Tridimensional Structural Analysis of Tau Isoforms Generated by Intronic Retention
Abstract
Background:
Tauopathies are a subset of neurodegenerative diseases characterized by abnormal tau inclusions. Recently, we have discovered a new, human specific, tau isoform termed W-tau that originates by intron 12 retention. Our preliminary data suggests this newly discovered W-tau isoform might prevent aberrant aggregation of other tau isoforms but is significantly downregulated in tauopathies such as Alzheimer's disease.
Objective:
To accurately predict, examine, and understand tau protein structure and the conformational basis for the neuroprotective role of W-tau.
Methods:
A tridimensional deep learning-based approach and in vitro polymerization assay was included to accurately predict, analyze, and understand tau protein structure and the conformational basis for the neuroprotective role of W-tau.
Results:
Our findings demonstrate: a) the predicted protein tridimensionality structure of the tau isoforms raised by intron retention and their comparison with the other tau isoforms; b) the interaction of W-tau peptide (from W-tau isoform) with other tau isoforms; c) the effect of W-tau peptide in the polymerization of those tau isoforms.
Conclusions:
This study supports the importance of the structure-function relationship on the neuroprotective behavior of W-tau inhibiting tau fibrillization in vitro.
INTRODUCTION
Tau protein is genetically encoded by the microtubule-associated protein tau (MAPT) gene located on chromosome 17q21.31 and is comprised of 16 exons: exons 0 and 1 encode the 5’ UTR of MAPT mRNA while exon 14 encodes for the 3’ UTR [1]. Exons 4a, 6, and 8 are skipped when tau is transcribed in the brain and are only found in the mRNA of peripheral tissues [2]. In the central nervous system (CNS), alternative splicing of exons 2, 3, and 10 result in six different tau isoforms that range from 352–441 residues in length [3–6]. The six CNS tau isoforms (0N4 R, 1N4 R, 2N4 R, 0N3 R, 1N3 R, 2N3 R) contain 0–2 N-terminal inserts and either 3 or 4 C-terminal microtubule binding repeats (MTBRs) with the 4 R variants having higher microtubule-binding affinity [7–9]. In the fetal brain, only the 0N3 R isoform is expressed while the adult brain possesses all six [10]. In a healthy adult brain, 4R- and 3R-tau are generally in equimolar quantities [7, 9] and divergence from this may be characteristic of tauopathies such as neurodegenerative frontotemporal dementias and Alzheimer’s disease (AD) [7, 11]. Despite tau being an intrinsically disordered protein (IDP) [12], the structural changes introduced during the alternative processing will have consequences on the functional versatility of tau [11]. Tau protein may acquire local structures such as α-helices, β-sheets, and polyproline-II helices in various sequence segments [13]. In this respect, a “paperclip” model for tau 3D structure has been suggested based on a fluorescence resonance energy transfer study [14], wherein the N-terminal, C-terminal and repeat domains are folded in such a manner that these regions approach each other [14, 15]. Unlike a well-folded protein whose structure restricts binding to only one type of ligand, tau is likely to adopt multiple conformations in a context-dependent manner [16].
Recently, a new tau isoform generated by retention of intron 12 of the human MAPT gene has been described [17]. Soon after the start of intron 12 of human MAPT, a stop codon appears, followed by a canonical polyadenylation sequence, resulting in the truncation of the protein at this point. Thus, this isoform differs from other human tau isoforms by lacking exon 13 of the MAPT gene and including an 18-amino-acid sequence corresponding to the translation of the retained fragment of the intron 12 in its place, at its carboxyl-terminal region, right after exon 12.1. The 18-residue sequence contains two tryptophan residues (W), an amino acid that cannot be found at any other location of the human tau sequence. Retention of the beginning of intron 12 and the truncation of exon 13, results in neuroprotective properties of W-tau isoforms such as a lower aggregation capacity or the ability to inhibit the polymerization of other tau isoforms [18].
In addition to W-tau isoforms, the tau11i isoform, generated by intron 11 retention is found elevated in the cortex of brains from AD patients. Tau11i is enriched in the sarkosyl-insoluble fraction in AD hippocampus and forms aggregates that colocalize weakly with 4R-tau fibril-like structure in AD temporal lobe [19]. Moreover, stably expressed Tau11i also shows weaker colocalization with α-tubulin of microtubule network in human mature cortical neurons [19].
The discovery of new tau isoforms generated by intron retention highlights the importance of the structure-function relationship. Moreover, recent developments in deep-learning-based methods improve protein structure prediction [20]. Protein structure prediction has been an active area of research for several decades, and theoretical methods have given insight into the structures of experimentally intractable proteins. The current availability of larger quantities of higher-quality structural data has resulted in improvements in training-data quality, and consequently in the accuracy of these predictive algorithms [20]. Deep learning-based methods have demonstrated high accuracy in protein structure prediction in the recent years. To date, the most accurate deep learning method is AlphaFold from Google’s DeepMind [21]. The accuracy of the method comes from the high accuracy of distance predictions. AlphaFold employs a convolutional neural network trained on protein structures from the Protein Data Bank. Given an input sequence and its multiple sequence alignment, it predicts pairwise distances and torsion angles between the residues. These distances are optimized using gradient descent minimization to obtain well-packed protein structures [20].
In the present work, tridimensional protein structure prediction of tau protein isoforms has been carried out using AlphaFold. To start untangling the structural basis of the neuroprotective role of W-tau we have resolved the tridimensional structure corresponding to the W-tau peptide using this new deep learning-based methodology, showing a compacted conformation form by two anti-parallel β-sheets. Furthermore, we have generated a modified version of the W-tau peptide in which consecutive cysteine residues are replaced by serine allowing us to study the potential structure-function relationship of the tau isoform generated by intron 12 retention,
MATERIALS AND METHODS
Heparin sodium salt from porcine intestinal mucosa was obtained from Sigma (H3393-100KU).
Synthesis of tau peptides
The tau peptide 317 – 335aa (1/2 R peptide) (317KVTSKCGSLGNIHHKPGGG335) was purcha-sed from Neosystem Laboratoire (Strasbourg, France). The W-tau peptide (KKVKGVGWVGCCPWVYGH) and its modification (KKVKGVGWVGSSPWVYGH) were obtained from Abyntek Biopharma S. L. (Parque Tecnológico de Bizkaia. Derio, Spain). All the peptides were dissolved in sterile Milli-Q distilled water.
3D protein structure assessment
A user-friendly interface for accessing AlphaFold2 has recently been made available through notebooks. We used the ColabFold notebook, whose structure prediction is powered by AlphaFold2 combined with RoseTTAFold and a fast, multiple sequence alignment generation stage using MMseqs2 [22]. Each isoform structure prediction was downloaded from ColabFold, and visualized with The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. For a correct visualization, constitutive exons of the isoforms were coloured in white, then each exon from the microtubule binding-core, exons 2 & 3 and intronic retention were performed with individual colors.
In vitro polymerization studies
Tau protein and its peptides were dissolved at a concentration of 10 mg/mL in distilled water, aliquoted, and immediately used or frozen to be used only once, to avoid several freezing/thawing cycles. The use of the synthetic peptide human tau 1/2 R as a polymerization element avoids the appearance of artifacts of purified microorganism proteins origin in the electron microscopic visualization. This peptide maintains a high polymerization capacity while obtaining a clear and precise view of the polymers formed in each variant [18]. Recombinant human tau 1/2 R peptide (10μg) was incubated in 10μL of Buffer A (0.1 MES (pH 6.4), 0.5 mM MgCl2, and 2 mM EGTA) and 0.5 M NaCl, in the absence or presence of different concentrations of heparin. For the 1/2 R tau peptide, the optimal peptide: heparin ratio was found to be 1:1 (mass/mass). To analyze the effect of W-tau peptides on 1/2 R tau peptide polymerization, W-tau peptides were added at final concentrations of 1, 0.1, and 0.01μg/μL. The reactions were allowed to proceed at room temperature for 7 days utilizing the drop vapor diffusion technique [23] before analysis. After 7 days of incubation at room temperature samples were visualized by electron microscopy.
Electron microscopy analysis
For the visualization of polymerization reactions samples were added to a Formvar (400 mesh) copper-coated grids ionized in BAE 120 Evaporator (Bal-Tec). Samples were adsorbed for 5 min, washed with milli Q water and left for 40 s in 2% uranyl acetate. Visualization was performed in JEM1400 Flash Transmission Electron Microscope (Jeol). Images were taken with a TemCam F416 (TVIPS) camera at a magnification of 20,000X. The extent of tau polymerization was analyzed using the image processing and analysis software Image J (Image J, NIH). Ten random fields from each condition were taken for image analysis. All analyses were performed in a blind manner. Each experiment was repeated at least three times.
Statistical analysis
For image analysis, the statistical significance was determined by One-way ANOVA followed by Dunnett’s multiple comparisons test. A p value of less than 0.05 was considered significant. Data represent the mean±SEM.
RESULTS
Taking advantage of the deep learning-based method AlphaFold the tridimensional structure prediction of the most representative tau isoforms found in the central nervous system were generated (Fig. 1). Tridimensional structure of tau isoforms was visualized by using the structure viewer software Pymol. Predicted protein structures were color coded by establishing a wavelength scale of the colors, assigning the shortest wavelength color to the first non-constitutive exon and continuing the scale of colors with the left non constitutive exons throughout the sequences of the isoforms.
Fig. 1
AlphaFold-predicted structure of tau. A) Diagram of MAPT gene and tau isoforms generated by alternative splicing. Tau isoforms abundantly expressed in the CNS are shown. B-D) AlphaFold-predicted tridimensional structure of tau isoforms including exons 2, 3, and 10 are shown. D) Predicted tridimensional structure of the longest isoform expressed in the CNS showing (inset) the distance between the carboxy- and amino-terminal segments of the protein. Exons are color coded and matched with diagram shown in A. The color scale would be as follows: violet for exon 9, blue purple for exon 11, blue for exon 12, sky blue for exon 2, green for exon 3, lime green for exon 10.
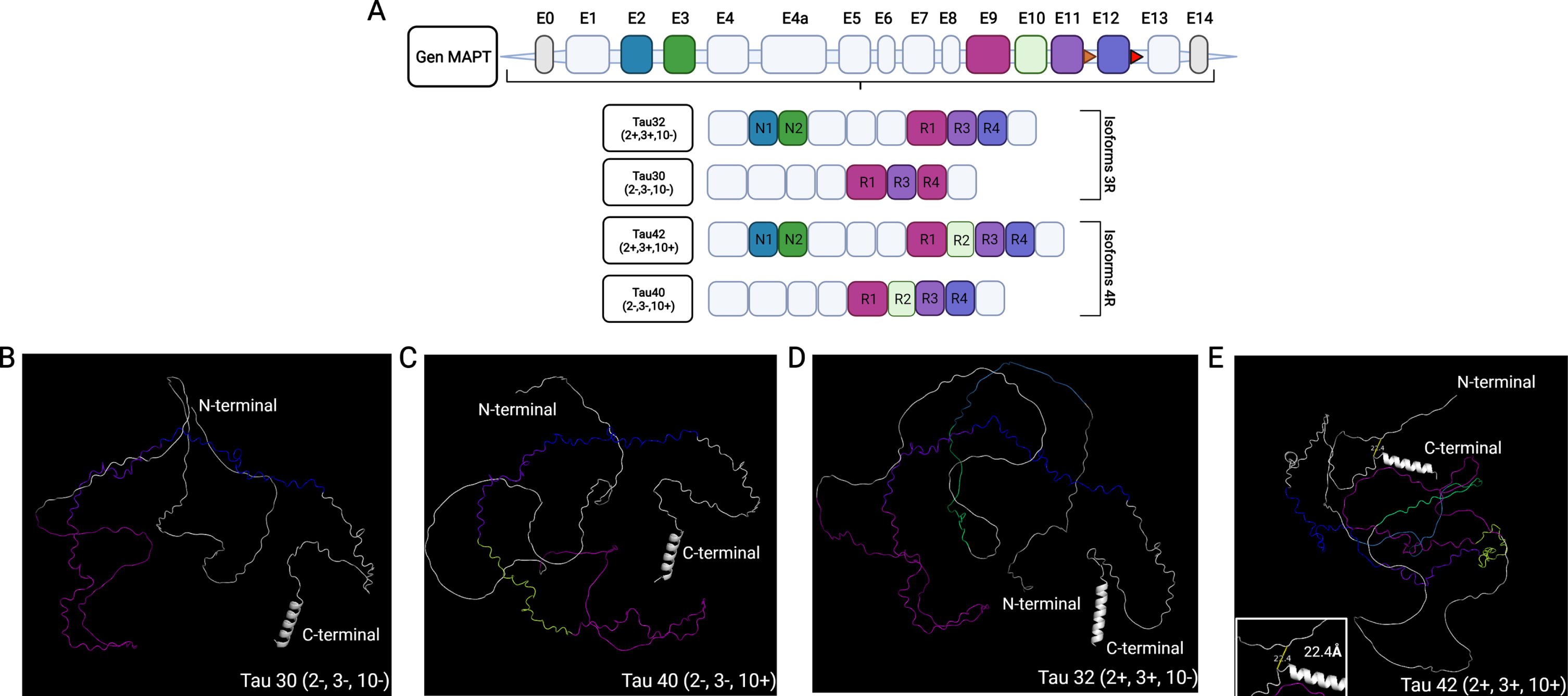
Comparing predicted tridimensional protein structures for Tau 30 (Fig. 1B) and Tau 40 isoforms (Fig. 1C) show an IDP structure which is not altered with the inclusion of exon 10 as seen in Tau 40 isoform (Fig. 1C). Remarkably, inclusion of exons 2 and 3 (Tau 32 and Tau 42 isoforms, Fig. 1D, E) show a more ordered tridimensional structure in which the N- and C-terminal segments of the protein are in close proximity. However, in regard to the spatial arrangement of the microtubule-binding domain, the inclusion of exons 2 and 3 does not change the external exposure of the protein sequences encoded by exons 9, 10, 11, and 12 (Fig. 1B-D). Figure 1E shows the three-dimensional structure prediction of the largest tau isoform found in the CNS, tau 42. It is important to mention this tridimensional protein structure prediction shows a distance of 22Å between the carboxy- and amino-terminal segments and a spatial orientation of the residues that conform these domains that corroborates the “paperclip” folding model previously proposed [13–15].
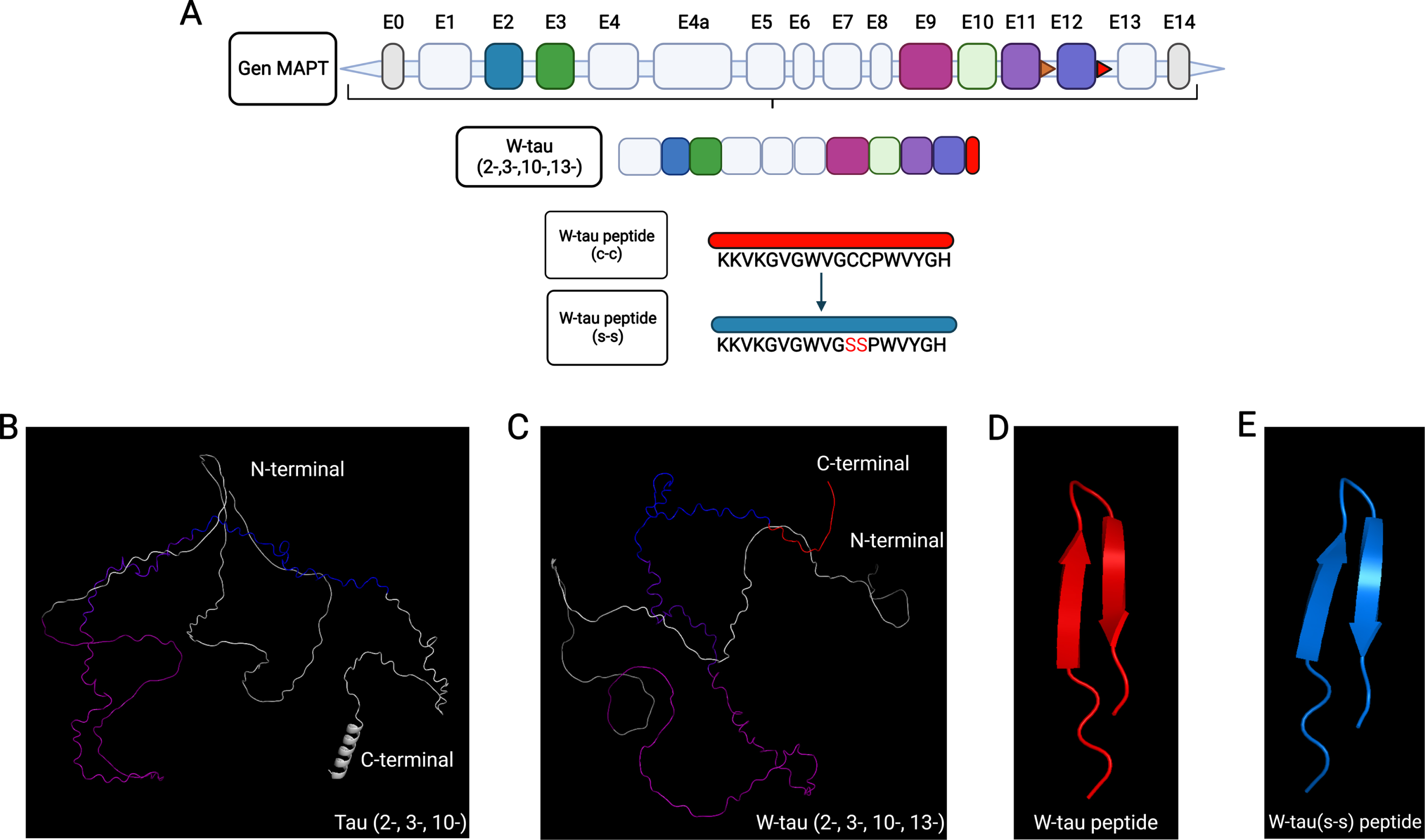
When comparing the predicted tridimensional structures of Tau 30 (Fig. 2B) and the recently discovered isoform generated by the retention of intron 12 (W-tau, Fig. 2C), the main observed change in the W-tau isoform is the loss of the alpha helix present in the carboxy-terminal domain (Fig. 2B, C). The intronic retention expressed in W-tau (Fig. 2C) does not show a particular ordered structure. Only a curved region due to the double cysteine present in its intronic sequence that gives a U-like structure due to the formation of a possible disulfide bridge is observed. This U-shape is exacerbated when only modelling the tridimensional structure of the peptide sequence corresponding to the expressed intron 12 of the W-tau isoform (Fig. 2D), showing a fully folded peptide in which two anti-parallel beta-sheet are found. Interestingly, the modification of the W-tau peptide sequence in which the double cysteine is replaced by a double serine (Fig. 2E) still shows a folded structure. Albeit it appears to have a different spatialorientation.
Fig. 2
AlphaFold-predicted structure of W-tau isoform. A) Diagram of MAPT gene and W-tau isoform generated by intron 12 retention. W-tau intronic peptide sequences are shown. B) AlphaFold-predicted tridimensional structure of tau isoform excluding exons 2, 3, and 10 is shown. C) Predicted tridimensional structure of the W-tau. D) Predicted peptide sequence expressed as a result of intron 12 retention (Red). E) Predicted peptide sequence expressed as a result of intron 12 retention with mutated cysteine residues (Blue). Exons are color coded and matched with diagram shown in A. The color scale would be as follows: violet for exon 9, blue purple for exon 11, blue for exon 12, and red for intronic retention.
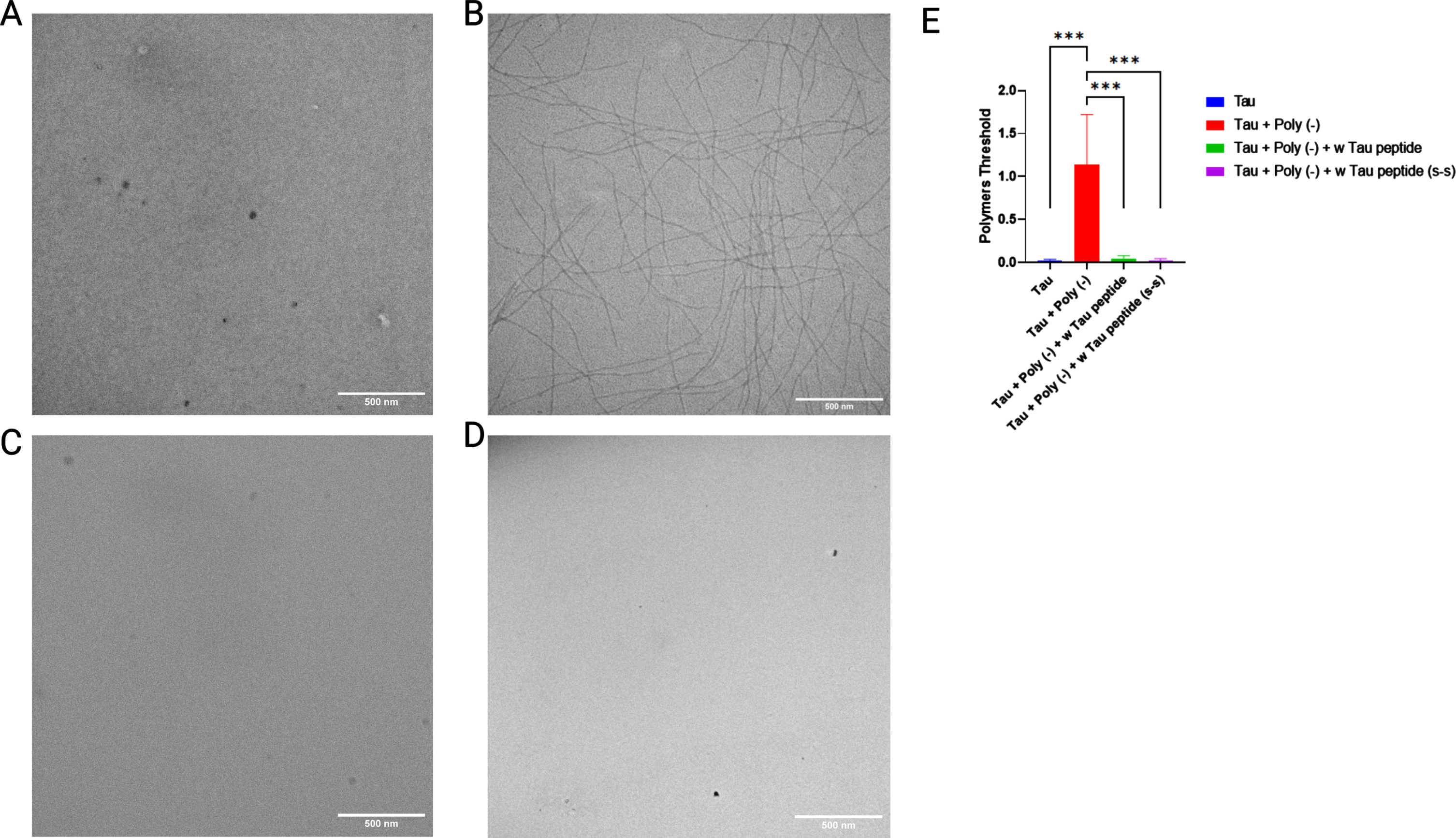
On the other hand, the main characteristic of this isoform generated by the retention of intron 12 is the inhibitory capacity on tau aggregation. We hypothesize one of the possible reasons for this property comes from the net positive charge provided by the sequence encoded by intron 12, which is the opposite charge of known polymerization inducers such as polyanions (e.g., heparin). To corroborate this hypothesis, the polymerization inhibitory capacity of the W-tau peptide in the presence of the polyanion heparin was assessed and confirmed (Fig. 3B, C, E). In addition, to analyze the effect that the double cysteine residues present in the W-tau peptide sequence could have on its polymerization inhibitory capacity due to their charge and reactivity, we tested the modified version of the W-tau peptide where cysteine residues are changed by serines residues. As shown in Fig. 3C-E, both peptides maintain the same tau polymerization inhibitory capacity.
Fig. 3
Mutated W-tau peptide inhibits polyanion-induced tau polymerization. Representative images of 1/2 R Tau polymerization in the absence (A) or presence (B) of heparin. B) Effect of increasing amounts of w-Tau peptide fragment (10aa) on 1/2 R Tau polymerization. C) Effect of W-tau peptide on 1/2 R Tau polymerization. D) Effect of mutant W-tau peptide on 1/2 R Tau polymerization. E) Image quantification of tau polymerization in (A-D). N = 10 per group; ***p < 0.001 by One-way ANOVA test. Data represent mean±SEM.
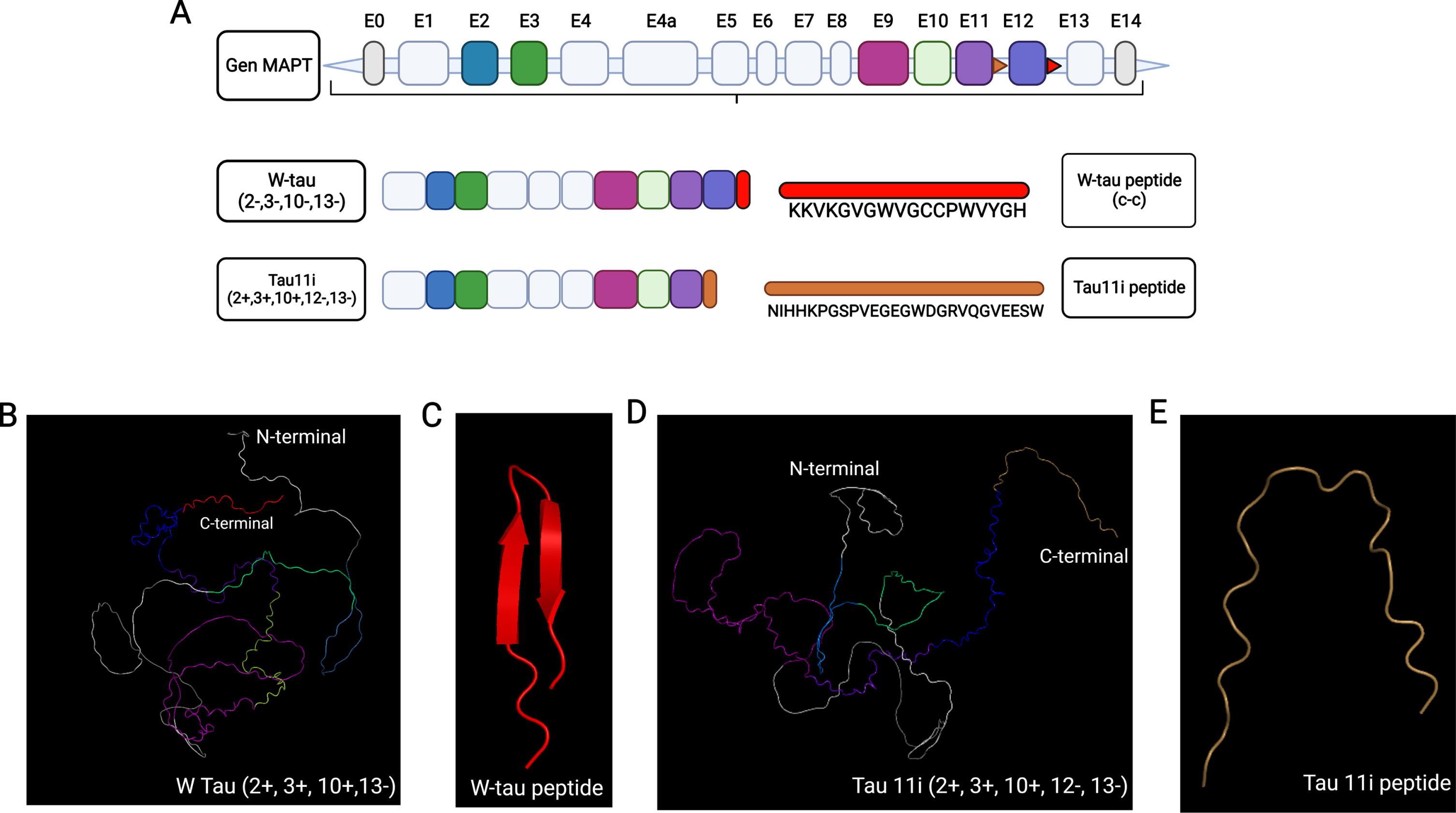
We recently have found the W-tau isoform, an isoform originated by retention of intron 12. Despite the truncation of exon 13, W-tau still maintains a “paperclip” like conformation (Figs. 2 and 4) and presents low self-aggregating capacity. In contrast, AlphaFold tridimensional structure prediction of Tau 11i, a tau isoform produced by intronic retention of exon 11 that accumulates in the hippocampus of AD brains, shows the N-terminal and C-terminal domains are distant from each other losing the “paperclip” conformation. Remarkably, both isoforms originated by intronic retention, W-tau and Tau11i (Fig. 4), differ in the net charge of their respective expressed intronic sequences, were overall charge of W-tau intronic sequence is positive and overall charge of Tau 11i intronic sequence is negative.
Fig. 4
AlphaFold-predicted structure of tau isoforms generated by intronic retention. A) Diagram of MAPT gene and tau isoform generated by intron 12 retention. W-tau and Tau11i intronic retention isoforms together with their respective expressed intronic sequences are shown. B) Predicted tridimensional structure of the W-tau including exons 2, 3, 10, and intron 12 retention. C) Predicted structure of peptide sequence expressed because of intron 12 retention (Red). D) Predicted tridimensional structure of the Tau11i. E) Predicted structure peptide sequence expressed because of intron 11 retention (Brown). The color scale would be as follows: violet for exon 9, blue purple for exon 11, sky blue for exon 2, green for exon 3, lime green for exon 10 and brown for intronic retention.
DISCUSSION
In this work, we have shown how by using the deep learning-based method AlphaFold we can start defining the tridimensional structure of the IDP Tau and its related intron retention isoforms.
We first compared the tridimensional structure of the main tau isoforms expressed in the human CNS (Fig. 1). Here we show tau isoforms were exon 2 and exon 3 are included present a greater degree of structuring, giving rise to the well-known “paperclip” conformation previously proposed for tau [13, 14]. It was shown previously, as judged by Fluorescence Resonance Energy Transfer, tau in solution adopts long range interactions between the repeat domain and the C terminus and between the N and C terminus (“paperclip” conformation) [14]. In solution, the N-terminal domain is close to the C-terminal tail where it causes FRET (21–24 Å) [15]. Strikingly, the AlphaFold predicted structures show a similar “paperclip” conformation presenting a similar distance between the C-terminal tail and the N-terminal domain (Fig. 1E).
Understanding the main structural differences of tau isoforms could give us a better understanding of the behavior of tau protein isoforms and inform us about the structure-function relationship. The charge difference along with the different structure-function relationship of these isoforms originated by intronic retention [17, 19] (Fig. 4) might explain their opposite behaviors in the CNS and supports a neuroprotective role for W-tau isoform.
ACKNOWLEDGMENTS
We thank Ms. Nuria de la Torre Alonso for technical and editorial assistance.
FUNDING
This work has been supported by grants from the Spanish Ministry of Science: PID2020-113204GB-I00 (F.H.) and PID2021-123859OB-100 from MCIN /AEI/10.13039/501100011033 / FEDER, UE (J.A.). The Centro de Biología Molecular Severo Ochoa (CBMSO) is a Severo Ochoa Center of Excellence (MICIN, award CEX2021-001154-S).
CONFLICT OF INTEREST
Jesus Avila is an Editorial Board Member of this journal but was not involved in the peer-review process nor had access to any information regarding its peer-review.
DATA AVAILABILITY
The datasets generated during the current study are available from the corresponding author upon reasonable request.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/ADR-230074.
REFERENCES
[1] | Muralidar S , Ambi SV , Sekaran S , Thirumalai D , Palaniappan B ((2020) ) Role of tau protein in Alzheimer’s disease: The prime pathological player. Int J Biol Macromol 163: , 1599–1617. |
[2] | Corsi A , Bombieri C , Valenti MT , Romanelli MG ((2022) ) Tau isoforms: Gaining insight into MAPT alternative splicing. Int J Mol Sci 23: , 15383. |
[3] | Andreadis A ((2005) ) Tau gene alternative splicing: Expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta 1739: , 91–103. |
[4] | Liu F , Gong CX ((2008) ) Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener 3: , 8. |
[5] | Wei ML , Andreadis A ((1998) ) Splicing of a regulated exon reveals additional complexity in the axonal microtubule-associated protein tau. J Neurochem 70: , 1346–1356. |
[6] | Zhou J , Yu Q , Zou T ((2008) ) Alternative splicing of exon 10 in the tau gene as a target for treatment of tauopathies. BMC Neurosci 9: (Suppl 2), S10. |
[7] | Gendron TF , Petrucelli L ((2009) ) The role of tau in neurodegeneration. Mol Neurodegener 4: , 13. |
[8] | Goedert M , Spillantini MG , Jakes R , Rutherford D , Crowther RA ((1989) ) Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3: , 519–526. |
[9] | Kosik KS , Orecchio LD , Bakalis S , Neve RL ((1989) ) Developmentally regulated expression of specific tau sequences. Neuron 2: , 1389–1397. |
[10] | Guo T , Noble W , Hanger DP ((2017) ) Roles of tau protein in health and disease. Acta Neuropathol 133: , 665–704. |
[11] | Takuma H , Arawaka S , Mori H ((2003) ) Isoforms changes of tau protein during development in various species. Brain Res Dev Brain Res 142: , 121–127. |
[12] | Wright PE , Dyson HJ ((2009) ) Linking folding and binding. Curr Opin Struct Biol 19: , 31–38. |
[13] | Gamblin TC ((2005) ) Potential structure/function relationships of predicted secondary structural elements of tau. Biochim Biophys Acta 1739: , 140–149. |
[14] | Jeganathan S , von Bergen M , Brutlach H , Steinhoff HJ , Mandelkow E ((2006) ) Global hairpin folding of tau in solution. Biochemistry 45: , 2283–2293. |
[15] | Jeganathan S , Hascher A , Chinnathambi S , Biernat J , Mandelkow EM , Mandelkow E ((2008) ) Proline-directed pseudo-phosphorylation at AT8 and PHF1 epitopes induces a compaction of the paperclip folding of Tau and generates a pathological (MC-1) conformation. J Biol Chem 283: , 32066–32076. |
[16] | Morris M , Maeda S , Vossel K , Mucke L ((2011) ) The many faces of tau. Neuron 70: , 410–426. |
[17] | Garcia-Escudero V , Ruiz-Gabarre D , Gargini R , Perez M , Garcia E , Cuadros R , Hernandez IH , Cabrera JR , Garcia-Escudero R , Lucas JJ , Hernandez F , Avila J ((2021) ) A new non-aggregative splicing isoform of human Tau is decreased in Alzheimer’s disease. Acta Neuropathol 142: , 159–177. |
[18] | Cuadros R , Perez M , Ruiz-Gabarre D , Hernandez F , Garcia-Escudero V , Avila J ((2022) ) Specific peptide from the novel W-Tau isoform inhibits tau and amyloid beta peptide aggregation in vitro. ACS Chem Neurosci 13: , 1974–1978. |
[19] | Ngian ZK , Tan YY , Choo CT , Lin WQ , Leow CY , Mah SJ , Lai MK , Chen CL , Ong CT ((2022) ) Truncated tau caused by intron retention is enriched in Alzheimer’s disease cortex and exhibits altered biochemical properties. Proc Natl Acad Sci U S A 119: , e2204179119. |
[20] | Singh A ((2020) ) Deep learning 3D structures. Nat Methods 17: , 249. |
[21] | Senior AW , Evans R , Jumper J , Kirkpatrick J , Sifre L , Green T , Qin C , Zidek A , Nelson AWR , Bridgland A , Penedones H , Petersen S , Simonyan K , Crossan S , Kohli P , Jones DT , Silver D , Kavukcuoglu K , Hassabis D ((2020) ) Improved protein structure prediction using potentials from deep learning. Nature 577: , 706–710. |
[22] | Mirdita M , Schutze K , Moriwaki Y , Heo L , Ovchinnikov S , Steinegger M ((2022) ) ColabFold: Making protein folding accessible to all. Nat Methods 19: , 679–682. |
[23] | Perez M , Valpuesta JM , Medina M , Montejo de Garcini E , Avila J ((1996) ) Polymerization of tau into filaments in the presence of heparin: The minimal sequence required for tau-tau interaction. J Neurochem 67: , 1183–1190. |



