Urinary Cytokines as Potential Biomarkers of Mild Cognitive Impairment and Alzheimer’s Disease: A Pilot Study
Abstract
Background:
Alzheimer’s disease (AD) is the most common form of dementia, accounting for 80% of all cases. Mild cognitive impairment (MCI) is a transitional state between normal aging and AD. Early detection is crucial, as irreversible brain damage occurs before symptoms manifest.
Objective:
This study aimed to identify potential biomarkers for early detection of AD by analyzing urinary cytokine concentrations. We investigated 37 cytokines in AD, MCI, and cognitively normal individuals (NC), assessing their associations with AD development.
Methods:
Urinary cytokine concentrations were measured in AD (n = 25), MCI (n = 25), and NC (n = 26) patients. IL6ST and MMP-2 levels were compared between AD and NC, while TNFRSF8, IL6ST, and IL-19 were assessed in AD versus MCI. Diagnostic models distinguished AD from NC, and in-silico analysis explored molecular mechanisms related to AD.
Results:
Significant perturbations in IL6ST and MMP-2 concentrations were observed in AD urine compared to NC, suggesting their potential as biomarkers. TNFRSF8, IL6ST, and IL-19 differed significantly between AD and MCI, implicating them in disease progression. Diagnostic models exhibited promising performance (AUC: 0.59–0.79, sensitivity: 0.72–0.80, specificity: 0.56–0.78) in distinguishing AD from NC. In-silico analysis revealed molecular insights, including relevant non-coding RNAs, microRNAs, and transcription factors.
Conclusion:
This study establishes significant associations between urinary cytokine concentrations and AD and MCI. IL6ST, MMP-2, TNFRSF8, IL6ST, and IL-19 emerge as potential biomarkers for early detection of AD. In-silico analysis enhances understanding of molecular mechanisms in AD. Further validation and exploration of these biomarkers in larger cohorts are warranted to assess their clinical utility.
INTRODUCTION
Alzheimer’s disease (AD) is a complex neurodegenerative disorder and the leading cause of dementia, resulting in memory loss, difficulty in thinking, and behavioral changes. The major neuropathological hallmarks include the accumulation of amyloid-β (Aβ) and neurofibrillary tangles in the brain. It is a growing global health concern with enormous implications for our society affecting approximately 50 million people worldwide. It is considered the fifth leading cause of death [1–3] in the United States. Mild cognitive impairment (MCI), is considered by many to be a transition state between cognitively normal and demented and globally affects 42% of those aged over 60 [4], with ∼7.5% developing dementia after one year following diagnosis. That number climbs to 15% by year two and 20% by year three [5].
The pathogenic process of AD is hypothesized to begin decades before clinical symptoms become apparent and irreversible brain damage has already occurred, emphasizing the need for early diagnoses when treatment options are believed to be most effective [6]. Current clinical modalities include invasive methods such as testing cerebrospinal fluid for levels of Aβ and tau [7] or using cost-prohibitive imaging techniques such as positron emission tomography [8]. Although with invasive methods, plasma/serum can be obtained from anuric patients and are less susceptible to bacterial contamination [9]; however, these methods can prove problematic as they are not readily available in every clinical setting, further highlighting the need for non-invasive and inexpensive strategies for diagnosing the disease in its infancy.
Increasing evidence suggests that inflammatory responses play a critical role in the pathogenesis of AD [10, 11]. Senile plaques and the neurofibrillary tangles bind to pattern recognition receptors on microglia and astrocytes, triggering an innate immune response, and the subsequent release of cytokines, chemokines, and other inflammatory mediators that are believed to contribute to disease progression and severity [12–14]. In this study, cytokine concentrations were measured in the urine of AD patients, MCI sufferers, and age and gender-matched cognitively normal controls (NC), to develop a non-invasive, biomarker tool for the early diagnosis of AD. Further, we employ an in-silico approach to provide insight into the inflammatory processes and how they relate to the onset and progression of dementia.
MATERIALS AND METHODS
Study design and sample preparation
We measured the concentration of 37 inflammatory cytokines in urine samples obtained from AD patients (n = 25), MCI sufferers (n = 25), and age and gender-matched cognitively normal controls (n = 26). The participants were recruited from Beaumont Health Outpatients. The diagnosis and evaluation of patients with AD and MCI sufferers was carried out by a fellowship-trained geriatrician according to the criteria of the National Institute of Neurological and Communicative Disorders and the Stroke (NINCDS) and Alzheimer’s Disease and Related Disorders Association (ADRDA) [15]. Participants underwent a focused history and physical examination (assessing motor strength and tone, the existence of a tremor, sensation, balance (Romberg), and gait) to include an exhaustive cognitive testing battery routinely utilized in the Geriatric Clinic at Beaumont health to include: Mini– Mental State Examination (MMSE), Saint Louis University Mental Status (SLUMS), clock drawing tasks (CLOX-I, and CLOX-II), trial making tests (Trails making A, and Trails making B), and Geriatric Depression Scale. The study was approved by the Beaumont Health Institutional Review Board (IRB# 2014-038) and all collection/testing parameters were carried out in accordance with the approved guidelines. The collection was completed in the early morning and all subjects were asked to refrain from eating, drinking, or smoking for at least 1 h before urine collection. Urine was collected in a polypropylene container, then aliquoted into an Eppendorf tube and stored at –80°C for further analysis.
Cytokine assay
Urine samples were defrosted and centrifuged at 1000× g for 5 min at 4°C and 50μl of the supernatant was analyzed according to the manufacturer’s protocol. In brief, using the Bio-Plex Pro™ Human Inflammation Panel 1, 37-Plex #171AL001M (BIO-RAD-USA), we measured 37 inflammatory cytokines from the TNF superfamily of proteins, the IFN family of proteins, Treg cytokines, and Matrix metalloproteinases (MMPs). While the methodology for the cytokine assay was done robustly, it should be noted that the limitation of the study is that only one technical replicate was acquired. This may have impacted the reliability and reproducibility of the results, and further studies with multiple technical replicates may be necessary to confirm our findings. The assay plates were read using a Luminex 200® analyzer system, and the raw concentration values for each inflammatory biomarker were calculated using the manufacturer’s software.
Univariate analysis
Before statistical analysis, any variable with >40% missing values were excluded from our analysis. Subsequently, the concentration for each cytokine was normalized using serially diluted internal calibrants, and standard curves were generated. The normality of the data was assessed by the Kolmogorov-Smirnov test of normality. Subsequently, using MetaboAnalyst (v 5.0) [16], an independent t-test and Mann Whitney U test were performed for all pair-wise comparisons for both parametric and non-parametric distributions, respectively. All demographic information was analyzed using the SPSS Statistics toolbox (ver 24.0).
Machine learning models
A total of 11 machine learning algorithms were evaluated in this study, including logistic regression, linear discriminant analysis, linear support vector machine (SVM), random forest, decision tree, xgboost, K-nearest Neighbor (knn), gaussian naïve bayes, and kernel SVM. Before examining the diagnostic performance of the models, each urinary cytokine was generalized log-transformed (glog) [17] and auto-scaled. Before performing pattern recognition, data from each group were analyzed using principal component analysis (PCA) to identify any potential outliers or systematic variation (p < 0.05). Subsequently, data were divided into training (60% of data) and testing (40% of data) sets. A Recursive Feature Elimination (RFE) method with Logistic Regression as the classifier was utilized to select the best predictor variables and eliminate redundancy in the variable space. Once the best set of markers was identified, model hyper-parameters [18] were optimized using 10-fold cross-validation. The trained models were assessed using the test set and a 10-fold CV method. Model performance was evaluated using the classification accuracy rate, the area under a receiver operating characteristic (AUROC) curve, and the identification of true discriminating features.
Pathway analysis
The differentially expressed inflammatory cytokines between NC and AD were categorized in FunRich software (https://www.funrich.org), an open access, functional enrichment, and network analysis tool [19]. The biological function was divided into four components: biological processes, transcription factors, cellular components, and molecular functions. The biological pathways and target miRNA for these cytokines were performed using the Enrichr software [20–22]. Differentially expressed cytokines (p < 0.05) between AD and NC were used for the pathway analysis. The miRtargetBase 2017 and targetScanmicroRNA2017 databases were employed to identify miRNAs linked to these cytokines. The LINCRNA database was used to identify corresponding Long intergenic non-coding RNA (LINCRNAs).
RESULTS
We measured the concentration of 37 inflammatory cytokines in the urine of AD patients (n = 25; mean age 81.44±4.56), MCI sufferers (n = 25; mean age 81.24±4.87), and age-, and gender-matched NCs (n = 26; mean age 80.65±5.87). Three cytokines (IL-29, MMP1, and MMP3) were excluded as they contained >40% missing values. A summary of each participant’s clinical and socio-demographic factors is reported in Supplementary Table 1A–C. The results reveal that education, gender, and age were not statistically different between the groups (p > 0.05) while SLUMS total scores, MMSE, CLOXs, and Trial Making Tests (TMT-Trails Making A, and Trails Making B were found to be statistically significantly different between the group as expected (p < 0.05).
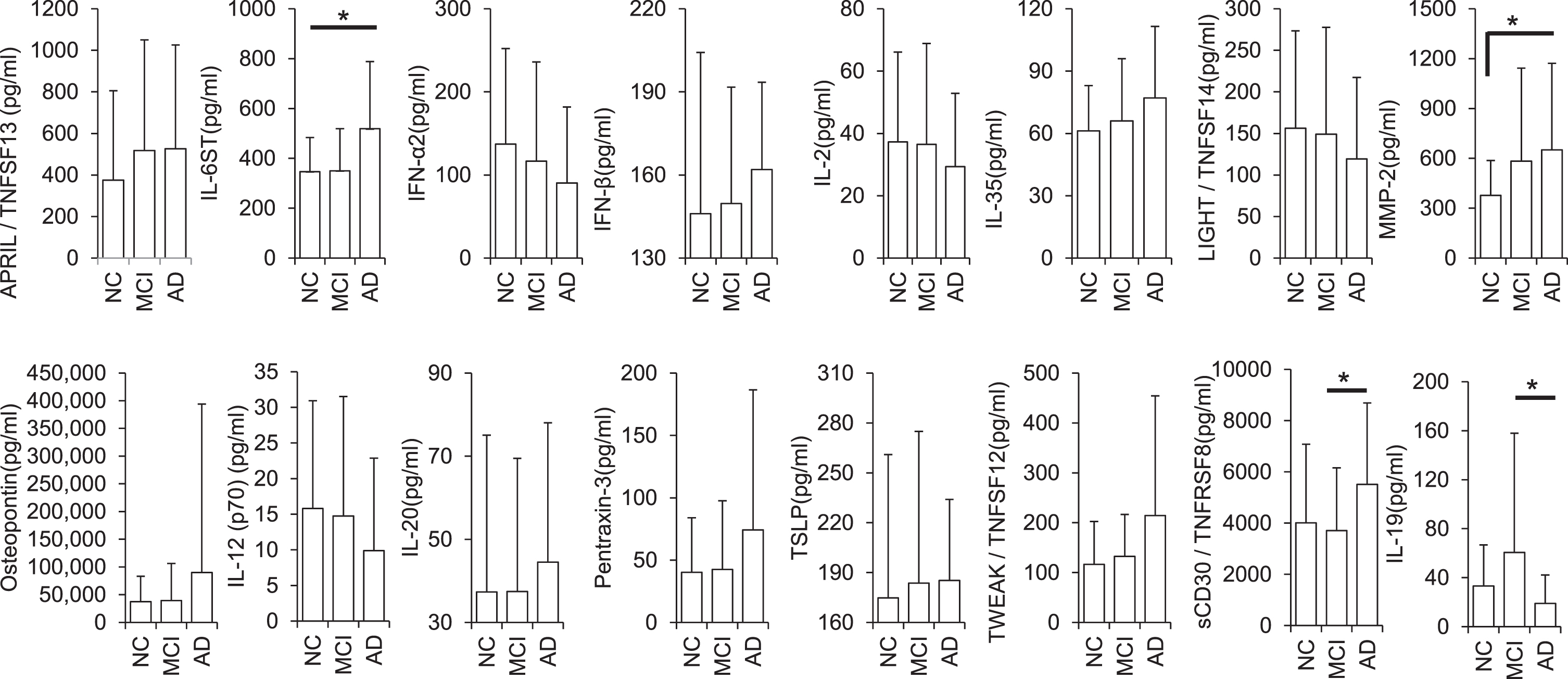
The univariate analysis comparing mean concentrations (pg/ml) of all inflammatory biomarkers between AD and MCI participants showed Tumor Necrosis Factor Receptor Superfamily Member 8 (TNFRSF8), Interleukin 6 Cytokine Family Signal Transducer (IL6ST/gp130), and Interleukin-19 (IL-19) to be differentially expressed. Contrastingly, while comparing the mean concentrations of all inflammatory biomarkers between NC versus AD, only IL6ST and Matrix metalloproteinase 2 (MMP-2) reached statistical significance. It is noteworthy that 41% of all cytokine concentrations observed exhibit either an upward or downward trend from NC to AD suggesting MCI a prodromal step before the onset of AD (Fig. 1, Supplementary Table 2).
Fig. 1
Differential expression of cytokines in AD, MCI, and normal control urine (pg/ml; *p < 0.05). Error bars represent standard deviations (SD).
Biomarker panels and machine learning models
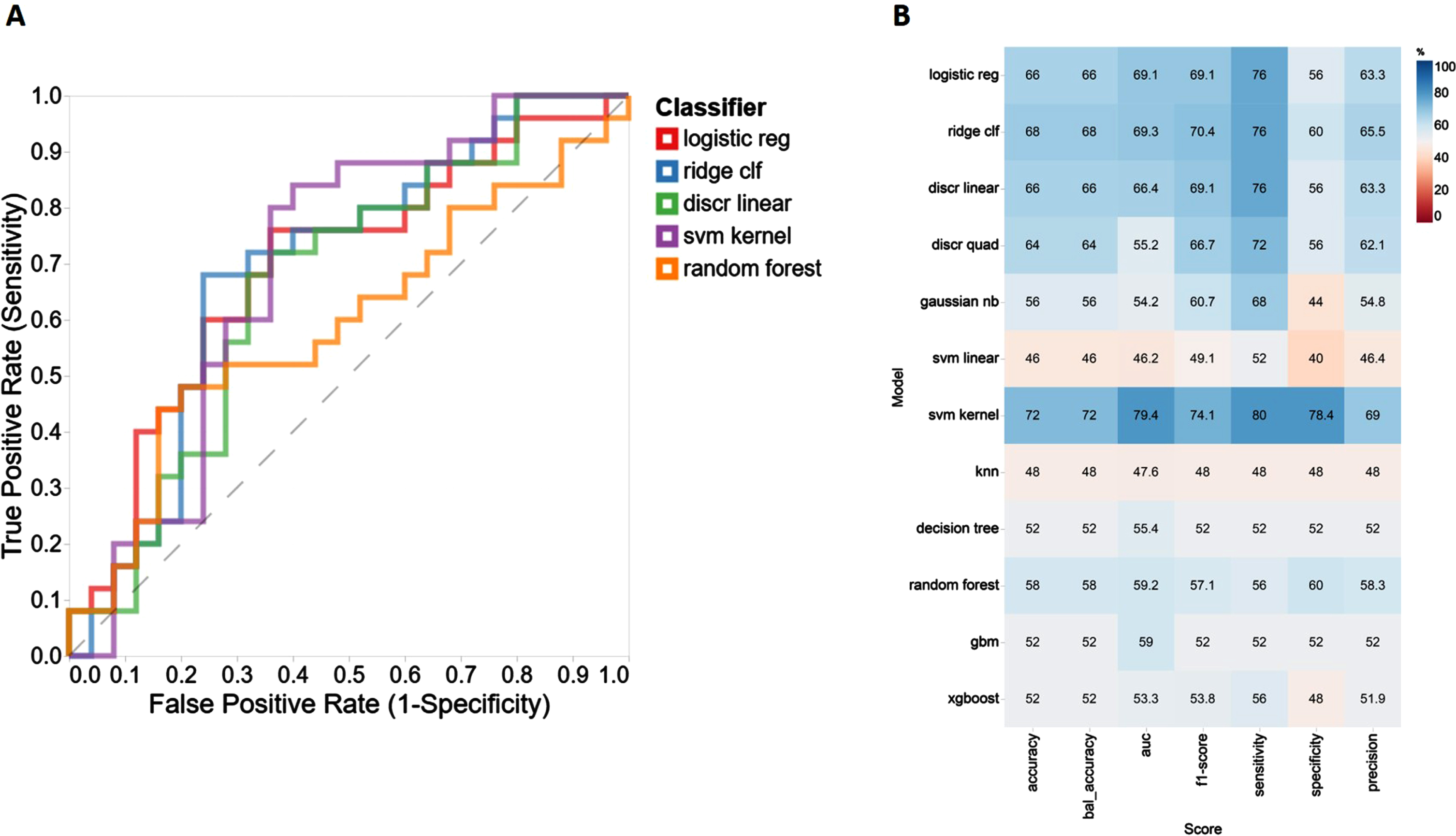
‘IFN-α2’, ‘IL-19’, ‘IL-12(p70)’, ‘IL-29/FN-λ1’, and ‘APRIL/TNFSF1’ were identified by RFE to be the most discriminative features when NC and AD cases were compared. In particular, K nearest neighbors algorithm (k = 5) with 200 oversampled points provided the best diagnostic outcome for all machine learning (ML) based models. The performance of 11 ML models used in this study was assessed by calculating a classification accuracy rate, AUROC, sensitivity, and specificity values. Based on these performance metrics, the best three classifiers diagnosing AD cases when compared to NC cases were found to be SVM kernel, logistic regression, and ridge clf (Fig. 2A) providing AUC values = 79.4, 69.3, and 69.1, respectively (Fig. 2B). When we compare MCI and NC individuals, ‘IFN-γ’, ‘IL-12(p40)’, ‘IFN-α2’, ‘IL-34’, and ‘MMP-2’ were found to be the most discriminative markers. Of the 11 different ML models we assessed for distinguishing MCI sufferers from NCs, the top three outperforming classifiers were found to be xgboost, knn, and gbm (Supplementary Figure 1A) providing AUC values = 78.6, 68.3, and 67.7, respectively (Supplementary Figure 1B). Finally, for the AD versus MCI comparison, of the 11 different ML models assessed and using RFE selected variables (IL-19, IFN-γ, IL-34, gp130/sIL-6Rβ, and sCD30/TNFRSF8) decision tree, svm kernel and logistic regression were found to be the most discriminative models (Supplementary Figure 2A) with AUC values of = 74.9, 65.4, and 65.3, respectively (Supplementary Figure 2B).
Fig. 2
Performance evaluation metrics for each ML-based model to include sensitivity, specificity, accuracy, and AUC using the RFE variable selection algorithm for distinguishing NC from AD and corresponding AUC curves top five models.
Pathway analysis
We report several biochemical pathways which may have been affected by the abnormal expression of the measured cytokines, and they include molecular functions, biological processes, biological pathways, and transcription factors when comparing AD cases with NC. Within the molecular function aspect, cytokine activity (p < 0.001) was identified as the most significantly different pathway when comparing AD and NC (Supplementary Figure 3A). In biological processes, the immune response pathway (p < 0.001) was the most significantly perturbed biochemical pathway (Supplementary Figure 3B) and in the biological pathway, validated transcriptional targets of the AP1 family (p = 0.017) and syndecan-2 mediated signaling events (p = 0.043) were identified as being significantly perturbed (Supplementary Figure 3C). Nuclear factor I C (NFIC) was the most significant transcription factor (TF) identified (p = 0.005) (Supplementary Figure 3D). We also identified several LINCRNAs whose expression could be modified by these cytokines. LINC01937 (p = 0.00001269) and LINC02376 (p = 0.00001269) were the top LINCRNAs we identified as being perturbed (Supplementary Table 3A). Subsequent Clustergram highlights MMP-2 as having significant co-expression with LINC01937 (p = 0.00001269), LINC01920 (p = 0.0004032), and LINC00687 (p = 0.0004032) (Supplementary Figure 4A).
Using an in-silico approach, we identified several miRNAs that may regulate these cytokines. FunRich provided gene hits on mmu-miRNAs; however, to maintain uniformity, we have converted all mmu-miRNAs to hsa-miRNAs based on the homolog search using Ingenuity Pathways Analysis [23]. Among the top 10 significant miRNAs (Supplementary Figure 4B), the human homolog of hsa-miR-503-3p (p = 0.00238) and hsa-miR-27a-3p (p = 0.0018) were the most significant. Of the top 10 miRNAs, hsa-miR-7152-3p (p = 0.015), hsa-miR-142a-3p (p = 0.020), hsa-miR-130a-3p (p = 0.025), and hsa-miR-4661-5p (p = 0.02) are regulated by IL6ST (Supplementary Table 3B). We also identified several miRNAs targeted by these cytokines. hsa-miR-4278 (p = 0.027) was the most significant target miRNA of IL6ST and MMP-2 (Supplementary Table 3C, Supplementary Figure 4C). Cytokines are widely known to modulate TF and affect molecular pathways. The in-silico analysis revealed an increase in the expression of several transcription factors, suggesting a possible regulatory role in our study (Supplementary Table 3D, (Supplementary Figure 4D). The pathway analysis further identified proteins MMP-2 and IL6ST in targeting the TF, FLI1.
DISCUSSION
We investigated the diagnostic utility of urinary cytokines for AD and MCI detection [24]. To the best of our knowledge, this is the first reported study measuring the level of inflammatory biomarkers in the urine of AD patients. Our results highlight significant differences in the concentrations of several urinary cytokines when we compare AD with NCs. Similarly, significant differences were found when we compared AD with MCI and NCs with MCI. Further, using the recorded concentration data we developed several ML models to differentiate both MCI and AD cases from controls and evaluated them based on classification accuracy rate, sensitivity, and specificity. It is worth noting that of 11 ML models evaluated herein, the decision tree repeatedly performed well.
We identified IL6ST and MMP-2 as significantly different between NC and AD. Our data show that MMP-2 levels are significantly elevated in AD compared to NC. MMP-2 is protective in AD as it plays a vital role in cell death, injury, and repair [25]. Elevated expression of MMP-2 is associated with Aβ induced neuronal cell death, a pathological hallmark of AD [26]. MMP-2 regulates many signaling molecules, including neuro-inflammation, synaptic dysfunction, and neuronal death [27]. It is known for remodeling the pericellular environment by regulating the cleavage of extracellular matrix proteins, cell surface components, neurotransmitter receptors, and growth factors [26]. Also, MMP-2 levels are positively correlated with MMSE scores in AD patients [28]. The interaction between Aβ and a receptor for advanced glycation end products (RAGE) activates an intracellular signaling cascade disrupting tight junction that leads to the breakdown of blood-brain barrier integrity. Aβ-RAGE-CaN-MMP cascade is critical for disruption and AD pathogenesis [29].
IL6ST is a signal transducer receptor shared by many cytokines, including interleukin 6 (IL-6), ciliary neurotrophic factor (CNTF), leukemia inhibitory factor (LIF), and oncostatin M (OSM) [30]. Also, IL6ST regulates metabolic, regenerative, and neural processes [30]. Other studies have shown that a variant (p.D358A, rs2228145) present in IL6R alters the ratio of IL6R in microglia resulting in elevated gene expression in late-onset AD [31]. Studies have shown that the IL-6 protein is involved in regulating the IL6ST gene [32]. A protein-protein complex consisting of IL-6 and IL6R increases the activation of the dimeric IL6ST protein [33]. Soluble IL6RA is also found to increase the antagonistic activity of soluble IL6ST protein [34]. Protein-protein binding interaction occurs between IL-6 and IL6ST [35]. IL-6 protein in the extracellular space is found to increase the activation of IL6ST protein in the plasma membrane [36]. IL6ST protein increases the neutralization of an active protein-protein complex consisting of IL-6 and soluble IL6R [32]. In cell assays, the IL-6 gene in JHOC5 cells is known to decrease the expression of IL6ST protein [37]. In Hela cells, IL-6 protein increases rapid translocation of IL6ST protein from cell surface to endosomal compartment [38], while in 293t cells, IL-6 protein increases polyubiquitination of IL6ST [38]. In our study, we observe elevated levels of TNFRSF8 and IL6ST in AD compared to MCI and NC; as such we hypothesize that elevated levels of IL6ST may contribute to the development of AD.
We identified TNFRSF8 (CD30), IL6ST, and IL-19 expression levels as significantly different between AD and MCI groups. TNFRSF8 is the proinflammatory cytokine mainly expressed by activated T cells and macrophages [39] and plays a vital role in cellular growth and survival [39–41]. TNFRSF8 has been associated with neuroinflammation [42] and could play a critical role in AD progression. The TNF gene at chromosome 6p21.3 has an AD association region [43–45]. The TNF haplotype TNF-308 2 and TNFa 2 polymorphisms are associated with increased transcriptional activity resulting in the overproduction of TNF receptors in AD [46]. An auto-amplified increased level of TNF-α can stimulate Aβ production and neuronal loss [47], and transduction of TNFRSF8 protein increases expression of TNF mRNA [48].
Immunosuppressive cytokine IL-19 has been positively correlated with AD pathogenesis at both upregulated and downregulated levels [49]; however, we observed downregulated expression of IL-19 in the urine of AD patients compared to MCI patients. This finding could be explained by microglial cells secreting IL-19 in the CNS [49]. Secreted IL-19 further regulates microglial function in an autocrine fashion and suppresses the production of pro-inflammatory cytokines IL-1β, IL-6, and TNF-α [49, 50]. However, under chronic inflammation, such as during AD pathogenesis, we have observed increased release of pro-inflammatory cytokines in the biofluids, which correlates with other studies [49]. This suggests that microglial cells may not secrete enough IL-19 resulting in lower levels of IL-19 in the urine of our AD sample cohort. Further studies are warranted to fully elucidate the relationship between the inflammatory stage and cytokine release in neurodegenerative disease while comparing the mean concentrations of all inflammatory biomarkers.
We also identified TNFSF13, IL6ST, IFN-α2, IFN-β, IL-2, IL-12, IL-20, IL-35, TNFSF14, MMP-2, Osteopontin, Pentraxin-3, TSLP, and TNFSF12 as intermediately expressed in MCI, as compared to AD and NC. Of all the measured cytokines, approximately 41% were recorded between the levels as observed in NCs and AD patients. This may suggest that MCI is a prodromal step before the onset of AD.
In silico analysis
miRNA and LINCRNA play an important role in neurite outgrowth, neuronal differentiation, synaptic plasticity, dendritic spine morphology [51, 52], and various cellular functions by interacting at the mRNA level resulting in the inhibition of protein synthesis [53]. In the immune system, miRNA regulates immune cell function in response to several stimuli [54], contributing to neural dysfunction in AD brain [51]. We identified several miRNAs and LINCRNAs that these inflammatory biomarkers could target. The miRNAs fall into two groups: a) miRNA that could be causal regulatory factors for cytokine expression, and b) Differentially expressed cytokines that target miRNAs. However, both groups are likely to play a critical role in AD pathogenesis [51]. Among regulatory miRNAs, hsa-miR-7152-3p, hsa-miR-142a-3p, hsa-miR-130a-3p, and hsa-miR-4661-5p are regulated by cytokine IL6ST. Hence, these miRNAs could prove to be promising therapeutic candidates for AD pathogenesis. Similarly, hsa-miR-4278 targets proinflammatory cytokine IL6ST and MMP-2. These miRNAs have not been previously reported for their role in AD pathogenesis; however, this novel finding requires further investigation.
Moreover, we identified LINC01937, LINC01920, and LINC00687 target MMP-2. As previously, these could prove to be promising therapeutic targets. For instance, the development of anti-oligonucleotide strategies to regulate these in those individuals suffering from AD.
Conclusion
This cross-sectional study measured cytokine levels in urine samples from AD, MCI, and cognitively normal subjects. Our study demonstrates the potential of inflammatory biomarkers, particularly cytokines in urine, as noninvasive diagnostic biomarkers of AD. Further, using an in-silico approach, we link the changes as observed in the dementia spectrum to the potential onset and pathogenesis of AD. There are several limitations to this study. First, the sample size used herein is modest, but we believe with a much larger sample set we would have the power to demonstrate their potential clinical utility. Second, systemic inflammatory diseases and comorbidities might affect the bladder and related urine conditions which have the potential to confound the results. Thus, additional studies are warranted to determine the importance of these changes, how inflammation is involved in AD development, and whether these targets can slow or stop the progression of the disease.
ACKNOWLEDGMENTS
We express our gratitude to the John and Marilyn Bishop Charitable Foundation, the Fred A. & Barbara M. Erb Foundation, and the Maibach family for their generous contributions. Their support has been invaluable in making this work possible.
FUNDING
This work was partly funded by the generous contribution made by the John and Marilyn Bishop Charitable Foundation and the Fred A. & Barbara M. Erb Foundation.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
DATA AVAILABILITY
The data supporting the findings of this study are available on request from the corresponding author.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/ADR-220081.
REFERENCES
[1] | ((2022) ) 2022 Alzheimer’s disease facts and figures. Alzheimers Dement 18: , 700–789. |
[2] | ((2021) ) 2021 Alzheimer’s disease facts and figures. Alzheimers Dement 17: , 327–406. |
[3] | Lane CA , Hardy J , Schott JM ((2018) ) Alzheimer’s disease. Eur J Neurol 25: , 59–70. |
[4] | Hu C , Yu D , Sun X , Zhang M , Wang L , Qin H ((2017) ) The prevalence and progression of mild cognitive impairment among clinic and community populations: A systematic review and meta-analysis. Int Psychogeriatr 29: , 1595–1608. |
[5] | Petersen RC , Lopez O , Armstrong MJ , Getchius TSD , Ganguli M , Gloss D , Gronseth GS , Marson D , Pringsheim T , Day GS , Sager M , Stevens J , Rae-Grant A ((2018) ) Practice guideline update summary: Mild cognitive impairment: Report of the Guideline Development, Dissemination, and Implementation Subcommittee of the American Academy of Neurology. Neurology 90: , 126–135. |
[6] | Rosenberg A , Solomon A , Soininen H , Visser PJ , Blennow K , Hartmann T , Kivipelto M ((2021) ) Research diagnostic criteria for Alzheimer’s disease: Findings from the LipiDiDiet randomized controlled trial. Alzheimers Res Ther 13: , 64. |
[7] | Jack CR Jr. , Bennett DA , Blennow K , Carrillo MC , Dunn B , Haeberlein SB , Holtzman DM , Jagust W , Jessen F , Karlawish J , Liu E , Molinuevo JL , Montine T , Phelps C , Rankin KP , Rowe CC , Scheltens P , Siemers E , Snyder HM , Sperling R ((2018) ) NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement 14: , 535–562. |
[8] | Hansson O ((2021) ) Biomarkers for neurodegenerative diseases. Nat Med 27: , 954–963. |
[9] | Schley G , Köberle C , Manuilova E , Rutz S , Forster C , Weyand M , Formentini I , Kientsch-Engel R , Eckardt KU , Willam C ((2015) ) Comparison of plasma and urine biomarker performance in acute kidney injury. PLoS One 10: , e0145042. |
[10] | Wang WY , Tan MS , Yu JT , Tan L ((2015) ) Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann Transl Med 3: , 136. |
[11] | Piancone F , La Rosa F , Marventano I , Saresella M , Clerici M ((2021) ) The role of the inflammasome in neurodegenerative diseases. Molecules 26: , 953. |
[12] | Dansokho C , Heneka MT ((2018) ) Neuroinflammatory responses in Alzheimer’s disease. J Neural Transm 125: , 771–779. |
[13] | Heneka MT , Carson MJ , El Khoury J , Landreth GE , Brosseron F , Feinstein DL , Jacobs AH , Wyss-Coray T , Vitorica J , Ransohoff RM , Herrup K , Frautschy SA , Finsen B , Brown GC , Verkhratsky A , Yamanaka K , Koistinaho J , Latz E , Halle A , Petzold GC , Town T , Morgan D , Shinohara ML , Perry VH , Holmes C , Bazan NG , Brooks DJ , Hunot S , Joseph B , Deigendesch N , Garaschuk O , Boddeke E , Dinarello CA , Breitner JC , Cole GM , Golenbock DT , Kummer MP ((2015) ) Neuroinflammation in Alzheimer’s disease. Lancet Neurol 14: , 388–405. |
[14] | Patterson ZR , Holahan MR ((2012) ) Understanding the neuroinflammatory response following concussion to develop treatment strategies. Front Cell Neurosci 6: , 58. |
[15] | McKhann G , Drachman D , Folstein M , Katzman R , Price D , Stadlan EM ((1984) ) Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34: , 939–944. |
[16] | Pang Z , Zhou G , Ewald J , Chang L , Hacariz O , Basu N , Xia J ((2022) ) Using MetaboAnalyst 5.0 for LC– HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat Protoc 17: , 1735–1761. |
[17] | Purohit PV , Rocke DM , Viant MR , Woodruff DL ((2004) ) Discrimination models using variance-stabilizing transformation of metabolomic NMR data. OMICS 8: , 118–130. |
[18] | Schratz P , Muenchow J , Iturritxa E , Richter J , Brenning A ((2019) ) Hyperparameter tuning and performance assessment of statistical and machine-learning algorithms using spatial data. Ecol Modell 406: , 109–120. |
[19] | Pathan M , Keerthikumar S , Ang CS , Gangoda L , Quek CY , Williamson NA , Mouradov D , Sieber OM , Simpson RJ , Salim A , Bacic A , Hill AF , Stroud DA , Ryan MT , Agbinya JI , Mariadason JM , Burgess AW , Mathivanan S ((2015) ) FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15: , 2597–2601. |
[20] | Chen EY , Tan CM , Kou Y , Duan Q , Wang Z , Meirelles GV , Clark NR , Ma’ayan A ((2013) ) Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: , 128. |
[21] | Xie Z , Bailey A , Kuleshov MV , Clarke DJB , Evangelista JE , Jenkins SL , Lachmann A , Wojciechowicz ML , Kropiwnicki E , Jagodnik KM , Jeon M , Ma’ayan A ((2021) ) Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1: , e90. |
[22] | Kuleshov MV , Jones MR , Rouillard AD , Fernandez NF , Duan Q , Wang Z , Koplev S , Jenkins SL , Jagodnik KM , Lachmann A , McDermott MG , Monteiro CD , Gundersen GW , Ma’ayan A ((2016) ) Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: , W90–W97. |
[23] | Li SC , Chan WC , Hu LY , Lai CH , Hsu CN , Lin WC ((2010) ) Identification of homologous microRNAs in 56 animal genomes. Genomics 96: , 1–9. |
[24] | Seol W , Kim H , Son I ((2020) ) Urinary biomarkers for neurodegenerative diseases. Exp Neurobiol 29: , 325–333. |
[25] | Miners JS , Baig S , Palmer J , Palmer LE , Kehoe PG , Love S ((2008) ) Abeta-degrading enzymes in Alzheimer’s disease. Brain Pathol 18: , 240–252. |
[26] | Wang XX , Tan MS , Yu JT , Tan L ((2014) ) Matrix metalloproteinases and their multiple roles in Alzheimer’s disease. Biomed Res Int 2014: , 908636. |
[27] | Hoogmartens J , Hens E , Engelborghs S , De Deyn PP , van der Zee J , Van Broeckhoven C , Cacace R , BELNEU consortium ((2021) ) Investigation of the role of matrix metalloproteinases in the genetic etiology of Alzheimer’s disease. Neurobiol Aging 104: , 105.e101–105.e106. |
[28] | Lim NK , Villemagne VL , Soon CP , Laughton KM , Rowe CC , McLean CA , Masters CL , Evin G , Li QX ((2011) ) Investigation of matrix metalloproteinases, MMP-2 and MMP-9, in plasma reveals a decrease of MMP-2 in Alzheimer’s disease. J Alzheimers Dis 26: , 779–786. |
[29] | Kook SY , Hong HS , Moon M , Ha CM , Chang S , Mook-Jung I ((2012) ) Aβ1-42-RAGE interaction disrupts tight junctions of the blood-brain barrier via Ca2+-calcineurin signaling. J Neurosci 32: , 8845–8854. |
[30] | Boulton TG , Stahl N , Yancopoulos GD ((1994) ) Ciliary neurotrophic factor/leukemia inhibitory factor/interleukin 6/oncostatin M family of cytokines induces tyrosine phosphorylation of a common set of proteins overlapping those induced by other cytokines and growth factors. J Biol Chem 269: , 11648–11655. |
[31] | Haddick PC , Larson JL , Rathore N , Bhangale TR , Phung QT , Srinivasan K , Hansen DV , Lill JR , Pericak-Vance MA , Haines J , Farrer LA , Kauwe JS , Schellenberg GD , Cruchaga C , Goate AM , Behrens TW , Watts RJ , Graham RR , Kaminker JS , van der Brug M ((2017) ) A common variant of IL-6R is associated with elevated il-6 pathway activity in Alzheimer’s disease brains. J Alzheimers Dis 56: , 1037–1054. |
[32] | Heinrich PC , Behrmann I , Müller-Newen G , Schaper F , Graeve L ((1998) ) Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J 334 (Pt 2): , 297–314. |
[33] | Stroud RM , Wells JA ((2004) ) Mechanistic diversity of cytokine receptor signaling across cell membranes. Sci STKE 2004: , re7. |
[34] | Heinrich PC , Behrmann I , Haan S , Hermanns HM , Müller-Newen G , Schaper F ((2003) ) Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 374: , 1–20. |
[35] | Schuliga M , Jaffar J , Harris T , Knight DA , Westall G , Stewart AG ((2017) ) The fibrogenic actions of lung fibroblast-derived urokinase: A potential drug target in IPF. Sci Rep 7: , 41770. |
[36] | O’Donoghue RJ , Knight DA , Richards CD , Prêle CM , Lau HL , Jarnicki AG , Jones J , Bozinovski S , Vlahos R , Thiem S , McKenzie BS , Wang B , Stumbles P , Laurent GJ , McAnulty RJ , Rose-John S , Zhu HJ , Anderson GP , Ernst MR , Mutsaers SE ((2012) ) Genetic partitioning of interleukin-6 signalling in mice dissociates Stat3 from Smad3-mediated lung fibrosis. EMBO Mol Med 4: , 939–951. |
[37] | Azar WJ , Christie EL ((2020) ) Noncanonical IL6 signaling-mediated activation of YAP regulates cell migration and invasion in ovarian clear cell cancer. Cancer Res 80: , 4960–4971. |
[38] | Tanaka Y , Tanaka N , Saeki Y , Tanaka K , Murakami M , Hirano T , Ishii N , Sugamura K ((2008) ) c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol Cell Biol 28: , 4805–4818. |
[39] | Mak TW , Saunders ME ((2006) ) 29 - Autoimmune Disease. In The Immune Response, MakTW, SaundersME, eds. Academic Press, Burlington, pp. 963–1023. |
[40] | Croft M ((2009) ) The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol 9: , 271–285. |
[41] | Yamashita M , Passegué E ((2019) ) TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell 25: , 357–372.e357. |
[42] | Yuan S , Li H , Xie J , Sun X ((2019) ) Quantitative trait module-based genetic analysis of Alzheimer’s disease. Int J Mol Sci 20: , 5912. |
[43] | Perry RT , Collins JS , Wiener H , Acton R , Go RC ((2001) ) The role of TNF and its receptors in Alzheimer’s disease. Neurobiol Aging 22: , 873–883. |
[44] | Collins JS , Perry RT , Watson B Jr , Harrell LE , Acton RT , Blacker D , Albert MS , Tanzi RE , Bassett SS , McInnis MG , Campbell RD , Go RC ((2000) ) Association of a haplotype for tumor necrosis factor in siblings with late-onset Alzheimer disease: The NIMH Alzheimer Disease Genetics Initiative. Am J Med Genet 96: , 823–830. |
[45] | Steffens DC , Garrett ME , Soldano KL , McQuoid DR , Ashley-Koch AE , Potter GG ((2020) ) Genome-wide screen to identify genetic loci associated with cognitive decline in late-life depression. Int Psychogeriatr. 10.1017/S1041610220001143. |
[46] | Kroeger KM , Steer JH , Joyce DA , Abraham LJ ((2000) ) Effects of stimulus and cell type on the expression of the -308 tumour necrosis factor promoter polymorphism. Cytokine 12: , 110–119. |
[47] | Koenigsknecht-Talboo J , Landreth GE ((2005) ) Microglial phagocytosis induced by fibrillar beta-amyloid and IgGs are differentially regulated by proinflammatory cytokines. J Neurosci 25: , 8240–8249. |
[48] | Muta H , Boise LH , Fang L , Podack ER ((2000) ) CD30 signals integrate expression of cytotoxic effector molecules, lymphocyte trafficking signals, and signals for proliferation and apoptosis. J Immunol 165: , 5105–5111. |
[49] | Horiuchi H , Parajuli B , Wang Y , Azuma YT , Mizuno T , Takeuchi H , Suzumura A ((2015) ) Interleukin-19 acts as a negative autocrine regulator of activated microglia. PLoS One 10: , e0118640. |
[50] | Sawada M , Suzumura A , Hosoya H , Marunouchi T , Nagatsu T ((1999) ) Interleukin-10 inhibits both production of cytokines and expression of cytokine receptors in microglia. J Neurochem 72: , 1466–1471. |
[51] | Angelucci F , Cechova K , Valis M , Kuca K , Zhang B , Hort J ((2019) ) MicroRNAs in Alzheimer’s disease: Diagnostic markers or therapeutic agents? Front Pharmacol 10: , 665. |
[52] | Zhou B , Li L , Qiu X , Wu J , Xu L , Shao W ((2020) ) Long non-coding RNA ANRIL knockdown suppresses apoptosis and pro-inflammatory cytokines while enhancing neurite outgrowth via binding microRNA-125a in a cellular model of Alzheimer’s disease. Mol Med Rep 22: , 1489–1497. |
[53] | Wahid F , Shehzad A , Khan T , Kim YY ((2010) ) MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta 1803: , 1231–1243. |
[54] | Chandan K , Gupta M , Sarwat M ((2019) ) Role of host and pathogen-derived MicroRNAs in immune regulation during infectious and inflammatory diseases. Front Immunol 10: , 3081. |




