Open science – towards reproducible research
Abstract
This paper presents an overview of several efforts towards reproducible research in the field of medical imaging and visualization. In the first section, the components of Open Science are presented: open access, open data and open source. In the second section, the challenges of open-science are described and potential solutions are mentioned. Finally, a discussion on the potential future of open science and reproducible research is introduced.
1.Introduction
For centuries, scientific publishing has been the driving mechanism for disseminating knowledge to scientific communities. The main role of publishing has been seen by many scientists as a way to “stand on the shoulders of giant” by reusing the methods and knowledge developed by peers in the common goal to advance Science.
While the act of publishing methods, results, and findings is critical to science, it has slowly become more of a requirement rather than a dissemination effort. For instance, in academia, researchers are primarily evaluated based on the number and the quality of their publications. Furthermore, in recent years, the overall cost of publishing has raised the level of entry for several institutions around the world. Finally, the content as well as the access of published materials has also been criticized by researchers who tried, and failed, to reproduce published recipes due to the lack of information and data.
For all these reasons, the Open Science movement has emerged in order to overcome these issues with “traditional scientific publishing.” This paper presents the key concepts of Open Science, as well as the challenges associated with this movement and the direction where Open Science is moving towards.
2.Open Science
To further understand how Open Science came to be, one must look at the concerns with traditional publishing.
First, publishing tends to be seen as a competition more than collaboration, especially in academia where the need to publish is critical. Second, traditional publishing is usually limited in content. In fact, often only written content along with figures can be associated with a given publication. This limitation is one of the sources of frustration for many researchers whom have been trying to reproduce published results without success. Third and last, traditional publishing is still a slow process from submission to publication with a long history of peer reviews and publisher establishment.
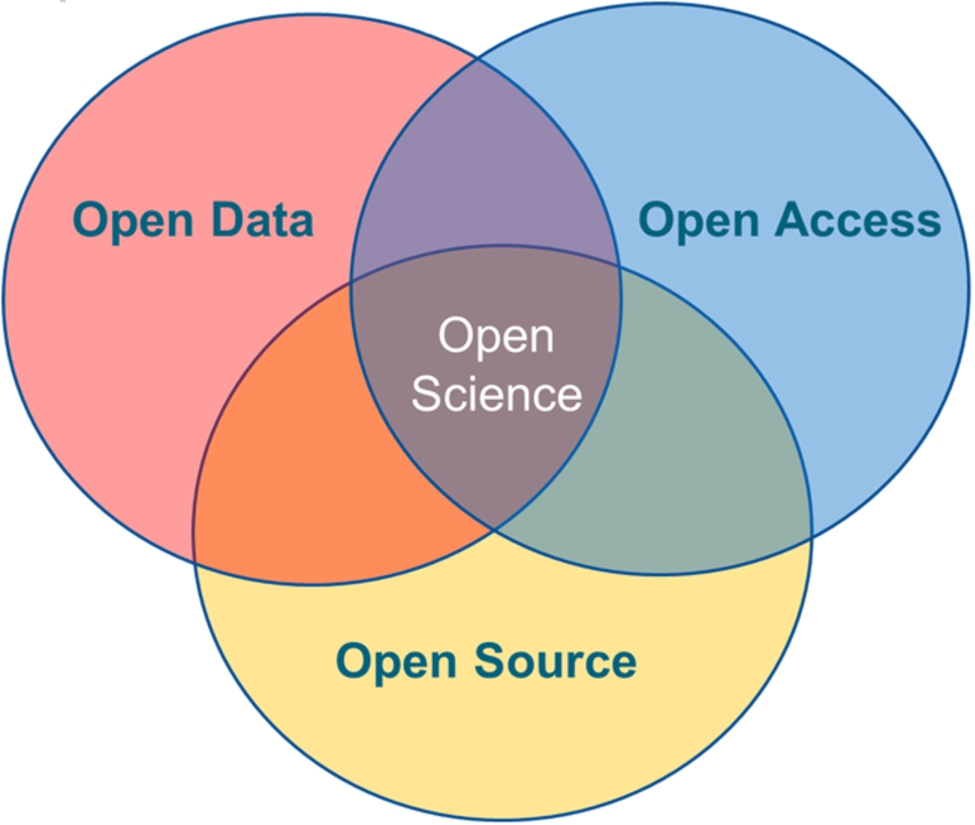
In the modern days of agile management, Open Science embraces the concept of agile publishing where the maximum number of information is disseminated quickly to the masses. Open Science currently encompasses three main concepts: Open Access, Open Data, and Open Source, as illustrated in Fig. 1. These concepts are described next.
Fig. 1.
Components of Open Science.
3.Open Access
Open Access publishing is mainly done online as a way to disseminate research free of all restrictions on access. One should note that most Open Access journals are usually free of many restrictions on use; however, some journals might restrict the usage, for instance, to non-commercial use.
Open Access requires a new business model for publishers as the revenue from publishing is not directly generated from readers. On the one hand, open access publishers have found creative ways to sustain these journals, either via advertisement, pay-per-use services around publications or by generating revenue exclusively from the authors. On the other hand, some Open Access journals, qualified as “delayed journals,” provide publications to their readers for free only after a period of time, meaning than recent publications have to be purchased.
4.Open Data
Another main component of Open Access is the notion of Open Data. As one can guess, the open data movement enables the dissemination of data in a free and open manner. The notion of open data was enabled by the emergence of (very) high speed internet access which allows to disseminate large datasets easily from the comfort of a personal computer. Scientists know that data is critical for any experiment and sharing data can significantly reduce the time of an experiment. Moreover, by making the data collection a collective effort, the quality as well as the quantity of the data is often improved.
However, the potential issues with disseminating data are numerous. First, the nature of the data is often diverse and usually depends on the scientific field, therefore the datasets are often very heterogeneous and their format, if not standardized, can be a bottleneck for reuse by other researchers. Second, datasets are becoming more and more massive which implies that either the data must be down-sampled in order to be shared, or the infrastructure, in terms of storage and bandwidth, must be upgraded to support a large amount of data. Third, the scientific communities are not only interested in the input data and final results, but they are also interested in intermediate data so that they can build upon already-processed datasets. And fourth, data sharing licenses have required some effort to be accepted and, as open source licenses, they are critical to regulate the dissemination and usage of such data.
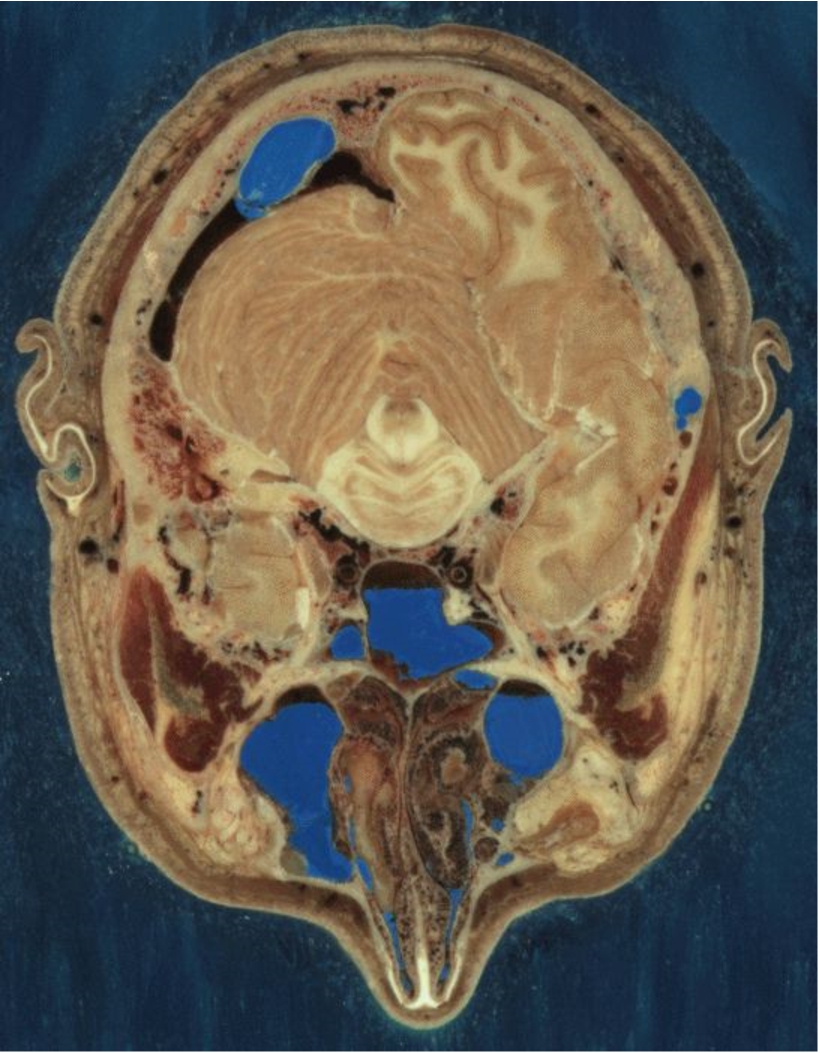
One example of Open Data is illustrated by the Visible Human Project [2] initiated by the National Library of Medicine (NLM) in the USA. This project disseminates openly the color cryosection CT, as illustrated in Fig. 2, and MRI scans of a former inmate whom gave his body to science. Upon his death, high dose CT scans of the whole body as well as high resolution (at the time) MRI scans were acquired. On top of these datasets, color cryosections (photographs) were generated by cutting the frozen body into thin slices. These datasets have become a standard for medical imaging research and have a tremendous value.
Fig. 2.
Slice of the Visible Human project.
A more recent example of Open Data for lung cancer research is illustrated by the Give a Scan [6] project initiated by the Lung Cancer Alliance in the USA. Give a Scan is the world’s first patient-powered open database for lung cancer research. The project currently host over seventy-six patients and provides the imaging data (CT scans) along with metadata information, which is critical for longitudinal studies and statistics. The datasets are accessible freely and are currently used in several research studies [5].
5.Open Source
The Open Source movement started back in the 1980s with the creation of the Free Software Foundation (FSF). The movement was strengthened in 1998 by the creation of the Open Source Initiative (OSI). The pillars of Open Source can be described by seven values: security, affordability, transparency, perpetuity, interoperability, flexibility, and localization. As the years have progressed, a variety of open source licenses have emerged making it somewhat complicated to understand the full extent of the license, but overall, permissive vs. non-permissive licenses allow both academia and the industry to make the most of open source software.
In the past decade, with more and more companies embracing and releasing open source software, the infrastructure for hosting, testing, and deploying open source tools have flourished. Such well-known pieces of the infrastructure include GitHub, gitlab, and iPython notebooks, among others.
In the medical imaging field, a well-known open source project, The Insight Toolkit (ITK) [3,7] was initiated in 2000 by the National Library of Medicine (NLM). After successfully initiating the Visible Human project, the question of processing the data was raised and in particular several universities and groups around the world started to implement their own, regretfully often non-interoperable, image processing software. NLM decided to fund the development of the Insight Toolkit in order to “standardize” the implementation and use of image processing in the medical field. ITK is an open source (BSD license) toolkit written in C
6.Open Science examples
There are many examples of open data, open source and open access projects which illustrate the concept of open science. In this section, two projects are presented.
The Insight Journal [4] was created in 2005 as an open access journal companion to the Insight Toolkit. The main idea of the journal is to bring agile programming to the publishing world. Agile publishing allows authors to publish instantaneously their finding while allowing reviewers to comment directly, without restrictions. Furthermore, based on open source and open data, the Insight Journal enforces reproducible science by running automatic testing upon submission. This reproducibility is shown as an automatic review by the testing system, letting users, developers and readers know if the submission is usable as is or not. The Journal is currently hosting over six hundred papers and has more than four thousand registered readers.
Elsevier’s Science Direct 3D data visualization project [1] was initiated in 2010 in order to bring 3D visualization to scientific publications. Through their collaboration with other publishers, Elsevier has enabled the use of their visualization technology as a tool for disseminating datasets and improving the reader’s overall experience. The project is currently hosting three-dimensional visualization for a large range of datasets: molecular data, cultural heritage, and medical images. More recently, the Virtual Microscope project allows authors to submit datasets acquired with confocal microscopes and visualize them online. These visualizations have been greatly welcomed by the scientific community.
7.Challenges of Open Science
The Open Science movement presents advantages as well as limitations. Based on current experiences and perspectives, one can see that Open Science is helping scientists in several ways. First, scientists can build on top of previous experiments, datasets, and software without starting from scratch. This allows researchers to allocate their resources and time to the performance actual science and not waste such precious resources attempting to replicate previous scientific findings. Second, knowledge is disseminated much more quickly, since the permissive licenses, along with the overall infrastructure, accelerates the sharing of data, publications, and software with other researchers. Third, by enabling collaboration, instead of competition, among scientists, better science is achieved.
It should be noted that Open Science also needs to be improved. For instance, the infrastructure required to share and deploy datasets and software is not free and usually is built and financially supported by large organizations and governments. Furthermore, Open Science requires the adoption of new technologies which can be disruptive, and the legal concerns about licenses can slow down the research. In addition, the security and privacy associated with Open Data and Open Source can be problematic and can require another level of legal and compliance review. Lastly, Open Science is bringing a disruptive change to the scientific publishing community, and the lack of credits (e.g., citations) can be problematic in academia where publish or perish is the rule. Also, the business models around Open Science for all stakeholders needs to be redefined.
8.The future of Open Science
As one can see, Open Science is fairly new and is expanding quickly; therefore, there is a need for a better education around Open Science and a need to better explain the pros and cons of the movement. Corporations and academia are starting to promote Open Science and educate the next generation of researchers about the concept. Along those lines, governments should incentivize, as it is currently done, the concept of Open Science and help promote it.
Regarding Open Data, the emergence of Data Papers which are defined as the scholarly publication of searchable metadata describing datasets, shows a clever way to bring Open Data into the more traditional publishing framework, therefore giving academic credits and citations to datasets. Additionally, publishers are starting to host not only papers, but large datasets and therefore are becoming the providers of data repositories. An important point is that scientific publishers have a larger role to play in Open Science and that collaboration between publishers and scientists is required to advance Open Science to the next level.
In parallel, the concept of Grand Challenges has emerged in recent years as a way to promote collaboration in a competitive way. The Grand Challenges in science are usually initiated by industrial partners who have access to challenging datasets and ask the best researchers to solve a given problem. The main idea is to provide a set of test data, have researchers submit their algorithm which is then run on sequestered datasets and evaluated against a metric that describes how well the algorithm has performed. While the notion of a Grand Challenge is not completely tied to Open Science, the infrastructure, along with the quick dissemination and collaboration of algorithms, is used for open challenges, such as hackathons.
One aspect of Open Science that has been left out in this paper is the notion of Open Standards. Open standards tend to bring ways to ensure interoperability in systems by proposing solutions in an open manner. In the coming decades, one can guess that open standards will become a larger part of Open Science.
9.Conclusion
This paper has presented the key concepts of Open Science along with some views on the future of this movement.
The current review of Open Science initiatives is calling for even more openness in science and several pushes in that direction have been triggered recently. Among them, two main actions have been promoted in the past years. The first action was initiated in 2015 to encourage scientists to publish negative results. New Negatives in Plant Science, Elsevier’s Open Access journal in biology, was specifically created to manage only negative results in plant sciences. Regretfully, this journal is no longer active, but the experiment demonstrated an interest in not only publishing great results, but also in publishing experiments that failed, so that researchers can avoid replicating unsuccessful experiments.
The second action deals with publishing the replication of previously published work. In some fields, such as chemistry, in which the experiments are complex and can often require very specific instrumentation, the replication of experiments is important in order to validate the reported findings. Regretfully, today authors and their publications must bring novelty to science, which is something that might change.
An overall action to enforce reproducibility at the journal level is necessary and, as it has been mentioned in this paper, would require that publishers update the current infrastructure to support reproducibility and that scientists and publishers collaborate to improve the ways in which scientific findings are published.
Acknowledgements
We would like to thank the National Federation of Abstracting and Information Services (NFAIS) for the opportunity to share our views on Open Science. We would also like to thank the many contributors to the Open Source and Open Data projects mentioned in this paper.
About the author
Julien Jomier is directing Kitware’s European office in Lyon, France. Prior to joining Kitware, Jomier was a Faculty Research Lecturer of Radiology at the University of North Carolina. In 2005, he started Kitware’s branch office in NC, where he led the MIDAS project, a system for collecting, processing, and distributing massive collections of data. Jomier is also leading the development of the Insight Journal, an electronic journal promoting Open Science.
References
[1] | I.J. Aalbersberg, P.C. Alvarez, J. Jomier, C. Marion and E. Zudilova-Seinstra, Bringing 3D Visualization Into the Online Research Article, Information Services and Use, IOS Press, (2014) . |
[2] | R.A. Banvard, The Visible Human Project© Image Data Set From Inception to Completion and Beyond, Proceedings CODATA 2002: Frontiers of Scientific and Technical Data, Track I-D-2, Medical and Health Data, Montréal, Canada, (2002) . Available at https://www.nlm.nih.gov/research/visible/visible_human.html. |
[3] | H.J. Johnson, M. McCormick and L. Ibanez, The ITK Software Guide: Design and Functionality, 4th edn, Kitware Inc., (2015) . |
[4] | Kitware, The Insight Journal. Available at http://www.insight-journal.org/. |
[5] | K. Krishnan, L. Ibanez, W. Turner, J. Jomier and R. Avila, An open-source toolkit for the volumetric measurement of CT lung lesions, Optics Express (2010). |
[6] | Lung Cancer Alliance, Give A Scan© – The first patient-powered open access database for lung cancer research. Available at www.giveascan.org. |
[7] | M. McCormick, X. Liu, J. Jomier, C. Marion and L. Ibanez, ITK: Enabling reproducible research and Open Science, Frontiers in Neuroinformatics ((2014) ). |




