Should the Treatment of Amyloidosis Be Personified? Molecular Mechanism of Amyloid Formation by Aβ Peptide and Its Fragments
Abstract
Aβ40 and Aβ42 peptides are believed to be associated with Alzheimer’s disease. Aggregates (plaques) of Aβ fibrils are found in the brains of humans affected with this disease. The mechanism of formation of Aβ fibrils has not been studied completely, which hinders the development of a correct strategy for therapeutic prevention of this neurodegenerative disorder. It has been found that the most toxic samples upon generation of fibrils are different oligomeric formations. Based on different research methods used for studying amyloidogenesis of Aβ40 and Aβ42 peptides and its amyloidogenic fragments, we have proposed a new mechanism of formation of amyloid fibrils. In accord with this mechanism, the main building unit for fibril generation is a ring-like oligomer. Association of ring-like oligomers results in the formation of fibrils of different morphologies. Our model implies that to prevent development of Alzheimer’s disease a therapeutic intervention is required at the earliest stages of amyloidogenesis—at the stage of formation of ring-like oligomers. Therefore, the possibility of a personified approach for prevention not only of Alzheimer’s disease development but also of other neurodegenerative diseases associated with the formation of fibrils is argued.
INTRODUCTION
Since the discovery of association of amyloids with different severe disorders, such as Alzheimer’s and Parkinson’s diseases, their research has grown. In the 1960s, mainly amyloids, for example, amyloid deposits in the brains of patients with Alzheimer’s disease, were studied. But since elaboration of the method for deciphering amino acid sequences of proteins and peptides [1] and identification of different compositions of amyloid formations the subject of studies has become the mechanism of formation of fibrils of definite proteins. As concerns Alzheimer’s disease, it is Aβ peptide where fibrils form aggregates (plaques) in human brain.
It has been established that the onset of fibril formation is connected with destabilization of the structure of monomer proteins, after which fibrils are formed according to the simplified scheme by generating oligomer aggregates from partially unfolded protein molecules. In the end fibril structures up to several micrometers long of about 10 nm in diameter appear. This process is common to all amyloid proteins and peptides; and of special interest is that average parameters of the length and diameter of fibrils remain mostly unchanged though dimensions of proteins vary greatly. Thus, it can be assumed that fibrils can be formed according to the common scheme and the mechanism of polymerization of destabilized proteins and peptides is a universal natural way of aggregation of partially unfolded proteins and peptides.
Much attention has been paid recently not only to fibrils but also to intermediate formations such as different oligomers upon generation of mature fibrils. This is explained by the fact that upon examination of toxicity it was found that proteins and peptides even in the form of dimers display toxicity to cell cultures [2].
It was reported that at early stages of fibril formation, circular ring-like particles with the diameter coinciding with that of mature fibrils were observed [3, 4]. Ring-like oligomers were observed in many amyloidogenic proteins. Moreover, the electron microscopy images presented in some papers clearly show that fibrils are constructed of ring-like oligomers [4, 5]. However, this does not attract special attention. In some papers it is reported that oligomer preparations have special peculiarities relating to fibril structures. These oligomers have some features specific for fibril formations, for example, the thioflavin T (ThT) binding and the presence of cross-β structure [6]. In addition, their toxicity is higher than that of mature fibrils. Therefore, it is proposed that this ring-like structure can form pores on the cell surface and change the viability of the cell [7]. This suggests that it is not amyloids themselves that are dangerous for the organism as a whole but namely different intermediate oligomer samples, because the presence of, for example, deposits of the Aβ peptide and at the same time the absence of any signs of Alzheimer’s disease were described. Accordingly the formation of fibrils can be considered as a defensive mechanism for reducing the number of oligomers in organism [8, 9]. Therefore studies of intermediate states of protein aggregation upon formation of fibrils are in the focus of attention not only for deciphering the mechanism of fibril formation, but also for elaboration of different approaches to cope with oligomerization, which will facilitate proposing novel methods for prophylactics and prevention of harsh amyloidogenic diseases.
The improvement of diagnostics methods and identifying more and more patients with Alzheimer’s disease have made foremost both the studies of this disorder and its revealing at the earliest stages. By the end of 2016, more than 70,000 research papers devoted to amyloidogenesis of Aβ peptide and its fragments were published. This may be one of the most dynamically developing directions in molecular biology, especially taking into account the significance of the influence of some amyloidogenic proteins on human health. However, no final mechanism of formation of fibrils by Aβ peptides has been proposed.
Studies of toxicity of all components of fibril formation (monomers, different oligomer formations, mature fibrils) and correspondingly elaboration of approaches that allow modifying all stages of this process have been greatly accentuated recently. They make it possible to come closer to the understanding of the toxicity mechanism of aggregates and designing corresponding therapeutic preparations capable of influencing the process of fibril formation at all stages. Moreover, fundamental studies of the morphology of mature amyloid fibrils under varying conditions of their formation (ionic conditions, temperature, pH and agitation) have shown that this process can be monitored.
Herein we propose one of the possible mechanisms of fibril formation based on the methods used for studying fibril generation by amyloid proteins. Our data allow us to propose that the main building unit for formation of fibrils may be a ring-like oligomer. Upon interaction with each other ring-like oligomers generate polymers in the form of fibrils. The proposed mechanism of fibril formation from ring-like oligomers suggests that it is required to search for ways to prevent generation of oligomer formations rather than of fibrils, i.e., to find reasons leading to destabilization of molecules of amyloidogenic proteins and peptides. Therefore of importance are studies of metabolism of the Aβ peptide and the search for its natural partners affecting its stability along with the search for conditions and ligands leading to its destabilization.
MATERIALS AND METHODS
Samples
Fragments Aβ16-25, Aβ31-40, and Aβ33-42 were synthesized chemically [10]. Synthetic samples of human Aβ40 (purity 95%, lot # 054M4822V) and Aβ42 (purity 95%, lot # SLBN3075V) peptides used in our experiments were purchased from Sigma-Aldrich. Purification of recombinant Aβ40 and Aβ42 was described in our paper [11].
Electron microscopy
All the samples were dissolved in DMSO (the final concentration 5%) then the buffer (50 mM Tris–HCl, pH7.5) was added (0.2–0.5 mg/ml). Prior to negatively staining, the concentration of the samples was adjusted to 0.1–0.2 mg/ml. A copper grid (400 mesh) coated with a formvar film (0.2% ) was mounted on a sample drop (10μl). After 5–10 min absorption, the grid with the preparation was negatively stained for 1.5–2.0 min with 1% (weight/volume) aqueous solution of uranyl acetate. The excess staining agent was removed with filter paper. The preparations were analyzed using a JEM-1200 EX transmission electron microscope at the accelerating voltage of 80 kV. Images were recorded on the Kodak electron image film (SO-163) at nominal magnification of 40,000–60,000.
X-ray analysis
The X-ray analysis of the preparations was conducted using a Microstar X-ray emitter with HELIOX optics equipped with a Platinum 135 CCD detector (X8 Proteum System, Bruker AXS). Cu Kα radiation (λ= 1.54 Å) was used (the sample was oriented perpendicular to the X-ray beam).
Limited proteolysis and mass spectrometry analysis
The isolated fibrils were treated with a mixture of proteases (trypsin, chymotrypsin, and proteinase K) for 8 h. The proteases were added so that the enzyme-to-peptide ratio was 1:25. To maintain the activity of proteinase K, CaCl2 was also added to the final concentration of 5 mM. Conditions of limited proteolysis were chosen so that the monomer molecules would be completely degraded while the fibril moiety would retain long fragments. The fragments set apart of the proteases were centrifuged and washed, and the remaining fragments included in fibril structures were pelleted once again and isolated. Then the isolated residual fibril structures were dried using a vacuum concentrator after which they were dissolved in a small volume of formic acid and thereafter in 10 mM ammonium acetate buffer. The obtained sets of peptides were separated by HPLC (EASY-nLC™ 1200 System, ThermoFisher Scientific, USA) and analyzed using a high-resolution mass spectrometer (Orbitrap Elite mass spectrometer, Thermo Scientific, Germany). Then the peptides were identified using the Peaks Studio 7.5 program (Bioinformatics Solutions Inc.), which allows not only to identify peptides but also to estimate their relative concentration in the probe.
Construction of molecular models of an amyloid fibril for Aβ peptide and its fragments
Structures 2M4J and 4UZR from the Protein Data Bank were taken as templates to construct an arrangement of β-sheets in square-like and ring-like manners. The dimension of these PDB structures corresponds to all parameters of oligomers observed in electron microscopy and X-ray analysis for Aβ40, Aβ42 and its fragments 31–40 and 33–42. The fitting of the desired amino acid sequences into the templates, modeling of oligomers, energy minimization and packing of oligomers were done by the YASARA program.
RESULTS AND DISCUSSION
Polymorphism and possible mechanism of formation of amyloid fibrils
Until recently it was believed that the assertion of the amyloid nature of proteins and peptides can be based on such research methods as birefringence upon Congo red staining, fluorescence spectroscopy (ThT binding), microscopic research (the existence of fibril morphology) and X-ray structure analysis (the presence of reflections characteristic of cross-β structure). Based on all such data it was concluded that fibrils observed using microscopic techniques (electron microscopy (EM), cryo-EM, and AFM) are amyloids [12, 13]. By the present time, data have been accumulated which show that ThT binds not to all amyloid fibrils [14] and not all amyloid fibrils have clear reflections specific of cross-β structure [15]. In addition, X-ray structure analyses reveal a large number of other reflections that are not always interpreted properly, which is explained by difficulties in their decoding [16–20]. This is also connected with the fact that at present the models of formation of amyloid fibrils cannot suggest an unambiguous answer to the question to what structure element a definite reflection can be ascribed. Electron microscopy, tandem mass spectrometry, and X-ray analyses have remained the main methods for studying amyloidogenesis. As a result of the accumulated experimental data, the role of analytical methods of bioinformatics as well as prediction of possible structure organization of amyloid fibrils increases. However, it is impossible to correctly interpret the organization of amyloid structures on the molecular level without a correct understanding of the mechanism(s) of formation of amyloid structures.
It is supposed that amyloid fibrils consist of filaments (from two to six) that can interact laterally with each other forming ribbons or bunches or intertwisting at different periods [21, 22]. In both cases, these formations generate bunches, bundles and large clusters of fibril aggregates at late stages of their formation. Polymorphism of amyloid fibrils is their characteristic property. The morphology of amyloid fibrils depends on conditions of their formation (ionic conditions, pH, temperature, and agitation), the producer, method of preparation (synthetic or recombinant) and different modifications of the amino acid sequence of peptides. Polymorphism is also observed under the same conditions. For example, it was demonstrated that Aβ40 peptide forms fibrils of different morphology under the same ionic conditions but upon agitation and without it. Agitation results mainly in the formation of fibrils twisted at different periods, and without agitation fibrils are prone to lateral association in ribbons of different lengths. It was also shown that fibrils obtained without agitation reveal high toxicity towards cell cultures [23, 24]. Therefore it is important to study all existing formations including oligomers at early stages of fibril generation and mature fibrils of different morphology.
In our studies of fibrillogenesis we analyzed both commercial preparations of Aβ42 and Aβ40 peptides manufactured by different producers [14] and genetically engineered preparations of these isoforms of the Aβ peptide [11, 25–27]. To this end, superproducer strains of corresponding peptides were obtained and techniques for their isolation and purification were developed [11, 25, 27]. The purity of all synthetic and recombinant Aβ42 and Aβ40 peptides was verified using the mass spectrometry analysis. Conditions of fibril formation were chosen and basic methods of their investigation were used: EM, X-ray structure analysis, fluorescence spectroscopy (ThT binding) and mass spectrometry. As concerns commercial preparations of Aβ42 peptide, we verified and compared synthetic samples of five producers including Anaspec, Invitrogen, Enzo, Sigma Aldrich, and SyntAssist for their applicability for studying the process of amyloidogenesis. It was found that of great importance is not only the producer but also the lot of the preparation [14]. Though all preparations form fibrils, their morphology and the rate of fibrillogenesis differ somewhat from each other. Our main recommendation is prerequisite verification of the purity of the Aβ peptide preparation (including even its presence in the packing) using the mass spectrometry analysis. Having analyzed reports available in PubMed (10,000 accessible papers on amyloidogenesis of Aβ peptide) we have found that Sigma Aldrich is the leader in marketing the synthetic preparation. However, it is also the most expansive preparation.
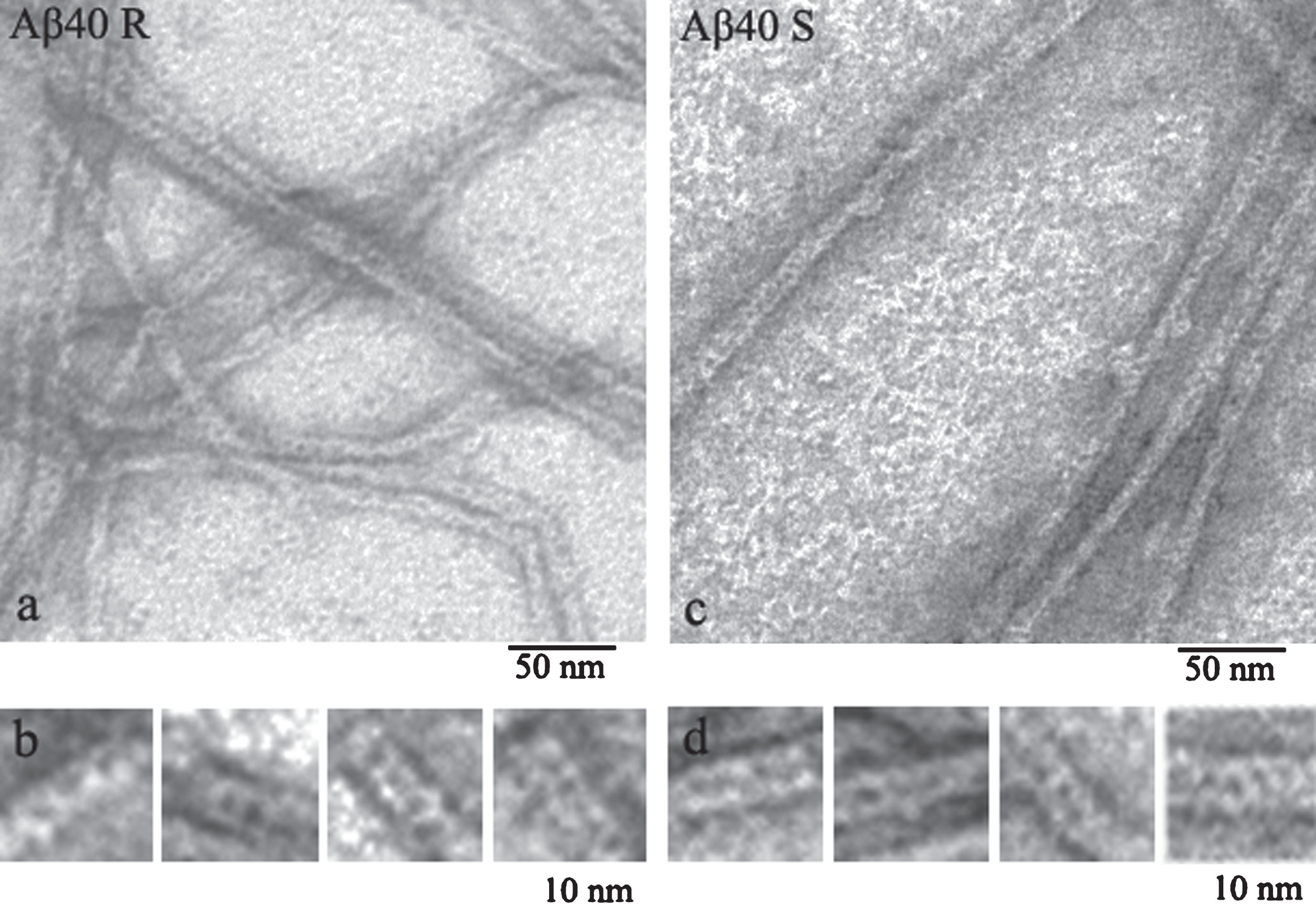
As commercial preparations of Aβ peptides are very expensive, we had to obtain recombinant Aβ40 and Aβ42 peptides and use them to analyze the process of amyloidogenesis. The comparison of the data on synthetic (Sigma Aldrich) and recombinant Aβ40 peptides showed that they have similar morphology that differs from morphology of recombinant and synthetic Aβ42 peptides (Fig. 1) [11, 27].
Fig.1
EM images of fragments of single fibrils formed by recombinant (a) and synthetic preparations of Aβ40 peptides (c) (R and S, respectively). Fragments of fibrils (b-R) and (d-S) at larger magnification.
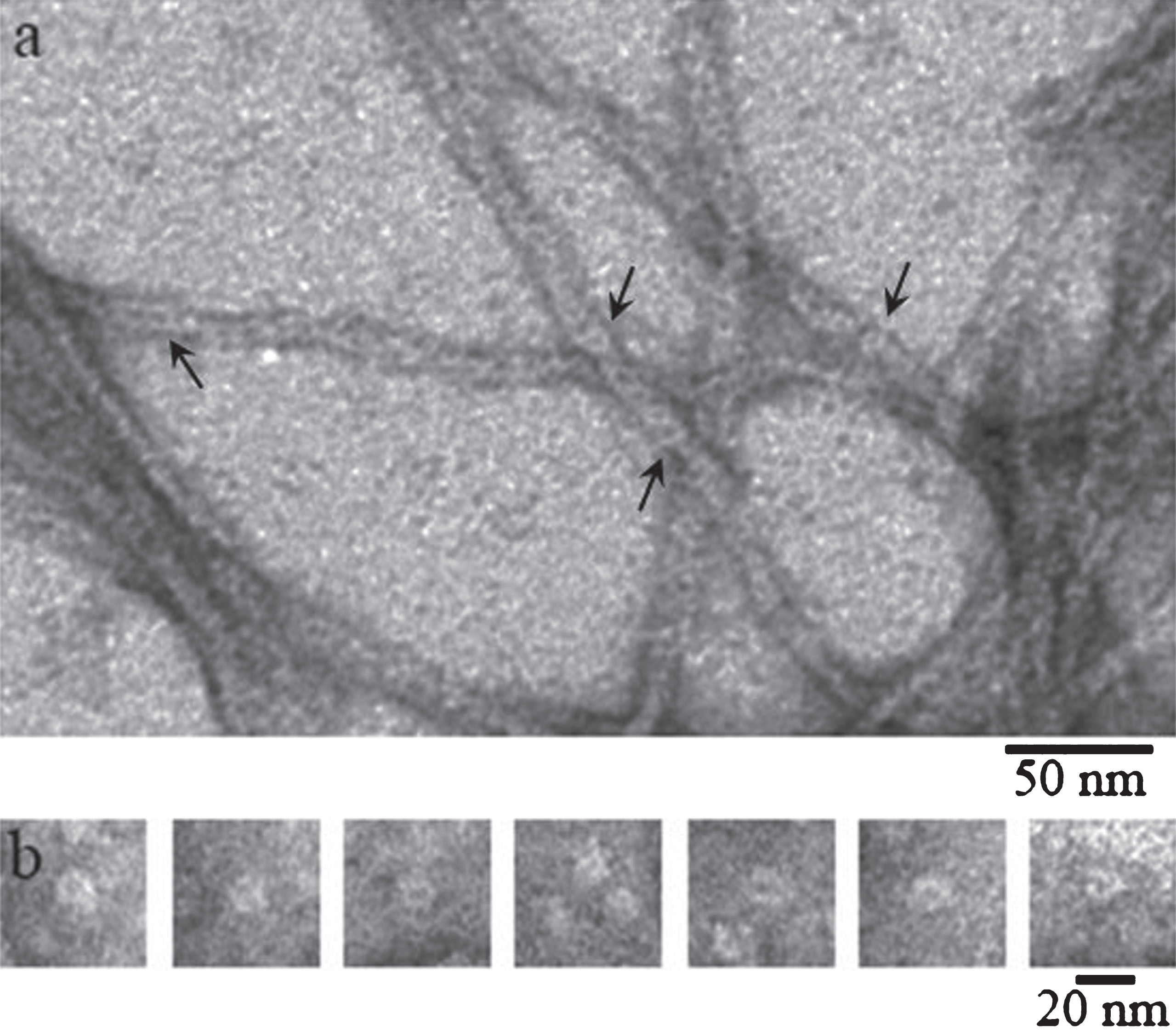
The use of fluorescence spectroscopy for studying the kinetics of fibrillogenesis of amyloid proteins and peptides has revealed a lag period, during which fibrils are prepared to grow and no inclusions of ThT are seen. Moreover, the presence of ThT also indicates active growth of fibrils. We have not succeeded in revealing a clearly defined and reproducible lag period for both synthetic and recombinant preparations of Aβ40 and Aβ42 peptides. But according to the EM data, even at the early period of fibrillogenesis it is possible to observe both oligomer rounded particles of about 8 nm in diameter (frequently as rings or rolls) and small (up to 50–100 nm in length) short fibrils of a similar diameter. By 8-h incubation the number of ring-like oligomers decreases and the length of fibrils increases (Fig. 2).
Fig.2
EM images of recombinant Aβ40 incubated at 25° (50 mM Tris_HCl, pH 7,5): (a) after 55 h incubation (the arrows indicate the ring-like oligomers); (b) gallery of separate ring-like oligomers (probable protofibril seeds) detected in the sample upon 8 h to 27 h incubation.
Upon further incubation of the preparations ring-like oligomers disappear and fibrils of about 5–10μm long and with the diameter of about 8 nm (the thinnest or single fibrils) appear. We observed considerable polymorphism of fibrils characterized by different forms and diameters as well as propensity to branching. Morphology of isoforms of Aβ peptides is different (Fig. 3). Aβ40 peptide has lateral association of fibrils with the formation of ribbons of different widths and ways of twisting (Fig. 3). Aβ42 peptide forms predominantly bunches and bundles of different diameters with rough surfaces of fibrils and tendency to their branching (Fig. 3).
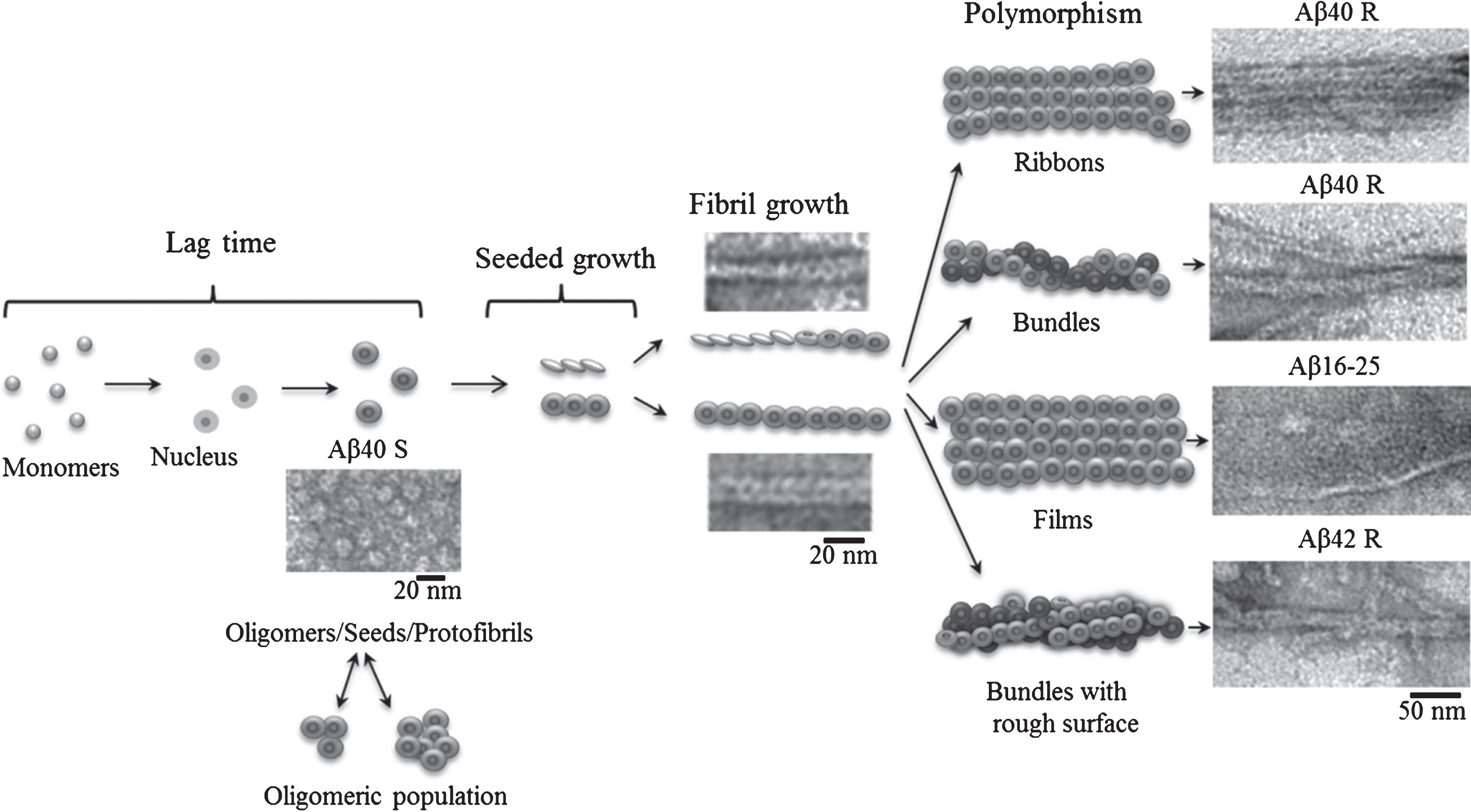
Fig.3
Schematic representation of the fibrillation process where the building block for fibril formation is a ring-like oligomer.
With a small magnification, it may seem that single fibrils of 8 nm in diameter consist of two filaments. This connects with features of EM [27]. But larger magnification shows that these fibrils consist of ring-like oligomers (Fig. 1).
Under such conditions, it is possible to watch a quite systematic packing of oligomers in single fibrils of Aβ40 peptide, while oligomers in Aβ42 peptide associate randomly so that by now single fibrils have rough surfaces. Similar fibrils are usually observed at the early period of fibril formation which is frequently reported by many authors studying the process of amyloidogenesis [3, 4, 28, 29]. If EM images of mature fibrils of rather high quality described in many papers are analyzed, it becomes clear that fibrils consist of ring-like structures [4, 5], although the authors do not mention this. Why? Maybe this is explained by the preconceived idea that fibrils are formed by a number of filaments (from two to six) [21, 22] passing along the entire axis of the fibril. The question arises: where do ring-like oligomers, seen in EM images at the early period of fibril formation, disappear? According to the simplified scheme, fibrils are formed in the following succession: destabilization of monomers, formation of oligomers from the monomers, and finally generation of mature fibrils (Fig. 3). The most inexplicable moment in the scheme is the transition from oligomers to mature fibrils. We propose that namely a ring-like oligomer is the main building block for formation of fibrils. As seen in Fig. 1, ring-like oligomers in a fibril interact by lateral sides with each other either in a ring-to-ring manner or slightly overlapping each other by their butt surfaces (like dominoes or a ladder). The diameter of a ring-like oligomer is about 8 nm which corresponds to the diameter of a single fibril, and the diameter of an internal hole in the ring is about 2 nm. An approximate height of the ring (a short hollow cylinder) is estimated by the fibril bend and is about 2 nm (the approximate diameter of a single fibril at its inflection point). The concept of a fibril consisting of two filaments can be explained by the fact that upon staining a preparation with uranyl acetate the stain covers not only the whole fibril, but it also fills all its cavities, which results in the formation of a fibril consisting of two filaments (with a small increase). Nonetheless, in some papers it is interpreted that fibrils consist of rounded oligomers. When studying the process of fibril formation by Aβ42 peptide with the EM method, Nielsen [3] demonstrated that at early stages of formation rounded oligomer particles are seen, which upon further incubation form mature fibrils via association with each other. A new mechanism of fibril formation was proposed upon analyzing generation of fibrils by α-synuclein [29]. According to this mechanism, a fibril is formed through association of the created oligomer granules that are a building block of the fibril (a double-concerted fibrillation model). The assembly of fibrils from oligomers was also demonstrated for lysozyme [30]. And finally in 2017 upon studying intermediates during fibril formation by Aβ peptide, Dean et al. [31] have come to a conclusion that oligomeric particles (dodecamers) form mature fibrils by interacting with each other.
To improve the quality of EM images that would provide more evidence for fibril formation from ring-like oligomeric structures, we used the method known in the literature as the Markham rotation techniques. It was first proposed by Markham with coworkers in 1963 [32] when they studied negatively stained viral particles using the electron microscopy method. The method can be applied to analyze objects with axial or linear symmetry with recurring structural elements poorly identified against the background. Image enhancement of recurring structural elements takes place as a result of multiple overlap with displacement of a semitransparent copy of the initial image. This accordingly leads to image averaging of an individual element and decreases the background “noise”. At present the idea of averaging is widely used also in contemporary software to enhance images for 3D reconstruction of macromolecules and their complexes based on the data of cryo-EM and EM images of negatively stained objects [33].
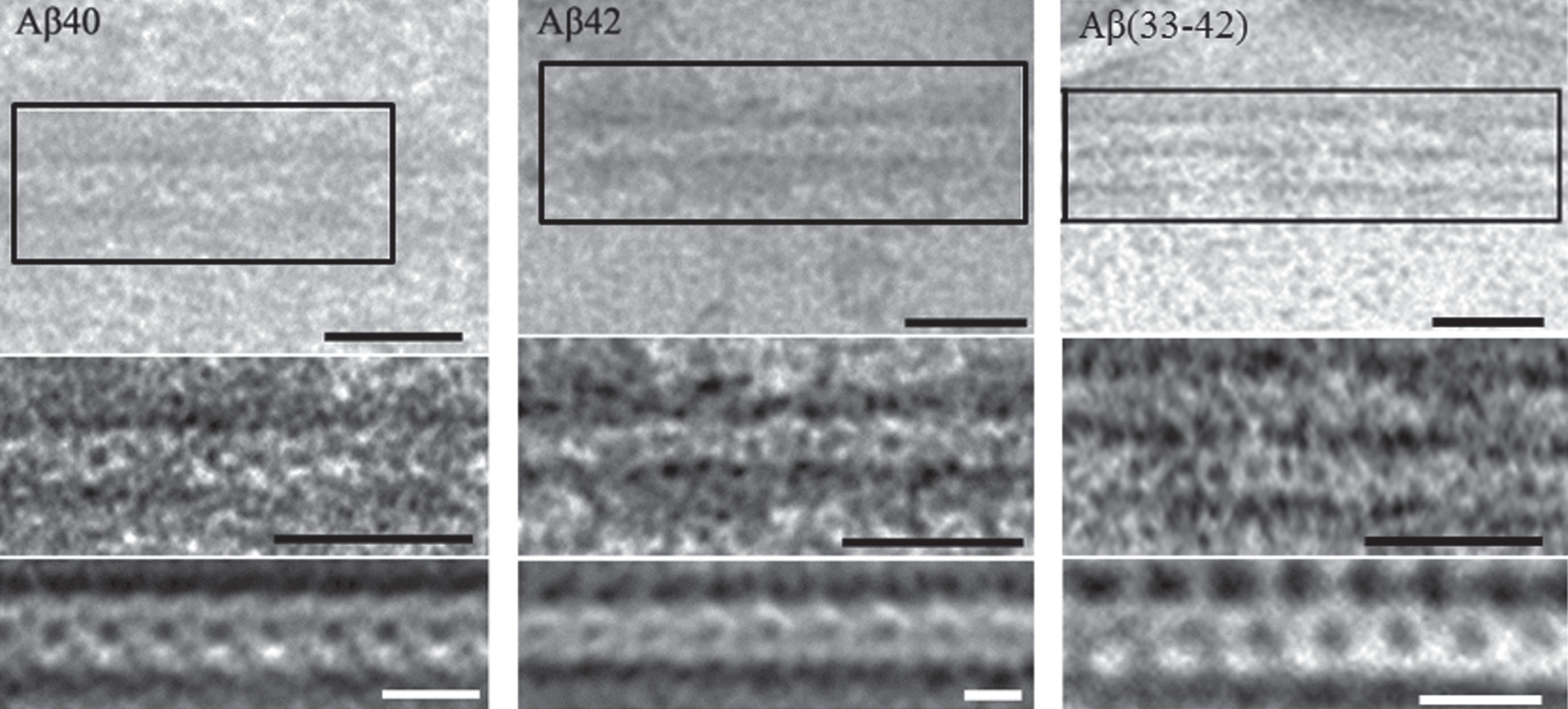
Figure 4 shows clearly the periodicity of overlapping ring-like structures, which supports the correctness of our assumption on the role of ring-like oligomers as main building blocks upon formation of amyloid structures. According to the data on enhancement of images of Aβ peptides, the average diameter of a ring-like oligomer is about 6–8 nm and the diameter of the cavity inside the ring is about 2 nm. As seen from Fig. 1, the diameter of a ring-like oligomer of Aβ is about 6–8 nm, and the diameter of the cavity inside the ring is about 2 nm.
Fig.4
EM images of fragments of fibrils formed by Aβ40, Aβ42 peptides and Aβ33–42 peptide fragment. Initial image of a fragment of a single fibril (top), enlarged detailed region of the fragment with enhanced contrast (middle), the result of applied the Markham method to better reveal morphological peculiarities of amyloid fibrils (bottom). Black labels correspond to 25 nm and white ones to 10 nm.
Two amyloidogenic fragments Aβ16-21 (KLVFFA) and Aβ32-36 (IGLMV) were predicted in the sequence of peptide Aβ42 [34]; and to study their amyloidogenic properties 10-membered fragments Aβ16-25, Aβ31-40, and Aβ33-42 were synthesized [10]. The last two fragments were synthesized to clarify the role of C-terminal amino acid residues in the morphology of fibrils, because earlier we determined that fibrils of Aβ40 and Aβ42 peptides have different morphologies [27]. The three fragments were studied using methods that are applied as a rule for analysis of formation of fibrils: ThT binding, EM, tandem mass spectrometry, X-ray analysis and structural modeling. All experiments were performed with concentrations of the preparations varying from 0.25 to 0.5 mg/ml in 5% DMSO, 50 mM Tris-HCl (pH 7.5) and incubation at 37°C. As shown by the fluorescence analysis, all the fragments interact with ThT, which reveals the presence of β-structure in them. But the shapes of the curves for different peptides dissimilar, which is evidence of various behavior of the preparations upon polymerization (see Supplementary Figure 1).
According to the data of EM analysis, the three peptides can self-assemble in polymer structures. In this case, Aβ31-40 peptide fragments aggregate forming ribbons and bundles, Aβ33-42 form bunches; and polymerization of Aβ16-25 peptide fragment is unusual for amyloidogenic fragments in the form of films (Fig. 5).
Fig.5
EM images of fibrils formed by Aβ42 peptide fragments. All experiments were performed at concentrations of the preparations 0.25–0.5 mg/ml in 5% DMSO, 50 mM Tris-HCl (pH 7.5) and incubation at 37°C for 24 h. The preparations were negatively stained with 1% aqueous solution of uranyl acetate. Bottom row: fragments of fields at large magnification. Arrows indicate ring-like oligomers with the outer diameter of about 6–7 nm and the inner diameter (hole) of about 2–3 nm.
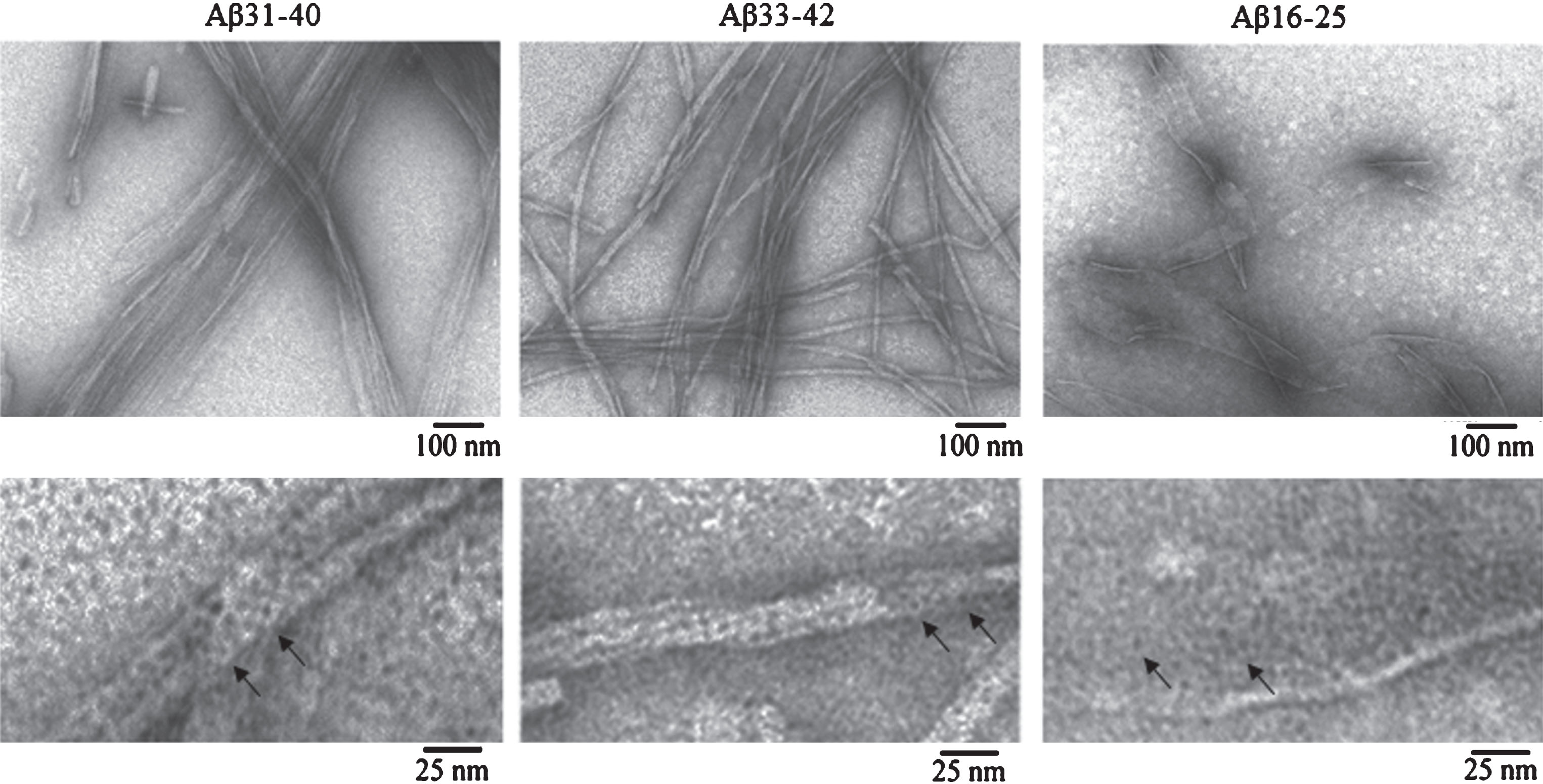
Under large magnification it is seen that all polymers consist of ring-like oligomers with the external diameter of about 6–7 nm, the internal diameter of about 2–3 nm, and in the bends of thinnest fibrils the diameter is about 3 nm, which allows determining an approximate height of the ring. Such oligomers can associate with each other by lateral sides or slightly overlap each other, forming ribbons in the lateral interaction of single fibrils and bundles of different diameters and twists (Aβ31-40); at irregular interaction of ring oligomers, bundles of different diameters with a rough surface are formed (Aβ33-42). In Aβ16-25 peptide fragment, oligomers associate side-by-side and form structures similar to honeycombs. It should be noted that the morphology of fibrils in Aβ31-40 and Aβ33-42 peptide fragments resembles that of fibrils of intact Aβ40 and Aβ42 peptides, respectively. It was demonstrated that the main difference between the Aβ40 and Aβ42 fibrils is the preferred formation of ribbons of different diameters and twists for Aβ40 and bunches of different diameters with rough surface and branching tendency for Aβ42 [14, 27]. The 2D reconstruction of Aβ33-42 fibrils also supports our assumption that fibrils consist of ring-like oligomers (Fig. 4).
We believe that it is easier to explain polymorphism inherent to all amyloid fibrils if a ring-like oligomer is taken as the basis for fibril construction. Depending on the amino acid sequence of protein or peptide and conditions of fibril formation, it is simpler to represent generation of single fibrils, their association in ribbons, bundles and bunches of different diameter and twisting, branching of fibrils and their rough surface.
X-ray structure analysis of amyloid fibrils of Aβ peptide and its fragments
At present the most popular idea on the structural organization of amyloid fibrils is that they consist of filaments [21, 22]. In turn, fibrils are formed of peptides which form β-sheets located parallel to the whole axis of a fibril at a distance of about 10 Å from each other. β-Sheets consist of regions packed as β-strands located perpendicular to the axis of a fibril/filament at a distance of about 4.7 Å from each other. This interpretation of the fibril structure is based on the data of X-ray structure analysis of preparations of amyloid fibrils and is called cross-β structure. This model of structural organization of amyloid fibrils was formed in the 2000s, and now it continues to be defined more exactly. The proposed model of the structure of amyloid fibrils of β-sheets was biased by the researches in which X-ray structure analysis was used (since 1935) to study some fibrillar proteins (fibroin, β-keratin, β-myosin, fibrin) as well as fibrils from globular denatured egg albumin. As a result, similar X-ray diffraction patterns were obtained [35]. The term “cross-β structure” was coined in 1968 [36] when intensive studies of amyloid formation from liver, kidney and spleen tissues of humans and experimental animals were performed. Using X-ray structure analysis, characteristic reflections were obtained: meridional of 4.75 Å and equatorial of 9.8 Å positioned perpendicular to each other (cross position). This interpretation of the structure of fibrils had an effect on the explanation of the EM data. Though the EM images of tissue sections in papers of 1960s showed ring-like oligomers the diameter of which coincided with that of a single fibril, they were interpreted frequently as cross sections of fibrils and it was concluded that fibrils consist of a number of filaments. Even when isolated preparations of tissues injured by amyloidosis began to be analyzed and some authors interpreted EM data from the point of view of fibril structure consisting of ring-like structures in form of a roll [28], the principal standpoint was the filament organization of fibrils.
In addition to the basic reflections ascribed to the cross-β structure, diffraction patterns of amyloid fibril preparations often contain a large number of other reflections. So far there has been no accurate interpretation of all reflections obtained with X-ray structure analysis of an amyloid preparation. It is notable however that a diffraction pattern can be affected by the orientation of amyloid fibrils, the concentration of the sample, its purity, source and technique of preparation. Data are available that well oriented preparations have a more complex pattern of clear reflections and yield more information on the structure [19]. Different explanations can be found in the literature concerning the presence of additional reflections for amyloid structures. Special attention is paid both to meridional and equatorial reflections [17, 18, 20]. An accurate interpretation of X-ray data requires not only the quality of the obtained diffraction patterns, but also the model of fibril structure on the molecular level.
All preparations of Aβ peptides and their fragments were studied using X-ray structure analysis. The obtained diffraction patterns of X-rays have more or less distinct reflections characteristic of cross-β structure, which is evidence of the amyloid nature of fibrils (Figs. 6 and 7). It should be underlined that X-ray pictures show a number of additional reflections in addition to the basic ones characteristic of amyloid reflections (β-strands at 4.7 Å and β-sheets at 11 Å). In the 1990s, additional reflections also attracted attention of researchers upon analysis of diffraction patterns. For example, when analyzing X-ray data at small angles for peptide Aβ1-40, Kirschner et al. [17] paid attention to the reflection at 53 Å and interpreted it so that either this reflection represents the object repeat along the fibril axis or the repeat upon the helix twisting (periodicity). Due to the complexity of interpretation of X-ray data that can be also a result of incorrect view on the structure of fibrils (infinite β-sheet along the whole axis of fibril), practically most part of reflections is not discussed in the following works on X-ray analysis of amyloid fibrils. But the reflections at 53 Å can be evidence of the repeat of some oligomer particles along the fibril axis. It should be mentioned that our experience in work with different conditions of sample preparation (ionic conditions) showed that attention should be paid to the solution in which the samples are prepared. Thus, we have found that, for example, Tris-HCl can have many reflections, part of which coincide with the reflections for amyloid structures [26].
Fig.6
X-ray diffraction patterns of fibrils formed by synthetic (Sigma) Aβ peptide: (a) schematic presentation of cross-β structure; (b) Aβ40 peptide; (c) Aβ42 peptide; (d) schematic representation of the arrangement of ring-like oligomers (the short tubular cylinders) in a fibril with the parameters corresponding to the reflections according to the X-ray data [17, 18]. Preparations were obtained in 50 mM Tris-HCl, pH 7.5, 48 h incubation at 37°C, 5 mg/ml.
![X-ray diffraction patterns of fibrils formed by synthetic (Sigma) Aβ peptide: (a) schematic presentation of cross-β structure; (b) Aβ40 peptide; (c) Aβ42 peptide; (d) schematic representation of the arrangement of ring-like oligomers (the short tubular cylinders) in a fibril with the parameters corresponding to the reflections according to the X-ray data [17, 18]. Preparations were obtained in 50 mM Tris-HCl, pH 7.5, 48 h incubation at 37°C, 5 mg/ml.](https://content.iospress.com:443/media/adr/2018/2-1/adr-2-1-adr180063/adr-2-adr180063-g006.jpg)
Fig.7
X-ray diffraction patterns of fibrils formed by the fragments of Aβ42 peptide. (a) Aβ16-25, (b) Aβ31-40, (c) Aβ33-40. Preparations were obtained in 50 mM Tris-HCl, pH 7.5, 48 h incubation at 37°C, 5 mg/ml. (d) Schematic representation of the arrangement of ring-like oligomers (short tubular cylinders) in a fibril with the parameters corresponding to the reflections according to the X-ray data [17, 59].
![X-ray diffraction patterns of fibrils formed by the fragments of Aβ42 peptide. (a) Aβ16-25, (b) Aβ31-40, (c) Aβ33-40. Preparations were obtained in 50 mM Tris-HCl, pH 7.5, 48 h incubation at 37°C, 5 mg/ml. (d) Schematic representation of the arrangement of ring-like oligomers (short tubular cylinders) in a fibril with the parameters corresponding to the reflections according to the X-ray data [17, 59].](https://content.iospress.com:443/media/adr/2018/2-1/adr-2-1-adr180063/adr-2-adr180063-g007.jpg)
Analysis of the literature data has allowed us to retrieve very important X-ray data obtained both for fragments of Aβ peptide and for Aβ40 itself. The authors of [17] obtained reflections that cannot be explained by the model accepted at present: the existence of “infinite” β-sheets formed along the axis of an amyloid fibril. This is an equatorial reflection at 49.2 Å corresponding to the distance of 55–57 Å, which is smaller than the fibril width (80–90 Å) and cannot be explained by lateral interaction of several fibrils. Another meridional reflection at 53 Å cannot be explained by the accepted model either. Kirschner et al. [17] proposed that this might take place only in three cases: 1) if the object is located recurringly along the fibril axis, 2) if fibrils are positioned stepwise, and 3) if fibrils are twisted. Unfortunately, the instrument used by us for measuring X-ray diffraction does not give reflections at small angles, i.e., does not produce large distances.
According to X-ray data, all three fragments of Aβ peptide have characteristic reflections for cross-β structure: equatorial at 8–10 Å and meridional at 4.5–4.8 Å, which is characteristic of amyloid fibrils. It should be mentioned that for Aβ31-40 and Aβ33-42 peptide fragments a large number of additional reflections is observed for amyloid fibrils. Additional reflections for amyloid fibrils are observed for many amyloid fibrils [17, 18] including undamaged Aβ40 and Aβ42 peptides [26]. The cross-β structure suggests that a fibril consists of β-sheets positioned parallel to the whole axis of the fibril at a distance of 8–12 Å from each other. The sheets are formed of β-strands perpendicular to the fibril axis and positioned at a distance of 4.5–4.8 Å from each other. But in this case how can we explain the presence of characteristic reflections for Aβ16-25 peptide fragment which forms nanofilms? A conclusion suggests itself that there can be another structure of fibrils and that the ring oligomers can have a cross-β in structure.
Identification of regions in the backbone of an amyloid fibril for Aβ peptide and its fragments
Fibrils obtained from Aβ40 and Aβ42 preparations were studied using the method of limited proteolysis combined with tandem mass spectrometry. Concentrations of a mixture of proteases (trypsin, chymotrypsin, and proteinase K) were chosen to perform proteolysis of regions not included in the backbone of an amyloid fibril. Then the peptides not included in the fibril backbone that remained non-cleaved were identified using tandem mass spectrometry. To determine and quantitatively estimate the contribution of each peptide, software PEAKS Studio 7.5 (Bioinformatics Solutions Inc.) was used. As a result of the studies, it was demonstrated that mainly C-terminal regions are protected from the action of proteases for Aβ40 [37], while for Aβ42 such are C-terminal and longer fragments (Fig. 8) [27].
Fig.8
Peptides detected after proteases treatment of amyloid fibrils formed by (a) Aβ40 peptide, (b) Aβ42, (c), Aβ33-42, (d) Aβ31-40, (e) Aβ16-25 peptide fragments and fragmentation spectra for the selected peptides.
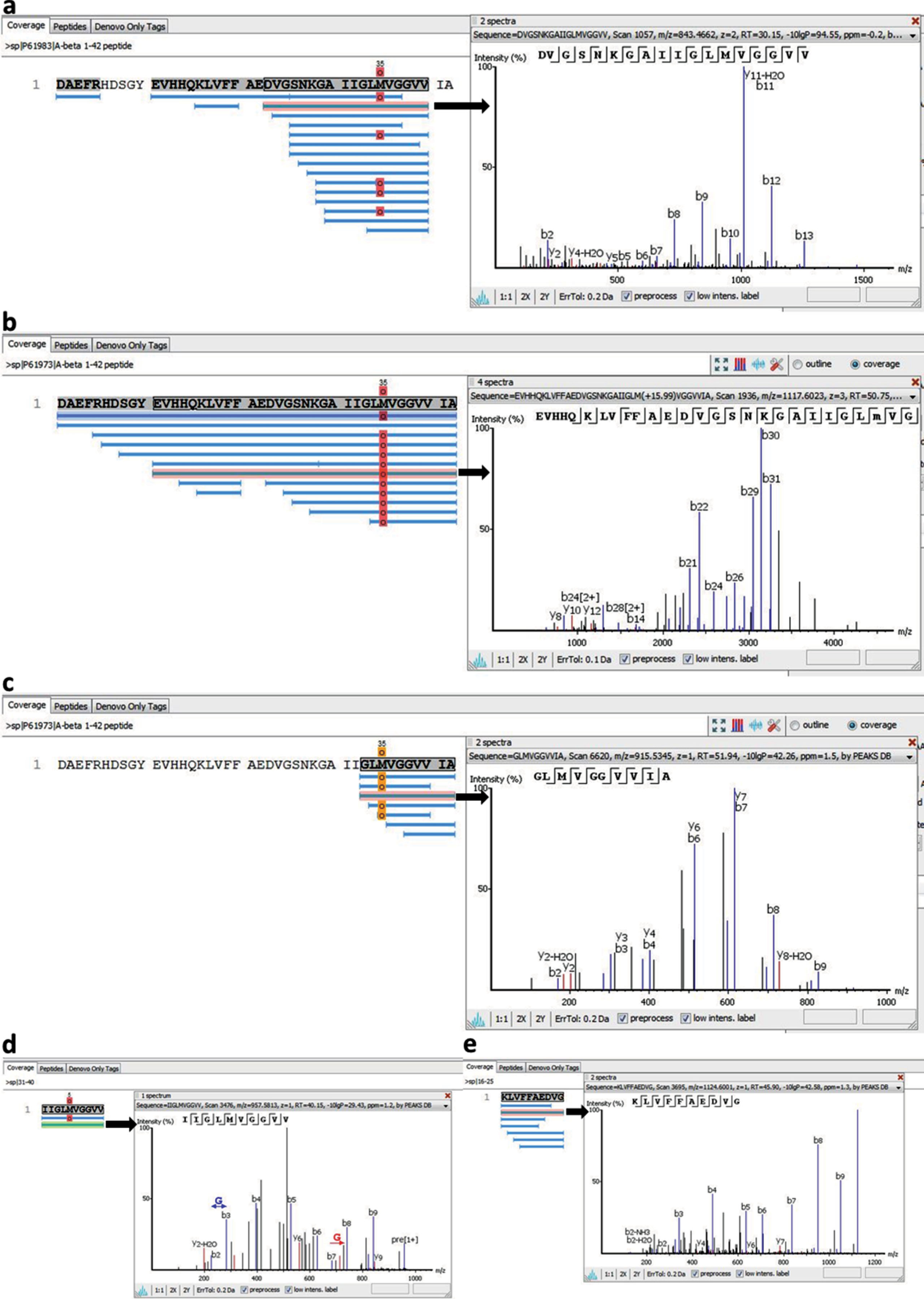
The data obtained agree with the results of the previous theoretical prediction of amyloidogenic regions of Aβ1-42 peptide made using software FoldAmyloid [34]. These are regions of amino acid residues 16–21 and 32–36 (Fig. 8). It was shown that regions of Aβ40 peptide including amino acid residues 16–22 and 22–40 are resistant to the action of proteases, i.e., are included in the backbone of an amyloid fibril. Our results correlate with the results of the theoretical prediction. In the case of Aβ42 peptide, some fragments accumulated after proteolysis correspond to predicted amyloidogenic regions of the sequence, are somewhat longer and protect the whole sequence. These data can be explained. According to the EM data, fibrils obtained for Aβ42 are the thickest in width, their width varying from 8 to 3 nm. Our model of generation of an amyloid fibril formed by docking of oligomeric structures suggests that amyloid fibrils for Aβ42 are formed as a result of irregular sticking of oligomeric structures (roughness of the surface) unlike fibrils formed for Aβ40, which are generated due to regular association of ring-like oligomers. The sticking of oligomers leads to stiffening of the amyloid structure and, as a consequence, to protection from the action of proteases. The limited proteolysis data demonstrate that the most protected from the proteolysis are the structures formed by Aβ31-40 peptide, which forms fibrils in the shape of bundles and ribbons (Fig. 8). The most susceptible to proteolysis is Aβ33-42, which forms uneven fibrils with a rough surface. The data indirectly support our model, because the side surfaces of the oligomers could be critical for their association.
3.4Molecular model of an amyloid fibril for Aβ peptide and its fragments
Based on the EM, mass spectrometry, theoretical and X-ray data we have proposed a molecular structure of an oligomer, which is the main building block of amyloid fibrils formed by Aβ40 and Aβ42 peptides and fragments of the latter (Fig. 9). To model an oligomer for a whole peptide we selected the structure obtained using solid phase NMR in the Laboratory of Prof. Tycko [38]. This model is based on a triangle structure (PDB 2M4J). In our experiments we did not observe such a triangle-like shape of amyloid fibrils; it rather resembled a square-like or ring-like structure. We developed a theory for computing folding nuclei based on the experimental data on the kinetics of changes in the involvement of monomers in an amyloid fibril, for example, the data on the change in the intensity of thioflavin fluorescence [39]. To compute the size of the primary nucleus for an amyloid fibril we used the data obtained in Prof. Dobson’s Laboratory [40, 41]. The nucleus of primary nucleation for a Aβ42 fibril consists of three monomers, whereas that for Aβ40 fibril consists of two monomers. Using the computed folding nuclei for amyloid fibrils and the experimental data, we proposed corresponding structures of oligomer particles—dodecamers consisting of three layers of tetramers. Such a dodecamer corresponds to the mass of 56 kDa. Interestingly, this structure contains two salt bridges: Arg5-Glu22 is formed between the structures and Asp23-Lys28 is formed within one of the structures. The dimension of this oligomer corresponds to the parameters observed using electron microscopy as well as to the X-ray data. This model allows explaining the meridional reflection at 53 Å and the equatorial reflection at 55 Å. The average height of the ring is about 3–4 nm. The ring consists of 12 monomer particles, and its height (side view, cylinder depth) shows that the height is formed by 3–4 layers of monomer particles. As a result, the ring contains up to 12 peptide monomers. Thus, this amount of the β-structure is quite enough for reflections characteristic of cross-β structure to be present in the diffraction patterns.
Fig.9
Schematic representation of a possible mechanism of the fibrils formed by (a) Aβ40 and Aβ42 peptides (the template was PDB 2M4J) and (b) its fragments Aβ31-40 and Aβ33-42 (the template was PDB 4UZR). (1) destabilized monomers; (2) fragment of oligomer (square-like primary oligomer for Aβ40 and Aβ42 and two β-sheets for fragments Aβ31-40 and Aβ33-42); (3) oligomer (the square-like oligomer composed of three primary oligomers for Aβ40 and Aβ42 and the ring-like oligomer composed of 12 β-sheets for fragments Aβ31-40 and Aβ33-42); (4) the interaction of oligomers in fibril.
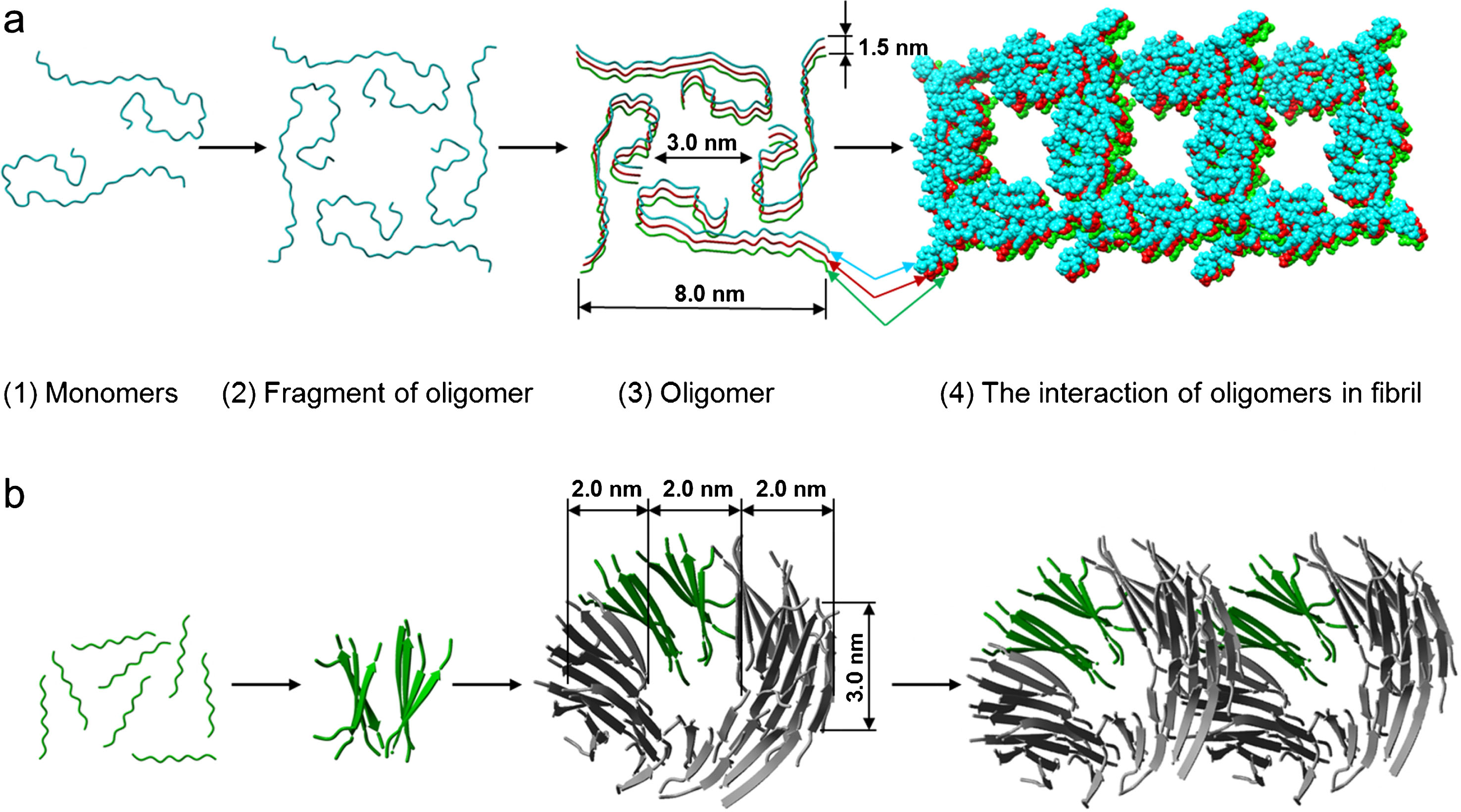
If a ring-like oligomer, which according to the EM data represents a short hollow cylinder with the external diameter of 6–7 nm, the internal diameter of 2–3 nm and the height of about 3 nm, is taken as a building block and structure modeling is used, it can be assumed for fragments of Aβ peptide that a ring-like oligomer consists of 48 peptide molecules forming 12 β-sheets layered in a circle (PDB 4UZR). The external diameter of this circle is about 6 nm and the internal about 2 nm. Upon association in a fibril this organization of a ring-like oligomer can produce a sufficient number of β-sheets and β-strands, so that the X-ray structure analysis would yield equivalent diffraction patterns for cross-β structure. In recent papers a special attention is paid namely to intermediates on the pathway of fibril formation, in particular, dodecamers which interact with other generated fibrils [31].
We believe that the model of fibril formation of ring oligomers, which interact with each other differently (ring-to-ring, ring-on-ring with some shift etc.), can give a better explanation to many properties such as strong polymorphism under the same conditions, fragmentation and branching inherent to amyloids. At the same time, the molecular structure of a ring-like oligomer can have distinctions for different short proteins and peptides as is observed for Aβ peptide and its fragments (Fig. 3).
Available structural models for amyloid fibrils of Aβ peptide and its fragments
By the moment, many models of structural organization of amyloid fibrils formed by Aβ peptide have been proposed. As a rule, all models are based on solid-state NMR spectra and rarely on cryo-EM data (see Table 1). Solid-state NMR is not a direct technique and is based on the use of distances obtained experimentally with special programs: the data are treated mathematically and a structure that would comply with all obtained parameters is deduced. The higher the number of such parameters is considered the less degenerated structure should be obtained. It is difficult to establish the uniqueness of assignments obtained by manual methods. Inasmuch as it is impossible to crystallize amyloid fibrils, their structure can be just deduced with the use of this method. However, there is a serious problem in structural studies of amyloid fibrils, i.e., obtaining of homogeneous preparations. Since amyloid fibrils are strongly polymorphous and their molecular structure depends greatly on conditions of fibrillogenesis, the obtaining of structure-homogeneous populations of fibrils in amounts sufficient for the method is an impediment for an unambiguous interpretation of the results. This may be the reason for the existence of a large number of models of structural organization of Aβ peptides (see Table 1).
Table 1
Model structures of monomer Aβ peptide in fibrils available in Protein Data Bank
| PDB, region Paper, method, region of investigated structure | 3D structure (1 model) | PDB, region Paper, method, region of investigated structure | 3D structure (1 model) |
| 2beg, WT, 17–42, Lührs et al., PNAS, 2005, [50] ssNMR 1–42 |  | 2lmn, WT, 9–40 Petkova et al., Biochemistry, 2006, [51] ssNMR 1–40 |  |
| 2lmp, WT, 9–40, Paravastu et al., PNAS, 2008, [52] ssNMR 1–40 |  | 2lmq WT, 9–40, Paravastu et al., PNAS, 2008, [52] ssNMR 1–40 |  |
| 2lnq, Iowa mutant: D23N 15–40, Qiang et al., PNAS, 2012, [53] ssNMR 1–40 |  | 2m4j, WT, 1–40, Lu et al., Cell, 2013, [38] ssNMR 1–40 |  |
| 2mvx, Osaka mutant: deletion of E22, 1–40, Schütz et al., Angew. Chem. Int. Ed. 2014, [54] ssNMR 1–40 |  | 2mxu WT, 11–42, Xiao et al., Nature Struct. Mol. Biol. 2015, [45] ssNMR 1–42 |  |
| 5aef, WT, 17–42, Schmidt et al., PNAS, 2015, [55] Cryo- EM, 5 Å 1–42 |  | 2mpz, D23N Iowa mutant, 15–40, Sgourakis et al., Structure, 2015, [56] ssNMR 1–40 |  |
| 2nao, WT, 1–42, Wälti et al., PNAS, 2016, [42] ssNMR 1–42 |  | 5kk3,WT, 11–42, Colvin et al., JACS, 2016, [43] ssNMR 1–42 |  |
| 2m4j, WT, 1–40, Dovidchenko et al., 2016, [11] modeling, 1–42 |  | 5oqv, WT, 1–42, Gremer et al., Science, 2017 [48] Cryo-EM, 4 Å, 1–42 | 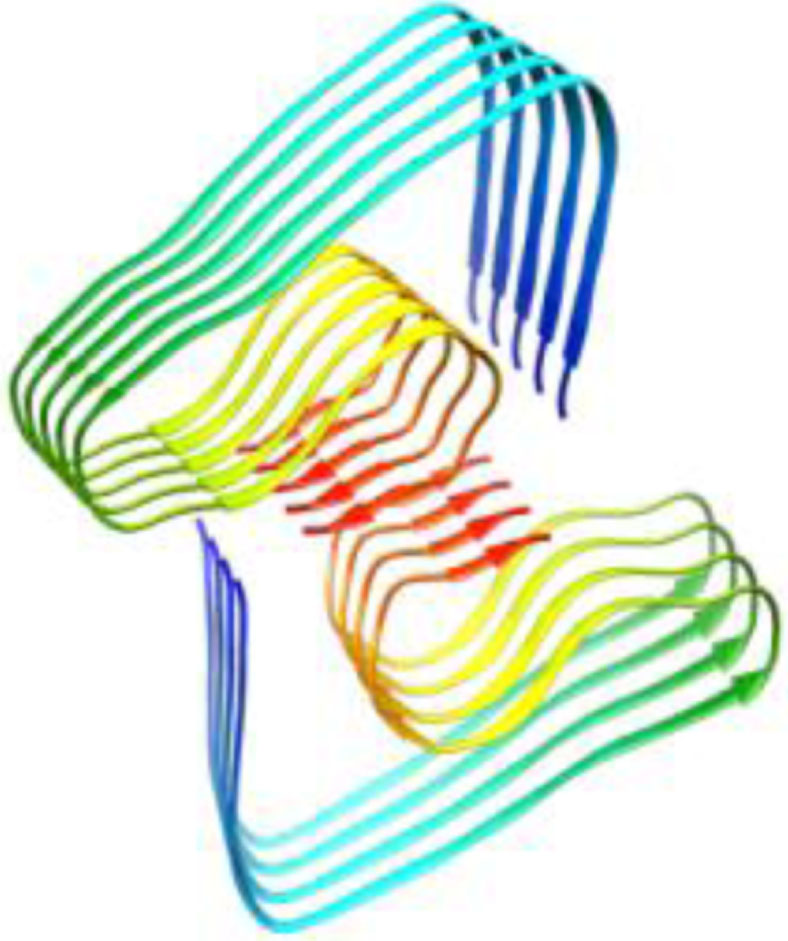 |
| 5w4i, de novo protein, 16–36, Kreutzer et al., Biochemistry, 2017, [57] X-ray, 2.026 Å 16–36 | 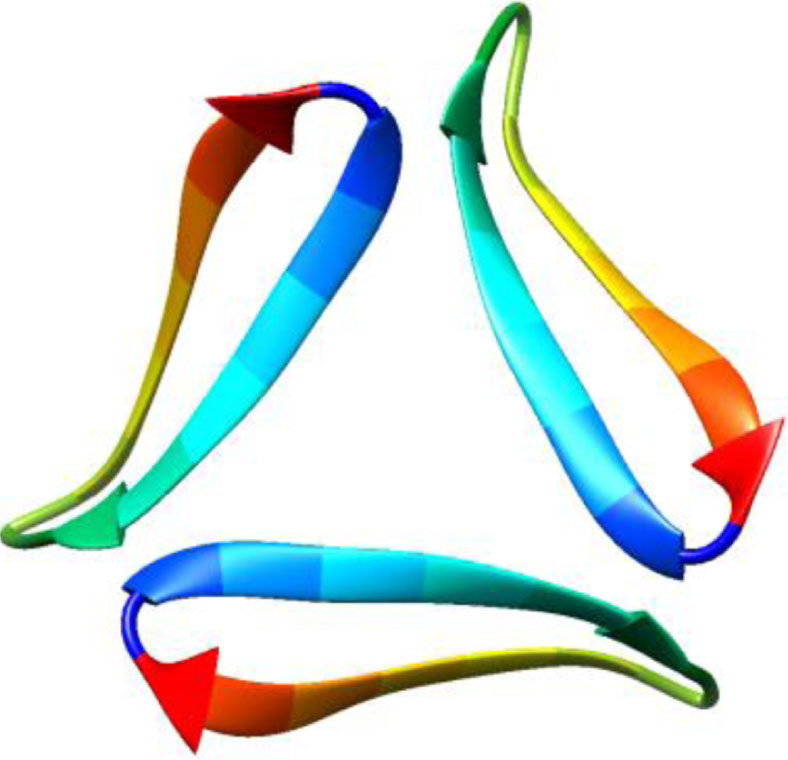 | 5how, de novo protein, 17–36, Kreutzer et al., JACS, 2016, [58] X-ray, 2.295 Å 17–36 | 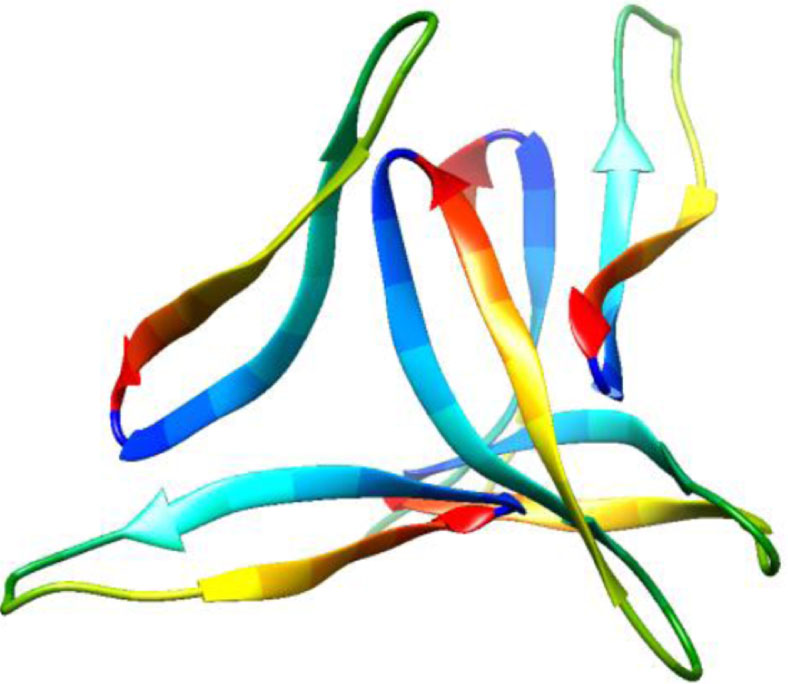 |
In 2016, based on solid-state NMR spectra two different research teams [42, 43] proposed a molecular structure for fibrils formed by Aβ42. Though conditions of sample preparation in these teams varied, the deduced structures of fibrils of Aβ42 appeared to be identical. As the authors of these publications believe, this structure of Aβ42 may be thermodynamically stable and is actively formed if a number of conditions are fulfilled. But in our opinion, the reason for deducing identical structures is the use of the same software to obtain a molecular structure using the distances found in the NMR measurements. Both research teams propose a model in which molecules of Aβ42 adopt an approximately S-shaped conformation consisting of short segments of β-regions interlinked by loops. Molecules are piled directly on each other along the direction of fibril growth thus forming strands of parallel cross-β structures in the register. Analogous to the structure of Aβ42 peptide, the same method was used to determine the structure of Aβ40 fibrils. However, in line with the obtained data, many details of the molecular conformation and interactions within and between molecules in fibrils formed by Aβ40 and Aβ42 vary greatly (Table 1). The authors of the mentioned papers think that these structural differences are caused by residues 41 and 42 and this prevents the formation of stable Aβ42 fibrils.
The main difference between the proposed models is that the C-terminal region of peptide is on the surface of Aβ42 fibrils, whereas in Aβ40 fibrils this region is embedded inside them. The authors of the above papers suppose that this difference in the structure can lead to dissimilar properties of amyloid fibrils: differences in interactions with neuron membranes, membrane-linked receptor proteins, dissimilar ways of stimulation of inflammation and different properties upon recognition of antibodies. However, these data do not agree with our data on accessibility of fibril regions to the action of three proteases. The data of mass spectroscopy analysis show that in both peptides the C-terminal region is hidden and inaccessible for binding with proteases. Moreover, the proposed different models of structural organization of fibrils formed by Aβ peptides are not compatible with the data published in 2017 [44], showing that seeds of Aβ42 peptide can accelerate the growth of Aβ40 fibrils and vice versa. This may be evidence of similarity of the basic structural block (oligomer) for both isoforms of Aβ peptide rather than of its divergence as was assumed previously [45–47].
The structure obtained using the cryo-EM data also raises several questions [48]. The analysis of an electron density slice shows that everything is not so unambiguous for obtaining the structure packing. But naturally it can be concluded that two monomers of Aβ peptide are insufficient for producing such a slice. The authors of the above papers demonstrate that upon choosing a certain parameter this model resembles greatly the structure suggested by Zhang et al. [49].
Conclusion
In spite of a large number of inconsistent theoretical and experimental data on the mechanism of formation of amyloid structures, in all probability there should exist general key moments in the process of their formation inherent to all proteins and peptides. A thorough analysis of the morphology of oligomeric particles and fibrils of the studied peptide preparations and the theoretical data on a possible mechanism of fibril generation have allowed us to propose a mechanism of polymerization of proteins and peptides in fibril formations. According to the EM data, at the initial period of fibril formation oligomeric particles of a ring-like structure are observed. Various studies of the mechanism of formation of amyloid fibrils contain schemes of their generation, where oligomers of different morphologies are invariably present; but nevertheless it is very infrequent that oligomers are considered as structural elements of fibrils. The most puzzling and unexplainable moment in the simplified scheme of formation of amyloid fibrils—destabilized monomer, oligomer, fibril—is transition from oligomers to mature fibrils. A detailed analysis of EM images of mature fibrils formed by Aβ40 and Aβ42 peptides and fragments of the latter was used by us as a basis for proposing a possible mechanism of amyloidogenesis. Destabilized monomers (in the case of Aβ peptide and α-synuclein we have already unstructured peptides) form ring-like oligomers which can be called protofibrils or basic building blocks of forthcoming fibrils. Ring-like oligomers have an external diameter similar to that of single fibrils. Interaction of complete building blocks (oligomers) leads to formation of fibrils. Ring-like oligomers in fibrils interact with each other by lateral sides, or more frequently slightly overlap each other by their front surfaces resembling dominoes or a “ladder”. When adsorbing single fibrils onto the formvar film, they can be twisted. This makes it possible to estimate the height of a ring-like oligomer in the bends and obtain its parameters. The average dimension of oligomers varies for each protein or peptide studied, but on the average the external diameter of the ring is about 6–8 nm, the internal diameter is about 2–3 nm (the hole in the ring) and the height of the ring-like oligomer (measured in the fibril bend) is about 3–4 nm.
Our X-ray data on preparations of Aβ peptides as well as the analysis of the literature data compelled us to study in more detail the possibility of formation of amyloid fibrils from β-sheets positioned along the whole fibril axis. The use of theoretical approaches (molecular modeling) has permitted us to propose a mechanism of formation and a molecular structure of a ring-like oligomer. Based on the totality of data, including information on proteolytic treatment of fibrils with the subsequent mass spectrometry analysis, we propose a new mechanism of generation of amyloid fibrils. This mechanism underlies the presumption that the basic building element of a fibril is a ring-like oligomer. However, the structural (molecular) organization (number of molecules, their packing, parameters) of ring-like oligomers of different proteins and peptides can vary. Formation of amyloid fibrils from ring-like oligomers simplifies the explanation of fibril polymorphism, fragmentation, branching and rough surfaces.
Correct representation of the structural organization of amyloid fibrils is a necessary condition for elaboration of strategic approaches upon developing methods of prevention of amyloidogenic diseases. An opinion can be found in the literature that when designing therapeutic preparations against amyloidosis a personified approach should be used, because fibril formations can differ morphologically in different patients. Therefore the way of formation of fibrils from oligomer structures, common for all fibrils discovered by us, could facilitate creation of general action preparations. But in our opinion, the basic attention should be focused on physiological, genetic and other reasons leading to destabilization of native molecules of proteins and peptides and triggering the process of fibrillogenesis. Destabilization of monomers depends on many factors, and this poses a lot of questions including the way of avoiding changes in the conformation of native proteins/peptides. As known, a number of mutations of Aβ peptides can accelerate the progression of Alzheimer’s disease, and therefore it is required to develop approaches for “treatment” of damaged proteins/peptides on the level of genes.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
ACKNOWLEDGMENTS
We are grateful to T.B. Kuvshinkina and Saikat Dutta Chowdhury for assistance in preparation of the manuscript, E.I. Grigorashvili and M.Yu. Suvorina for their help in obtaining mass spectrometry images, A.D. Nikulin for obtaining X-ray images. Research was supported by facilities of United Pushchino Center for Electron Microscopy (No 507648) and mass spectrometry. The studies were supported by the Russian Science Foundation (grant numbers 14-14-00536 and 18-14-00321).
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: 10.3233/ADR-180063.
REFERENCES
[1] | Glenner GG , Terry W , Harada M , Isersky C , Page D ((1971) ) Amyloid fibril proteins: Proof of homology with immunoglobulin light chains by sequence analyses. Science 172: , 1150–1151. |
[2] | Finder VH , Glockshuber R ((2007) ) Amyloid-beta aggregation. Neurodegener Dis 4: , 13–27. |
[3] | Nielsen EH , Nybo M , Svehag SE ((1999) ) Electron microscopy of prefibrillar structures and amyloid fibrils. Methods Enzymol 309: , 491–496. |
[4] | Lashuel HA , Hartley DM , Petre BM , Wall JS , Simon MN , Walz T , Lansbury PT ((2003) ) Mixtures of wild-type and a pathogenic (E22G) form of Abeta40 in vitro accumulate protofibrils, including amyloid pores. J Mol Biol 332: , 795–808. |
[5] | Paravastu AK , Qahwash I , Leapman RD , Meredith SC , Tycko R ((2009) ) Seeded growth of beta-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci U S A 106: , 7443–7448. |
[6] | Dobson CM ((2004) ) Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol 15: , 3–16. |
[7] | Quist A , Doudevski I , Lin H , Azimova R , Ng D , Frangione B , Kagan B , Ghiso J , Lal R ((2005) ) Amyloid ion channels: A common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A 102: , 10427–10432. |
[8] | Mal’tsev AV , Galzitskaia OV ((2010) ) Formation and participation of nano-amyloids in pathogenesis of Alzheimer’s disease and other amyloidogenic diseases. Biomed Khim (Russia) 56: , 624–638. |
[9] | Maltsev AV , Bystryak S , Galzitskaya OV ((2011) ) The role of β-amyloid peptide in neurodegenerative diseases. Ageing Res Rev 10: , 440–452. |
[10] | Selivanova OM , Surin AK , Ryzhykau YL , Glyakina AV , Suvorina MY , Kuklin AI , Rogachevsky VV , Galzitskaya OV ((2018) ) To be fibrils or to be nanofilms? Oligomers are building blocks for fibril and nanofilm formation of fragments of Aβ Peptide. Langmuir ACS J Surf Colloids 34: , 2332–2343. |
[11] | Dovidchenko NV , Glyakina AV , Selivanova OM , Grigorashvili EI , Suvorina MY , Dzhus UF , Mikhailina AO , Shiliaev NG , Marchenkov VV , Surin AK , Galzitskaya OV ((2016) ) One of the possible mechanisms of amyloid fibrils formation based on the sizes of primary and secondary folding nuclei of Aβ40 and Aβ42. J Struct Biol 194: , 404–414. |
[12] | Chiti F , Dobson CM ((2006) ) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75: , 333–366. |
[13] | Chiti F , Dobson CM ((2009) ) Amyloid formation by globular proteins under native conditions. Nat Chem Biol 5: , 15–22. |
[14] | Suvorina MY , Selivanova OM , Grigorashvili EI , Nikulin AD , Marchenkov VV , Surin AK , Galzitskaya OV ((2015) ) Studies of polymorphism of amyloid-β 42 Peptide from different suppliers. J Alzheimers Dis 47: , 583–593. |
[15] | Selivanova OM , Suvorina MY , Dovidchenko NV , Eliseeva IA , Surin AK , Finkelstein AV , Schmatchenko VV , Galzitskaya OV ((2014) ) How to determine the size of folding nuclei of protofibrils from the concentration dependence of the rate and lag-time of aggregation. II. Experimental application for insulin and LysPro insulin: Aggregation morhology, kinetics, and sizes of nuclei. J Phys Chem B 118: , 1198–1206. |
[16] | Koltun WL , Waugh DF , Bear RS ((1954) ) An X-Ray Diffraction investigation of selected types of insulin fibrils. J Am Chem Soc 76: , 413–417. |
[17] | Inouye H , Fraser PE , Kirschner DA ((1993) ) Structure of beta-crystallite assemblies formed by Alzheimer beta-amyloid protein analogues: Analysis by x-ray diffraction. Biophys J 64: , 502–519. |
[18] | Malinchik SB , Inouye H , Szumowski KE , Kirschner DA ((1998) ) Structural analysis of Alzheimer’s beta(1-40) amyloid: Protofilament assembly of tubular fibrils. Biophys J 74: , 537–545. |
[19] | Makin OS , Serpell LC ((2005) ) X-ray diffraction studies of amyloid structure. Methods Mol Biol Clifton NJ 299: , 67–80. |
[20] | Sunde M , Serpell LC , Bartlam M , Fraser PE , Pepys MB , Blake CC ((1997) ) Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol 273: , 729–739. |
[21] | Jiménez JL , Nettleton EJ , Bouchard M , Robinson CV , Dobson CM , Saibil HR ((2002) ) The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci U S A 99: , 9196–9201. |
[22] | Close W , Neumann M , Schmidt A , Hora M , Annamalai K , Schmidt M , Reif B , Schmidt V , Grigorieff N , Fändrich M ((2018) ) Physical basis of amyloid fibril polymorphism. Nat Commun 9: , 699. |
[23] | Petkova AT , Leapman RD , Guo Z , Yau W-M , Mattson MP , Tycko R ((2005) ) Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science 307: , 262–265. |
[24] | Petkova AT , Ishii Y , Balbach JJ , Antzutkin ON , Leapman RD , Delaglio F , Tycko R ((2002) ) A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci U S A 99: , 16742–16747. |
[25] | Grigorashvili EI , Selivanova OM , Dovidchenko NV , Dzhus UF , Mikhailina AO , Suvorina MY , Marchenkov VV , Surin AK , Galzitskaya OV ((2016) ) Determination of size of folding nuclei of fibrils formed from recombinant Aβ(1-40) peptide. Biochemistry (Moscow) 81: , 538–547. |
[26] | Selivanova OM , Grigorashvili EI , Suvorina MY , Dzhus UF , Nikulin AD , Marchenkov VV , Surin AK , Galzitskaya OV ((2016) ) X-ray diffraction and electron microscopy data for amyloid formation of Aβ40 and Aβ42. Data Brief 8: , 108–113. |
[27] | Selivanova OM , Surin AK , Marchenkov VV , Dzhus UF , Grigorashvili EI , Suvorina MY , Glyakina AV , Dovidchenko NV , Galzitskaya OV ((2016) ) The mechanism underlying amyloid polymorphism is opened for Alzheimer’s disease amyloid-β peptide. J Alzheimers Dis 54: , 821–830. |
[28] | Benditt EP , Eriksen N ((1966) ) Amyloid. 3. A protein related to the subunit structure of human amyloid fibrils. Proc Natl Acad Sci U S A 55: , 308–316. |
[29] | Bhak G , Lee J-H , Hahn J-S , Paik SR ((2009) ) Granular assembly of alpha-synuclein leading to the accelerated amyloid fibril formation with shear stress. PloS One 4: , e4177. |
[30] | Hill SE , Robinson J , Matthews G , Muschol M ((2009) ) Amyloid protofibrils of lysozyme nucleate and grow via oligomer fusion. Biophys J 96: , 3781–3790. |
[31] | Dean DN , Das PK , Rana P , Burg F , Levites Y , Morgan SE , Ghosh P , Rangachari V ((2017) ) Strain-specific fibril propagation by an Aβ dodecamer. Sci Rep 7: , 40787. |
[32] | Markham R , Frey S , Hills GJ ((1963) ) Methods for the enhancement of image detail and accentuation of structure in electron microscopy. Virology 20: , 88–102. |
[33] | Ryazantsev S , Tischenko V , Nguyen C , Abramov V , Zav’yalov V ((2013) ) Three-dimensional structure of the human myeloma IgG2. PLoS One 8: , e64076. |
[34] | Garbuzynskiy SO , Lobanov MY , Galzitskaya OV ((2010) ) FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics 26: , 326–332. |
[35] | Astbury WT , Dickinson S , Bailey K ((1935) ) The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J 29: , 2351–2360.1. |
[36] | Eanes ED , Glenner GG ((1968) ) X-ray diffraction studies on amyloid filaments. J Histochem Cytochem 16: , 673–677. |
[37] | Surin AK , Grigorashvili EI , Suvorina MY , Selivanova OM , Galzitskaya OV ((2016) ) Determination of regions involved in amyloid fibril formation for Aβ(1-40) peptide. Biochemistry (Moscow) 81: , 762–769. |
[38] | Lu J-X , Qiang W , Yau W-M , Schwieters CD , Meredith SC , Tycko R ((2013) ) Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell 154: , 1257–1268. |
[39] | Dovidchenko NV , Finkelstein AV , Galzitskaya OV ((2014) ) How to determine the size of folding nuclei of protofibrils from the concentration dependence of the rate and lag-time of aggregation. I. Modeling the amyloid protofibril formation. J Phys Chem B 118: , 1189–1197. |
[40] | Cohen SIA , Linse S , Luheshi LM , Hellstrand E , White DA , Rajah L , Otzen DE , Vendruscolo M , Dobson CM , Knowles TPJ ((2013) ) Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci U S A 110: , 9758–9763. |
[41] | Meisl G , Yang X , Hellstrand E , Frohm B , Kirkegaard JB , Cohen SIA , Dobson CM , Linse S , Knowles TPJ ((2014) ) Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc Natl Acad Sci U S A 111: , 9384–9389. |
[42] | Wälti MA , Ravotti F , Arai H , Glabe CG , Wall JS , Böckmann A , Güntert P , Meier BH , Riek R ((2016) ) Atomic-resolution structure of a disease-relevant Aβ(1-42) amyloid fibril. Proc Natl Acad Sci U S A 113: , E4976–4984. |
[43] | Colvin MT , Silvers R , Ni QZ , Can TV , Sergeyev I , Rosay M , Donovan KJ , Michael B , Wall J , Linse S , Griffin RG ((2016) ) Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J Am Chem Soc 138: , 9663–9674. |
[44] | Tran J , Chang D , Hsu F , Wang H , Guo Z ((2017) ) Cross-seeding between Aβ40 and Aβ42 in Alzheimer’s disease. FEBS Lett 591: , 177–185. |
[45] | Xiao Y , Ma B , McElheny D , Parthasarathy S , Long F , Hoshi M , Nussinov R , Ishii Y ((2015) ) Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol 22: , 499–505. |
[46] | Pauwels K , Williams TL , Morris KL , Jonckheere W , Vandersteen A , Kelly G , Schymkowitz J , Rousseau F , Pastore A , Serpell LC , Broersen K ((2012) ) Structural basis for increased toxicity of pathological aβ42:aβ40 ratios in Alzheimer disease. J Biol Chem 287: , 5650–5660. |
[47] | Cukalevski R , Yang X , Meisl G , Weininger U , Bernfur K , Frohm B , Knowles TPJ , Linse S ((2015) ) The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation. Chem Sci 6: , 4215–4233. |
[48] | Gremer L , Schölzel D , Schenk C , Reinartz E , Labahn J , Ravelli RBG , Tusche M , Lopez-Iglesias C , Hoyer W , Heise H , Willbold D , Schröder GF ((2017) ) Fibril structure of amyloid-β(1-42) by cryo-electron microscopy. Science 358: , 116–119. |
[49] | Zhang R , Hu X , Khant H , Ludtke SJ , Chiu W , Schmid MF , Frieden C , Lee J-M ((2009) ) Interprotofilament interactions between Alzheimer’s Abeta1-42 peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci U S A 106: , 4653–4658. |
[50] | Lührs T , Ritter C , Adrian M , Riek-Loher D , Bohrmann B , Döbeli H , Schubert D , Riek R ((2005) ) 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci U S A 102: , 17342–17347. |
[51] | Petkova AT , Yau W-M , Tycko R ((2006) ) Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry (Mosc) 45: , 498–512. |
[52] | Paravastu AK , Leapman RD , Yau W-M , Tycko R ((2008) ) Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci U S A 105: , 18349–18354. |
[53] | Qiang W , Yau W-M , Luo Y , Mattson MP , Tycko R ((2012) ) Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci U S A 109: , 4443–4448. |
[54] | Schütz AK , Vagt T , Huber M , Ovchinnikova OY , Cadalbert R , Wall J , Güntert P , Böckmann A , Glockshuber R , Meier BH ((2015) ) Atomic-resolution three-dimensional structure of amyloid β fibrils bearing the Osaka mutation. Angew Chem Int Ed Engl 54: , 331–335. |
[55] | Schmidt M , Rohou A , Lasker K , Yadav JK , Schiene-Fischer C , Fändrich M , Grigorieff N ((2015) ) Peptide dimer structure in an Aβ(1-42) fibril visualized with cryo-EM. Proc Natl Acad Sci U S A 112: , 11858–11863. |
[56] | Sgourakis NG , Yau W-M , Qiang W ((2015) ) Modeling an in-register, parallel “iowa” aβ fibril structure using solid-state NMR data from labeled samples with rosetta. Struct Lond Engl 1993 23: , 216–227. |
[57] | Kreutzer AG , Spencer RK , McKnelly KJ , Yoo S , Hamza IL , Salveson PJ , Nowick JS ((2017) ) A Hexamer of a Peptide Derived from Aβ16-36. Biochemistry (Mosc) 56: , 6061–6071. |
[58] | Kreutzer AG , Hamza IL , Spencer RK , Nowick JS ((2016) ) X-ray crystallographic structures of a trimer, dodecamer, and annular pore formed by an Aβ17-36 β-hairpin. J Am Chem Soc 138: , 4634–4642. |
[59] | Perutz MF , Finch JT , Berriman J , Lesk A ((2002) ) Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci U S A 99: , 5591–5595. |




