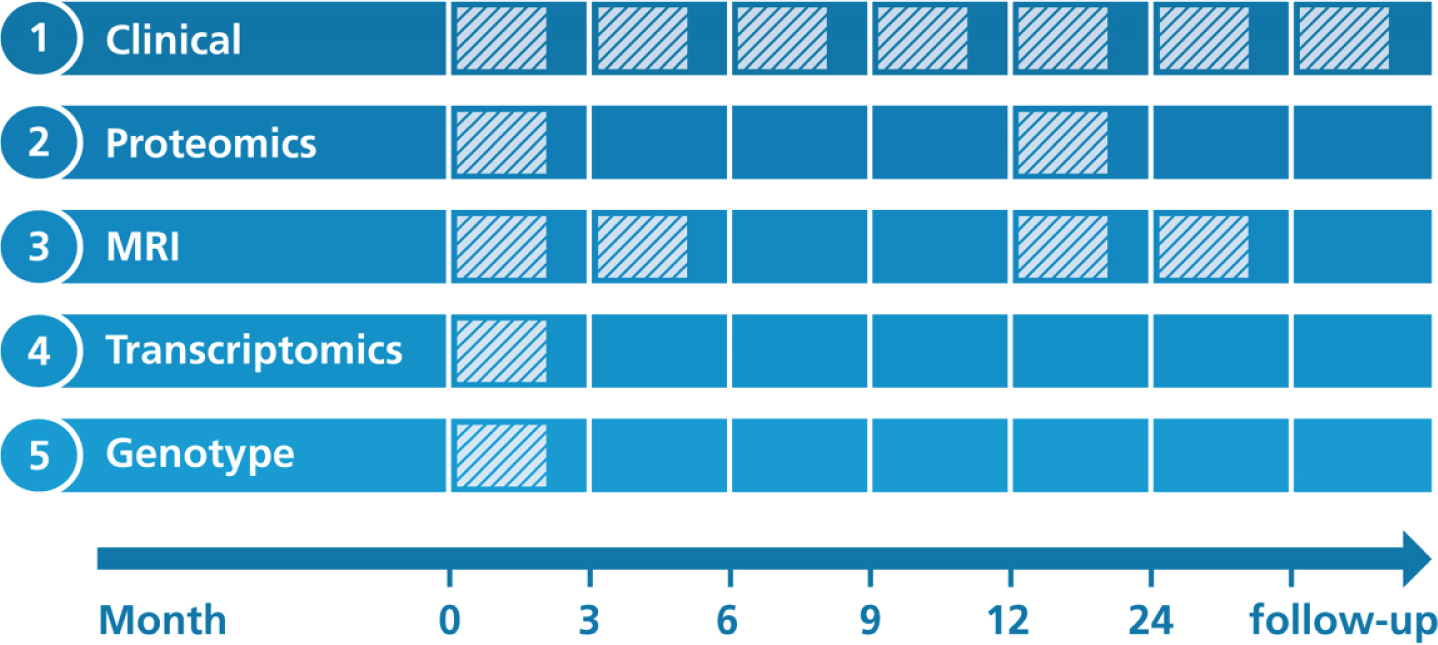
AddNeuroMed’s study protocols were designed to be at least partially compatible with ADNI [6]. Figure 1 illustrates when data collection was performed for each modality.
Fig. 1
Overview on longitudinal data collection per modality. Proteomics, Proteomic data from blood plasma. Transcriptomics, Transcriptomic data from blood plasma. MRI, Structural magnetic resonance imaging.
Clinical assessments
At each participant’s visit throughout the study, a broad collection of neurocognitive and psychological assessments were performed, including the MMSE, CDR, GDS (Geriatric Depression Scale), NPI (Neuropsychiatric Inventory), ADAS-Cog (Alzheimer’s Disease Assessment Scale-Cognitive Subscale), ADCS-ADL (Alzheimer’s Disease Cooperative St-udy Activities of Daily Living Scale), the full CE-RAD battery [12], the Hachinski Ischemic Score, and the Webster Rating Scale. The frequency with which assessments were made varied between diagnostic groups. During the first year, AD cases completed assessments every three months and annual follow-up visits afterwards. MCI patients and healthy individuals from AddNeuroMed, as well as all participants from the ART and DCR cohorts, were assessed regularly every twelve months.
Proteomics
Proteomic data were measured in blood plasma using a Slow Off-rate Modified Aptamer (SOMAmer)-based array called ‘SOMAscan’ (SomaLogic, Inc, Boulder, Colorado). Data collection was performed at baseline and again one year into the study. Details on data acquisition are presented in Kiddle et al. [13] and Sattlecker et al. [14]. In brief, using chemically altered nucleotides the protein signal is turned into a nucleotide signal that can be measured using microarrays. Per sample 8μL plasma were required and levels of 1,001 distinct proteins were measured. An in-depth description of the array technology can be found in Gold et al. [15].
Genotyping
AddNeuroMed participants were genotyped in three batches. For batch one, the Illumina HumanHap610-Quad Beadchip was used, while batches two and three were processed using the Illumina HumanOmniExpress-12 v1.0. More information can be found in the method section of Loudursamy et al. [16] and Proitsi et al. [17]. All genotyping was performed at the Centre National de Génotypage in France.
Transcriptomics
Blood samples for the collection of gene expression data were taken at study baseline. Transcriptional profiling was performed in two batches using the Illumina HumanHT-12 v3 (batch one) and v4 (batch two) Expression BeadChip kits. Original raw data can be found in GEO1. Preprocessed raw data files, as well as post quality control, batch corrected expression values, are distributed via Synapse. The processed data underwent background correction, log base two transformation and all values were robust spline normalized [18]. Outlying samples were excluded. Batch correction was performed using ComBat [19]. All data were subset to probes that could reliably be detected in at least 80%of samples in at least one diagnostic group. More details on the processing of the data is explained in Voyle et al. [18].
Magnetic resonance imaging
1.5 Tesla T1-weighted MRI images were taken at three different timepoints throughout the study (Month 0, 3, 12). The first 3-month interval was exp-licitly chosen to contrast the 6-month MRI follow-up of ADNI and thereby evaluate if 3 months could potentially be enough to observe substantial changes in brain structure. Protocols for imaging were aligned to the ADNI study. Details on the AddNeuroMed MRI data acquisition have been described in Simmons et al. [8, 20]. ANMerge provides access to collected raw images as well as processed brain vol-umes and cortical thickness calculated using FreeSurfer 5.3 and 6.0.
Data preprocessing
As a first step, manual investigation of all raw AddNeuroMed data files was inevitable to assess the availability and state of each data type. To avoid irreproducible changes to the data, we did not alter any entry manually but relied on programming for each data changing step.
We tried to build the most informative and complete, yet minimally complex, version of AddNe-uroMed possible. Therefore, we carefully selected variables from the raw data for inclusion into AN-Merge. To limit the number of variables in ANMerge, we only included total scores of clinical assessments in the new ANMerge files instead of listing all sub-scores and individual answers. Variables not considered for inclusion into ANMerge, such as the test subscores, are accessible through the additionally provided raw data.
Not all participants from the DCR and ART cohorts underwent data collection in the course of AddNeuroMed. However, since clinical assessments between the original AddNeuroMed study and DCR were largely overlapping, we decided to include all DCR participants into ANMerge, even if they lacked other modalities apart from clinical data. From the ART cohort, only those individuals who had been assessed in at least one modality next to the clinical data were included in order to reduce sparsity in the resulting tables.
In the original AddNeuroMed data, modality spe-cific data tables lacked interoperability because distinct patient identifiers were used for many of them. Additionally, only the visit numbers were reported instead of the actual months in study. This was mis-leading due to differences in assessment intervals between diagnostic groups (e.g., visit 2 for healthy and MCI participants corresponds to visit 5 of AD patients). Information which is not subject to change (e.g., APOE genotype) was only reported at baseline which led to sparsity in follow-up visit entries. Furthermore, to increase interoperability not only within AddNeuroMed itself but also to other data resources, we mapped variable names to public data-base identifiers wherever possible. Finally, we en-riched ANMerge with data previously not available in the Synapse version. Among others, we added missing diagnoses and clinical assessment scores as well as months in study as an unambiguous time scale.
RESULTS
Overview on data
The resulting ANMerge dataset comprises four data modality specific subtables, genotype data in PLINK format and one combined table providing all preprocessed information as one. Respectively, one subtable was created for clinical data, proteo-mics, FreeSurfer calculated MRI features, and gene expression values. Next to diagnosis and clinical ass-essments, the clinical subtable also provides participants demographics, family history, and medication data.
In total, the dataset comprises information on 1,702 patients, out of which 773, 665, and 264 originated from the AddNeuroMed, DCR, and ART cohorts, respectively (Table 1). Data on 4,585 individual participant visits are reported. At study baseline, 512 participants had been diagnosed with AD, 397 with MCI, and 793 were non-cognitively impaired individuals. Table 1 describes the average characteristics of each diagnosis group at baseline. On average, cognitively affected individuals (i.e., MCI and AD) in ANMerge were 77 years old at baseline, completed 9.7 years of full-time education and 59%of them were female. Healthy individuals averaged to an age of 74.5 years, underwent 12.3 years of education and 59%are female. During study runtime 48 and 11 healthy participants converted to MCI and AD respectively. Out of all patients diagnosed with MCI at baseline 70 converted to AD.
Table 1
Summary statistics describing the ANMerge dataset at baseline
| Diagnosis | N | ANM | DCR | ART | Age (SD) | Female % | Education (SD) | APOE ∈4 positive % |
| CTL | 793 | 266 | 423 | 104 | 74.5 (6.4) | 59 | 12.3 (4.3) | 25 |
| MCI | 397 | 247 | 89 | 61 | 76.0 (6.5) | 55 | 10.0 (4.3) | 40 |
| AD | 512 | 260 | 153 | 99 | 78.6 (7.2) | 63 | 9.4 (4.3) | 54 |
| Total | 1702 | 773 | 665 | 264 | 76.4 (6.9) | 59 | 10.9 (4.5) | 39 |
N, Number of participants with the corresponding diagnosis; ANM, Number of participants originally from the AddNeuroMed study; DCR, Number of participants originally from the DCR study; ART, Number of participants originally from ART study; CTL, Healthy control participants; SD, Standard deviation.
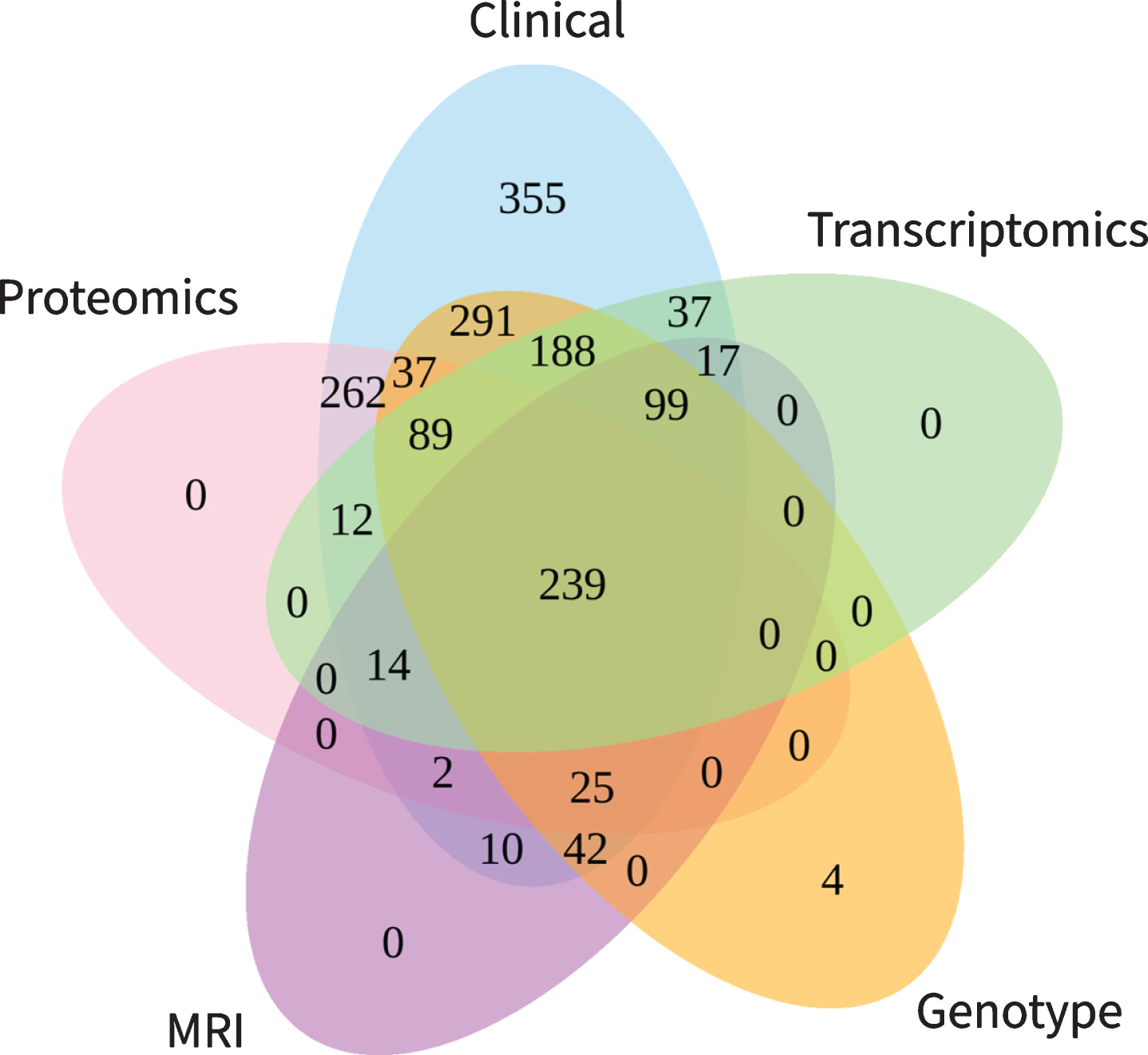
Not every study participant took part in data collection of all modalities. For our evaluation, we considered participants as represented in a modality if at least one modality specific variable was measured. This implies that not necessarily all variables of that modality were available for a given participant (e.g., an individual listed in the clinical table might have MMSE scores but no ADAS-Cog). We found that clinical data is reported for all 1,702 participants, while MRI, proteomic, gene expression, and genotype data were collected for subsets of several hundred participants each (Table 2 ‘Participants’). Figure 2 demonstrates the number of patients assessed across multiple modalities. In total, 239 participants have been assessed with regard to all five data modalities. By reducing the number of modalities included into an analysis, subsequently the number of available participants rises. For example, when conducting a multimodal study using transcriptomic, genotype and clinical variables data from 614 participants would be available. Focusing only on genotype and clinical data yields 1,010 analyzable subjects.
Table 2
Number of assessed variables and participants per modality subtables
| Modality | Participants | Variables |
| Clinical | 1,702 | 40 |
| Proteomics | 680 | 1,016 |
| MRI | 453 | 136 |
| Gene expression | 709 | 56,701 |
| Genotype | 1,014 | 789,470 |
Fig. 2
Participant overlap across modalities. The numbers illustrate the number of participants with available information for the intersection of the respective modalities.
All in all, data on more than 800,000 variables are reported in ANMerge. 40 of them correspond to the clinical modality, 56,701 originate from gene expression analysis, 136 are MRI variables, and 1,016 were assessed in blood proteomics (Table 2 ‘Variables’).
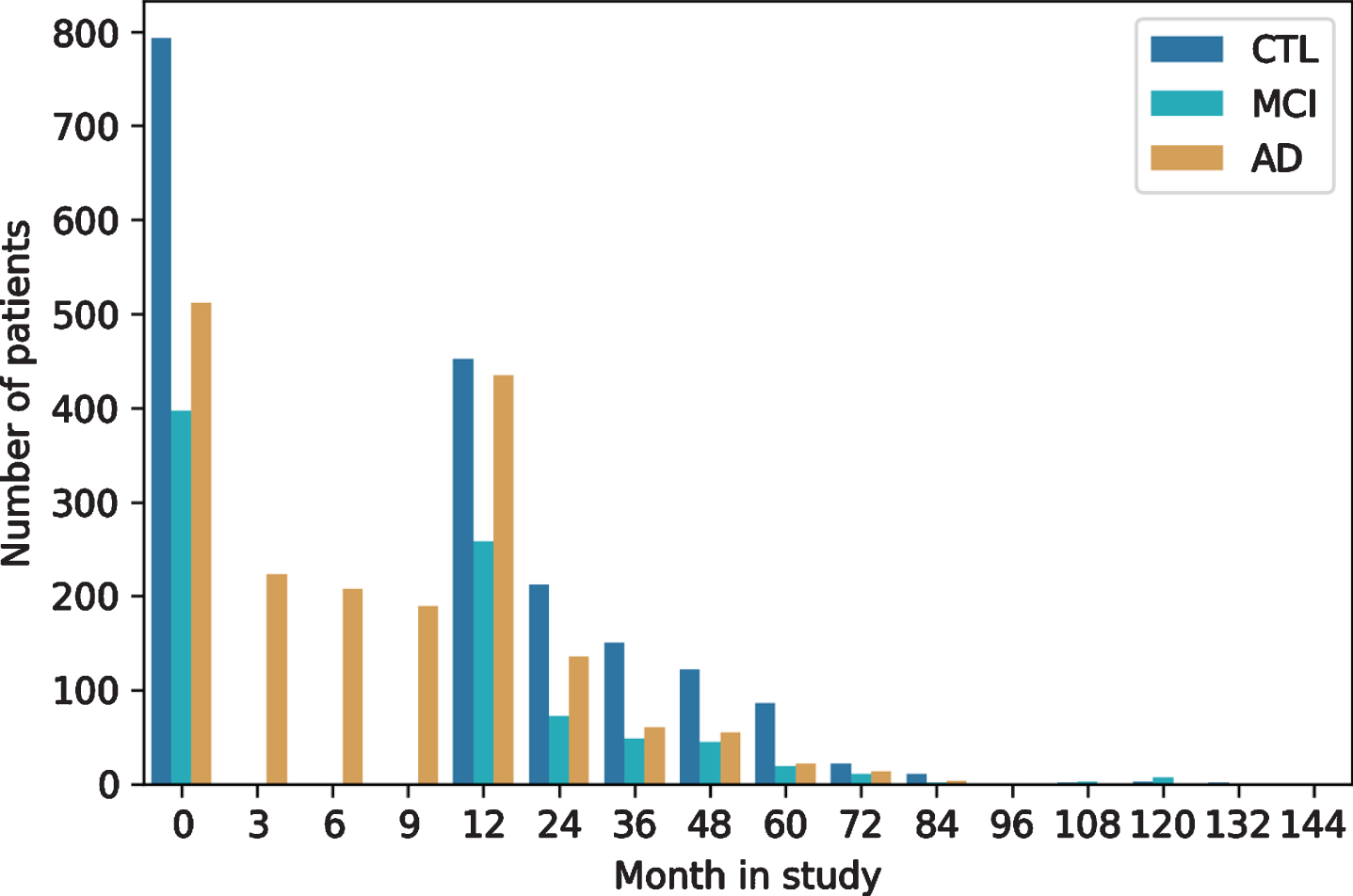
As with most clinical studies, AddNeuroMed exh-ibits a declining number of participants over study runtime (Fig. 3). For most patients (n = 1,136) at least one additional visit 12 months after baseline is available in the data. The drop of AD patients at month 3 to 9 is explained by the fact that only AD cases recruited in the original AddNeuroMed study had three monthly visits during the first year, while ART and DCR assessed all patients annually. The longest follow-up exhibited in the data spanned 12 years.
Fig. 3
Longitudinal follow-up and patient drop-out throughout study runtime per diagnosis group. CTL, healthy controls; MCI, mild cognitive impaired participants; AD, Alzheimer’s disease patients.
Data after preprocessing
The new ANMerge dataset is divided into mod-ality specific subtables which makes unimodal ana-lysis straightforward. During the preprocessing of AddNeuroMed we addressed multiple issues dete-cted in the original data. The previous version of AddNeuroMed was indexed using distinct patient identifiers across its modalities, thereby impeding mul-timodal analysis due to missing internal interoperability. Standard data integration techniques like table joins were impossible. By mapping all present identifiers to a unique one, we enabled inter-modality interoperability such that tables can now easily be analyzed together. Additionally, we provide a new identifier mapping file which helps to map the un-ified identifiers to the raw data for backwards compatibility. To increase interoperability also beyond ANMerge itself, we mapped variable names to pub-lic database identifiers. For example, proteomic variables are now also given as UniProt identifiers, genotype data is encoded as rs-numbers, and gene expression probes as Illumina IDs [21]. All of these identifiers can be easily mapped to other resources and be enriched with information from public data-bases. Instead of relying on the misleading reported visit numbers, in ANMerge we added an unambiguous time scale (months in study) to patient entries to make longitudinal follow-up easier to understand. Information that will stay permanent (e.g., APOE ɛ4 status) throughout study runtime is now reported at every visit for that respective patient, not only at baseline. Multiple issues found in the data (e.g., typos and erroneous entries) have been corrected.
Although proteomic and transcriptomic data, for example, were presented for some DCR and ART participants in the previous AddNeuroMed version, no corresponding clinical data was available, including important information like participant diagnosis. ANMerge now has all available clinical data for the two associated cohorts, which critically increases the amount of actionable information in the dataset.
Accessing ANMerge
ANMerge and the underlying data are available under https://doi.org/10.7303/syn22252881. To ensure data privacy, a straight-forward data access application has to be completed. During this access application, researchers are asked to 1) register a Synapse account, 2) have all collaborators who will access the data sign a data use certificate (DUC), 3) provide a brief research proposal (1–3 paragraphs), and 4) agree that the appropriate citation of ANMerge will be used. By signing the DUC, applicants confirm that the planned study underwent ethical review. If successful, access approval is granted within approximately 14 days.
DISCUSSION
In this work, we presented ANMerge, a longitudinal multimodal AD cohort dataset that we made accessible to the research community. Since the most time-consuming part about data analysis is often the preprocessing of data, we believe that the cumulative time save, achieved by sharing readily preprocessed datasets, can lead to faster global scientific advancement. Additionally, by describing the characteristics of the dataset in detail, we aim to enable researchers to evaluate on first sight if ANMerge is suited for their analysis.
Establishing reliable results through external validation on independent cohorts is of utmost importance, especially when dealing with high complex diseases like AD. Up to date, and to the best of our knowledge, the vast majority of data-driven appro-aches in AD relied solely on ADNI data. To validate discoveries made in ADNI on other datasets, a high overlap in measured variables is a prerequisite. Previously, we could demonstrate that despite evident differences to ADNI, ANMerge is a viable validation dataset [22].
Providing clean, preprocessed datasets is a key prerequisite to enable any data-driven AD research. However, small cohort studies, for example conducted in single hospitals, often lack the resources to provide such readily preprocessed data. In an era where data re-use beyond the initial study itself becomes increasingly important, we believe that adequate data preprocessing and sharing should resemble a planned position in the initial funding proposal for all cohort studies.
Limitations
While AddNeuroMed collected a valuable dataset, it still has some noteworthy limitations. The main limitation of the data is that the amyloid status of participants is unknown. No positron emission tomography (PET) imaging was performed and cerebrospinal fluid markers were not assessed. This difference to the ADNI data could partially explain the comparably lower number of citations of the original AddNeuroMed data.
As in many clinical cohort datasets, missing data is a considerable issue in AddNeuroMed. Not every patient was involved in the assessment of every data modality and within a modality not necessarily all variables were measured for each patient.
Compared to ADNI, AddNeuroMed lacks comprehensive documentation. Retrospectively searching for study procedures and protocols of an already concluded, older cohort study proved to be very difficult. The original study website is not available anymore and exhaustive study protocols were not findable. However, we tried to address this limitation by collecting and assembling all available information and links in this publication. While the original AddNeuroMed dataset provided descriptive data dictionaries for most clinical variables, we extent the documentation by meaningful connections of other modalities to public databases (e.g., UniProt or dbSNP) by mapping their variable names to appropriate identifiers wherever possible.
The genotype and transcriptomic data presented in ANMerge was acquired in two separate batches of participants. This implies that the data can be subject to systematic batch effects and appropriate adjustments should be made [23].
Conclusion
Over the last years, the AD field witnessed a fortunate shift to a more accessible and comprehensible data culture. New studies such as PREVENT-AD [24] and EPAD [25] recently joined the ranks of ADNI, DIAN [26], and others by making their data accessible to third party researchers. Currently running studies, for example the Deep Frequent Phenotype Study [27], already emphasized that the collected data will be published. On the metadata-level, projects such as EMIF [28] and ROADMAP [29] aimed at aiding researchers to understand the datasets in our field by providing comprehensive metadata resources. This shift in the AD data landscape toward increasingly accessible and understandable datasets marks an important development to facilitate data-driven research in the dementia domain.
By publishing ANMerge, we want to contribute to a culture of data sharing in AD research and follow the open science paradigm. Participation in observational clinical cohort studies represents an immense investment by volunteering patients and healthy individuals. They undergo extensive and sometimes intrusive repeated measurements, most of the time without any direct benefit for the individuals themselves, with the ultimate aim to contribute to disease research. We believe that it is an ethical imperative to honor their investment by enabling their data to be used for generating the most societal benefit possible.
Availability of data and materials
All data are available under: https://doi.org/10.7303/syn22252881
ACKNOWLEDGMENTS
The authors of this article wanted to thank every participant and professional involved in AddNeuroMed, DCR, and ART data collection.
This project has received partial support from the Innovative Medicines Initiative Joint Undertaking “AETIONOMY” under grant agreement #115568, resources of which are composed of financial con-tributions from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution.
This project was partially funded by an MRC Mental Health Data Pathfinder Award to the University of Oxford (MC_PC_17215), and supported by the NIHR Oxford Health Biomedical Research Centre.
The original AddNeuroMed study was supported by InnoMed (Innovative Medicines in Europe), an Integrated Project funded by the European Union of the Sixth Framework program priority FP6-2004-LIFESCIHEALTH-5, Life Sciences, Genomics, and Biotechnology for Health.
Additional support for data collection was granted by the Alzheimer’s Research Trust and from the NIHR Specialist Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust and King’s College London, Institute of Psychiatry, London, United Kingdom.
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/20-0948r2 ).
REFERENCES
[1] | Sperling RA , Jack CR Jr , Aisen PS ((2011) ) Testing the right target and right drug at the right stage. Sci Transl Med 3: , 111cm. |
[2] | Morgan AR , Touchard S , Leckey C , O’Hagan C , Nevado-Holgado AJ ; NIMA Consortium, Barkhof F , Bertram L , Blin O , Bos I , Dobricic V , Engelborghs S , Frisoni G , Frölich L , Gabel S , Johannsen P , Kettunen P , Kłoszewska I , Legido-Quigley C , Lleó A , Martinez-Lage P , Mecocci P , Meersmans K , Molinuevo JL , Peyratout G , Popp J , Richardson J , Sala I , Scheltens P , Streffer J , Soininen H , Tainta-Cuezva M , Teunissen C , Tsolaki M , Vandenberghe R , Visser PJ , Vos S , Wahlund LO , Wallin A , Westwood S , Zetterberg H , Lovestone S , Morgan BP ; Annex: NIMA–Wellcome Trust Consortium for Neuroimmunology of Mood Disorders and Alzheimer’s Disease ((2019) ) Inflammatory biomarkers in Alzheimer’s disease plasma. Alzheimers Dement 15: , 776–787. |
[3] | Mueller SG , Weiner MW , Thal LJ , Petersen RC , Jack CR , Jagust W , Trojanowski JQ , Toga AW , Beckett L ((2005) ) Ways toward an early diagnosis in Alzheimer’s disease: The Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimers Dement 1: , 55–66. |
[4] | Whitwell JL , Wiste HJ , Weigand SD , Rocca WA , Knopman DS , Roberts RO , Boeve BF , Petersen RC , Jack CR Jr ; Alzheimer Disease Neuroimaging Initiative ((2012) ) Comparison of imaging biomarkers in the Alzheimer Disease Neuroimaging Initiative and the Mayo Clinic Study of Aging. Arch Neurol 69: , 614–622. |
[5] | Fröhlich H , Balling R , Beerenwinkel N , Kohlbacher O , Kumar S , Lengauer T , Maathuis MH , Moreau Y , Murphy SA , Przytycka TM , Rebhan M , Röst H , Schuppert A , Schwab M , Spang R , Stekhoven D , Sun J , Weber A , Ziemek D , Zupan B ((2018) ) From hype to reality: Data science enabling personalized medicine. BMC Med 16: , 150. |
[6] | Lovestone S , Francis P , Strandgaard K ((2007) ) Biomarkers for disease modification trials–the innovative medicines initiative and AddNeuroMed. J Nutr Health Aging 11: , 359–361. |
[7] | Hye A , Lynham S , Thambisetty M , Causevic M , Campbell J , Byers HL , Hooper C , Rijsdijk F , Tabrizi SJ , Banner S , Shaw CE , Foy C , Poppe M , Archer N , Hamilton G , Powell J , Brown RG , Sham P , Ward M , Lovestone S ((2006) ) Proteome-based plasma biomarkers for Alzheimer’s disease. Brain 129: , 3042–3050. |
[8] | Simmons A , Westman E , Muehlboeck S , Mecocci P , Vellas B , Tsolaki M , Kłoszewska I , Wahlund LO , Soininen H , Lovestone S , Evans A , Spenger C ; AddNeuroMed Consortium ((2009) ) MRI measures of Alzheimer’s disease and the AddNeuroMed study. Ann N Y Acad Sci 1180: , 47–55. |
[9] | McKhann G , Drachman D , Folstein M , Katzman R , Price D , Stadlan EM ((1984) ) Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34: , 939–944. |
[10] | Petersen RC ((2004) ) Mild cognitive impairment as a diagnostic entity. J Intern Med 256: , 183–194. |
[11] | Hye A , Riddoch-Contreras J , Baird AL , Ashton NJ , Bazenet C , Leung R , Westman E , Simmons A , Dobson R , Sattlecker M , Lupton M , Lunnon K , Keohane A , Ward M , Pike I , Zucht HD , Pepin D , Zheng W , Tunnicliffe A , Richardson J , Gauthier S , Soininen H , Kłoszewska I , Mecocci P , Tsolaki M , Vellas B , Lovestone S ((2014) ) Plasma proteins predict conversion to dementia from prodromal disease. Alzheimers Dement 10: , 799–807. |
[12] | Morris JC , Heyman A , Mohs RC , Hughes JP , van Belle G , Fillenbaum G , Mellits ED , Clark C ((1989) ) The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer’s disease. Neurology 39: , 1159–1165. |
[13] | Kiddle SJ , Sattlecker M , Proitsi P , Simmons A , Westman E , Bazenet C , Nelson SK , Williams S , Hodges A , Johnston C , Soininen H , Kłoszewska I , Mecocci P , Tsolaki M , Vellas B , Newhouse S , Lovestone S , Dobson RJ ((2014) ) Candidate blood proteome markers of Alzheimer’s disease onset and progression: A systematic review and replication study. J Alzheimers Dis 38: , 515–531. |
[14] | Sattlecker M , Kiddle SJ , Newhouse S , Proitsi P , Nelson S , Williams S , Johnston C , Killick R , Simmons A , Westman E , Hodges A , Soininen H , Kłoszewska I , Mecocci P , Tsolaki M , Vellas B , Lovestone S ; AddNeuroMed Consortium, Dobson RJ ((2014) ) Alzheimer’s disease biomarker discovery using SOMAscan multiplexed protein technology. Alzheimers Dement 10: , 724–734. |
[15] | Gold L , Ayers D , Bertino J , Bock C , Bock A , Brody EN , Carter J , Dalby AB , Eaton BE , Fitzwater T , Flather D , Forbes A , Foreman T , Fowler C , Gawande B , Goss M , Gunn M , Gupta S , Halladay D , Heil J , Heilig J , Hicke B , Husar G , Janjic N , Jarvis T , Jennings S , Katilius E , Keeney TR , Kim N , Koch TH , Kraemer S , Kroiss L , Le N , Levine D , Lindsey W , Lollo B , Mayfield W , Mehan M , Mehler R , Nelson SK , Nelson M , Nieuwlandt D , Nikrad M , Ochsner U , Ostroff RM , Otis M , Parker T , Pietrasiewicz S , Resnicow DI , Rohloff J , Sanders G , Sattin S , Schneider D , Singer B , Stanton M , Sterkel A , Stewart A , Stratford S , Vaught JD , Vrkljan M , Walker JJ , Watrobka M , Waugh S , Weiss A , Wilcox SK , Wolfson A , Wolk SK , Zhang C , Zichi D ((2010) ) Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5: , e15004. |
[16] | Lourdusamy A , Newhouse S , Lunnon K , Proitsi P , Powell J , Hodges A , Nelson SK , Stewart A , Williams S , Kloszewska I , Mecocci P , Soininen H , Tsolaki M , Vellas B , Lovestone S ; AddNeuroMed Consortium, Dobson R , Alzheimer’s Disease Neuroimaging Initiative ((2012) ) Identification of cis-regulatory variation influencing protein abundance levels in human plasma. Hum Mol Genet 21: , 3719–3726. |
[17] | Proitsi P , Lupton MK , Velayudhan L , Newhouse S , Fogh I , Tsolaki M , Daniilidou M , Pritchard M , Kloszewska I , Soininen H , Mecocci P , Vellas B ; Alzheimer’s Disease Neuroimaging Initiative, Williams J ; GERAD1 Consortium, Stewart R , Sham P , Lovestone S , Powell JF ((2014) ) Genetic predisposition to increased blood cholesterol and triglyceride lipid levels and risk of Alzheimer disease: A Mendelian randomization analysis. PLoS Med 11: , e1001713. |
[18] | Voyle N , Keohane A , Newhouse S , Lunnon K , Johnston C , Soininen H , Kloszewska I , Mecocci P , Tsolaki M , Vellas B , Lovestone S , Hodges A , Kiddle S , Dobson RJ ((2016) ) A pathway based classification method for analyzing gene expression for Alzheimer’s disease diagnosis. J Alzheimers Dis 49: , 659–669. |
[19] | Johnson WE , Li C , Rabinovic A ((2007) ) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: , 118–127. |
[20] | Simmons A , Westman E , Muehlboeck S , Mecocci P , Vellas B , Tsolaki M , Kłoszewska I , Wahlund LO , Soininen H , Lovestone S , Evans A , Spenger C ((2011) ) The AddNeuroMed framework for multi-centre MRI assessment of Alzheimer’s disease: Experience from the first 24 months. Int J Geriatr Psychiatry 26: , 75–82. |
[21] | Du P , Kibbe WA , Lin SM ((2008) ) lumi: A pipeline for processing Illumina microarray. Bioinformatics 24: , 1547–1548. |
[22] | Birkenbihl C , Emon MA , Vrooman H , Westwood S , Lovestone S ; AddNeuroMed Consortium, Hofmann-Apitius M , Fröhlich H , Alzheimer’s Disease Neuroimaging Initiative ((2020) ) Differences in cohort study data affect external validation of artificial intelligence models for predictive diagnostics of dementia - lessons for translation into clinical practice. EPMA J 11: , 367–376. |
[23] | Benito M , Parker J , Du Q , Wu J , Xiang D , Perou CM , Marron JS ((2004) ) Adjustment of systematic microarray data biases. Bioinformatics 20: , 105–114. |
[24] | Tremblay-Mercier J , Madjar C , Das S , Dyke SO , Étienne P , Lafaille-Magnan M , Bellec P , Collins DL , Rajah MN , Bohbot VD , Leoutsakos J , Iturria-Medina Y , Kat J , Hoge RD , Gauthier S , Chakravarty MM , Poline J , Rosa-Neto P , Villeneuve S , Evans AC , Poirier J , Breitner JCS , the PREVENT-AD Research Group ((2020) ) Creation of an open science dataset from PREVENT-AD, a longitudinal cohort study of pre-symptomatic Alzheimer’s disease. bioRxiv; doi: https://doi.org/10.1101/2020.03.04.976670 |
[25] | Solomon A , Kivipelto M , Molinuevo JL , Tom B , Ritchie CW EPAD Consortium ((2019) ) European Prevention of Alzheimer’s Dementia Longitudinal Cohort Study (EPAD LCS): Study protocol. BMJ Open 8: , e021017. |
[26] | Morris JC , Aisen PS , Bateman RJ , Benzinger TL , Cairns NJ , Fagan AM , Ghetti B , Goate AM , Holtzman DM , Klunk WE , McDade E , Marcus DS , Martins RN , Masters CL , Mayeux R , Oliver A , Quaid K , Ringman JM , Rossor MN , Salloway S , Schofield PR , Selsor NJ , Sperling RA , Weiner MW , Xiong C , Moulder KL , Buckles VD ((2012) ) Developing an international network for Alzheimer research: The Dominantly Inherited Alzheimer Network. Clin Investig (Lond) 2: , 975–984. |
[27] | Koychev I , Lawson J , Chessell T , Mackay C , Gunn R , Sahakian B , Rowe JB , Thomas AJ , Rochester L , Chan D , Tom B , Malhotra P , Ballard C , Chessell I , Ritchie CW , Raymont V , Leroi I , Lengyel I , Murray M , Thomas DL , Gallacher J , Lovestone S ((2019) ) Deep and Frequent Phenotyping study protocol: An observational study in prodromal Alzheimer’s disease. BMJ Open 9: , e024498. |
[28] | Oliveira JL , Trifan A , Bastião Silva LA ((2019) ) EMIF Catalogue: A collaborative platform for sharing and reusing biomedical data. Int J Med Inform 126: , 35–45. |
[29] | Gallacher J , de Reydet de Vulpillieres F , Amzal B , Angehrn Z , Bexelius C , Bintener C , Bouvy JC , Campo L , Diaz C , Georges J , Gray A , Hottgenroth A , Jonsson P , Mittelstadt B , Potashman MH , Reed C , Sudlow C , Thompson R , Tockhorn-Heidenreich A , Turner A , van der Lei J , Visser PJ , ROADMAP Consortium ((2019) ) Challenges for optimizing real-world evidence in Alzheimer’s disease: The ROADMAP Project. J Alzheimers Dis 67: , 495–501. |




