Studies on Copper and Aβ1-16-Induced Conformational Changes in CAG/CTG Trinucleotide Repeats Sequence
Abstract
DNA conformation and stability are critical for the normal cell functions, which control many cellular processes in life, such as replication, transcription, DNA repair, etc. The accumulation of amyloid-β peptide (Aβ) and Copper (Cu) are the etiological factors for neurodegenerative diseases and hypothesized that they can cause DNA instability. In the current investigation, we studied copper and Aβ1-16 induced conformation and stability changes in CAG/CTG sequences and found alterations from B-DNA to altered B-conformation. Further, the interaction of the copper and Aβ1-16 with CAG/CTG sequences was studied by molecular docking modeling and results indicated that the interaction of copper and Aβ1-16 was through the hydrogen bond formation between adenine, guanine, and cytocine. This study illustrates the role of the copper and Aβ1-16 in modulating the DNA conformation and stability.
INTRODUCTION
DNA contains all the genetic information for cellular function and plays an important role in the life process [1]. DNA is polymorphic in nature that adopts different conformations like B, Z, C DNA, and also alterations in the geometry of the double helix, including the depth and width of the major and minor grooves, is variable in different conformations [1]. The repetitive DNA sequences like triplets are found to be dispersed throughout eukaryotic genomes and trinucleotide repeat regions in the genome have been shown to be unstable and polymorphic [1]. CAG trinucleotide repeats, a unique class of repeats that forms hairpin secondary conformations, and this alters DNA replication and repair [2]. Trinucleotide repeats namely CAG, CTG, CCG sequences are linked to various neurological disorders including Huntington’s disease and myotonic dystrophy [3–5]. It is hypothesized that genome containing long stretches of CAG repeats in the N-terminal region of the huntingtin protein, associated with neuronal cell death [6–9]. Further, studies evidenced that polypeptides such as amyloid-β (Aβ), tau, and prion have been shown directly interacting with several metal ions in vitro and undergo conformational change [10, 11]. Earlier studies from our lab showed that, Aβ and metals bind to DNA and induce conformational change from B-DNA to an altered B-DNA and Ψ-DNA, which are one of the pathological features of Alzheimer’s disease (AD) [12]. The conformational change from B-DNA to altered B-DNA/Z-DNA/Ψ-DNA plays a critical role in the DNA instability of the genome and associated cellular dysfunctions in brain disorders [13–15]. There are limited reports on Aβ and copper (Cu) induced conformational changes in DNA. The current studies focused on Cu and Aβ induced conformational changes in triplet CAG/CTG sequences to get an insight for their role in neurodegenerative disorders.
MATERIALS AND METHODS
Repeats
(CAG)5/(CTG)5 sequence were purchased from MWG Biotech GmbH and dissolved the DNA with the MilliQ water. Further dilutions of DNA were prepared with 5 mM Tris-HCl (pH of 7.4) buffer for the circular dichroism (CD) and UV studies.
Copper chloride (CuCl2.2H2O)
Copper chloride dihydrate (Cu) was purchased from Merck Schuchard, and was used without purification. Stock solution (50 mM) was prepared in milliQ water and used for the interaction studies with DNA and also with peptides. The required dilutions were prepared as and when necessary for the experiments.
Aβ1-16 peptide
Aβ1-16 fragment with the sequence DAEFRHDSGYEVHHQK having molecular weight 1956 Da was purchased from M/s USV Peptides and used without further purification.
Circular dichroism studies
CD is used to explore the conformations of nucleic acids. The Cu and Aβ1-16 induced conformational changes of (CAG)5 and (CTG)5 were recorded using a Jasco J-715 spectropolarimeter at 25°C. Quartz cuvetts with path length of 1 mm were used for spectral recording in the range between 200 nm to 320 nm with sampling interval at every 1 nm.The spectra recorded were average of four repetitive scans in the absence and presence of different concentrations of the Cu and Aβ1-16.
UV/Vis absorption studies
The electronic absorption studies of (CAG)5/ (CTG)5 and its binding ability to Cu, Aβ1-16, were investigated with a Jasco V-530. Spectrophotometer (Jasco, Japan) equipped with a peltier temperature controller. (CAG)5/(CTG)5 samples were dissolved in Tris–HCl buffer (5 mM, pH 7.4) in the absence and presence of Cu and Aβ1-16 were recorded between 220 nm to 320 nm with a matched set of 1 cm path length quartz cuvettes.
Thermal denaturation studies of CD and UV
Thermodynamic studies of (CAG)5 and (CTG)5 in the absence and presence of Cu were measured on a Jasco J-715 spectropolarimeter connected with Model PTC-348WI, Peltier type temperature control system. Samples were recorded in the wavelength between 200 nm and 320 nm by varying the temperature of 20°C to 100°C.
UV- Thermal melting studies of (CAG)5 repeats in 10 mM HEPES buffer (pH 7.4) in the absence and presence of Cu were measured with Jasco V-530 spectrophotometer equipped with Jasco-ETC-505T temperature controller and cell holder that permits temperature control. Absorbance and Tm was measured at 253 nm from 25°C to 95°C, with a heating rate at 1.0°C/min. For each optically monitored transition Tm of (CAG)5 was determined as the transition midpoint.
Molecular docking studies
Considering the results obtained in vitro study and to have a better understanding, it was assumed of more significance to perform molecular docking studies to identify the mode of interactions of the Cu and Aβ1-16 with DNA- CAG and CTG repeats, which further correlate both in silico and in vitro results. The Docking approaches were carried out using discovery studio 3.5 modelling program (DS 3.5); GOLD (Genetic Optimization for Ligand Docking) and Cdocker, A charmm-based molecular dynamics (md) simulated-annealing-based algorithm and a conventional molecular mechanics force field. Initial work involved the Crystal structure retrieval and preparation. The crystal structures, PDBID: 4YN6 of RNA model r (CAG) motif repeat sequence (UUGGGCCAGCAGCAGGUCC)2 and CTG repeat (ATGCTGCAT) PDBID:1MNV retrieved from the Research collaboration for structural bioinformatics Protein Data Bank (RCSB PDB) were used in this study. Crystallization water molecules were removed from the DNA molecules and are optimized for docking by the preparation and function edit Nucleic Acid tool of by DS 3.5. Similarly, the Cu (II) ion (PubChem CID:27099) retrieved form Pubchem database and the 3D coordinates of Aβ1-16 fragment was taken from the NMR structures elucidated [16]. The DNA, ligand molecules; metal ion and Aβ1-16 were minimized prior to docking.by using CHARMm force field with potential energy (kcal/mol) of –978.23719. DNA is kept rigid while the ligands are treated as fully flexible and a final minimization step is used to refine the docked possess. The docking tool “GOLD” employs genetic algorithm to search the rotational flexibility of receptor hydrogen’s and ligand conformational flexibility.
The active site with a 6Å radius sphere was defined by selecting define and edit binding site function of DS 3.5. A set of top 10 solutions was explored for favorable and stable interactions. Gold Score function was used to evaluate the docking poses. The individual binding poses of each ligand were identified and their interactions with the DNA were studied. The best and most energetically favorable conformation of each ligand was selected and the interactions tabulated. To further illustrate, the influence of Cu and Aβ1-16 binding effect on conformation in B-DNA structure, Structural overlay approaches was carried between native DNA form and a docked Aβ-Cu bound DNA complexes. The outcomes and orientation of the nucleotide bases/DNA axis and the influences of the DNA major and minor grooves were further evaluated.
RESULTS
Interaction of DNA or nucleotide sequences with Cu and Aβ1-16
CD Studies on the effect of Cu and Aβ1-16 on the conformation of (CAG)5/(CTG)5 sequence
CD spectra’s (CAG)5 and (CTG)5 repeats were monitored in the absence and presence of Cu and Aβ1-16 peptides. The spectra depicted in Figs. 1 and 2 showed typical B-DNA conformation with a characteristic positive band at 275 nm for CAG and 285 for CTG sequence attribute to the base stacking and a negative band at 252 nm (CAG) and 258 nm (CTG) due to the right-handed helicity [17, 18]. Substantial changes in the CD spectra were observed upon interaction with Cu (250 μM) to CAG, the spectra shifted to higher wavelengths exhibited the largest change in the positive band to 288 nm (14 nm) and negative band to 261 nm (9 nm) as shown in Fig. 1. Similar results were noticed in (CTG) with Cu as depicted in Fig. 2. The results indicate that, DNA sequences exhibited the marked conformational changes from B-DNA to altered B-DNA conformation upon Cu binding.
Fig.1
Circular dichroism studies showing the conformational change of (CAG)5 in the presence of Copper (Cu). (CAG)5 alone (a), (CAG)5 with 250 μM Cu (b), Cu alone (c).
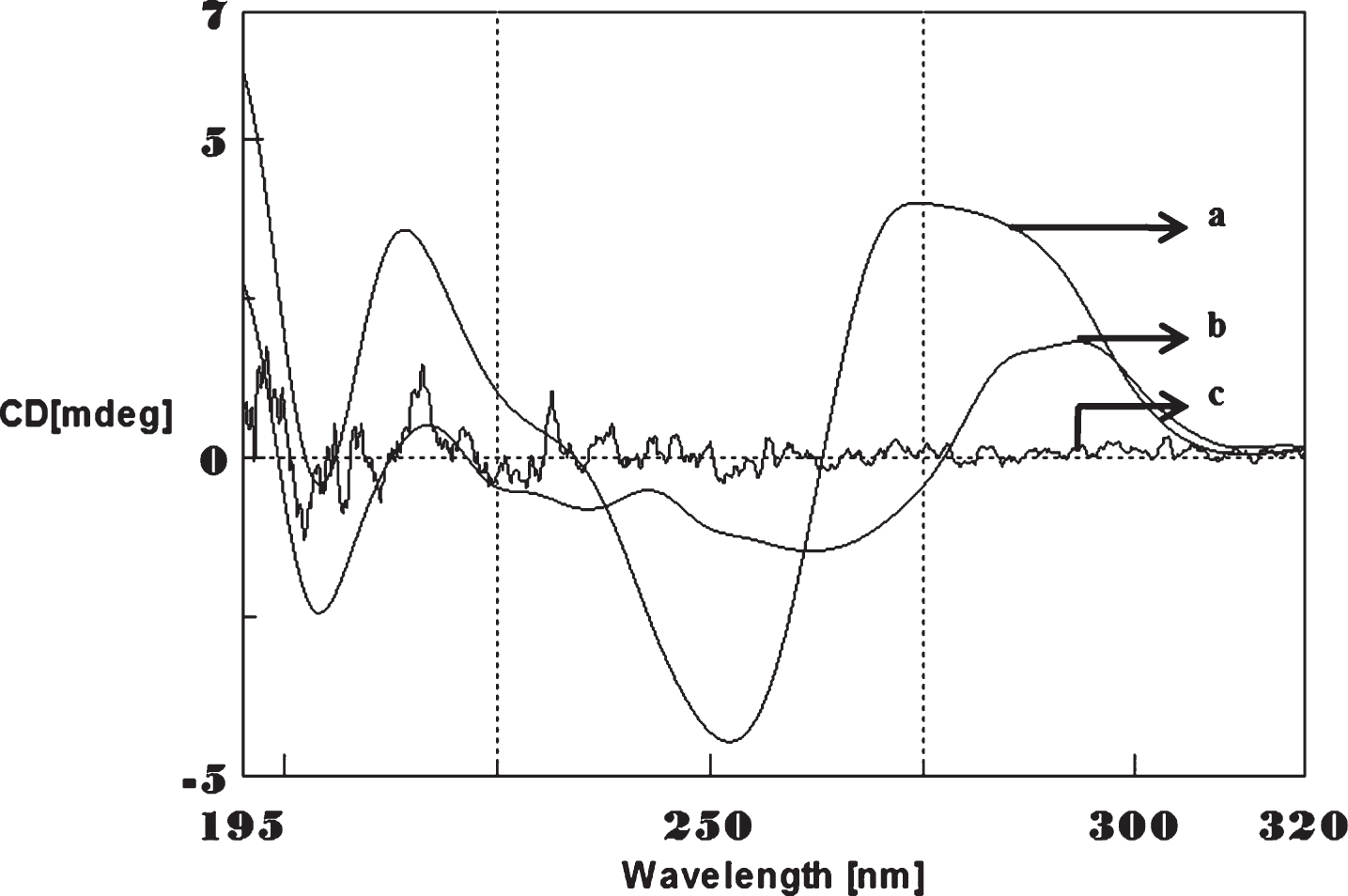
Fig.2
Circular dichroism studies showing the conformational change of (CTG)5 in the presence of Cu. (CTG)5 alone (a), (CTG)5 with 250 μM Cu (b), Cu alone (c).
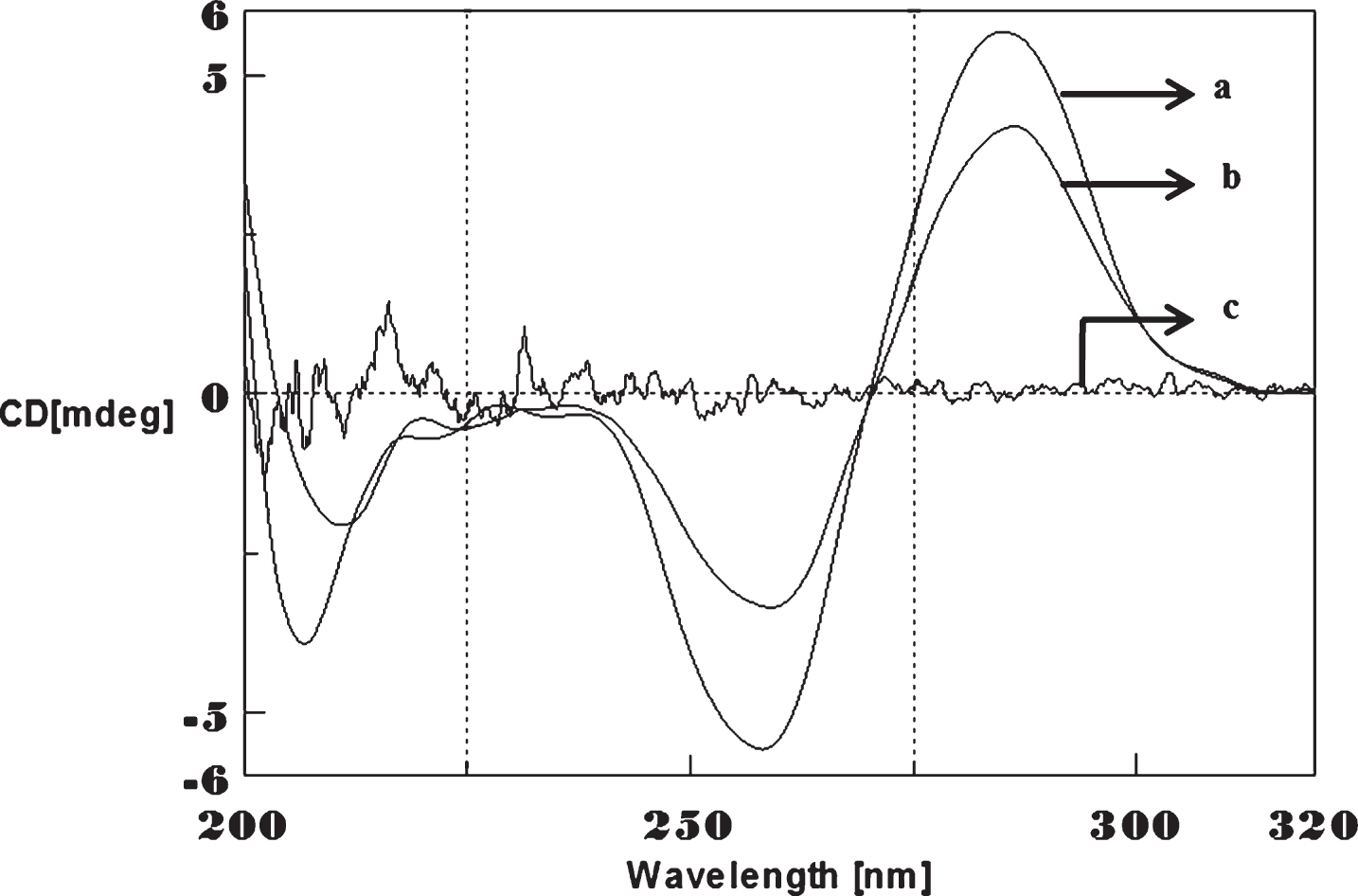
To determine whether Aβ1-16 interaction influences DNA conformation, we measured CD spectra of CAG and CTG sequences in the absence and presence of Aβ1-16. B-form DNA with two typical bands at 276 nm and 252 nm in CAG shown in Fig. 3 and 285 nm and 258 nm in CTG represented in Fig. 4 in the absence of Aβ peptide. In the presence of Aβ, the 250 nm and 272 nm bands diminished. The spectral variations in (CAG)5 and (CTG)5 sequence DNA is due to the interaction of Cu and Aβ1-16 peptide to a specific binding site and may push the strands of DNA apart to accommodate the incoming Cu and Aβ1-16 peptide. The decrease in the magnitude of both positive and negative band could be related to an increase in the winding angle of the DNA helix may be due to the super helical tension caused by Cu and Aβ1-16 [19, 20]. The binding of Cu to DNA causes the perturbation of B-DNA and alter the base stacking pattern in the DNA structures of each strand resulting in unwinding of the helices. This process leads to altered B-DNA conformations. These results suggested the effect of interaction of Cu and Aβ1-16 peptide into base stacking and a decreased right handedness of the DNA. Together, the changes indicate that there was a DNA conformational transition during incubation with Aβ as well as Cu, and also indicate that the secondary conformation of DNA was disturbed. Earlier reports indicated DNA binding property of Aβ and synuclein leading to the modulation of DNA topology [21, 22]. There were reports on the nuclear localization of Aβ in AD brain and a possible mechanism for Aβ playing a role in DNA conformational change [23]. The present study provides a clue on the mechanism of Aβ1-16 and Cu binding to triplet DNA and altering conformation.
Fig.3
Circular dichroism studies showing the conformational change of (CAG)5 in the presence of Aβ1-16 peptide. (CAG)5 alone (a), (CAG)5 with Aβ peptide (0.2 mM) (b), Aβ alone (c).
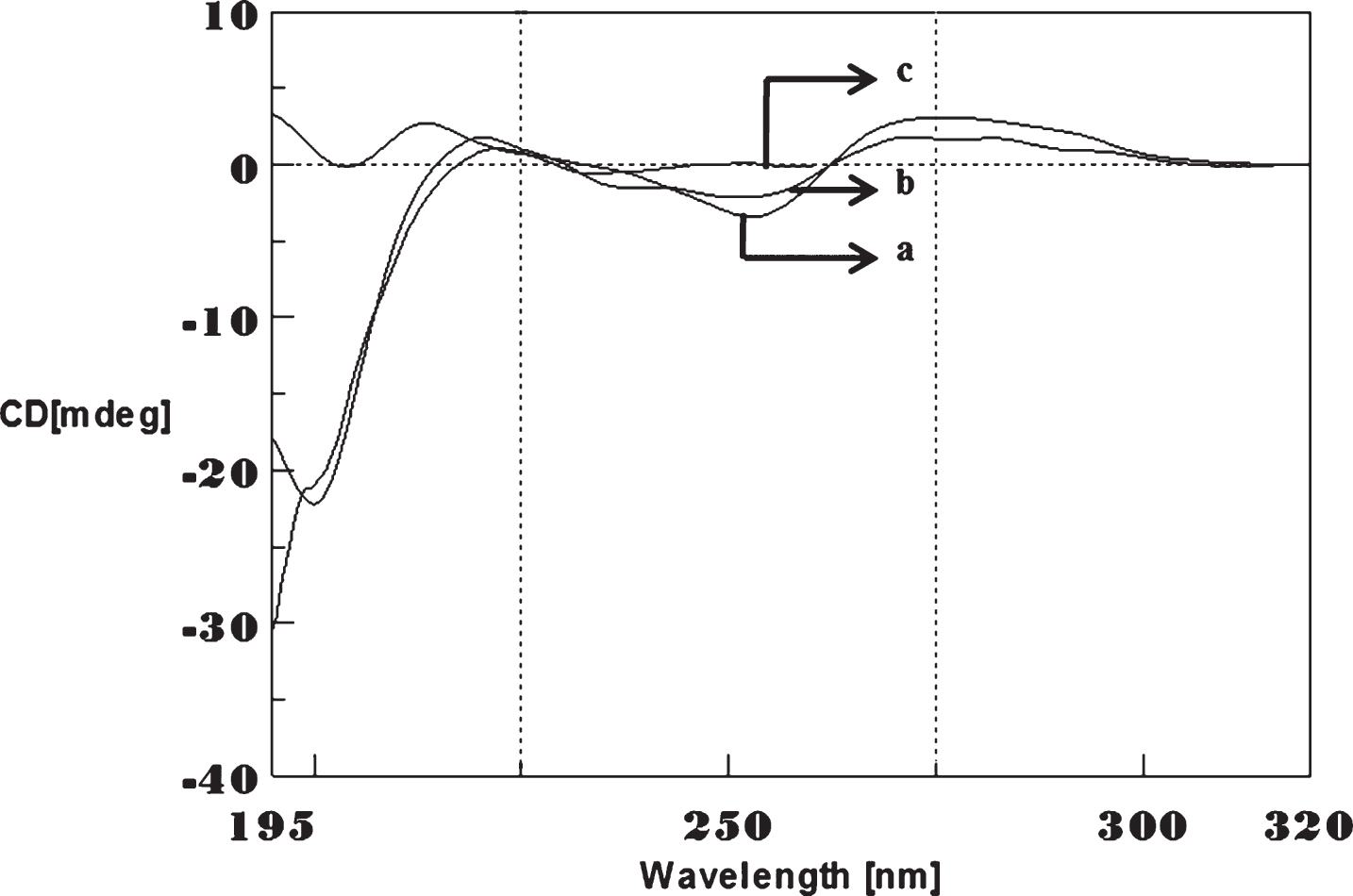
Fig.4
Circular dichroism studies showing the conformational change of (CTG)5 in the presence of Aβ1-16 peptide. (CTG)5 alone (a), (CTG)5 with Aβ peptide (0.2 mM) (b), Aβ alone (c).
UV Absorption studies on the effect of Cu and Aβ1-16 on the conformation of (CAG)/(CTG)5
Interaction of Cu with (CAG)5 and (CTG)5 sequences were monitored by UV absorption spectrum. Figure 5 showed the maximum absorbance of (CAG)5 sequence at 253 nm, there was a negligible change in absorbance upon addition of Cu at lower concentration. At higher concentration of Cu, there was an increase in the absorbance (hyperchromicity) with a peak shift to lower wavelength to at 244 nm (blue shift) as a function of Cu concentration. Similar changes were also noticed in (CTG)n with Cu (Fig. 6). The spectral changes may be attributed to a strong interaction between π–π* electron cloud of the interaction of Cu with DNA base pairs.
Fig.5
UV absorbance studies of (CAG)5 in the presence of Cu. (CAG)5 alone (a), (CAG)5 with 250 μM Cu (b).
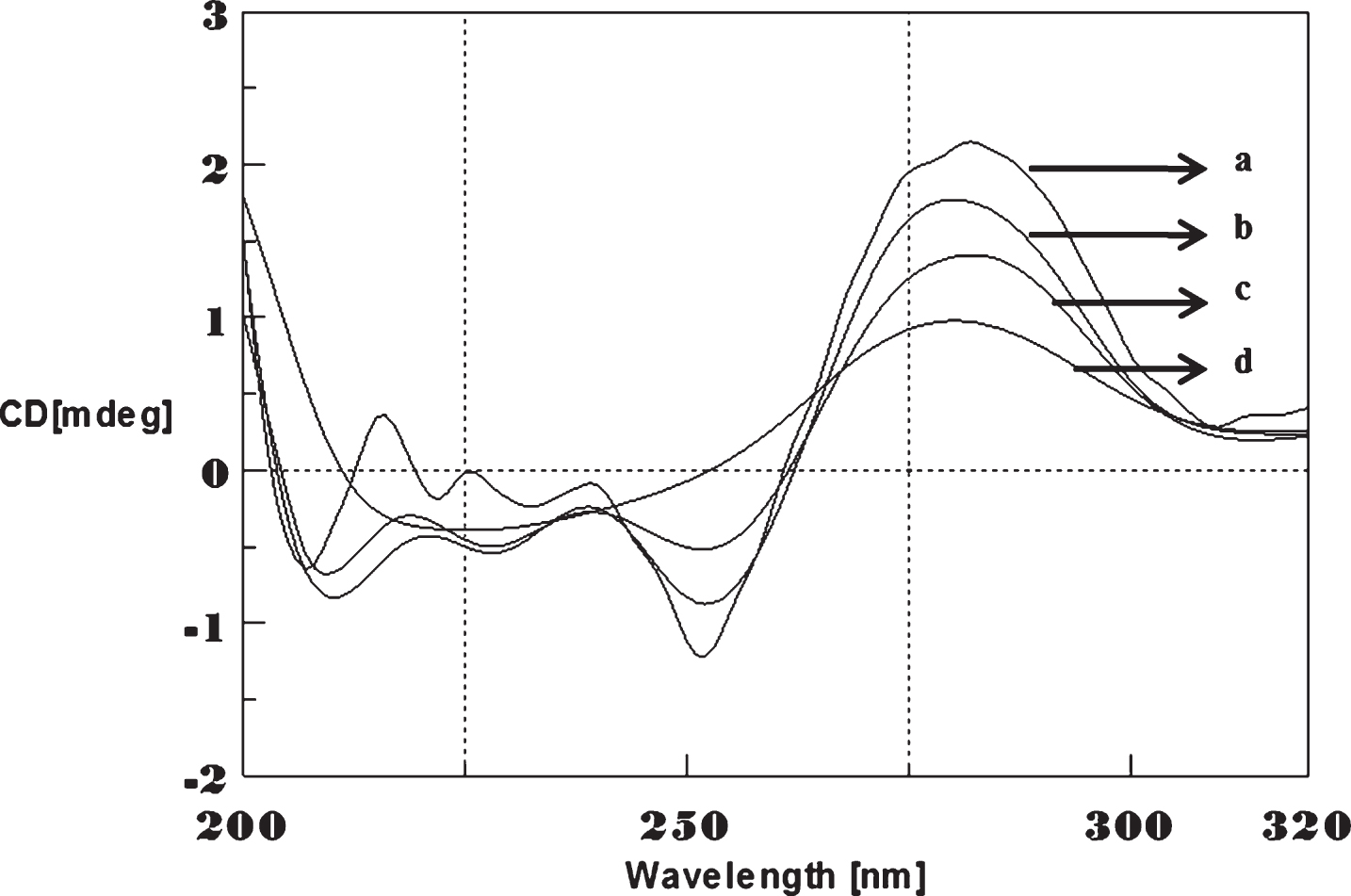
Fig.6
UV absorbance studies of (CTG)5 in the presence of Cu. (CTG)5 alone (a), (CTG)5 with 250 μM Cu (b).
The Absorption spectra of (CAG)5 and (CTG)5 sequence were monitored by the addition of varying concentrations of Aβ and found that there were significant changes in the absorbance at 260 nm, supporting Aβ and DNA interaction (Figs. 7 and 8). The changes observed in the spectra of DNA reveals that the Aβ interacts with DNA and the changes in conformation may be due to the interaction between the electronic state of the intercalating Aβ peptide with that of the DNA bases [23, 24]. The hypochromism along with red shift observed in the spectra may be due to the interaction of Aβ to the DNA through interacalation resulting electronic perturbations and twist the helix(winding) in DNA to accommodate the incoming Aβ peptide.
Fig.7
UV absorbance studies of (CAG)5 in the presence of Aβ. CAG alone (a), CAG-Aβ (0.2 mM) (b).
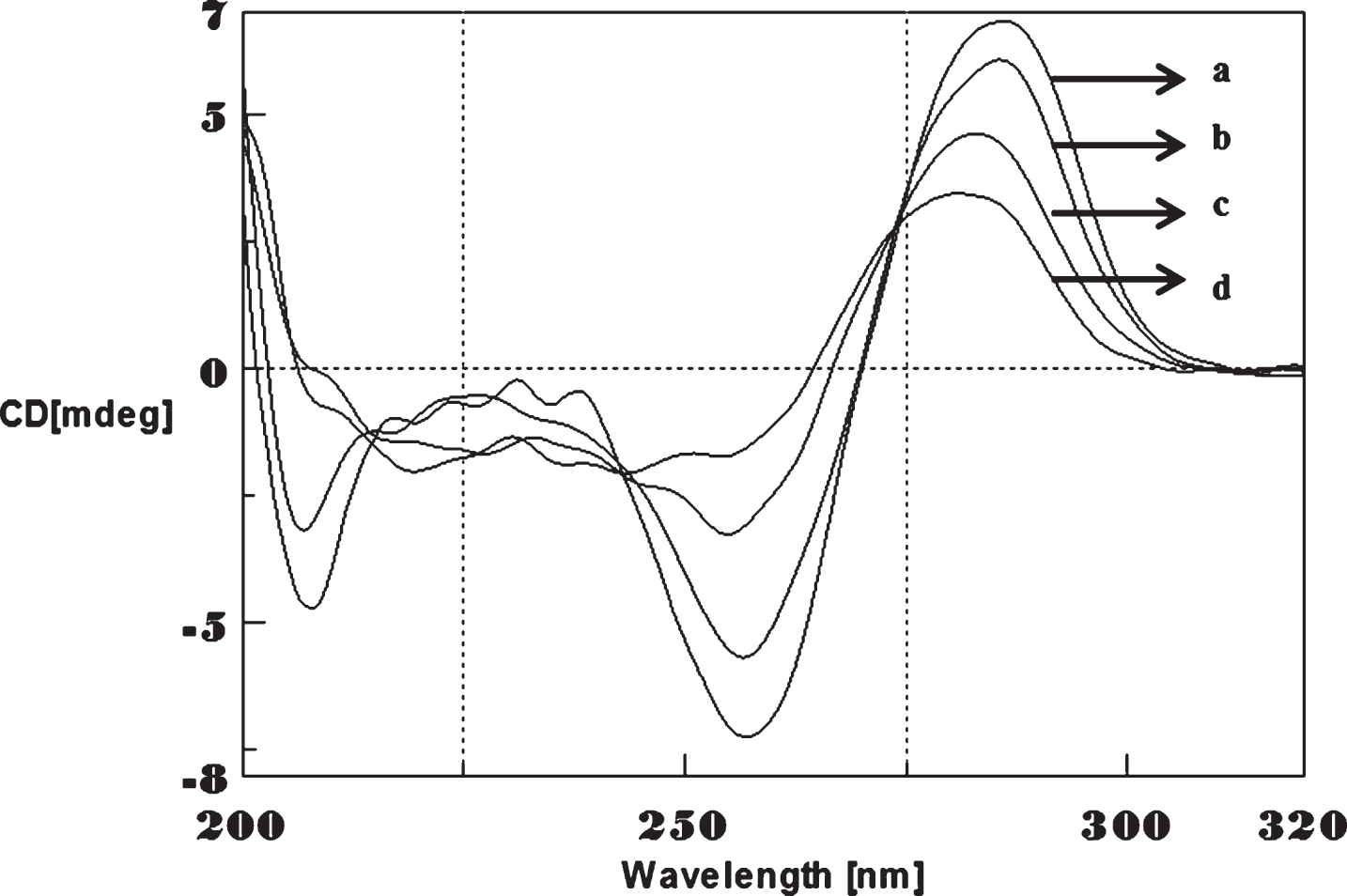
Fig.8
UV absorbance studies of (CTG)5 in the presence of Aβ1-16. CTG alone (a), CTG-Aβ (0.2 mM) (b).
Effect of temperature on the conformation changes of (CAG)/CTG sequence
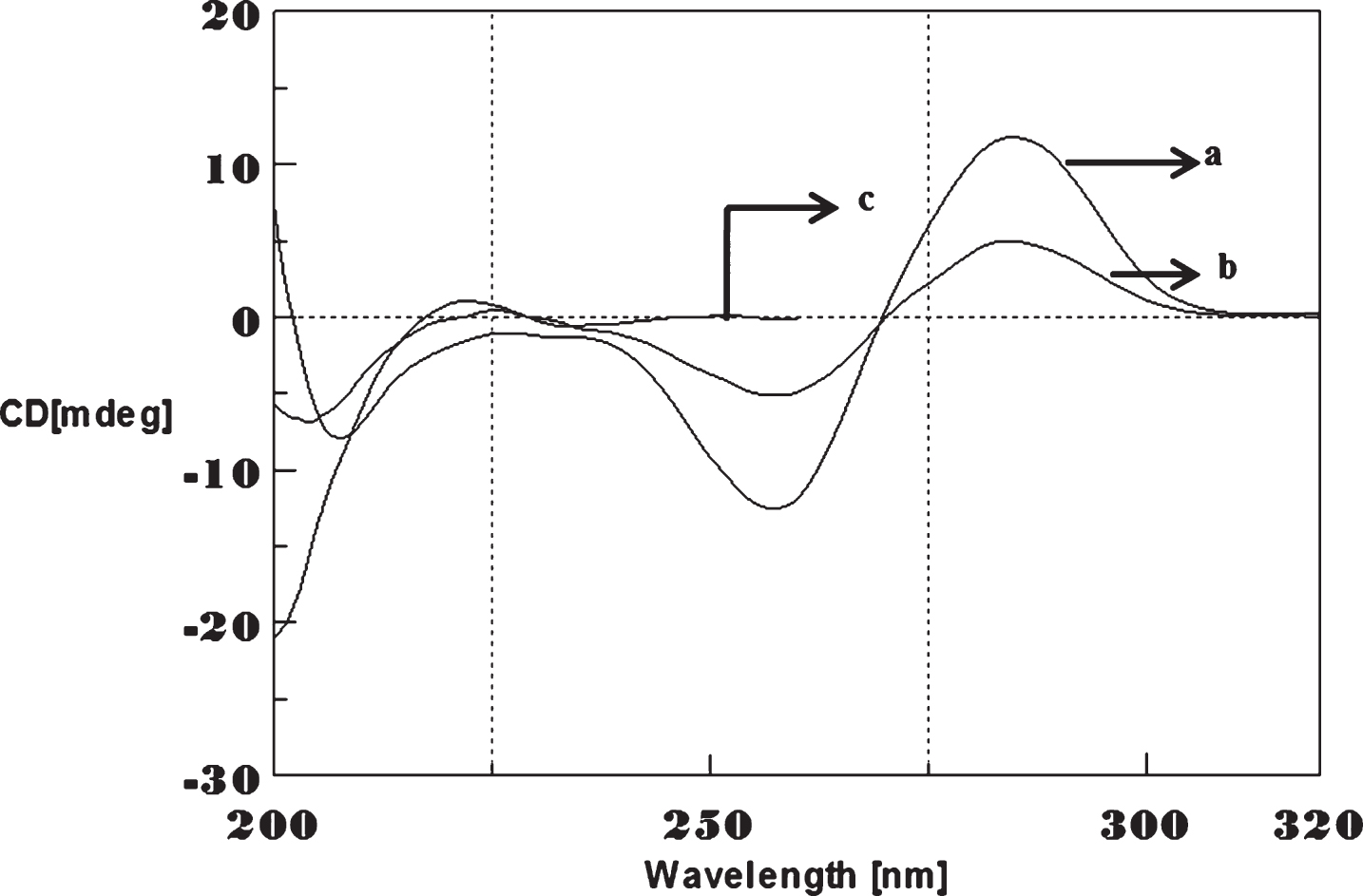
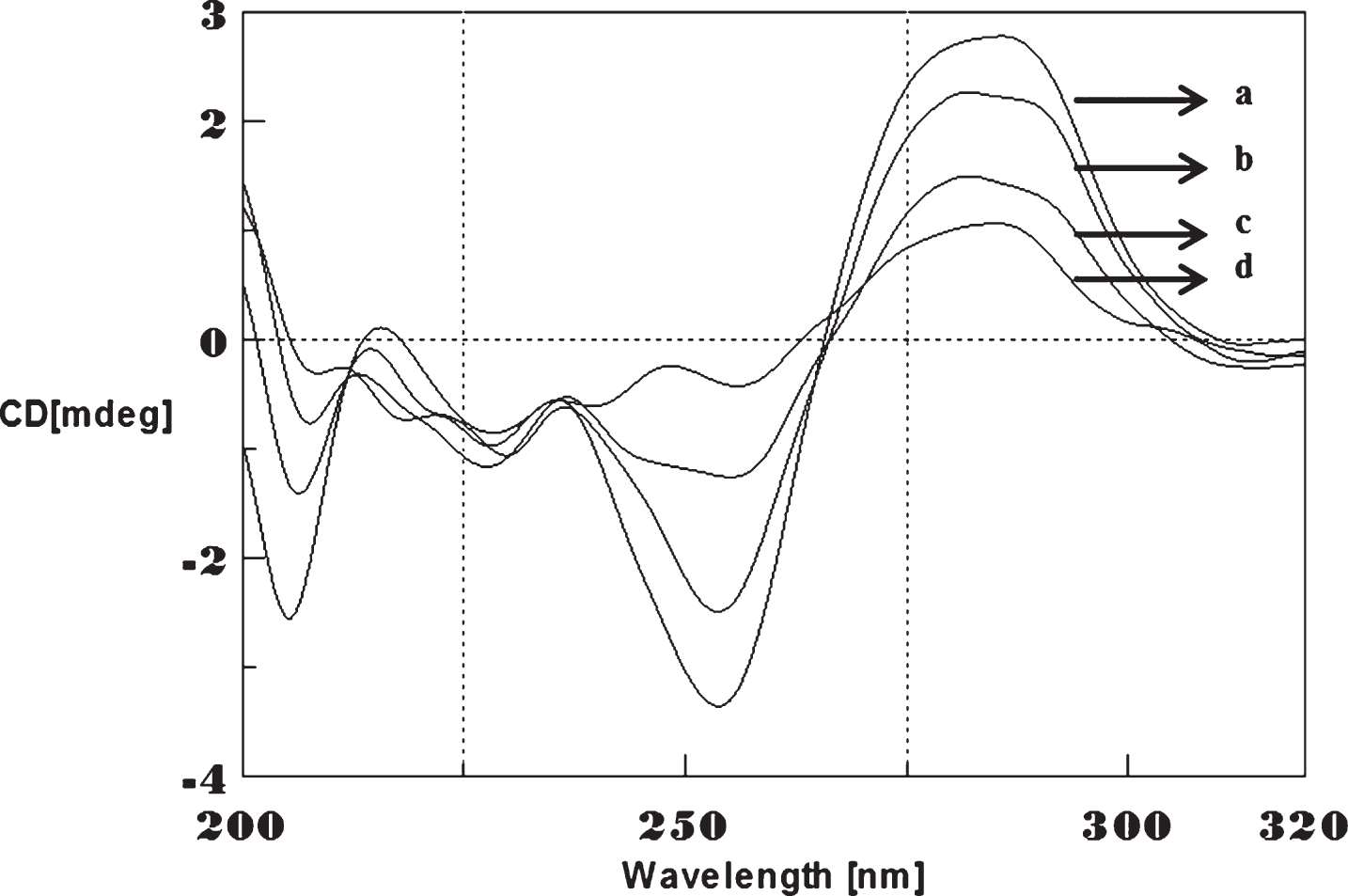
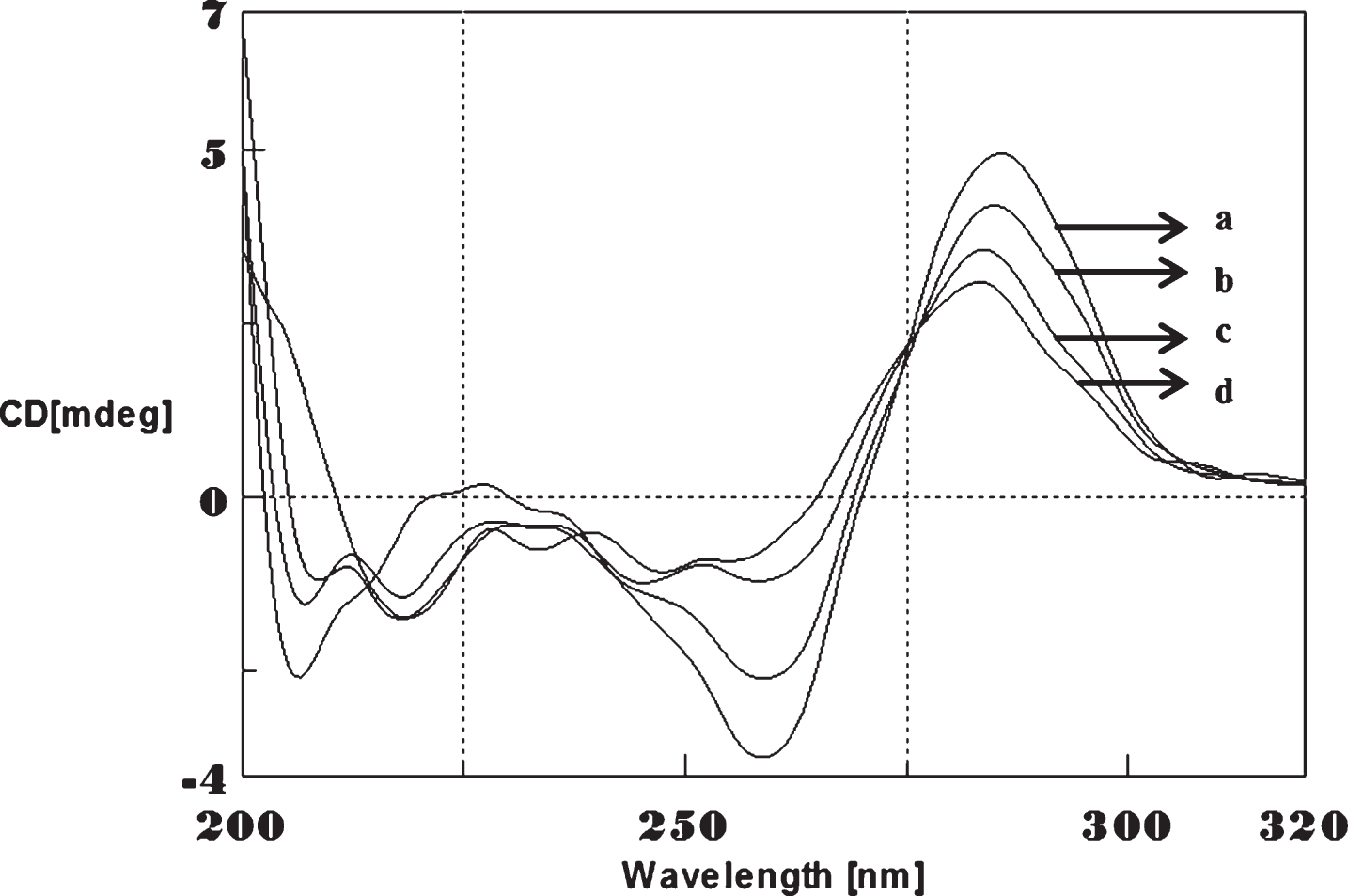
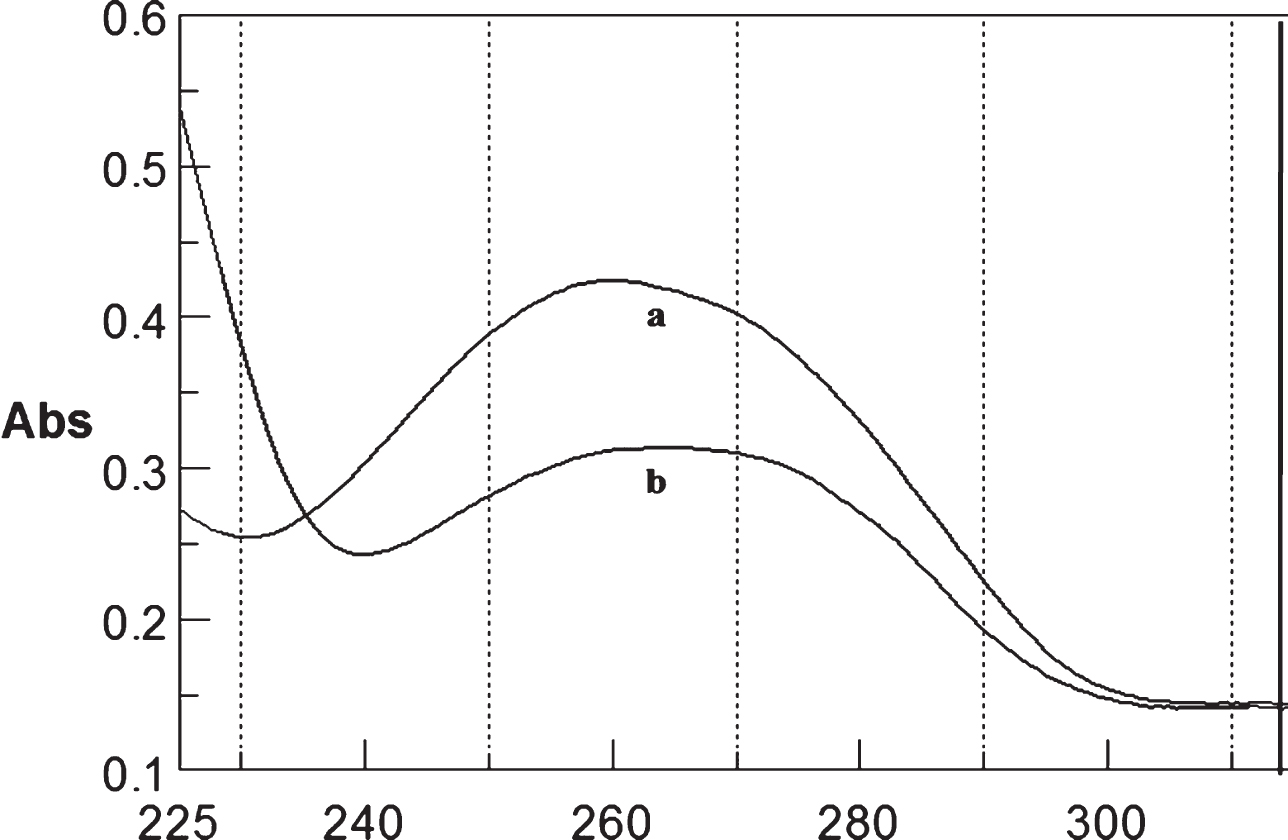
To determine the effect of temperature on the DNA conformation and stability, we measured CD spectra of CAG and CTG sequences in the absence and presence of Cu from 20°C to 100°C. CD spectra of (CAG) show characteristics of B-DNA (Figs. 9–12). The decrease in the CD bands with complete loss of the negative band with the redshift provided evidence for the disruption of a base-paired structure as the temperature increased. It was saturated to at 80°C. In the presence of 50 μM Cu, similar decrease in the positive and negative bands with the redshift noticed and was saturated to at 70°C (Fig. 11). The effect of temperature on (CTG) spectrum was shown in Fig. 12. As temperature increased, there was a decrease in intensity of positive band and complete loss of the negative band with the blue shift was noticed. Similar pattern of changes were also observed with 50 μM Cu and an isodichroic point was observed at 265 nm. The result indicates that both the bands are sensitive to temperature and the conformational change showed cooperative and biphasic transition. The melting profile provides insight into the physical attributes of the (CAG)5 and (CTG)5sequence. At high temperature, the hydrogen bonds between the two complementary DNA strands are broken, and disturb the secondary conformation. The melting of strands (Tm) occurs over a narrow temperature range depends strongly on the percentage of the stronger hydrogen bonding present between G-C base pairs, therefore, DNA with a higher G-C content will have a higher Tm [25, 26].
Fig.9
Circular dichroism studies showing the conformational change of (CAG)5 as a function of temperature. 20 (a), 40 (b), 60 (c), and 80°C (d).
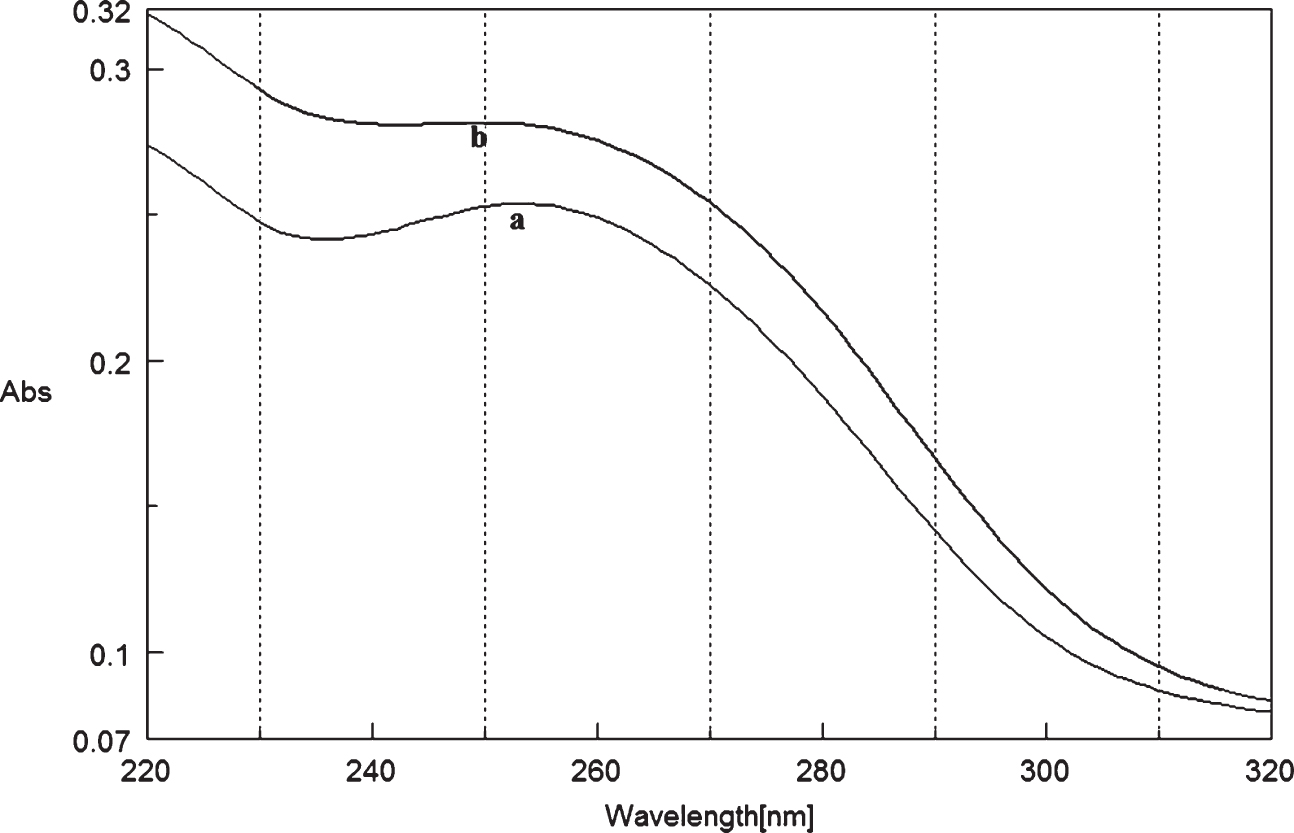
Fig.10
Circular dichroism studies showing the conformational change of (CAG)5 in the presence of 50 μM Cu as a function of temperature. 20 (a), 40 (b), 60 (c), and 80°C (d).
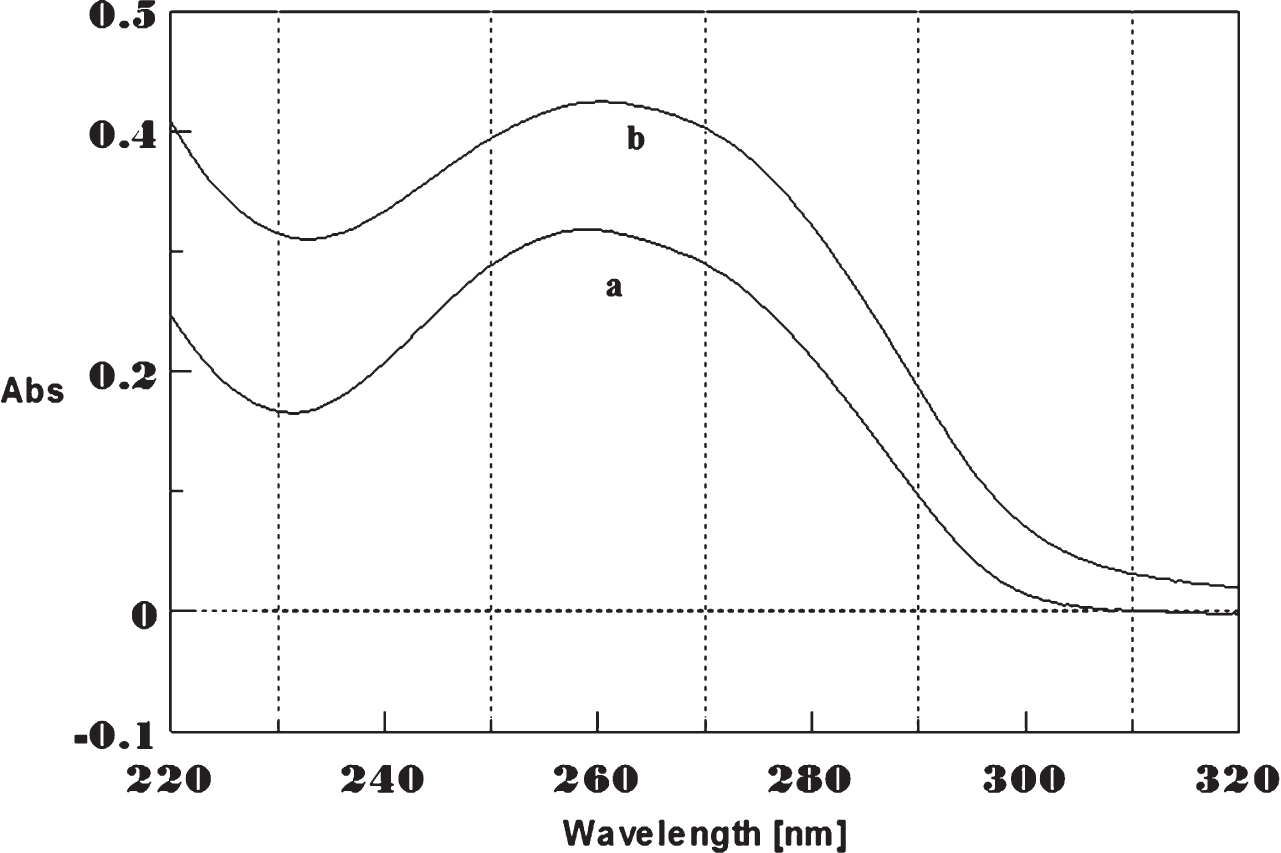
Fig.11
Circular dichroism studies showing the conformational change of (CTG)5 as a function of temperature. 20 (a), 40 (b), 60 (c), and 80°C (d).
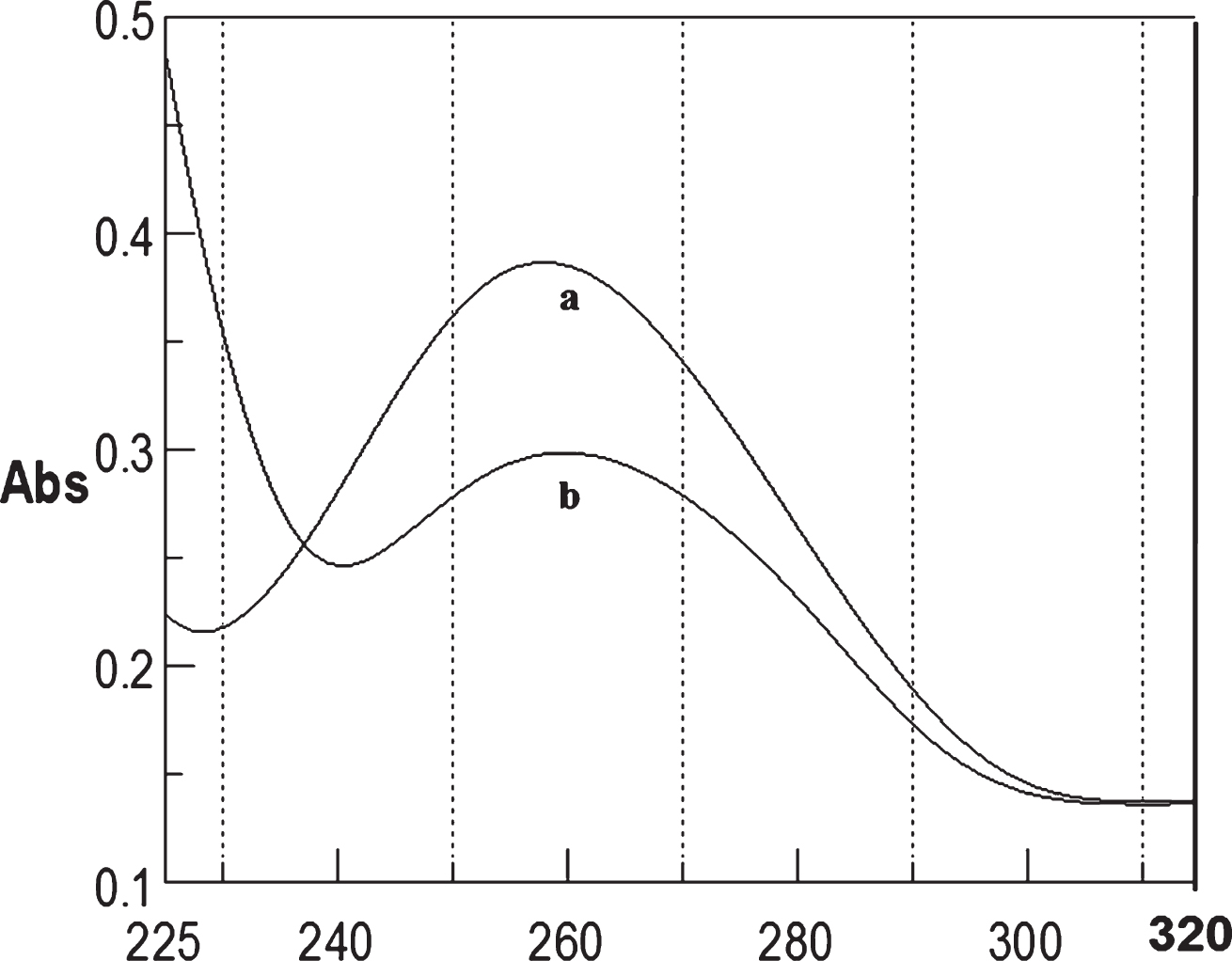
Fig.12
Circular dichroism studies showing the conformational change of (CTG)5 in the presence of 50 μM Cu as a function of temperature. 20 (a), 40 (b), 60 (c), and 80°C (d).
The results of UV absorption spectroscopy showed the Tm of (CAG) sequence as 70°C under experimental conditions. Addition of 500 nM Cu increased the Tm to 73°C, and in the presence of 50 μM, it increased to 80.6°C. However, at 100 μM Cu, Tm decreased to 78.4°C). The increase in Tm at lower concentration of Cu has been attributed to the stabilization of the DNA conformation by binding to phosphate backbone of the DNA, and decrease in Tm at higher concentrations of Cu may be due to the binding of Cu to the bases of (CAG)5 sequence and breaking the hydrogen bonds that held between the G-C bases of the complimentary strands of DNA. DNA-Cu (II) complex is sensitive to higher temperatures and involves complex binding sites between strands and is strong enough to compete effectively and the secondary conformations of each strand are destroyed due to unwinding of the helices. Cu ion perturbs the B-form residues in the base composition of DNA, resulting in destabilization of the DNA double helix [27]. The Tm data of both CD and UV studies suggest that Cu ions bind to both phosphate and nucleotide bases of DNA, unwind the two strands at high temperature, and lower the stability.
Docking studies
The systematic molecular docking techniques used in the study may possibly provide a clue toward understanding the specific binding site of the DNA for Cu and Aβ1-16. To determine the most possible binding site, interaction mode and binding affinity, docking studies have been performed on Cu and Aβ1-16 with DNA- CAG repeat and CTG- DNA repeats. As shown in Figs. 12 and 14 (Tables 1–4), the complexes interact with DNA through partial intercalation at the major groove. The Docking results have been established that, Aβ1-16 and Cu have favorable DNA-CAG repeat binding affinities across major groove, with a resulting relative binding energy of –31.9763 kJ mol–1 through, CYT7, ADE 8, GUA9, CYT10, ADE11, GUA12, CYT13 of A chain, indicating that there are certain hydrogen bonding interaction between the complexes and DNA by forming total of 8 Hydrogen bonds with amino acids. Similarly, 11 hydrogen bonds are formed with CYT7, ADE8, GUA9, CYT10, ADE11, GUA12, CYT13, ADE14 of B chain. The more negative binding energy shows more potent DNA-binding affinity of the Aβ1-16. Likewise, interaction of Cu specifically with guanine contributes to the changes in the stability of the DNA molecule. Hence, formation of certain hydrogen-bonding interaction and more number of hydrogen bond interactions with nitrogenous bases of B-DNA- (CAG) confirm the interaction of Aβ1-16 in presence of Cu. Similarly, docking studies of CTG- DNA repeat (ATGCTGCAT) [PDBID: 1MNV] with Aβ1-16 in the presence of Cu, as evident from the results (Fig. 14 and Tables 3 and 4), that the CTG-DNA repeats also exhibit the promising interaction with the Aβ1-16 and Cu. To further illustrate the influence of Cu and Aβ1-16 and their binding effect on conformation in B-DNA structure, the structural overlay was carried out for both the CAG and CTG DNA repeats between native DNA and docked Cu Aβ bound DNA complexes. As the overlay shown in Figs. 13II and 14II depicts that, the base pairs show significant rotations/deviations upon binding of Aβ1-16 and Cu, while compared to the native form of DNA. The circled nucleotides (Figs. 13 and 14) exhibit the shift in position of the nucleotides. On the other hand, from Fig. 13II, Guanine residues reveal the interaction of Cu ions by forming covalent coordination bonds preferentially to N7 position of guanine and there was no indication of binding of Cu to phosphate oxygen has been observed.
Fig.13
I) best docked conformations of Aβ1-16 and Cu with RNA model r (CAG) motif repeat sequence of (UUGGGCCAGCAGCAGGUCC)2, [PDBID: 4YN6]. II) Structural overlay between native DNA form and a docked Aβ-Cu bound DNA complexes. The encircled nucleotide bases displays the orientation of nucleotide base pairs/significant rotations across the base pairs/ base stacking pattern of Bound (dark orange) and Unbound forms of DNA helices (Green) indicating the shift in the position upon binding of the Aβ-Cu representing the changes in the altered B-DNA conformations.
![I) best docked conformations of Aβ1-16 and Cu with RNA model r (CAG) motif repeat sequence of (UUGGGCCAGCAGCAGGUCC)2, [PDBID: 4YN6]. II) Structural overlay between native DNA form and a docked Aβ-Cu bound DNA complexes. The encircled nucleotide bases displays the orientation of nucleotide base pairs/significant rotations across the base pairs/ base stacking pattern of Bound (dark orange) and Unbound forms of DNA helices (Green) indicating the shift in the position upon binding of the Aβ-Cu representing the changes in the altered B-DNA conformations.](https://content.iospress.com:443/media/adr/2017/1-1/adr-1-1-adr170027/adr-1-adr170027-g013.jpg)
Fig.14
Docking of CTG repeat (ATGCTGCAT) with Aβ1-16 and Cu [PDBID: 1MNV]. II) Structural overlay between Native DNA form and docked Aβ-Cu bound DNA complexes. The encircled nucleotide bases display the orientation of nucleotide base pairs/significant rotations across the base pairs. Base stacking pattern of bound (lemon Yellow) and unbound forms of DNA helices (Green) are showing the shift in the position upon binding of the Aβ-Cu representing the changes in the altered B-DNA conformations.
![Docking of CTG repeat (ATGCTGCAT) with Aβ1-16 and Cu [PDBID: 1MNV]. II) Structural overlay between Native DNA form and docked Aβ-Cu bound DNA complexes. The encircled nucleotide bases display the orientation of nucleotide base pairs/significant rotations across the base pairs. Base stacking pattern of bound (lemon Yellow) and unbound forms of DNA helices (Green) are showing the shift in the position upon binding of the Aβ-Cu representing the changes in the altered B-DNA conformations.](https://content.iospress.com:443/media/adr/2017/1-1/adr-1-1-adr170027/adr-1-adr170027-g014.jpg)
Thus, our Insilco results furthermore confirm and support the perturbation of B-DNA and alter the base stacking pattern in the DNA structures of each strand resulting in altered B-DNA conformations. The binding of Cu to DNA probably causes disturbed complementary interactions of DNA. Thus, our Insilco results are in consistent with the DNA-binding properties as evidenced by CD and UV-Vis Spectroscopic studies.
Table 1
CAG Repeat and Aβ1-16 amino acid interaction data across A chain
| Sl NO | Nucleotides in DNA A chain interacting with DNA repeats | Aβ1-16 amino acids distance in A0 | Hydrogen Bonds |
| 1 | CYT7 | ARG5 | 18.5 |
| 2 | ADE8 | GLN15 | 2.473 |
| 3 | GUA9 | GLN15 | 2.528 |
| GLN11 | 3.431 | ||
| 4 | CYT10 | VAL12 | 2.827 |
| 5 | ADE11 | HIS14 | 4.889 |
| 6 | GUA12 | HIS13 | 4.500 |
| 7 | CYT13 | ASP1 | 5.861 |
Table 2
CAG Repeat and Aβ1-16 amino acid interaction data across B chain
| Sl No | Nucleotides in DNA B chain interacting with DNA repeats | Aβ1-16 amino acids | Hydrogen Bonds distance in A0 |
| 1 | CYT7 | ASP1 | 4.080 |
| 2 | ADE8 | GLU3 | 3.197 |
| 3 | GUA9 | HIS13 | 3.722 |
| GLN6 | 2.066 | ||
| 4 | CYT10 | ASP7 | 3.163 |
| VAL12 | 3.960 | ||
| 5 | ADE11 | HIS14 | 4.889 |
| GLY9 | 2.327 | ||
| 6 | GUA12 | VAL12 | 2.110 |
| 7 | CYT13 | GLU11 | 3.886 |
| 8 | ADE14 | GLU11 | 3.999 |
Table 3
CTG repeat and Aβ1-16 amino acid interaction data across A chain
| Sl NO | Nucleotides in DNA A chain | Aβ1-16 amino acids interacting with DNA repeats | Hydrogen Bonds distance in A0 |
| 1 | CYT4 | GLN15 | 3.071 |
| 2 | THY5 | TYR10 | 5.450 |
| 3 | GUA6 | GLU3 | 2.715 |
| GLN3 | 3.083 | ||
| 4 | CYT7 | GLU3 | 3.667 |
Table 4
CTG repeat and Aβ1-16 amino acid interaction data across B chain
| Sl NO | Nucleotides in DNA B chain | Aβ1-16 amino acids interacting with DNA repeats | Hydrogen Bonds distance in A0 |
| 1 | CYT13 | ASP1 | 2.743 |
| 2 | THY14 | TYR10 | 4.956 |
| 3 | GUA15 | HIS14 | 4.185 |
| GLN3 | 3.083 | ||
| 4 | CYT7 | GLU3 | 3.667 |
DISCUSSION
The B-DNA conformation is the normal conformation of DNA in cells and is attributed with a majority of biological functions and change in the conformation will alter the major and minor groove properties [28, 29] and thus alters minor transcription binders leading to altered gene expression [30, 31].
Cu is an essential metal for normal CNS development and function. However, excess Cu possesses toxicity through interaction with DNA and proteins [32]. Cells in the specific regions of brain, the basal ganglia and parts of the cortex are very sensitive to the effects of the DNA change [33]. Several observations implicated that accumulation of Cu in the brain can modulate DNA stability/other physiological events, may possibly lead to brain pathology [10, 34–39]. In this study, we provide evidence that, conformational change of (CAG) and (CTG) induced by Cu and Aβ1-16, may have relevance to the genomic biology of neurodegeneration.
Cu ions have a strong base-affinity and binds to G-C bases, specifically bind to N7 of guanine and N3 of cytosine of (CAG)/(CTG) sequence which was confirmed by docking studied. Coordination of the Cu(II) with N7 of guanine, which approaches the outer sphere of the DNA more closely in the Cu(II)-DNA complex and was related to the positions of the atom that is most susceptible to oxidative damage due to its lowest oxidation potential (dG 1,29 V, dA 1.42 V, dC 1.6 V, dT 1.7 V). Cu perturbs the hydrogen bonds rotate one of the bases around its glycosidic bond to bring the bonding position of the other base by a minor tilting of the bases in the adjacent base pairs. This causes unwinding of double helix and alters the angle of rotation of helix axis, the decrease in the pitch of the helix with a greater degree of twisting of the chain may affect sugar puckering, and the stacking patterns resulting changes in structural transition from a B-geometry to intermediate conformation of the altered B-DNA conformation. Our research gains the importance in showing the relationship between Cu and Aβ in altering DNA topology.
Conclusion
The Cu and Aβ interacts with CAG and CTG sequences and induce changes in conformation and stability of DNA and may have a role in genome biology in neurodegeneration.
CONFLICT OF INTEREST
All the authors hereby declare that they do not have any financial or any other conflict of interest related to the work in this publication.
ACKNOWLEDGMENTS
K.S. Rao is thankful for the SNI grant of SENACYTPanama, and the Melo Brain Grant for supporting the research project. K.S. Rao Jayanth is grateful for the Internship award by Sham Foundation-India. All authors thank the Chair, Molecular Biophysics Unit, IISc, Bangaore, India, for support.
REFERENCES
[1] | Richard GF , Kerrest A , Dujon B ((2008) ) Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 72: , 686–727. |
[2] | Orr HT , Zoghbi HY ((2007) ) Trinucleotide repeat disorders. Annu Rev Neurosci 30: , 575–621. |
[3] | Brouwer JR , Willemsen R , Oostra BA ((2009) ) Microsatellite repeat instability and neurological disease. Bioessays 31: , 71–83. |
[4] | Mirkin SM ((2007) ) Expandable DNA repeats and human disease. Nature 447: , 932–940. |
[5] | Zuccato C , Valenza M , Cattaneo E ((2010) ) Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev 90: , 905–981. |
[6] | Vonsattel JP , Myers RH , Stevens TJ , Ferrante RJ , Bird ED , Richardson EP Jr ((1985) ) Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol 44: , 559–577. |
[7] | Hedreen JC , Peyser CE , Folstein SE , Ross CA ((1991) ) Neuronal loss in layer V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett 133: , 257–261. |
[8] | Langbehn DR , Brinkman RR , Falush D , Paulsen JS , Hayden MRA ((2004) ) New model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet 65: , 267–277. |
[9] | Andrew SE , Goldberg YP , Kremer B , Telenius H , Theilmann J , Adam S , Starr E , Squitieri F , Lin B , Kalchman MA ((1993) ) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet 4: , 398–403. |
[10] | Savelieff MG , Lee S , Liu Y , Lim MH ((2013) ) Untangling amyloid-β, tau, and metals in Alzheimer’s disease. ACS Chem Biol 8: , 856–865. |
[11] | Azimi S , Rauk A ((2011) ) On the involvement of copper binding to the n-terminus of the amyloid beta peptide of Alzheimer’s disease: A computational study on model systems. Int J Alzheimers Dis 2011: , 539762. |
[12] | Suram A , Rao KS , Latha KS , Viswamitra MA ((2002) ) First evidence to show the topological change of DNA from B-DNA to Z-DNA conformation in the hippocampus of Alzheimer’s brain. Neuromolecular Med 2: , 289–297. |
[13] | Bacolla A , Wells RD ((2009) ) Non-B DNA conformations as determinants of mutagenesis and human disease. Mol Carcinog 48: , 273–285. |
[14] | Wells RD ((1996) ) Molecular basis of genetic instability of trinucleotide repeats. J Biol Chem 271: , 2875–2878. |
[15] | Hegde ML , Gupta VB , Anitha M , Harikrishna T , Shankar SK , Muthane U , Subba Rao K , Jagannatha Rao KS ((2006) ) Studies on genomic DNA topology and stability in brain regions of Parkinson’s disease. Arch Biochem Biophys 449: , 143–156. |
[16] | Narayan P , Krishnarjuna B , Vishwanathan V , Jagadeesh Kumar D , Babu S , Ramanathan KV , Easwaran KR , Nagendra HG , Raghothama S ((2013) ) Does aluminium bind to histidine? An NMR investigation of amyloid β12 and amyloid β16 fragments. Chem Biol Drug Design 82: , 48–59. |
[17] | Ivanov VI , Minchenkova IE , Schyolkina AK , Poletayev AI ((1973) ) Different conformations of double stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers 12: , 89–110. |
[18] | Hou MH , Robinson H , Gao YG , Wang AH ((2002) ) Crystal structure of actinomycin D bound to the CTG triplet repeat sequences linked to neurological diseases. Nucleic Acids Res 30: , 4910–4917. |
[19] | Hou MH , Lin SB , Yuann JM , Lin WC , Wang AH , Kan Ls L ((2001) ) Effects of polyamines on the thermal stability and formation kinetics of DNA duplexes with abnormal structure. Nucleic Acids Res 29: , 5121–5128. |
[20] | Eichhorn GL , Clark P ((1965) ) Interactions of metal ions with polynucleotides and related compounds v. The unwinding and rewinding of DNA strands under the influence of copper (II) ions. Proc Nat Acad Sci U S A 53: , 586–593. |
[21] | Hegde ML , Rao KS ((2007) ) DNA induces folding in alpha-synuclein: Understanding the mechanism using chaerone property of osmolytes. Arch Biochem Biophys 464: , 57–69. |
[22] | Hegde ML , Vasudevaraju P , Rao KS ((2010) ) DNA induced folding/fibrillation of alpha-synuclein: New insights inarkinson’s disease. Front Biosci 15: , 418–436. |
[23] | Hegde ML , Anitha S , Latha KS , Mustak MS , Stein R , Ravid R , Rao KS ((2004) ) First evidence for helical transitions in supercoiled DNA by amyloid beta peptide (1-42) and aluminum: A new insight in understanding Alzheimer’s disease. J Mol Neurosci 22: , 19–31. |
[24] | Zou XH , Ye BH , Li H , Zhang QL , Chao H , Liu JG , Ji LN , Li XY ((2001) ) The design of new molecular “light switches” for DNA. J Biol Inorg Chem 6: , 143–150. |
[25] | Zhang QI , Liu JG , Chao H , Xue GQ , Ji LN ((2001) ) DNA binding and photocleavage studies of cobalt(III) polypyridyl complexes: [Co(phen)2IP]3+ and [Co(phen)2 PIP]3+. J Inorg Biochem 83: , 49–55. |
[26] | Marmur J , Doty P ((1962) ) Determination of the base composition of DNA from its thermal denaturation temperature. J Mol Biol 5: , 109–118. |
[27] | Zimmer C , Venner H ((1970) ) Influence of copper (II) ions on the denaturation and spectral properties of DNA and methylated DNA. Eur J Biochem 15: , 40–47. |
[28] | Eichhorn GL ((1962) ) Metal ions as stabilizers or destabilizers of the DNA structure. Nature 194: , 474–475. |
[29] | Watson JD , Crick FH ((1953) ) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171: , 737–738. |
[30] | Ghosh A , Bansal MA ((2003) ) Glossary of DNA structures from A to Z. Acta Crystallogr D Biol Crystallogr 59: , 620–626. |
[31] | Hinoi E , Balcar VJ , Kuramoto N , Nakamichi N , Yoneda Y ((2002) ) Nuclear transcription factors in the hippocampus. Prog Neurobiol 68: , 145–165. |
[32] | Gaeta A , Hider RC ((2005) ) The crucial role of metal ions in neurodegeneration: The basis for a promising therapeutic strategy. Br J Pharmacol 146: , 1041–1059. |
[33] | Rosas HD , Salat DH , Lee SY , Zaleta AK , Hevelone N , Hersch SM ((2008) ) Complexity and heterogeneity: What drives the ever-changing brain in Huntington’s disease? Ann N Y Acad Sci 1147: , 196–205. |
[34] | Perera WS , Hooper NM ((2001) ) Ablation of the metal ion-induced endocytosis of the prion protein by disease-associated mutation of the octa repeat region. Curr Biol 11: , 519–523. |
[35] | Bush AI ((2000) ) Metals and neuroscience. Curr Opin Chem Biol 4: , 184–191. |
[36] | Bush AI ((2003) ) The metallobiology of Alzheimer’s disease. Trends Neurosci 26: , 207–214. |
[37] | Fox JH , kama JA , Lieberman G , Chopra R , Dorsey K , Chopra V , Volitkis I , Cherny RA , Bush AI , Hersch S ((2007) ) Mechanisms of copper ion mediated Huntington’s disease progression. PLoS One 2: , e334. |
[38] | Pérez P , Flores A , Santamaría A , Ríos C , Galván-Arzate S ((1996) ) Changes in transition metal contents in rat brain regions after in vivo quinolinate intrastriatal administration. Arch Med Res 27: , 449–452. |
[39] | Hands SL , Mason R , Sajjad MU , Giorgini F , Wyttenbach A ((2010) ) Metallothioneins and copper metabolism are candidate therapeutic targets in Huntington’s disease. Biochem Soc Trans 38: , 552–558. |



