Tocotrienol Rich Fraction Supplementation Modulate Brain Hippocampal Gene Expression in APPswe/PS1dE9 Alzheimer’s Disease Mouse Model
Abstract
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder characterized by loss of memory and other cognitive abilities. AD is associated with aggregation of amyloid-β (Aβ) deposited in the hippocampal brain region. Our previous work has shown that tocotrienol rich fraction (TRF) supplementation was able to attenuate the blood oxidative status, improve behavior, and reduce fibrillary-type Aβ deposition in the hippocampus of an AD mouse model. In the present study, we investigate the effect of 6 months of TRF supplementation on transcriptome profile in the hippocampus of APPswe/PS1dE9 double transgenic mice. TRF supplementation can alleviate AD conditions by modulating several important genes in AD. Moreover, TRF supplementation attenuated the affected biological process and pathways that were upregulated in the AD mouse model. Our findings indicate that TRF supplementation can modulate hippocampal gene expression as well as biological processes that can potentially delay the progression of AD.
INTRODUCTION
Alzheimer’s disease (AD) is defined by a chronic progressive neurodegenerative disorder that is characterized by brain deterioration leading to progressive impairment of intellectual, cognition, behavior, and memory [1]. Accumulation and aggregation of amyloid-β (Aβ) peptide in brain tissue is known to be the main etiological factor for AD [2, 4]. Although there is still some controversy, the amyloid cascade hypothesis formulated in the early 1990s has been a dominant model for AD pathogenesis [5, 6]. This hypothesis suggested that an increment of abnormal extracellular Aβ level in the brain can cause aggregation of Aβ into β-sheet-rich structures [7]. Increasing evidence shows that Aβ and hyperphosphorylated tau protein alteration involved at the cellular and molecular levels contribute to neurodegeneration in AD [8–11]. The hippocampus brain region is the most affected region with AD. Many neurobiological alterations in the hippocampus have been found to be associated with cognitive impairment [12–17].
Numerous studies have highlighted the potential of tocotrienol rich fraction (TRF) as the active compounds that assist in preventing many diseases including neurodegenerative disease and are thus worthy of further scientific investigation. TRF is a different mixture of vitamin E that consists of a combination of α-, β-, γ-, and δ-tocotrienol and α-tocopherol [18]. TRF can be found in a variety of plants and vegetable oils (soybean, corn, sesame, peanuts, and cotton) while tocotrienols are also available in oil from rice, barley, wheat germ, and rye, as well as palm oil [19, 20]. TRF is claimed to possess high antioxidant [21–25], anticancer [26–30], anti-aging [31, 32], and anti-diabetic [33, 34] properties, show neuroprotective effects [35–39], attenuate the blood oxidative status [40], and improve behavior and reduce Aβ deposition in the hippocampus [41, 42]. Therefore, in the present study, we sought to determine the effects of TRF on the hippocampus in an AD mouse model and describe its potential for management of AD.
MATERIALS AND METHODS
Chemicals
TRF (Golden Tri™ E 70) was purchased from Sime Darby (Kuala Lumpur, Malaysia) and consisted of 24% α-tocopherol (αT), 27% α-tocotrienol (αT3), 4% β-tocotrienol (βT3), 32% γ-tocotrienol (γT3), and 14% δ-tocotrienol (δT3) in every gram of TRF.
Animals
Double transgenic male mouse B6C3-Tg (APPswe, PS1dE9) 85Dbo/Mmjax with C57BL/6J genetic background was purchased from The Jackson Laboratory (Bar Harbor, ME, USA). The AD mouse model expressed the mutant amyloid precursor protein (Mo/HuAPP659swe) and mutant presenilin 1 (PS1dE9), both of which were associated with the early-onset of AD [43]. All mice were placed in a specific-pathogen-free condition and maintained individually in polycarbonate cages on a 12:12 h light-dark cycle with light started at 7:00 am and 24 h ventilation. All equipment and materials such as cages, corn cobs, food pellets, and drink containers were sterilized using UV light and autoclaved before use. This study was approved by the UKM Animal Ethics Committee (FP/BIOK/2015/HANAFI/9-DEC./716-DEC.-2015;FEB.-2017) and all animal works were conducted in accordance with national and international guidelines.
Supplementation and sample collection
Nine-month-old animals were divided into a wild type (WT) group (n = 4), control group of AD transgenic mice (n = 4), and AD transgenic mice receiving TRF supplementation (200 mg/kg, n = 4). TRF dosage was based on the previously published work [40, 44, 45]. Animals received daily supplementation via oral gavages for 6 months. At the end of the treatment period, all mice were sacrificed by cervical dislocation and the hippocampus was collected using brain matrix for microarray analysis.
RNA extraction and purification
Total RNA was extracted from the brain hippocampus region using Tri Reagent (Sigma Aldrich) according to manufacturer’s guidelines and RNA clean-up was performed using RNeasy spin columns (Qiagen, USA). Total RNA from each sample was quantified by the NanoDrop 1000 Spectrophotometer (Wilmington, DE) and RNA integrity was assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, USA) using RNA 6000 Nano LabChip ® kit.
Microarray and pathway analysis
For microarray analysis, GeneChip mouse transcriptome assay 1.0 (MTA 1.0) from Affymetrix, Inc. was used to detect the expression profiles of mRNA which contains 23,000 mouse mRNAs. The raw data was analyzed using Expression Console (EC) from Affymetrix, Inc. The gene expression was analyzed using Partek Genomics Suite (Partek GS). Significantly, differentially expressed genes (DEG) were identified by analysis of variance (ANOVA) and false discovery rate (FDR) that of less than 5%. Then, the DEG with≥1.5-fold change was defined as up regulation or down regulation. Principal component analysis (PCA) was performed to show the distinction between samples and hierarchical clustering was carried out to distinguish the mRNA expression profile between the control and the treatment group. Pathway analysis was performed to explore the significant pathway of the DEG according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) using the software Transcriptome Analysis Console (TAC) from Affymetrix, Inc.
Quantitative real time RT-PCR (qRT-PCR)
Randomized genes were validated via quantitative real time PCR (qRT-PCR) to confirm the results of microarray analysis. Total RNA (100 ng) was isolated using Tri Reagents (Sigma Aldrich). The one-step RT-PCR kit was obtained from Qiagen using the manufacturer’s protocol. Briefly, total RNA was mixed with a cocktail of RT and PCR reaction buffers, including 400μM of each dNTP, 1.5 mM MgCl2, and 1μM of the gene-specific primer pairs, Gapdh: 5’ TGGTGAAGCAGGCATCTGAG -3’(sense), 5’ TGGTGTTGAAGTCGCAGGAG-3’ (antisense) and a one-step RT-PCR enzyme mix (Omniscript and Sensiscript RTs and HotStartTaq DNA polymerase). The thermal cycler conditions consisted of RT at 50°C for 30 min, the initial PCR activation step at 95°C for 15 min, and a three-step cycling (total number of cycles 40), denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min. The aliquots (10μl) of the reaction were then run in 2% agarose gels for visualization of the products.
Statistical analysis
Data was expressed as mean±SEM and was determined using GraphPad Prism 5 Software (GraphPad Software, San Diego, CA, USA). p value≤0.05 was considered statistically significant.
RESULTS
Principle component analysis
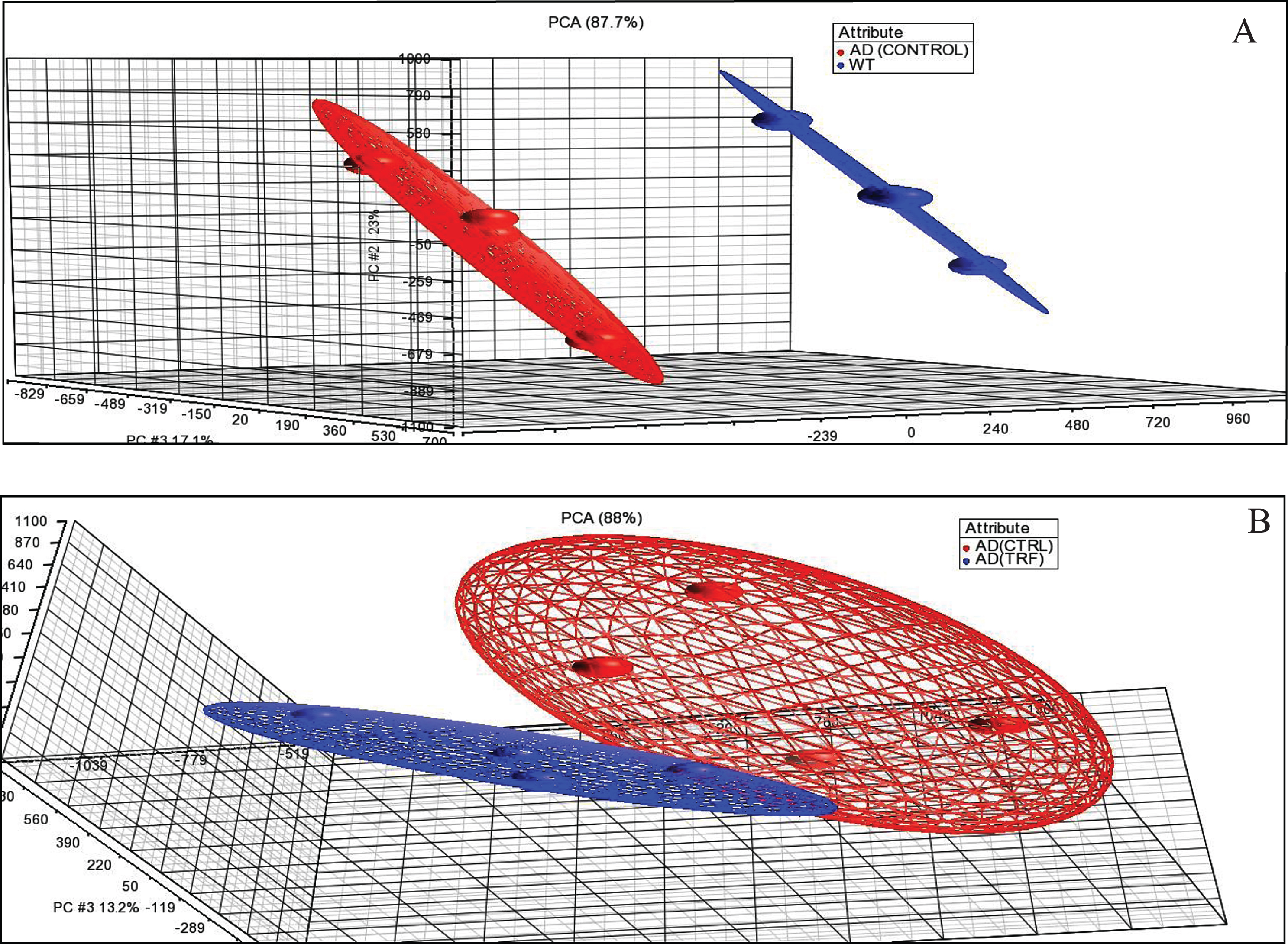
Through the PCA analysis, the sample expression profile was in similar condition as the other groups. This PCA showed the samples that were in the same group gather closer to each other than the samples in different groups. Figure 1A shows the PCA between samples of control group of AD with WT group. Figure 1B shows the PCA between samples with 6-month supplementation of TRF and the control group with AD. They showed a similar expression profile when compared to others in the same group and this unity of PCA implies that all variance is common or shared.
Fig. 1.
Principal component analysis (PCA). A) AD (Control) group compared to wild type (WT) group. B) AD (TRF) group compared to AD (Control) group.
Hierarchical clustering analysis
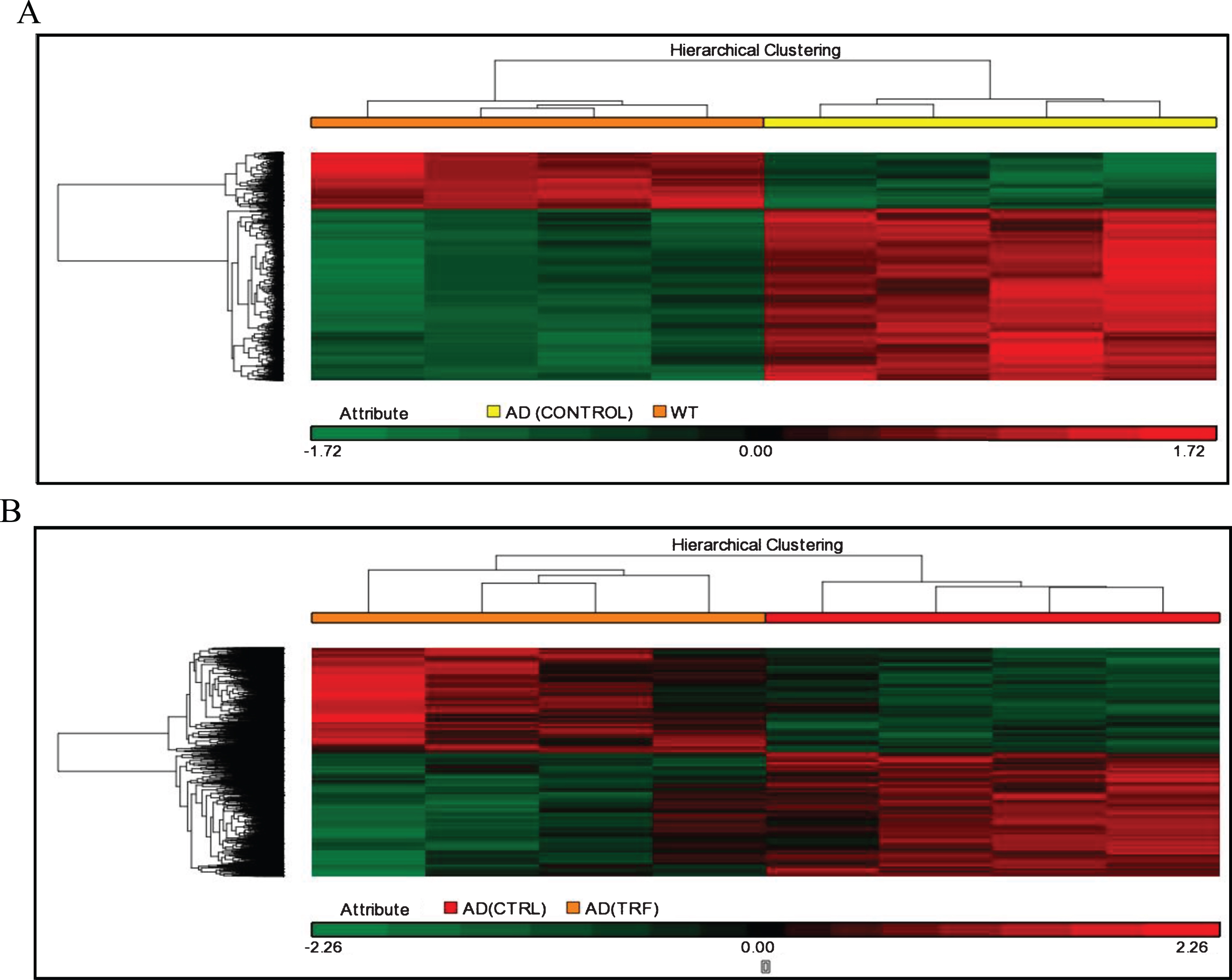
The next analysis involved analysis of hierarchical clusters based on the group using Person Centered metric distance with centroid network regulation over the collection of filtered genes based on expression. The unsupervised hierarchical clustering analysis was used to investigate the possibility of identifying different groups based on their molecular expression profile rather than morphological features. The separated cluster indicates that the datasets were significantly different between groups. Hierarchical clustering in the control group of AD compared to the WT group and in the group of TRF compared to the control group of AD showed that most of those with a similar expression profile were joined together to form a group (Fig. 2A, B).
Fig. 2.
Hierarchical clustering analysis. A) AD (Control) group compared to wild type (WT) group. B) AD (TRF) group compared to AD (Control) group.
Differential expression genes
Once the sample passed the above quality control, the steps to identify different gene expressions were performed. Through a one-way ANOVA test at p < 0.05 with fold change > ±1.5, the significant yield of DEGs was 36,323 genes for comparison between the control group of AD and the WT group. While 6,679 genes were regulated between TRF group and control group of AD in hippocampus (Table 1). A total of 20 lists of the highest gene that significantly regulated in the comparable group using ANOVA statistical analysis of one way at p < 0.05 and multiply variables > ±1.5 was adjusted. Table 2 showed the list of top 20 regulated genes between the control group of AD and the WT group. Table 3 shows the list after a 6-month supplementation of TRF in the hippocampus brain region compared to the control group of AD. Among the top 20 genes in the hippocampus, gene analysis of the AD mouse model exhibited downregulated genes of Sorbs1(p = 1.61E-04; FC = –19.342) and Unc80 (p = 6.82E-05; FC = –7.365), and upregulated genes of Lpcat2 (p = 1.53E-06; FC = 3.211) and Havcr2 (p = 4.96E-06; FC = 2.394). In contrast, TRF supplementation significantly upregulated the hippocampal gene expression of Slc24a2 (p = 4.84E-04;FC = 1.535), Exo1(p = 1.58E-04;FC = 2.107), and Enox1(p = 5.27E-04;FC = 2.079) but downregulated Pla2g4a (p = 1.61E-03; FC = –1.747), Tfap2b (p = 8.13E-04; FC = –2.622), and Oip5 (p = 9.73E-04;FC = –2.134) genes.
Table 1
Comparison of transcriptome changes in hippocampus
| Differentially expressed genes | Upregulated genes | Downregulated genes | |
| AD (Control) versus WT | 36,323 | 26,710 | 9613 |
| AD (TRF) versus AD (Control) | 6679 | 3064 | 3615 |
Table 2
List of top 20 regulated genes of AD (Control) compared to wild type (WT) group
| Group/No. | AD (Control) versus WT | |||||
| Upregulated | Downregulated | |||||
| Gene Symbol | p-value | FC | Gene Symbol | p-value | FC | |
| 1 | Tbc1d31 | 3.50E-07 | 1.79141 | Mut | 2.67E-06 | –1.50786 |
| 2 | Zfp426 | 6.87E-07 | 2.21531 | Grem2 | 1.67E-05 | –1.56731 |
| 3 | Lpcat2 | 1.53E-06 | 3.21152 | Ncor2 | 2.67E-05 | –1.70539 |
| 4 | Havcr2 | 4.96E-06 | 2.39496 | Zswim4 | 3.80E-05 | –5.67512 |
| 5 | Rpl26 | 5.35E-06 | 1.81951 | Fer1l6 | 4.81E-05 | –2.25303 |
| 6 | Htr7 | 7.27E-06 | 1.74825 | Plekhg5 | 5.23E-05 | –1.82099 |
| 7 | Clcn1 | 7.88E-06 | 1.57086 | Tbc1d8 | 5.33E-05 | –2.17651 |
| 8 | Idh1 | 4.49E-05 | 1.63123 | Ncoa5 | 5.89E-05 | –1.50567 |
| 9 | Wdr17 | 8.77E-06 | 1.68763 | Copz2 | 6.44E-05 | –4.5788 |
| 10 | Mgat5 | 1.40E-05 | 1.72288 | Unc80 | 6.82E-05 | –7.36583 |
| 11 | Calcrl | 1.96E-05 | 1.573 | Nfkbid | 8.58E-05 | –1.75874 |
| 12 | Dnajc27 | 2.14E-05 | 1.55246 | Cd72 | 9.06E-05 | –1.90163 |
| 13 | Pgap1 | 2.34E-05 | 3.44235 | Tectb | 9.49E-05 | –2.34042 |
| 14 | Hpgds | 2.43E-05 | 3.22743 | Neurod4 | 9.53E-05 | –1.52969 |
| 15 | Uba3 | 2.49E-05 | 1.94474 | Nav1 | 9.65E-05 | –1.64426 |
| 16 | Aif1 | 2.60E-05 | 2.49363 | Fnta | 0.000146 | –5.1144 |
| 17 | Vps35 | 2.79E-05 | 1.8234 | Mink1 | 0.000149 | –1.63021 |
| 18 | ATM | 3.70E-05 | 2.22508 | Sorbs1 | 0.000161 | –19.3424 |
| 19 | Atp7a | 4.16E-05 | 2.08218 | Iqgap2 | 0.000168 | –5.65503 |
| 20 | Slc14a1 | 4.20E-05 | 1.59435 | Syne1 | 0.000173 | –5.5011 |
Table 3
List of top 20 regulated genes after 6 months supplementation of TRF
| Group/No. | AD (TRF) versus AD (Control) | |||||
| Upregulated | Downregulated | |||||
| Gene Symbol | p-value | FC | Gene Symbol | p-value | FC | |
| 1 | Lekr1 | 2.42E-05 | 1.73768 | Irx6 | 8.13E-05 | –1.52715 |
| 2 | Edc4 | 3.26E-05 | 1.63423 | Cyth2 | 0.00011 | –1.97821 |
| 3 | Marco | 4.52E-05 | 2.17188 | Spns3 | 0.000122 | –1.89363 |
| 4 | Fam189b | 7.25E-05 | 1.98532 | Brip1 | 0.000246 | –1.52473 |
| 5 | Epb41 | 0.000144 | 1.7128 | Usp37 | 0.000301 | –1.79451 |
| 6 | Exo1 | 0.000158 | 2.10773 | Trdn | 0.000431 | –2.04844 |
| 7 | Sik3 | 0.000198 | 1.50578 | Grin1 | 0.00046 | –1.63866 |
| 8 | Poc1b | 0.000263 | 1.51857 | Bpi | 0.000707 | –1.76336 |
| 9 | Otoa | 0.000429 | 1.67172 | Als2cl | 0.000709 | –1.6858 |
| 10 | Sik2 | 0.000431 | 1.50532 | Tfap2b | 0.000813 | –2.62277 |
| 11 | Cacna1i | 0.000441 | 1.90664 | Col16a1 | 0.000893 | –1.63068 |
| 12 | Wac | 0.000448 | 1.7586 | Sag | 0.000937 | –1.87902 |
| 13 | Pds5a | 0.000471 | 1.58806 | Ckmt1 | 0.000937 | –1.54843 |
| 14 | Slc24a2 | 0.000484 | 1.53536 | Oip5 | 0.000973 | –2.13417 |
| 15 | Enox1 | 0.000527 | 2.07988 | Ust | 0.001076 | –1.83303 |
| 16 | Chst2 | 0.000603 | 1.90556 | Pkp3 | 0.001338 | –2.14347 |
| 17 | Itgal | 0.000674 | 1.86292 | Polr1a | 0.001463 | –1.80335 |
| 18 | Muc5b | 0.000744 | 1.50761 | Lama4 | 0.001567 | –1.54965 |
| 19 | Ccnb2 | 0.000799 | 1.69749 | Pla2g4a | 0.001614 | –1.7476 |
| 20 | Tsta3 | 0.000828 | 1.58963 | Rnf17 | 0.001621 | –1.51101 |
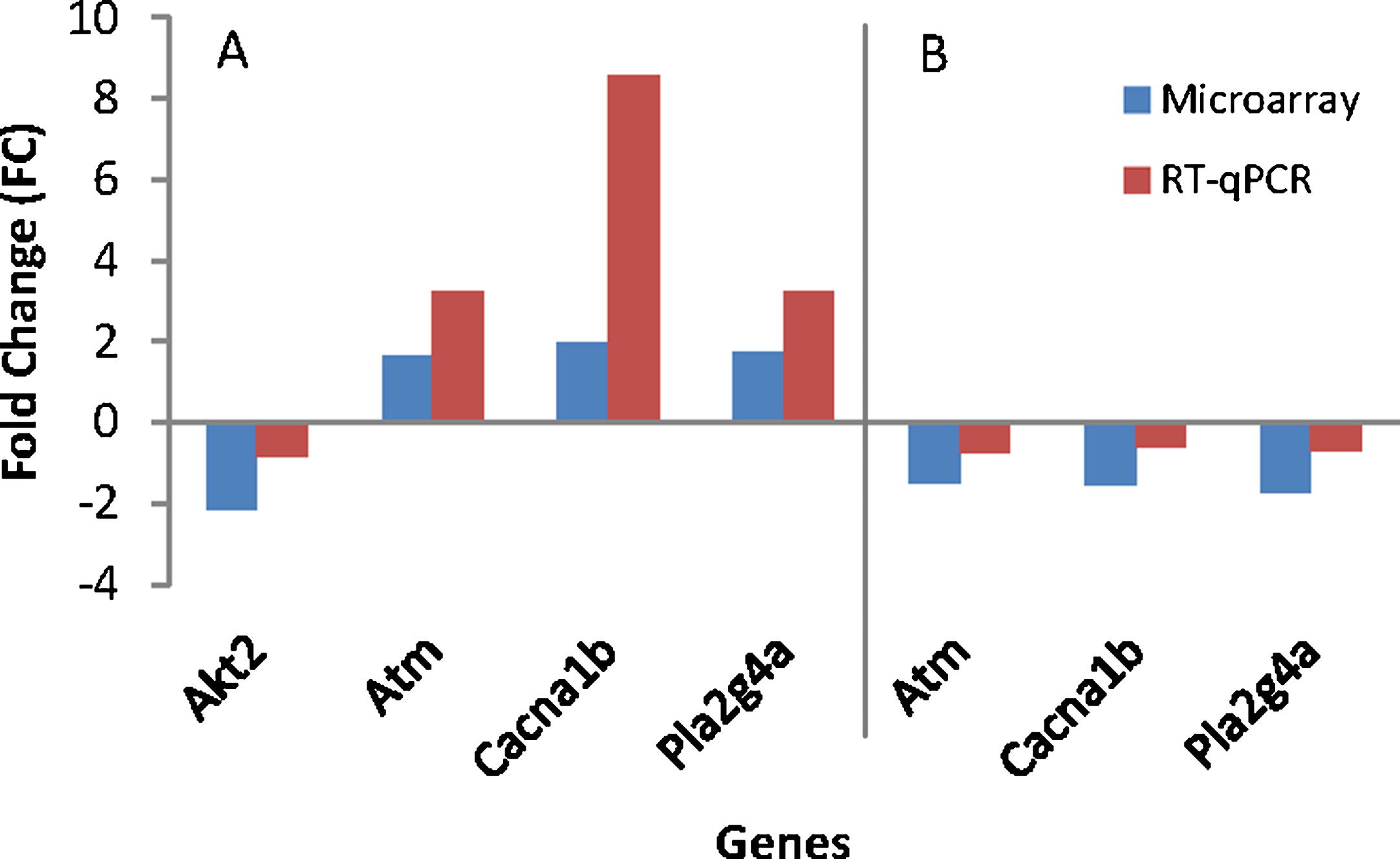
To assess differences between treatments, the effect from the study was evaluated by comparing outcomes for the treated and the untreated group. Table 6 shows discrimination of modulated genes between the AD (Control) and the WT group and also between AD (TRF) and AD (Control). The RT-qPCR analysis confirmed the four-gene expression found to significantly difference in microarray (Fig. 4). These genes include randomly selected Akt2, Atm, Cacna1b, and Pla2g4a. All these genes showed the same expression in both techniques of microarray and RT-qPCR.
Biological process or pathway analysis
According to biological process or pathway analysis, KEGG pathway analysis (p < 0.05, fold change > 1.2) revealed that TRF supplementation significantly downregulated the genes in the hippocampus that are involved in biological processes and pathways such as mRNA processing, Focal Adhesion-PI3K-Akt-mTOR signaling, EGFR1 signaling, p53 signaling, T cell receptor signaling, TNF-alpha NF-kB signaling, Alzheimer’s disease, and MAPK signaling pathways. It is important to note that most of the biological processes and pathways affected by TRF were upregulated in the AD mouse model (Tables 4 and 5). Figure 3 shows an Alzheimer’s disease pathway with regulated genes after a 6-month supplementation of TRF.
Table 4
List of significant biological process or pathways of AD (Control) compared to Wild Type (WT) group in hippocampus brain region. Symbol (↓) indicated the fold change of downregulated genes and symbol of (↑) indicated the fold change of upregulated genes
| Biological process/ Pathways | Total | ↑ | Up list (↑) | ↓ | Down list (↓) |
| Focal Adhesion-PI3K-Akt-mTOR-Signaling Pathway | 18 | 16 | Itgam,Osmr,Itgav,Itgb1,Itgb5,Angpt1,Fgf14,Hgf,Fgfr2, Kit,Pdgfra,Gsk3b,Casp9,Slc2a3,Hmgcr,Acaca | 2 | Ifna4,Chrm1 |
| EGFR1 Signaling Pathway | 13 | 10 | Stat1,Stat2,Eps15,Fos, Rps6ka3,Ctnnd1,Pak1, Casp9,Rps6ka5,Smad2 | 3 | Elk1,Ndufa13,Ptk2b |
| MAPK Signaling Pathway | 13 | 11 | Tgfbr1,Mapk6,Map4k4,Map2k4,Stmn1,Rps6ka3, FosMapk9,Pak1,Casp9,Map3k5 | 2 | Elk1,Dusp4 |
| B Cell Receptor Signaling Pathway | 12 | 8 | Blnk,Casp9,Fcgr2b,Gsk3b, Atp2b4,Ccnd2,Stat1,Rasgrp3 | 4 | Elk1,Ptk2b,Pip5k1b,Dusp4 |
| IL-6 Signaling Pathway | 8 | 7 | Il6st,Stat1,Map2k4,Fos,Casp9,Ppp2r2a,Gsk3b | 1 | Ptk2b |
| Alzheimer’s Disease | 12 | 9 | App,Ncstn,Mme,Lpl,Casp9,Capn2,Gsk3b, Nae1,Casp12 | 3 | Grin2a,Cacna1c,Bad |
| T Cell Receptor Signaling Pathway | 7 | 6 | Stat1,Pak1,Tubb5,Dlg1,Fos,Stk39 | 1 | Ptk2b |
| TNF-alpha NF-kB Signaling Pathway | 7 | 6 | Psmd12,Psmd1,Stat1,Gsk3b,Ktn1,Rps6ka5 | 1 | Iqgap2 |
| Wnt Signaling Pathway and Pluripotency | 7 | 7 | Fzd4,Gsk3b,Mapk9,Ccnd2,Apc,Ctnnd1,Ppp2r2a | – | – |
| p53 Signaling | 5 | 5 | Ccnd2,Casp9,Rrm2b,Sesn3,Ccng1 | – | – |
| Oxidative Stress | 4 | 4 | Fos,Maoa,Gsr,Gpx1 | – | – |
| Parkinson’s Disease Pathway | 3 | 3 | Lrrk2,Mapk11,Casp9 | – | – |
Table 5
List of significant biological process or pathways after 6 months supplementation of TRF on Alzheimer’s disease mouse model in hippocampus brain region. Symbol (↓) indicated the fold change of downregulated genes and symbol of (↑) indicated the fold change of upregulated genes
| Biological process/ Pathways | Total | ↑ | Up list (↑) | ↓ | Down list (↓) |
| mRNA Processing | 13 | 3 | Pcbp2,Pcbp3,Rnu12 | 10 | Snrpb2,Cdc40,Son,Rbm7,Adarb2,Rbmx,Tardbp,Ttc14,Ddx5,Eif4a2 |
| B Cell Receptor Signaling Pathway | 6 | 1 | Ccna2 | 5 | Actr2,Actr3,Arpc2,Nck1,Ppp3ca |
| Focal Adhesion-PI3K-Akt-mTOR-Signaling Pathway | 5 | 2 | Ifna7,Il2 | 3 | Osmr,Tek,Pfkfb2 |
| EGFR1 Signaling Pathway | 3 | – | – | 3 | Nck1,Ralb,Snca |
| p53 Signaling | 3 | – | – | 3 | Siah1b,Ccng2,Ppm1d |
| T Cell Receptor Signaling Pathway | 2 | – | – | 2 | Nck1,Skap2 |
| TNF-alpha NF-kB Signaling Pathway | 2 | – | – | 2 | Hdac2,Iqgap2 |
| Alzheimer’s Disease | 6 | 2 | Cacna1c,Bad | 4 | Ppp3ca,Snca,Nae1,Casp12 |
| Wnt Signaling Pathway NetPath | 1 | – | – | 1 | Fzd1 |
| MAPK Signaling Pathway | 1 | – | – | 1 | Ppp3ca |
| Wnt Signaling Pathway | 1 | – | – | 1 | Fzd1 |
| Parkinson’s Disease Pathway | 1 | – | – | 1 | Snca |
Table 6
Selected identified genes for discrimination between AD (Control) and Wild Type (WT) group and between AD (TRF) and AD (Control). ↑, Upregulated; ↓, Downregulated
| Genes | Description | AD (Control) versus WT | AD (TRF) versus AD (Control) | ||
| p | Fold change | p | Fold change | ||
| App | Amyloid beta precursor protein | 0.011 | ↑(1.82) | 0.353 | ↓(–1.02) |
| Adam10 | A disintegrin and metallopeptidase domain 10 | 0.407 | ↑(1.24) | 0.221 | ↓ (–1.23) |
| Adam17 | A disintegrin and metallopeptidase domain 17 | 0.006 | ↑(1.37) | 0.316 | ↓(–1.35) |
| Psen1 | Presenilin 1 | 0.046 | ↑(1.17) | 0.404 | ↑(1.02) |
| Psen2 | Presenilin 2 | 0.647 | ↑(1.04) | 0.874 | ↓(–1.04) |
| Psenen | Presenilin enhancer gamma secretase subunit | 0.459 | ↑(1.19) | 0.397 | ↑(1.36) |
| Ncstn | Nicastrin | 0.024 | ↑(1.61) | 0.574 | ↓(–1.14) |
| Aph1a | Aph1 homolog A, gamma secretase subunit | 0.133 | ↑(1.14) | 0.453 | ↓(–1.11) |
| Bace1 | Beta-site APP cleaving enzyme 1 | 0.238 | ↑(1.15) | 0.607 | ↑(1.11) |
| Abca7 | ATP-binding cassette, sub-family A (ABC1), member 7 | 0.748 | ↑(1.03) | 0.180 | ↑(1.08) |
| Ache | Acetylcholinesterase | 0.065 | ↑(2.25) | 0.425 | ↓(–1.07) |
| Bin1 | Bridging integrator 1 | 0.516 | ↑(1.17) | 0.575 | ↓(–1.09) |
| Mapt | Microtubule-associated protein tau | 0.537 | ↑(1.14) | 0.914 | ↑(1.06) |
| Sorl1 | Sortilin-related receptor, LDLR class A repeats-containing | 0.310 | ↑(1.40) | 0.349 | ↓(–1.32) |
| Sptbn4 | Spectrin beta, non-erythrocytic 4 | 0.455 | ↑(1.09) | 0.546 | ↓(–1.04) |
| Apoe | Apolipoprotein E | 0.040 | ↑(1.18) | 0.545 | ↓(–1.09) |
Fig. 3.
Alzheimer’s disease pathway with regulated genes after 6 months’ supplementation of TRF. *The green box indicates downregulated genes and the red box indicates upregulated genes.
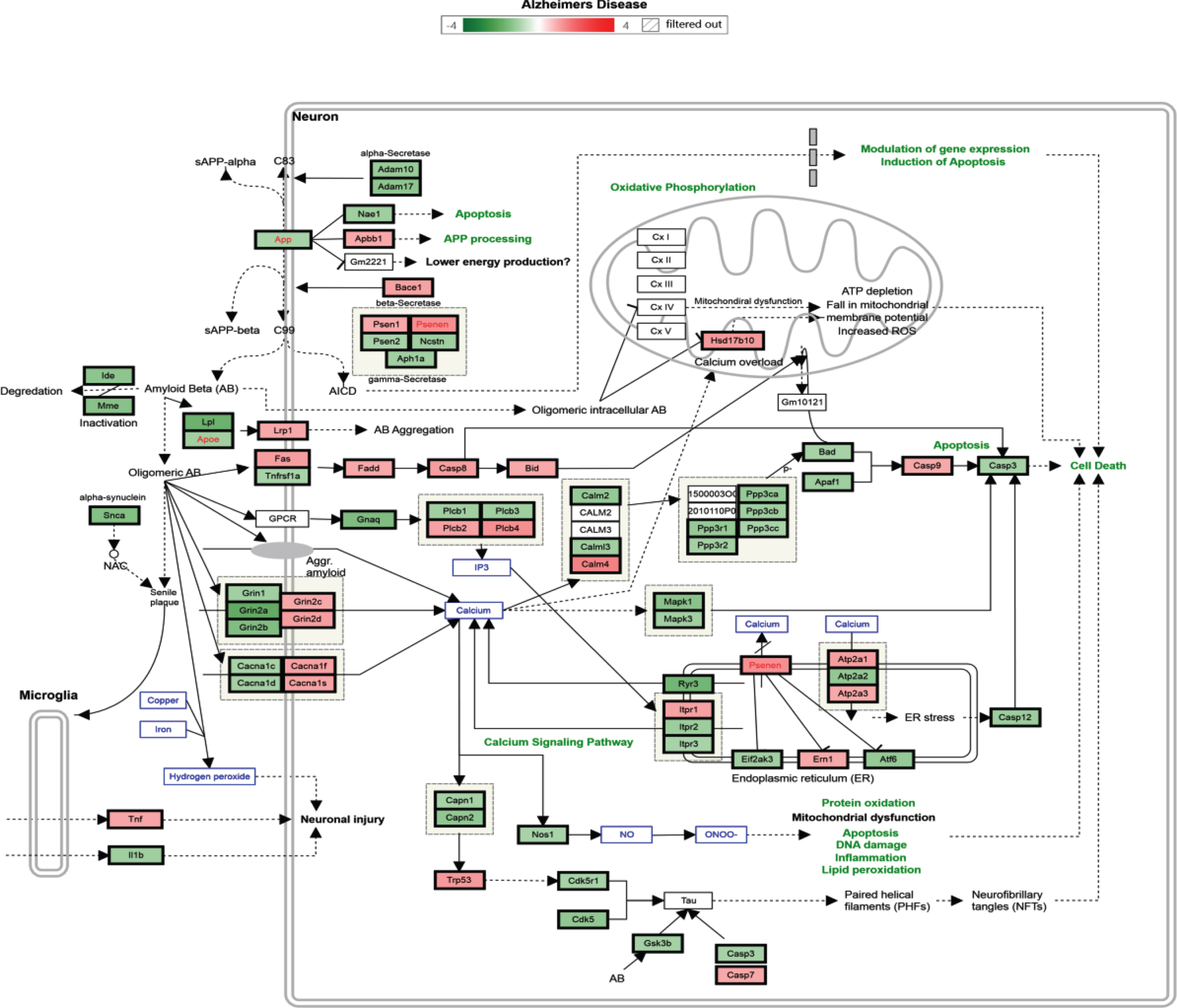
Fig. 4.
Gene’s validation using RT-qPCR. A) AD (Control) group compared to wild type (WT) group. B) AD (TRF) group compared to AD (Control) group.
DISCUSSION
Neuronal gene profiling studies have an enormous impact on our understanding of the pathology of several neurodegenerative diseases, including AD, Parkinson’s disease, schizophrenia, multiple sclerosis, and Huntington’s disease by providing identification of novel regulators of neuronal gene expression, gene functions, and also by defining the biological process or pathways that interplay to promote the neurodegenerative diseases. TRF has gained great attention as a potential therapeutic agent to combat or delay neurodegenerative disease due to its wide biological properties. From this microarray data, we suggest that most of the regulated genes were linked to AD in the hippocampus. We also found that the susceptible genes and pathways in AD were alleviated through 6 months of TRF supplementation.
Regulated genes of AD control group parallel to AD condition
Among regulated genes, Sorbs1 (Sorbin and SH3 domain containing 1) are reported to be important in the development of neuromuscular synapse and activation of nerve growth factors in neuronal differentiation [46, 47] and Unc80 (Unc-80, NALCN activator) that are involved in neuron excitability were inhibited in AD (Control). While Lpcat2 (Lysophosphatidylcholine acyltransferase 2) genes are involved in cell inflammation and neuropathic pain [49, 50] and Havcr2 (Hepatitis A virus cellular receptor 2) genes related to tumors [51] increased. Additionally, elevation of biological processes and pathways of Focal Adhesion-P13k-Akt-mTOR signaling pathway, MAPK signaling pathway, Alzheimer’s disease pathway, TNF-alpha NF-kB signaling pathway, p53 signaling, oxidative stress, and Parkinson’s disease pathway showed that regulated genes were parallel to the conditions found in AD [52–55].
TRF supplementation alleviated the AD conditions
Being rich in so many key benefits, research has uncovered that the most top-modulated genes by TRF are involved in alleviating several neurological diseases. After 6 months of TRF supplementation, the gene of Slc24a2 (Solute carrier family 24 [sodium/potassium/ calcium exchanger], member 2) that plays a physiological role in neuronal plasticity, acts as a neuroprotective during ischemic brain injury, and is also associated with biological processes critical to vascular dementia has been upregulated. Slc24a2 is the most abundant member of the calcium/cation antiporter superfamily of transport protein that present in the brain and important in Ca (2+) homeostasis [56–58]. A study on Slc24a2 knockout mice caused deficits in motor learning and spatial working memory in the hippocampus [59]. Therefore, TRF supplementation potentially improves neuroprotective protein and cognition in the AD mouse model.
The exo1 (Exonuclease 1) gene is important in the cleavage step of DNA mismatch repair. Studies on mice reported that inactivation of Exo1 caused defects in DNA, which then increased the susceptibility to cancer and infertility [60]. Exo1 was also proven to act as a critical mediator of survival during DNA double-stranded break repair in hematopoietic stem and progenitor cells [61]. Exo1 is also required to activate the reaction response to S (N) 1 DNA methylating agents [62]. Thus, TRF supplementation helps to stabilize the DNA process.
While Enox1 (Ecto-NOX disulfide-thiol exchanger 1) is involved in the pathway of electron transport of plasma membrane and the activity of hydroquinone oxidation (NADH) and also in conversion of disulfide-thiol protein [63], Nuclear-1 respiratory factor (NRF-1), associated in neurite growth in neuroblastoma cells, positively controls the Enox1 in human IMR-32 cells neuroblastoma and primary mice cortical neurons [64]. Thus, speculation can be made that TRF supplementation acts as an antioxidant that helps to stabilize the activity of NADH and disulfide-thiol.
For the significantly downregulated gene of Pla2g4a (Phospholipase A2, group IVA [cytosolic, calcium-dependent]), this gene plays a crucial role in neurological disease pathology as Pla2g4a is reported to play a role in elevated AβPP protein expression induced by aggregated Aβ1 - 42 in cortical neurons and in signal events leading to AβPP induction. The protein encoded by this gene is a member of the phospholipase A2 group IV family. Via protein kinase A pathway, accumulation of Aβ induced elevation of AβPP protein expression mediated by activation of Pla2g4a, Pge2 release, and Creb [65]. As known, oxidative stress and inflammation are important factors contributing to pathophysiology, overexpression of Pla2g4a leads to microglial activation and causes a neuroinflammatory condition in the brain [66]. Thus, further studies on targeted inhibition of Pla2g4a will be valuable in investigating the therapeutic potential for AD.
For the gene of Tfap2b (Transcription factor AP-2 beta), research by Rossello et al. in 2012, reported that AP-2b regulates Aβ protein stimulation of apolipoprotein E transcription in astrocytes. AP-2b contributed to the elevation of Aβ induction in ApoE overflow [67]. In addition, AP-2b is a genetic candidate in the cause of adipocyte hypertrophy and associated with abnormal adipocyte in obesity [68]. Thus, TRF supplementation helps in the reduction of Aβ induction in ApoE overloaded.
An increased level of Oip5 (Opa interacting protein 5) was found during the G1 phase of cell proliferation and upon cell cycle exit in quiescence, senescence, and differentiation. Studies also reported that this gene leads to subsequent cell death [69]. Oip5 also promotes accumulation of pre-mature and mature adipocytes and leads to adipose hyperplasia [70]. Upon TRF supplementation, this gene was suppressed, which helps to alleviate the adverse events above.
TRF supplementation attenuated the affected biological process or pathway in AD
Malfunction in signaling of cellular and molecular networks is the root cause disturbing the signaling processes and leading to production of disturbance or abnormal proteins involved in neurodegenerative disease. The signaling pathway itself was inappropriate in an AD brain, causing extensive neuronal and synaptic degradation with an active inflammatory response, apoptosis, and ultimately loss of function of neurons and brain tissue [71–73].
We performed biological process and pathway analysis to forecast the biological function of modulated genes and potential roles of the differentially expressed genes after 6 months of TRF supplementation in an AD mouse model. When using biological process and pathway analysis, we discovered that these regulated genes were mainly associated with mRNA processing followed by B cell receptor signaling pathway, focal adhesion-PI3K-Akt-mTOR signaling, EGFR1 signaling, p53 signaling, T cell receptor signaling, TNF-alpha NF-kB signaling, Alzheimer’s disease pathway, Wnt signaling pathway, MAPK signaling pathway, and Parkinson’s disease pathway. For the mRNA processing gene of Pcbp2 (Poly (rc) binding protein 2), this gene showed promising therapeutic effects by TRF as this gene may increase the p73 expression. p73 is a member of the p53 family tumor suppressors, plays a critical role in tumor suppression and neuronal development. Increased expressions of p73 subsequently slow down the ROS production and cellular senescence [74]. A study by Zhang and his colleagues in 2016 also showed that overexpression of Pcbp2 rescued the inflammation by Meis1 gene effects in Akt-mTOR pathway [75].
In addition, the mutation of Tardbp (TAR DNA binding protein) gene caused neurological disease of amyotrophic lateral sclerosis and there were supporting studies showing that the inhibition of Tardbp mitochondrial localization blocked its neuronal toxicity. In addition, suppression of Tardbp alleviated motor-coordinative and cognitive dysfunction, reduced neuroinflammation, prevented neuronal loss, and restored mitochondrial functions [76, 77]. Thus, TRF supplementation is able to improve neuronal functions in an AD mouse model.
The regulated pathway of focal adhesion-PI3K-Akt-mTOR signaling pathway, TNF-alpha NF-kB signaling pathway, T cell receptor signaling pathway, MAPK signaling pathway, and Wnt signaling pathway have been well studied in the literature for their complicated impact on inflammation and AD. Deviation from strict control of the above pathways has been implicated in the progression of AD. Neuroinflammation plays critical roles in the pathogenesis of AD. PI3K-Akt-mTOR signaling pathway is often associated in crosstalk with mTOR and NF-kB that were reported to play a vital role in neurodegenerative disease by regulating cell growth, proliferation, and differentiation [78–80]. TNF-α that secreted mainly in microglial cells in response to the abnormal accumulation of Aβ protein give the chronic response by elevating its level in the AD brain as well as in the brain of transgenic mouse models of AD [81–84]. Several pro-inflammatory mediators or cytokines are commanded by transcription factor NF-κB. NF-κB that stimulates microglials is a ubiquitous transcription factor found in almost all animal cell types. NF-κB regulates the expression of many cytokines and chemokines, such as interferons, interleukins, lymphokines, and tumor necrosis factors [85]. Inhibition of NF-κB signaling became the popular approach among researchers in order to prevent or delay the onset of many diseases. Many natural products or polyphenol from plants targeted the NF-κB pathway to modulate the pro-inflammatory mediators such as curcumin, resveratrol, capsaicin, and apigenin [86–88].
Mitogen-activated protein kinases (MAPKs) are serine-threonine kinases that mediate intracellular signaling associated with a variety of cellular responses and biological process in reaction to a stress signal. MAPK family consists of extracellular signal-regulated kinase (ERK), p38, and c-Jun NH2-terminal kinase (JNK). In AD patients, vulnerable neurons activate MAPK pathways and are involved in pathogenesis of AD [89–91]. The Wnt signaling pathway is a protein network associated with embryo development and cancer progression [92–94]. The Wnt pathway believed to be involved in promoting the survival, proliferation, differentiation, and migration of cells in many different types of tissues, including nervous tissue, as well as in synapse formation in the nervous system. Regulation of β-Catenin levels is a major step in Wnt signaling [95]. Dysfunction of the Wnt signaling pathway is implicated in the pathophysiology of neuronal degeneration of AD [96, 97]. Thus, TRF supplementation potentially causes suppression of neuroinflammation in an AD mouse model.
Oxidative stress has been implicated in the pathogenesis of AD [98, 99]. Previous studies suggested there was involvement of p53 in degenerating neurons in AD. p53 levels are largely regulated in response to injury by changes in protein degradation. Data on the expression of p53 found a significant increase of p53 levels in both MCI and AD [100]. Abundant p53-immunoreactive neurites and glial cell processes also appeared to be associated with neurogenerative diseases [101]. Hence p53 inhibition may protect neuron cells from neurotoxicity insults in AD [102]. From the result, TRF supplementation may inhibit the activity of p53 signaling, consequently relieving the oxidative stress in AD.
EGRF1 signaling was downregulated by TRF supplementation. EGF receptors are reportedly involved in activating pathways that promote cell proliferation, independence, migration, epithelial tissue distinction, fibroblasts, and endothelial cells in which EGF biological activity depends on its binding on receptors [103]. In squamous oral carcinoma (OSCC) cells, EGFR is reportedly overexpressed and associated with tumor invasion and metastasis and deterioration of patient survival rate [104, 105]. Deficiency of EGFR is reported to inhibit tumor cell migration and suppress EMT (epithelial-to-mesenchymal transition) in human ovarian cancer cells [106]. In addition, Sylvester et al. (2001) also demonstrated that tocotrienol was more potent than tocopherol in preventing mitogenesis in normal mammary epithelial cells (extracted from BALB / c pregnant-mid mice) by reducing EGF-receptor activity (EGFR) [107]. From the result, TRF supplementation may ameliorate the effect of antagonizing EGRF1 signaling.
Moreover, TRF supplementation modulated the biological process of AD by deregulated genes of Ppp3ca, SNCA, Nae1, Casp12. Protein phosphatase 3, catalytic subunit, and alpha isoform (Ppp3ca) are confirmed to interact with NF-κB-inducing kinase. This interaction activates NF-κB pathways, thereby regulating a wide variety of immune system functions. Calcineurin is a serine/threonine protein phosphatase including a catalytic subunit (CnA) and regulatory subunit (CnB), which participates in calcium ion-dependent signal transduction pathways. Calcineurin activation also promotes apoptosis of glomerular podocytes in diabetic nephropathy. In Parkinson’s disease, activation of calcineurin and NFAT dependent pathway are involved in α-synuclein induced degeneration of midbrain dopaminergic neurons [108–111].
SNCA alpha-synuclein is a member of the synuclein family, which also includes beta- and gamma-synuclein. Synucleins are abundantly expressed in the brain and alpha- and beta-synuclein inhibit phospholipase D2 selectively. SNCA play causal roles in the pathogenesis of Parkinson disease that lead to neural degeneration [112, 113]. SNCA peptide is also reported to be a major component of amyloid plaques in the brains of patients with AD [114–116].
NEDD8 Activating Enzyme E1 Subunit 1 (Nae1) also known as amyloid-β Precursor Protein-Binding Protein 1 (APPBP1) is a protein encoded by this gene, which binds to AβPP. It is reported to play a role in the pathogenesis of AD. This protein is required for cell cycle progression through the S/M checkpoint. Mutation of this gene marked in disruption in the proliferation of fetal neural stem cells [117]. A study on transgenic mice that express Swedish/Dutch/Iowa mutant AβPP in the brain showed that processing of β-secretase is accelerated and the Aβ peptide production in AD brain is enhanced [118–120].
Caspases 12 (Casp12) is a cysteine protease that cleaves C-terminal aspartic acid residues on their substrate molecules. Caspase-12 antagonizes the inflammasome and NF-κB [121]. Casp12 often is associated with the progression of AD because it promotes the neuronal death through apoptosis. Casp12 reported mediation of endoplasmic reticulum-specific apoptosis and cytotoxicity by aggregation of Aβ. In addition, neuronal cells treated with caspase inhibitors are resistant to attack by Aβ [122–124].
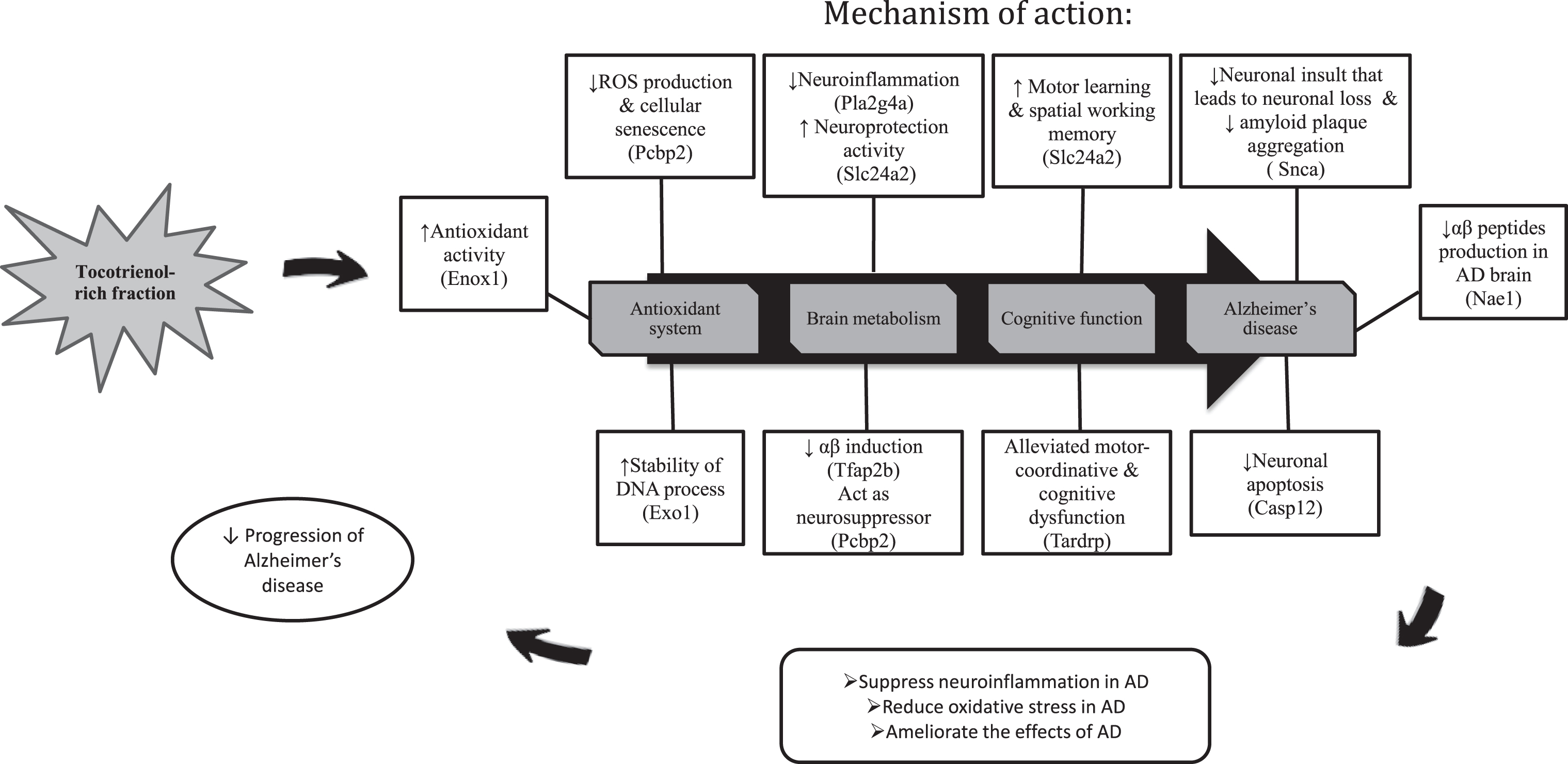
In a nutshell, the results were astounding and the therapeutic effects of TRF were uncovered through transcriptome profiling in the hippocampus of AβPPswe/PS1dE9 double transgenic mice. TRF successfully halted the AD conditions by modulating several genes and also attenuated the affected biological process and pathways in AD (Fig. 5). Therefore, we proposed that TRF supplementation could be a promising therapeutic agent for delaying the progression of AD.
Fig. 5.
The summary on the effects of TRF supplementation on the gene expression, biological process and pathways in the hippocampus of AD mouse model.
ACKNOWLEDGMENTS
This study was supported by Geran Universiti Penyelidikan (GUP) from Universiti Kebangsaan Malaysia (GUP-2015-045) and Long-Term Research Grant Scheme (LRGS) from the Ministry of Higher Education, Malaysia (LRGS/BU/2012/UKM-UKM/K/04).
Authors’ disclosures available online (https://www.j-alz.com/manuscript-disclosures/18-0496r1).
REFERENCES
[1] | Grand JH , Caspar S , MacDonald SW ((2011) ) Clinical features and multidisciplinary approaches to dementia care. J Multidiscip Healthc 4: , 125–147. |
[2] | Hardy JA , Higgins GA ((1992) ) Alzheimer’s disease: The amyloid cascade hypothesis. Science 256: , 184–185. |
[3] | Rosenberg RN ((2005) ) Translational research on the way to effective therapy for Alzheimer disease. Arch Gen Psychiatry 62: , 1186–1192. |
[4] | Veerhuis R ((2011) ) Histological and direct evidence for the role of complement in the neuroinflammation of AD. Curr Alzheimer Res 8: , 34–58. |
[5] | Hardy J , Allsop D ((1991) ) Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol Sci 12: , 383–388. |
[6] | Selkoe DJ , Hardy J ((2016) ) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8: , 595–608. |
[7] | Ding F , Borreguero JM , Buldyrey SV , Stanley HE , Dokholyan NV ((2003) ) Mechanism for the α-helix to ß-hairpin transition. Proteins 53: , 220–228. |
[8] | Wang W-X , Rajeev BW , Stromberg AJ , Ren N , Tang G , Huang Q , Rigoutsos I , Nelson PT ((2008) ) The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of ß-site amyloid precursor protein-cleaving enzyme 1. JNeurosci 28: , 1213–1223. |
[9] | Takeda S , Sato N , Rakugi H , Morishita R ((2011) ) Molecular mechanisms linking diabetes mellitus and Alzheimer disease: Beta-amyloid peptide, insulin signaling, and neuronal function. Mol Biosyst 7: , 1822–1827. |
[10] | Sorrentino G , Bonavita V ((2007) ) Neurodegeneration and Alzheimer’s disease: The lesson from tauopathies. Neurol Sci 28: , 63–71. |
[11] | Cortes N , Andrade V , Guzman-Martinez L , Estrella M , Maccioni RB ((2018) ) Neuroimmune tau mechanisms: Their role in the progression of neuronal degeneration. Int J Mol Sci 19: , 956. |
[12] | Nowotny P , Kwon JM , Goate AM ((2001) ) Alzheimer disease. In: eLS. John Wiley & Sons Ltd, Chichester. http://www.els.net [doi: 10.1038/npg.els.0000228] |
[13] | Munder T , Pfeffer A , Schreyer S , Guo J , Braun J , Sack I , Steiner B , Klein C ((2018) ) MR elastography detection of early viscoelastic response of the murine hippocampus to amyloid ß accumulation and neuronal cell loss due to Alzheimer’s disease. J Mag Reson Imaging 47: , 105–114. |
[14] | Gallagher M , Bizon JL , Hoyt EC , Helm KA , Lund PK ((2003) ) Effects of aging on the hippocampal formation in a naturally occurring animal model of mild cognitive impairment. Exp Gerontol 38: , 71–77. |
[15] | Fellgiebel A , Wille P , Muller MJ , Winterer G , Scheurich A , Vucurevic G , Schmidt LG , Stoeter P ((2004) ) Ultrastructural hippocampal and white matter alterations in mild cognitive impairment: A diffusion tensor imaging study. Dement Geriatr Cog Disord 18: , 101–108. |
[16] | Duyckaerts C , Potier M-C , Delatour B ((2008) ) Alzheimer disease models and human neuropathology: Similarities and differences. Acta Neuropathol 115: , 5–38. |
[17] | Bizon JL , Helm KA , Han JS , Chun HJ , Pucilowska J , Lund PK , Gallagher M ((2001) ) Hypothalamic-pituitary-adrenal axis function and corticosterone receptor expression in behaviourally characterized young and aged Long-Evans rats. Eur J Neurosci 14: , 1739–1751. |
[18] | Chin S-F , Hamid NAA , Latiff AA , Zakaria Z , Mazlan M , Yusof YAM , Karim AA , Ibahim J , Hamid Z , Ngah WZW ((2008) ) Reduction of DNA damage in older healthy adults by Tri E Tocotrienol supplementation. Nutrition 24: , 1–10. |
[19] | Eitenmiller RR , Lee J ((2004) ) Vitamin E: Food chemistry, composition, and analysis. CRC Press. |
[20] | Sen CK , Khanna S , Roy S ((2007) ) Tocotrienols in health and disease: The other half of the natural vitamin E family. Mol Aspects Med 28: , 692–728. |
[21] | Taib IS , Budin SB , Ghazali AR , Jayusman PA , Louis SR , Mohamed J ((2015) ) Palm oil tocotrienol-rich fraction attenuates testicular toxicity induced by fenitrothion via an oxidative stress mechanism. Toxicol Res 4: , 132–142. |
[22] | Sen CK , Khanna S , Roy S ((2006) ) Tocotrienols: Vitamin E beyond tocopherols. Life Sci 78: , 2088–2098. |
[23] | McIntyre BS , Briski KP , Gapor A , Sylvester PW ((2000) ) Antiproliferative and apoptotic effects of tocopherols and tocotrienols on preneoplastic and neoplastic mouse mammary epithelial cells (44544). Proc Soc Exp Biol Med 224: , 292–301. |
[24] | Gugliandolo A , Bramanti P , Mazzon E ((2017) ) Role of vitamin E in the treatment of Alzheimer’s disease: Evidence from animal models. Int J Mol Sci 18: , 2504. |
[25] | Chin S-F , Ibahim J , Makpol S , Hamid NAA , Latiff AA , Zakaria Z , Mazlan M , Yusof YAM , Karim AA , Ngah WZW ((2011) ) Tocotrienol rich fraction supplementation improved lipid profile and oxidative status in healthy older adults: A randomized controlled study. Nutr Metab 8: , 42. |
[26] | Zhang J-S , Zhang S-J , Li Q , Liu Y-H , He N , Zhang J , Zhou P-H , Li M , Guan T , Liu J-R ((2015) ) Tocotrienol-rich fraction (TRF) suppresses the growth of human colon cancer xenografts in Balb/C nude mice by the Wnt pathway. PLoS One 10: , e0122175. |
[27] | Srivastava JK , Gupta S ((2006) ) Tocotrienol-rich fraction of palm oil induces cell cycle arrest and apoptosis selectively in human prostate cancer cells. Biochem Biophys Res Commun 346: , 447–453. |
[28] | Rahman AA , Makpol S , Jamal R , Harun R , Mokhtar N , Ngah WZW ((2014) ) Tocotrienol-rich fraction,[6]-gingerol and epigallocatechin gallate inhibit proliferation and induce apoptosis of glioma cancer cells. Molecules 19: , 14528–14541. |
[29] | Hafid SRA , Radhakrishnan AK , Nesaretnam K ((2010) ) Tocotrienols are good adjuvants for developing cancer vaccines. BMC Cancer 10: , 5. |
[30] | Hafid SRA , Chakravarthi S , Nesaretnam K , Radhakr-ishnan AK ((2013) ) Tocotrienol-adjuvanted dendritic cells inhibit tumor growth and metastasis: A murine model of breast cancer. PLoS One 8: , e74753. |
[31] | Makpol S , Yeoh TW , Ruslam FAC , Arifin KT , Yusof YAM ((2013) ) Comparative effect of Piper betle, Chlorella vulgaris and tocotrienol-rich fraction on antioxidant enzymes activity in cellular ageing of human diploid fibroblasts. BMC Complement Altern Med 13: , 210. |
[32] | Makpol S , Durani LW , Chua KH , Yusof M , Anum Y , Ngah W , Zurinah W ((2011) ) Tocotrienol-rich fraction prevents cell cycle arrest and elongates telomere length in senescent human diploid fibroblasts. J Biomed Biotechnol (2011) . |
[33] | Lee H , Lim Y ((2018) ) Tocotrienol-rich fraction supplementation reduces hyperglycemia-induced skeletal muscle damage through regulation of insulin signaling and oxida-tive stress in type 2 diabetic mice. J Nutr Biochem 57: , 77–85. |
[34] | Budin SB , Othman F , Louis SR , Bakar MA , Das S , Mohamed J ((2009) ) The effects of palm oil tocotrienol-rich fraction supplementation on biochemical parameters, oxidative stress and the vascular wall of streptozotocin-induced diabetic rats. Clinics 64: , 235–244. |
[35] | Musa I , Khaza'ai H , Mutalib MSA , Yusuf F , Sanusi J , Chang SK ((2017) ) Effects of oil palm tocotrienol rich fraction on the viability and morphology of astrocytes injured with glutamate. Food Biosci 20: , 168–177. |
[36] | Mazlan M ((2010) ) Comparison of the effects of α-tocopherol and γ-tocotrienol against oxidative stress in two different neuronal cultures. Sains Malays 39: ,145–156. |
[37] | Khanna S , Parinandi NL , Kotha SR , Roy S , Rink C , Bibus D , Sen CK ((2010) ) Nanomolar vitamin E α-tocotrienol inhibits glutamate-induced activation ofphospholipase A2 and causes neuroprotection. J Neurochem 112: ,1249–1260. |
[38] | Elsy B , Khan AA , Maheshwari V ((2017) ) Neuroprotective effects of d-á-tocotrienol rich fraction on crushed sciatic nerve in diabetic rats. Eur J Pharm Med Res 4: , 489–500. |
[39] | Aan GJ , Adi MM , Hairi HA ((2013) ) Tocotrienol rich fraction (TRF) increases viability of senescent fibroblast. Res Updates Med Sci 1: , 37–40. |
[40] | Damanhuri H , Rahim NA , Nasri W , Tan J , Makpol S , Ngah W , Mazlan M , Tooyama I ((2016) ) Tocotrienol-rich fraction supplementation modulates antioxidant enzymes activity and reduces DNA damage in APP-swe/PS1dE9 Alzheimer’s disease mouse model (suplementasi fraksi kaya tokotrienol memodulasi aktiviti enzim antioksidan dan mengurangkan kerosakan DNA pada APPswe/PS1dE9 model mencit penyakit Alzheimer). Sains Malays 45: , 1363–1370. |
[41] | Ibrahim NF , Yanagisawa D , Durani LW , Hamezah HS , Damanhuri HA , Ngah W , Zurinah W , Tsuji M , Kiuchi Y , Ono K ((2017) ) Tocotrienol-rich fraction modulates amyloid pathology and improves cognitive function in AßPP/PS1 mice. J Alzheimers Dis 55: , 597–612. |
[42] | Durani LW , Hamezah HS , Ibrahim NF , Yanagisawa D , Nasaruddin ML , Mori M , Azizan KA , Damanhuri HA , Makpol S , Ngah W ((2018) ) Tocotrienol-rich fraction of palm oil improves behavioral impairments and regulates metabolic pathways in AßPP/PS1 mice. J Alzheimers Dis 64: , 249–267. |
[43] | Jankowsky JL , Slunt HH , Ratovitski T , Jenkins NA , Copeland NG , Borchelt DR ((2001) ) Co-expression of multiple transgenes in mouse CNS: A comparison of strategies. Biomol Eng 17: , 157–165. |
[44] | Taridi NM , Abd Rani N , Abd Latiff A , Wan Ngah WZ , Mazlan M ((2014) ) Tocotrienol rich fraction reverses age-related deficits in spatial learning and memory in aged rats. Lipids 49: , 855–869. |
[45] | Mazlan M , Hamezah HS , Taridi NM , Jing Y , Liu P , Zhang H , Ngah W , Zurinah W , Damanhuri HA ((2017) ) Effects of aging and tocotrienol-rich fraction supplementation on brain arginine metabolism in rats. Oxi Med Cell Longev 2017: , 6019796. |
[46] | Limpert AS , Karlo JC , Landreth GE ((2007) ) Nerve growth factor stimulates the concentration of TrkA within lipid rafts and extracellular signal-regulated kinase activation through c-Cbl-associated protein. Mol Cell Biol 27: , 5686–5698. |
[47] | Hallock PT , Chin S , Blais S , Neubert TA , Glass DJ ((2016) ) Sorbs1 and-2 interact with CrkL and are required for acetylcholine receptor cluster formation. Mol Cell Biol 36: , 262–270. |
[48] | Lu B , Su Y , Das S , Wang H , Wang Y , Liu J , Ren D ((2009) ) Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature 457: , 741. |
[49] | Shindou H , Shiraishi S , Tokuoka SM , Takahashi Y , Harayama T , Abe T , Bando K , Miyano K , Kita Y , Uezono Y ((2017) ) Relief from neuropathic pain by blocking of the platelet-activating factor-pain loop. FASEB J 31: , 2973–2980. |
[50] | Morimoto R , Shindou H , Oda Y , Shimizu T ((2010) ) Phosphorylation of lysophosphatidylcholine acyltransferase 2 at Ser34 enhances platelet-activating factor production in endotoxin-stimulated macrophages. J Biol Chem 285: , 29857–29862. |
[51] | Gujar R , Maurya N , Yadav V , Gupta M , Arora S , Khatri N , Sen P ((2016) ) c-Src suppresses dendritic cell antitumor activity via T cell Ig and mucin protein-3 receptor. J Immunol 197: , 1650–1662. |
[52] | Wang Q , Fan H , Liu Y , Yin Z , Cai H , Liu J , Wang Z , Shao M , Sun X , Diao J ((2014) ) Curcumin enhances the radiosensitivity in nasopharyngeal carcinoma cells involving the reversal of differentially expressed long non-coding RNAs. Int J Oncol 44: , 858–864. |
[53] | Munoz L , Ammit AJ ((2010) ) Targeting p38 MAPK pathway for the treatment of Alzheimer’s disease. Neuropharmacology 58: , 561–568. |
[54] | Maccioni RB , Rojo LE , Fernandez JA , Kuljis RO ((2009) ) The role of neuroimmunomodulation in Alzheimer’s disease. Ann N Y Acad Sci 1153: , 240–246. |
[55] | Braidy N , Essa MM , Poljak A , Selvaraju S , Al-Adawi S , Manivasagm T , Thenmozhi AJ , Ooi L , Sachdev P , Guillemin GJ ((2016) ) Consumption of pomegranates improves synaptic function in a transgenic mice model of Alzheimer’s disease. Oncotarget 7: , 64589. |
[56] | Cuomo O , Pignataro G , Gala R , Boscia F , Tortiglione A , Molinaro P , Renzo GD , Lytton J , Annunziato L ((2007) ) Involvement of the potassium-dependent sodium/calcium exchanger gene product NCKX2 in the brain insult induced by permanent focal cerebral ischemia. Ann N Y Acad Sci 1099: , 486–489. |
[57] | Cuomo O , Gala R , Pignataro G , Boscia F , Sec-ondo A , Scorziello A , Pannaccione A , Viggiano D , Adornetto A , Molinaro P ((2008) ) A critical role for the potassium-dependent sodium-calcium exchanger NCKX2 in protection against focal ischemic brain damage. J Neurosci 28: , 2053–2063. |
[58] | Zhang Y , Sharma S , Lytton J ((2015) ) Anatomical evidence for a non-synaptic influence of the K+-dependent Na+/Ca2+-exchanger, NCKX2, on hippocampal plasticity. Neuroscience 310: , 372–388. |
[59] | Li X-F , Kiedrowski L , Tremblay F , Fernandez FR , Periz-zolo M , Winkfein RJ , Turner RW , Bains JS , Rancourt DE , Lytton J ((2006) ) Importance of K+-dependent Na+/Ca2+-exchanger 2, NCKX2, in motor learning and memory. J Biol Chem 281: , 6273–6282. |
[60] | Wei K , Clark AB , Wong E , Kane MF , Mazur DJ , Parris T , Kolas NK , Russell R , Hou H , Kneitz B ((2003) ) Inactiva-tion of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev 17: , 603–614. |
[61] | Desai A , Qing Y , Gerson SL ((2014) )Exonuclease 1 is a critical mediator of survival during DNA double strand break repair in nonquiescent hematopoietic stem and progenitor cells. Stem Cells 32: , 582–593. |
[62] | Izumchenko E , Saydi J , Brown KD ((2012) ) Exonuclease 1 (Exo1) is required for activating response to S N 1 DNA methylating agents. DNA Repair (Amst) 11: , 951–964. |
[63] | Jiang Z , Gorenstein NM , Morre DM , Morre DJ ((2008) ) Molecular cloning and characterization of a candidate human growth-related and time-keeping constitutive cell surface hydroquinone (NADH) oxidase. Biochemistry 47: , 14028–14038. |
[64] | Wang J-L , Tong C-W , Chang W-T , Huang A-M ((2013) ) Novel genes FAM134C, C3orf10 and ENOX1 are regulated by NRF-1 and differentially regulate neurite outgrowth in neuroblastoma cells and hippocampal neurons. Gene 529: , 7–15. |
[65] | Sagy-Bross C , Kasianov K , Solomonov Y , Braiman A , Friedman A , Hadad N , Levy R ((2015) ) The role of cytoso-lic phospholipase A2 a in amyloid precursor protein induction by amyloid beta1-42: Implication for neurodegeneration. J Neurochem 132: , 559–571. |
[66] | Chuang DY , Simonyi A , Kotzbauer PT , Gu Z , Sun GY ((2015) ) Cytosolic phospholipase A 2 plays a crucial role in ROS/NO signaling during microglial activation through the lipoxygenase pathway. J Neuroinflammation 12: , 199. |
[67] | Rossello XS , Igbavboa U , Weisman GA , Sun GY , Wood WG ((2012) ) AP-2ß regulates amyloid beta-protein stimulation of apolipoprotein E transcription in astrocytes. Brain Res 1444: , 87–95. |
[68] | Tao Y , Maegawa H , Ugi S , Ikeda K , Nagai Y , Egawa K , Nakamura T , Tsukada S , Nishio Y , Maeda S ((2006) ) The transcription factor AP-2ß causes cell enlargement and insulin resistance in 3T3-L1 adipocytes. Endocrinology 147: , 1685–1696. |
[69] | Naetar N , Hutter S , Dorner D , Dechat T , Korbei B , Gotz-mann J , Beug H , Foisner R ((2007) ) LAP2α-binding protein LINT-25 is a novel chromatin-associated protein involved in cell cycle exit. J Cell Sci 120: , 737–747. |
[70] | Inoue K , Maeda N , Mori T , Sekimoto R , Tsushima Y , Mat-suda K , Yamaoka M , Suganami T , Nishizawa H , Ogawa Y ((2014) ) Possible involvement of Opa-interacting protein 5 in adipose proliferation and obesity. PLoS One 9: , e87661. |
[71] | Rossor M ((1993) ) Molecular pathology of Alzheimer’s disease. J Neurol Neurosurg Psychiatry 56: , 583. |
[72] | Mitra A , Dey B ((2013) ) Therapeutic interventions in Alzheimer disease. Neurodegenerative Diseases Uday Kishore, IntechOpen, pp. 291–317. Available from: https://www.intechopen.com/books/neurodegenerative-diseases/therapeutic-interventions-in-alzheimer-disease |
[73] | Castellani RJ , Zhu X , Lee H-G , Smith MA , Perry G ((2009) ) Molecular pathogenesis of Alzheimer’s disease: Reductionist versus expansionist approaches. Int J Mol Sci 10: , 1386–1406. |
[74] | Ren C , Zhang J , Yan W , Zhang Y , Chen X ((2016) ) RNA-binding protein PCBP2 regulates p73 expression and p73-dependent antioxidant defense. J Biol Chem 291: , 9629–9637. |
[75] | Zhang Y , Si Y , Ma N ((2016) ) Meis1 promotes poly (rC)-binding protein 2 expression and inhibits angiotensin II-induced cardiomyocyte hypertrophy. IUBMB Life 68: , 13–22. |
[76] | Wang W , Wang L , Lu J , Siedlak SL , Fujioka H , Liang J , Jiang S , Ma X , Jiang Z , Da Rocha EL ((2016) ) The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med 22: , 869. |
[77] | Wang W , Arakawa H , Wang L , Okolo O , Siedlak SL , Jiang Y , Gao J , Xie F , Petersen RB , Wang X ((2017) ) Motor-coordinative and cognitive dysfunction caused by mutant TDP-43 could be reversed by inhibiting its mitochondrial localization. Mol Ther 25: , 127–139. |
[78] | Zhao M , Zhou A , Xu L , Zhang X ((2014) ) The role of TLR4-mediated PTEN/PI3K/AKT/NF-kB signaling pathway in neuroinflammation in hippocampal neurons. Neuroscience 269: , 93–101. |
[79] | Kumar S , Patel R , Moore S , Crawford DK , Suwanna N , Mangiardi M , Tiwari-Woodruff SK ((2013) ) Estrogen receptor ß ligand therapy activates PI3K/Akt/mTOR signaling in oligodendrocytes and promotes remyelination in a mouse model of multiple sclerosis. Neurobiol Dis 56: , 131–144. |
[80] | Gu Q , Zhai L , Feng X , Chen J , Miao Z , Ren L , Qian X , Yu J , Li Y , Xu X ((2013) ) Apelin-36, a potent peptide, protects against ischemic brain injury by activating the PI3K/Akt pathway. Neurochem Int 63: , 535–540. |
[81] | Ruan L , Kang Z , Pei G , Le Y ((2009) ) Amyloid deposition and inflammation in APPswe/PS1dE9 mouse model of Alzheimer’s disease. Curr Alzheimer Res 6: , 531–540. |
[82] | Park KM , Bowers WJ ((2010) ) Tumor necrosis factor-alpha mediated signaling in neuronal homeostasis and dysfunction. Cell Signal 22: , 977–983. |
[83] | Latta CH , Brothers HM , Wilcock DM ((2015) ) Neuroinflammation in Alzheimer’s disease; a source of heterogeneity and target for personalized therapy. Neuroscience 302: , 103–111. |
[84] | Grammas P , Ovase R ((2001) ) Inflammatory factors are elevated in brain microvessels in Alzheimer’s disease. Neurobiol Aging 22: , 837–842. |
[85] | Figuera-Losada M , Rojas C , Slusher BS ((2014) ) Inhibition of microglia activation as a phenotypic assay in early drug discovery. J Biomol Screen 19: , 17–31. |
[86] | Singh S , Natarajan K , Aggarwal BB ((1996) ) Capsaicin (8-methyl-N-vanillyl-6-nonenamide) is a potent inhibitor of nuclear transcription factor-kappa B activation by diverse agents. J Immunol 157: , 4412–4420. |
[87] | Kundu JK , Shin YK , Kim SH , Surh Y-J ((2006) ) Resveratrol inhibits phorbol ester-induced expression of COX-2 and activation of NF-kB in mouse skin by blocking IkB kinase activity. Carcinogenesis 27: , 1465–1474. |
[88] | Abe Y , Hashimoto S , Horie T ((1999) ) Curcumin inhibition of inflammatory cytokine production by human peripheral blood monocytes and alveolar macrophages. Pharmacol Res 39: , 41–47. |
[89] | Zhu X , Lee H-G , Raina AK , Perry G , Smith MA ((2002) ) The role of mitogen-activated protein kinase pathways in Alzheimer’s disease. Neurosignals 11: , 270–281. |
[90] | Kim EK , Choi E-J ((2010) ) Pathological roles of MAPK signaling pathways in human diseases. Biochim Biophys Acta 1802: , 396–405. |
[91] | Kaminska B ((2005) ) MAPK signalling pathways as molecular targets for anti-inflammatory therapy—from molecular mechanisms to therapeutic benefits. Biochim Biophys Acta 1754: , 253–262. |
[92] | Cook D , Fry MJ , Hughes K , Sumathipala R , Woodgett JR , Dale TC ((1996) ) Wingless inactivates glycogen synthase kinase-3 via an intracellular signalling pathway which involves a protein kinase C. EMBO J 15: , 4526–4536. |
[93] | Manoukian AS , Woodgett JR ((2002) ) Role of glycogen synthase kinase-3 in cancer: Regulation by Wnts and other signaling pathways. Adv Cancer Res 84: , 203–229. |
[94] | Fuchs SY , Ougolkov AV , Spiegelman VS , Minamoto T ((2005) ) Oncogenic ß-catenin signaling networks in colorectal cancer. Cell Cycle 4: , 1522–1539. |
[95] | Jope RS , Johnson GV ((2004) ) The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sciences 29: , 95–102. |
[96] | Inestrosa NC , Toledo EM ((2008) ) The role of Wnt signaling in neuronal dysfunction in Alzheimer’s disease. Mol Neurodegener 3: , 9. |
[97] | Caricasole A , Copani A , Caruso A , Caraci F , Iacovelli L , Sortino MA , Terstappen GC , Nicoletti F ((2003) ) The Wnt pathway, cell-cycle activation and ß-amyloid: Novel therapeutic strategies in Alzheimer’s disease? Trends Pharmacol Sci 24: , 233–238. |
[98] | Feng Y , Wang X ((2012) ) Antioxidant therapies for Alzheimer’s disease. Oxid Med Cell Longev 2012: ,472932. |
[99] | E Abdel Moneim A ((2015) ) Oxidant/antioxidant imbalance and the risk of Alzheimer’s disease. Curr Alzheimer Res 12: , 335–349. |
[100] | Cenini G , Sultana R , Memo M , Butterfield DA ((2008) ) Elevated levels of pro-apoptotic p53 and its oxidative modification by the lipid peroxidation product, HNE, in brain from subjects with amnestic mild cognitive impairment and Alzheimer’s disease. J Cell Mol Med 12: , 987–994. |
[101] | Sohn Y , Ganju N , Wands J ((1998) ) P53-and CD95-associated apoptosis in neurodegenerative diseases. Lab Invest 78: , 401–411. |
[102] | Morrison RS , Kinoshita Y , Johnson MD , Guo W , Garden GA ((2003) ) p53-dependent cell death signaling in neurons. Neurochem Res 28: , 15–27. |
[103] | Bernardes VF , Gleber-Netto FO , Sousa SF , Silva TA , Abreu MHNG , Aguiar MCF ((2011) ) EGF in saliva and tumor samples of oral squamous cell carcinoma. App Immunohistochem Mol Morphol 19: , 528–533. |
[104] | Dai W , Li Y , Zhou Q , Xu Z , Sun C , Tan X , Lu L ((2014) ) Cetuximab inhibits oral squamous cell carcinoma invasion and metastasis via degradation of epidermal growth factor receptor. J Oral Pathol Med 43: , 250–257. |
[105] | Berlanga-Acosta J , Gavilondo-Cowley J , Lopez-Saura P , González-López T , Castro-Santana MD , Lopez-Mola E , Guillen-Nieto G , Herrera-Martinez L ((2009) ) Epidermal growth factor in clinical practice-a review of its biological actions, clinical indications and safety implications. Int Wound J 6: ,331–346. |
[106] | Yue P , Zhang X , Paladino D , Sengupta B , Ahmad S , Holloway RW , Ingersoll SB , Turkson J ((2012) ) Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene 31: , 2309. |
[107] | Sylvester P , McIntyre B , Gapor A , Briski K ((2001) ) Vitamin E inhibition of normal mammary epithelial cell growth is associated with a reduction in protein kinase Cα activation. Cell Prolif 34: , 347–357. |
[108] | Wang L , Chang J-H , Paik S-Y , Tang Y , Eisner W , Spurney RF ((2011) ) Calcineurin (CN) activation promotes apoptosis of glomerular podocytes both in vitro and in vivo. Mol Endocrinol 25: , 1376–1386. |
[109] | Shinzawa M , Konno H , Qin J , Akiyama N , Miyauchi M , Ohashi H , Miyamoto-Sato E , Yanagawa H , Akiyama T , Inoue J-i ((2015) ) Catalytic subunits of the phosphatase calcineurin interact with NF-κB-inducing kinase (NIK) and attenuate NIK-dependent gene expression. Sci Rep 5: , 10758. |
[110] | Luo J , Sun L , Lin X , Liu G , Yu J , Parisiadou L , Xie C , Ding J , Cai H ((2014) ) A calcineurin-and NFAT-dependent pathway is involved in α-synuclein-induced degeneration of midbrain dopaminergic neurons. Hum Mol Genet 23: , 6567–6574. |
[111] | Li H , Rao A , Hogan PG ((2011) ) Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol 21: , 91–103. |
[112] | Mullin S , Schapira A ((2013) ) α-Synuclein and mitochon-drial dysfunction in Parkinson’s disease. Mol Neurobiol 47: , 587–597. |
[113] | Bendor JT , Logan TP , Edwards RH ((2013) ) The function of α-synuclein. Neuron 79: , 1044–1066. |
[114] | Ueda K , Fukushima H , Masliah E , Xia Y , Iwai A , Yoshi-moto M , Otero D , Kondo J , Ihara Y , Saitoh T ((1993) ) Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci USA 90: , 11282–11286. |
[115] | Trojanowski JQ , Goedert M , Iwatsubo T , Lee VM ((1998) ) Fatal attractions: Abnormal protein aggregation and neuron death in Parkinson’s disease and Lewy body dementia. Cell Death Differ 5: , 832. |
[116] | Goedert M , Spillantini MG , Del Tredici K , Braak H ((2013) ) 100 years of Lewy pathology. Nat Rev Neurol 9: , 13. |
[117] | Joo Y , Ha S , Hong BH , Kim Ja , Chang KA , Liew H , Kim S , Sun W , Kim JH , Chong YH , Suh YH , Kim HS ((2010) ) Amyloid precursor protein binding protein-1 modulates cell cycle progression in fetal neural stem cells. PLoS One 5: , e14203. |
[118] | Citron M , Oltersdorf T , Haass C , McConlogue L , Hung AY , Seubert P , Vigo-Pelfrey C , Lieberburg I , Selkoe DJ ((1992) ) Mutation of the ß-amyloid precursor protein in familial Alzheimer’s disease increases ß-protein production. Nature 360: , 672–674. |
[119] | Mullan M , Crawford F , Axelman K , Houlden H , Lilius L , Winblad B , Lannfelt L ((1992) ) A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of ß-amyloid. Nat Genet 1: , 345–347. |
[120] | Davis J , Xu F , Deane R , Romanov G , Previti M , Zeigler K , Zlokovic BV , Van Nostrand WE ((2004) ) Early-onset and robust cerebral microvascular accumulation of amyloid ß-protein in transgenic mice expressing low levels of a vasculotropic Dutch/Iowa mutant form of amyloid ß-protein precursor. J Biol Chem 279: , 20296–202306. |
[121] | Labbe K , Miu J , Yeretssian G , Serghides L , Tam M , Finney CA , Erdman LK , Goulet M-L , Kain KC , Stevenson MM ((2010) ) Caspase-12 dampens the immune response to malaria independently of the inflammasome by targeting NF-kB signaling. J Immunol 185: , 5495–5502. |
[122] | Nakagawa T , Zhu H , Morishima N , Li E , Xu J , Yankner BA , Yuan J ((2000) ) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxic-ity by amyloid-ß. Nature 403: , 98. |
[123] | Mattson MP ((2000) ) Apoptosis in neurodegenerative disorders. Nat Rev Mol Cell Biol 1: , 120–129. |
[124] | Hitomi J , Katayama T , Taniguchi M , Honda A , Imaizumi K , Tohyama M ((2004) ) Apoptosis induced by endoplasmic reticulum stress depends on activation of caspase-3 via caspase-12. Neurosci Lett 357: , 127–130. |




