Influence of APOE Genotype on Mortality and Cognitive Impairment
Abstract
While many studies have examined the associations between APOE genotype and mortality, findings have often been conflicting and it remains unclear whether APOE genotype affects longevity. Using selected individuals from the Manchester arm of the Brains for Dementia Research programme and University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age, we investigated relationships between APOE genotype and age at death in both cognitively normal and cognitively impaired individuals. Results indicated that carrying the APOE ɛ4 allele led to a reduced chance in an individual reaching 80+ years and remaining cognitively healthy. Conversely, APOE ɛ2 carriers tended to live longer and remain cognitively normal. These findings add to the evidence that APOE genotype influences longevity, especially in cognitively impaired individuals who carry the APOE ɛ4 allele.
INTRODUCTION
The Apolipoprotein E (APOE) gene has been strongly implicated in Alzheimer’s disease (AD) and in longevity. APOE has three polymorphic forms, ɛ2, ɛ3, and ɛ4, which have a frequency in the general worldwide population of 8.4%, 77.9%, and 13.7% [1]. Carriers of the APOE ɛ4 allele are at a higher risk of AD [2] whereas those who carry the APOE ɛ2 allele are thought to be protected from the disease (relative to APOE ɛ3 and APOE ɛ4 carriers) [3]. Studies of older individuals have found that the frequency of APOE ɛ4 is lower and APOE ɛ2 is higher than in the general population [4].
Many studies have examined the associations between APOE genotype and mortality with often conflicting findings. Genome-wide association studies have linked APOE with longevity [5]. It has been reported that carriers of the APOE ɛ4 allele die earlier than expected [6], whereas other studies find earlier age at death only in those with the APOE ɛ4/ɛ4 genotype [7]. A study looking at three independent cohorts from Italy, Spain, and Japan concluded that longevity was negatively associated with the ɛ4 allele and positively associated with the ɛ2 allele [8]. Similarly, another study reported that extreme longevity was negatively associated with APOE ɛ4/ɛ4, ɛ3/ɛ4, and ɛ2/ɛ4 genotypes whereas there was a positive association between longevity and the APOE ɛ2/ɛ3 genotype [9]. However, associations between age at death and APOE ɛ4 allele(s) were not found in other studies based on individuals over the age of 85 [10, 11], nor were they found when attempting to correlate APOE isoforms, age, and levels of blood cholesterol [12].
In the present study, we have investigated relationships between APOE genotype and age at death in both cognitively normal and cognitively impaired individuals recruited in the Manchester arm of the Brains for Dementia Research (BDR) programme and University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age (UMLCHA). We focused principally on those individuals with AD pathology (without concomitant or secondary pathologies) and those with pathology considered normal for age. The main aims of the study were to assess the interactions between APOE genotype and age group at death (79 years and under, 80–89 years, and 90 years and over) and investigate the impact of cognitive impairment on any associations found.
MATERIALS AND METHODS
The present study combines the UMLCHA and the Manchester arm of the BDR cohorts. Details concerning clinical characteristics and neuropathological features of these cohorts have been presented by the authors elsewhere [13–15].
The study was approved by Manchester Brain Bank Management Committee (REC reference 19/NE/0242). Under conditions agreed with the Research Ethics Committee, The Manchester Brain Bank can supply data to researchers, without any requirement for researchers to apply individually to the REC for approval.
For BDR, participants underwent cognitive assessments either via telephone interview or via a visit to the participant’s home. Details of cognitive assessments have been previously described [12].
For UMLCHA, cognitive status at death was assigned using a combination of last modified Telephone Instrument for Cognitive Status (TICSm) score, patient notes obtained via the participants’ general practitioner, cause of death as recorded on the death certificate and information gained from the Brain Bank Coordinator (PT).
Neuropathological assessment
Postmortem assessment of the individuals in these cohorts has been previously described [14, 15]. Consensus criteria were used to establish the presence and stage of neurodegenerative diseases and cerebrovascular pathology.
For the purpose of this study, we excluded all cases where the primary neuropathological diagnosis was not AD and also excluded AD cases where there was any concomitant or secondary pathology (other than cerebral amyloid angiopathy or small vessel disease). We included cases of primary age-related tauopathy, aging-related tau astrogliopathy, and limbic-predominant age-related TDP-43 encephalopathy due to the fact that they are relatively common age-related pathologies.
The two ongoing cohorts currently comprise 290 subjects. After applying the above exclusion criteria, a total of 182 participants (84 BDR and 98 UMLCHA) were considered eligible for the present study (Supplementary Table 1). These were then stratified into groups based on age at death: 79 years and under, 80–89 years, and 90 years and over.
APOE genotyping
DNA was extracted from frozen brain tissue using REDExtract-N-Amp™ Tissue PCR Kit (Sigma) or from blood (UMLCHA –3 cases). The APOE genotype was determined using routine polymerase chain reaction (PCR) methods [16].
Statistical analysis
Pearson’s Chi-squared test was used to compare demographic features. This test was also used to analyze whether there were differences between proportions of APOE ɛ4 and APOE ɛ2 carriers in the various age at death groups. T-test was used to distinguish between differences in mean age at death for APOE genotypes both overall and stratified by cognitive status.
A p value of <0.05 was considered significant for all tests.
RESULTS
Demographics
Demographic information, stratified by age group at death, can be found in Table 1. There were differences in sex ratio when comparing those 79 years and under with those 80–89 years (χ2 = 4.25; p = 0.039) and with those 90 years and over (χ2 = 4.78; p = 0.029) with males appearing more frequently in the 79 years and under group and females over represented in the 80–89 years and 90 years and over groups. However, there were no differences in the sex ratio between those 80–89 years and those 90 years and over (χ2 = 0.03; p = 0.869).
Table 1
Basic demographics of the 182 eligible participants split by age group at death
| Age group at death | ||||||
| 79 years and under | 80–89 years | 90 years and over | ||||
| N | % | N | % | N | % | |
| Sex | ||||||
| Male | 23 | 56.1 | 26 | 36.1 | 24 | 34.8 |
| Female | 18 | 43.9 | 46 | 63.9 | 45 | 65.2 |
| Cognitive status | ||||||
| Normal | 16 | 39.0 | 37 | 51.4 | 45 | 65.2 |
| Impaired | 25 | 61.0 | 35 | 48.6 | 24 | 34.8 |
The proportion of cognitively impaired individuals was lower in the 90 years and over age group when compared with the 79 years and under age group (χ2 = 7.14; p = 0.008). No other differences in cognitive status were apparent between the age groups.
APOE genotype distribution
The frequency and distribution of APOE genotype, stratified by cognitive status, can be found in Table 2. The most common APOE genotype was 3/3 (53.2%) and the least common was 2/2 (1.1%). When analyzing all eligible participants, regardless of cognitive status, we found that those with APOE 2/3 genotype had a later age at death compared to those with APOE 3/4 genotype (p = 0.004) and APOE 4/4 (p < 0.001). Mean age at death was also significantly later in those with APOE 2/4 genotype when compared with those with APOE 3/4 genotype (p = 0.026). Similarly, those with APOE 3/3 genotype died at a later age compared to those with APOE 4/4 genotype (p = 0.008). When stratifying the participants based on cognitive status, there were no differences in mean age at death when comparing cognitively normal and cognitively impaired individuals of the same genotype.
Table 2
Mean (±SD) age at death for each APOE genotype stratified by cognitive status
| APOE genotype | N | Age at death (Mean±SD) | |
| Cognitively normal | Cognitively impaired | ||
| 2/2 | 2 | 81.0±4.2 | n/a |
| 2/3 | 17 | 90.5±6.2 | 86.7±3.2 |
| 2/4 | 4 | 79.0±14.1 | 73.0±19.8 |
| 3/3 | 97 | 86.8±9.1 | 86.1±8.6 |
| 3/4 | 50 | 85.2±11.1 | 82.8±9.1 |
| 4/4 | 12 | n/a | 79.3±7.2 |
Effects of APOE ɛ4 and APOE ɛ2 alleles on age at death
The proportion of cases exhibiting one or more APOE ɛ4 and APOE ɛ2 allele(s) and allele frequencies of APOE ɛ4 and APOE ɛ2 alleles are shown in Fig. 1A and B.
Fig. 1
Frequencies and proportions of APOE ɛ4 and APOE ɛ2, stratified by age group at death and cognitive status (White –79 years and under; Grey –80 to 89 years; Black –90 years and older). A) The proportion of all eligible cases exhibiting at least one APOE ɛ4 or APOE ɛ2 allele, stratified by age group at death. B) Allele frequencies of APOE ɛ4 and APOE ɛ2 allele for all eligible cases stratified by age group at death. C, D) Allele frequencies of APOE ɛ4 and APOE ɛ2 allele stratified by age group at death and cognitive status; Panel C shows cognitively normal individuals and Panel D shows cognitively impaired individuals.
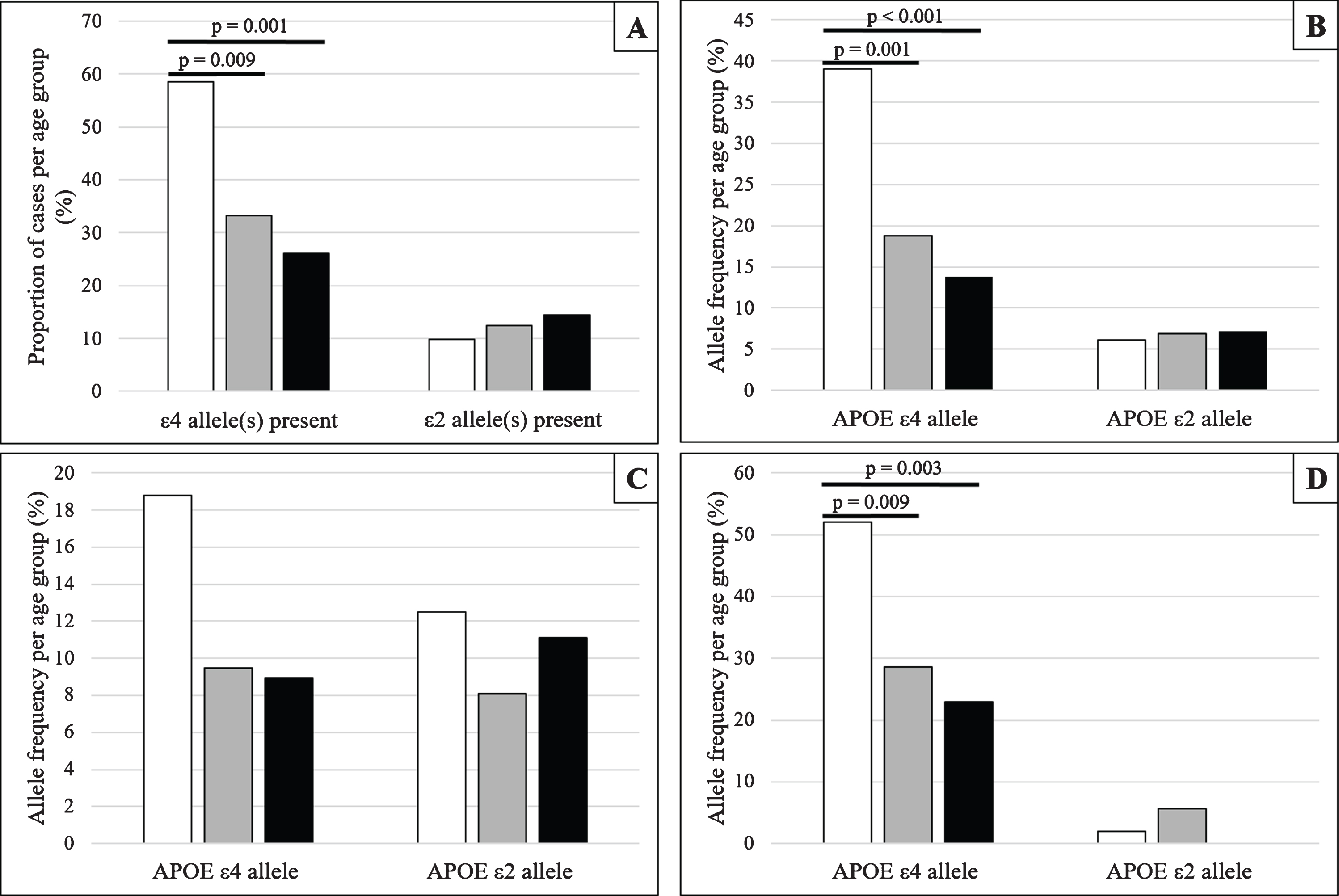
When analyzing all eligible participants, regardless of cognitive status, we found a significantly greater proportion of APOE ɛ4 carriers in the 79 years and under age group compared with those in the 80–89 years group (χ2 = 6.791; p = 0.009) and also the 90 years and over age group (χ2 = 11.472; p = 0.001). In addition, the APOE ɛ4 appeared more frequently in the 79 years and under age group when compared with the 80–89 years age group (χ2 = 11.133; p = 0.001) and the 90 years and over age group (χ2 = 18.425; p < 0.001). Although there was a trend for those who carry APOE ɛ2 to be found in the older age groups, and for the frequency of the APOE ɛ2 allele to be greater in the older age groups, these were not statistically significantly different.
Similar analyses were conducted after stratifying cases by cognitive status (Fig. 1C, D). In cognitively normal individuals, frequencies for APOE ɛ4 and APOE ɛ2 alleles were not found to be statistically different between the age groups. However, in those considered cognitively impaired at death, the frequency of APOE ɛ4 allele was greater in the 79 and under age group when compared with the 80–89 years age group (χ2 = 6.773; p = 0.009) and the 90 years and over age group (χ2 = 8.814; p = 0.003). No such differences were found between the 80–89 years and 90 years and over age groups or in APOE ɛ2 allele frequency for any of the age groups.
Investigation into relationships between severity of small vessel disease in the basal ganglia and presence of APOE ɛ4 allele(s), APOE ɛ2 allele(s), age group at death and cognitive impairment at death yielded no significant associations.
DISCUSSION
The importance of APOE genotype in AD and longevity has long been known but remains to be clearly understood. Here, using a carefully selected group of individuals with autopsy confirmed AD and also those considered (both cognitively and neuropathologically) normal for their age we found that an individual carrying APOE ɛ4 has a reduced chance of reaching 80+ years while avoiding cognitive impairment. Thus, it can be concluded that the effects of APOE ɛ4 on reducing longevity are strongest when cognitive impairment is also present. In contrast, there was a (non-significant) trend that suggested that carrying APOE ɛ2 increases the chances of living longer and avoiding cognitive impairment (unless an individual is APOE ɛ2/ɛ4). The lack of statistical significance could be due to the low numbers of APOE ɛ2 carriers in the study.
Although a large number of previous studies have found similar results [5–9], a smaller proportion of studies have shown conflicting findings [10–12]. A large, recent review of 12 cohorts [17] sought to use meta-analyses to assist in deducing the influence of APOE on longevity. Here, they concluded that, when compared to the ɛ3 allele, the ɛ4 allele was associated with a shorter lifespan and the ɛ2 allele was associated with a longer lifespan. Other meta-analyses have come to similar conclusions [18] as have studies examining the effects of APOE on the longevity of those with Down syndrome (where AD pathology is almost ubiquitous in aged cases) [19] leading to the assumption that our findings are in line with what has previously been found.
A main strength of the present study is that it uses autopsy verified cases of AD and also confirms no (or little) pathology in the cognitively normal individuals. Many prior studies have used other methods of deducing disease state at death, such as death certification, which can lead to errors in assigning cognitive status as under-reporting of dementia in clinical practice is a known phenomenon [20].
Alongside the robust neuropathological work-up, the study uses clinical information regarding cognitive impairment from a number of varied sources rather than simply relying on one measure of dementia (e.g., Mini-Mental State Examination). This ensures that the cognitive status of the individuals is as accurate as possible.
A general limitation of the study is that the recruitment strategies for both BDR and UMLCHA could introduce bias. They are both self-selecting and only cover specific geographical areas in the UK. Future studies might perhaps include the whole BDR cohort which may better reflect society in the UK. Another possible limitation is that the study combined two cohorts which had different methods of collecting data. However, previous studies on these cohorts have shown that, although different in their approach, they both provide a robust clinical diagnosis which matches well to the neuropathological diagnosis [15].
In conclusion, our data show that carrying the APOE ɛ4 allele leads to an increased chance of cognitive impairment and an early death. Conversely, carrying the APOE ɛ2 allele may lead to a longer life and better odds of remaining cognitively normal. These findings provide further evidence that APOE genotype is associated with mortality, especially in those who find themselves to be cognitively impaired.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
ACKNOWLEDGMENTS
University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age was funded by Medical Research Council, Economic and Social Research Council, The Wellcome Trust (grant reference number 003889) and Unilever PLC.
The work of Manchester Brain Bank is supported by Alzheimer’s Research UK and Alzheimer’s Society through the Brains for Dementia Research (BDR) Programme.
We also thank Daniel du Plessis and Piyali Pal for their help and assistance with neuropathology.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/ADR-200203.
REFERENCES
[1] | Farrer LA , Cupples LA , Haines JL , Hyman B , Kukull WA , Mayeux R , Myers RH , Pericak-Vance MA , Risch N , van Duijn CM ((1997) ) Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium, JAMA 278: , 1349–1356. |
[2] | Strittmatter WJ , Roses AD ((1996) ) Apolipoprotein E and Alzheimer’s disease, Annu Rev Neurosci 19: , 53–77. |
[3] | Safieh M , Korczyn AD , Michaelson DM ((2019) ) ApoE4: An emerging therapeutic target for Alzheimer’s disease, BMC Med 17: , 64. |
[4] | Lewis SJ , Brunner EJ ((2004) ) Methodological problems in genetic association studies of longevity-the apolipoprotein E gene as an example, . Intl J Epidemiol 33: , 962–970. |
[5] | Joshi PK , Pirastu N , Kentistou KA , Fischer K , Hofer E , Schraut KE , Clark DC , Nutile T , Barnes CLK , Timmers PRHJ , Shen X , Gandin I , McDaid AF , Hansen TF , Gordon SD , Giulianini F , Boutin TS , Abdellaoui A , Zhao W , Medina-Gomez C , Bartz TM , Trompet S , Lange LA , Raffield L , van der Spek A , Galesloot TE , Proitsi P , Yanek LR , Bielak LF , Payton A , Murgia F , Concas MP , Biino G , Tajuddin SM , Seppälä I , Amin N , Boerwinkle E , Børglum AD , Campbell A , Demerath EW , Demuth I , Faul JD , Frod I , Gialluisi A , Gögele M , Graff M , Hingorani A , Hottenga J , Hougaard DM , Hurme MA , Ikram MA , Jylhä M , Kuh D , Ligthart L , Lill CM , Lindenberger U , Lumley T , Mägi R , Marques-Vidal P , Medland SE , Milani L , Nagy R , Ollier WER , Peyser PA , Pramstaller PP , Ridker PM , Rivadeneira F , Ruggiero D , SAba Y , Schmidt R , Schmidt H , Slagboom PE , Smith BH , Smith JA , Sotoodehnia N , Steinhagen-Thiessen e , van Rooij FJA , Verbeek AL , Vermeulen SH , Vollenweider P , Wang Y , Werge T , whitfield JB , Zonderman AB , Lehtimäki T , Evans MK , Pirastu M , Fuchsberger C , Bertram L , Pendleton N , Kardia SLR , Ciullo M , Becker DM , Wong A , Psaty BM , van Duijn CM , Wilson JG , Jukema JW , Kiemeney L , Uitterlinden AG , Franceschini N , North KE , Weir DR , Metspalu A , Boomsma DI , Hayward C , Cahsman D , Martin NG , Sattar N , Campbell H , Esko T , Kutalik Z , Wilson JF ((2017) ) Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity, Nat Commun 8: , 910. |
[6] | Tilvis RS , Strandberg TE , Juva K ((1998) ) Apolipoprotein E phenotypes, dementia and mortality in a prospective population sample, J Am Geriatr Soc 46: , 712–715. |
[7] | Juva K , Verkkoniemi A , Viramo P , Polvikoski T , Kainulainen K , Kontula K , Sulkava R ((2000) ) APOE epsilon4 does not predict mortality, cognitive decline, or dementia in the oldest old, Neurology 54: , 412–415. |
[8] | Garatachea N , Emanuele E , Calero M , Fuku N , Arai Y , Abe Y , Murakami H , Miyachi M , Yvert T , Verde Z , Zea MA , Venturini l , Santiago C , Santo-Lozano A , Rodr íguez-Romo G , Ricevuti G , Hirose N , Rábano A , Lucia A ((2014) ) ApoE gene and exceptional longevity: Insights from three independent cohorts, Exp Gerontol 53: , 16–23. |
[9] | Garatachea N , Marín PJ , Santos-Lozano A , Sanchis-Gomar F , Emanuele E , Lucia A ((2014) ) The ApoE gene is related with exceptional longevity: A systematic review and meta-analysis, Rejuvenation Res 18: , 3–13. |
[10] | Skoog I , Hesse C , Aevarsson O , Landahl S , Wahlström J , Fredman P , Blennow K ((1998) ) A population study of APOE genotype at the age of 85: Relation to dementia, cerebrovascular disease, and mortality, J Neurol Neurosurg Psychiatry 64: , 37–43. |
[11] | Tschanz JT , Corcoran C , Skoog I , Khachaturian AS , Herrick J , Hayden KM , Welsh-Bohmer KA , Calvert T , Norton MC , Zandi P , Breitner JCS , ((2004) ) Cache County Study Group Dementia: The leading predictor of death in a defined elderly population (The Cache County Study), Neurology 62: , 1156–1162. |
[12] | Wang J , Shi L , Zou Y , Tang J , Cai J , Wei Y , Qin J , Zhang Z ((2018) ) Positive association of familial longevity with the moderate-high HDL-C concentration in Bama aging Study, Aging (Albany NY) 10: , 3528–3540. |
[13] | Rabbitt PMA , McInnes L , Diggle P , Holland F , Bent N , Abson V , Pendleton N , Horan M ((2004) ) The University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age, 1983 through 2003, Aging Neuropsychol Cogn 11: , 245–279. |
[14] | Robinson AC , Davidson YS , Horan MA , Pendleton N , Mann DMA ((2018) ) Pathological correlates of cognitive impairment in The University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age, J Alzheimers Dis 64: , 483–496. |
[15] | Robinson AC , Chew-Graham S , Davidson YS , Horan MA , Roncaroli F , Minshull J , du Plessis D , Pal P , Payton A , Pendleton N , Mann DMA ((2020) ) A comparative study of pathological outcomes in The University of Manchester Longitudinal Study of Cognition in Normal Healthy Old Age and Brains for Dementia Research Cohorts, J Alzheimers Dis 73: , 619–632. |
[16] | Wenham PR , Price WH , Blandell G ((1991) ) Apolipoprotein E genotyping by one-stage PCR, Lancet 337: , 1158–1159. |
[17] | Revelas M , Thalamuthu A , Oldmeadow C , Evans T.-J , Armstrong NJ , Kwok JB , Brodaty H , Schofield PR , Scott RJ , Sachdev PS , Attia JR , Mather KA ((2018) ) Review and meta-analysis of genetic polymorphisms associated with exceptional human longevity, Mech Ageing Dev 175: , 24–34. |
[18] | Sebastiani P , Gurinovich A , Nygaard M , Sasaki T , Sweigart B , Bae H , Andersen SL , Villa F , Atzmon G , Christensen K , Arai Y , Barzilai N , Puca A , Christiansen L , Hirose N , Perls TT ((2019) ) APOE alleles and extreme human longevity, J Gerontol A Biol Sci Med Sci 74: , 44–51. |
[19] | Royston MC , Mann D , Pickering-Brown S , Owen F , Perry R , Raghavan R , Khin-Nu C , Tyrer S , Day K , Crook R , Hardy J , Roberts GW ((1994) ) Apolipoprotein E epilson 2 allele promotes longevity and protects patients with Down’s syndrome from dementia, Neuroreport 5: , 2583–2585. |
[20] | Chodosh J , Petitti DB , Elliott M , Hays RD , Crooks VC , Reuben DB , Galen Buckwalter J , Wenger N ((2004) ) Physician recognition of cognitive impairment: Evaluating the need for improvement, J Am Geriatr Soc 52: , 1051–1059. |




