Towards new tools for bioresource use and sharing
Abstract
Data sharing institutional incentives exist but no practical tools to implement such policy are in place. The BRIF (Bioresource Research Impact Factor) initiative was conceived as a possible way to fill this gap. It is an ongoing initiative that encompasses considerations and actions from various stakeholders (researchers, funders, industrials, editors) towards (i) standardized identification schemes and reporting for better visibility and tracing of bioresources on the web; (ii) incentive policies from hosting institutions; (iii) creation of tools allowing follow-up of their use.
Tracing the use of bioresources is the first step in this process. For this purpose we have published the CoBRA (Citation of BioResources in journal Articles) guideline that standardizes the way bioresources are referred to in academic literature. We have launched the Open Journal of Bioresources in close collaboration with the open access publisher Ubiquity Press allowing both the resources to be cited and authors to get metrics on reuse and impact. Meanwhile, we have started developing new better adapted metrics; a first list of relevant parameters to take into account in the impact measure of bioresources has been provided.
The tools proposed here foster easier access to samples and associated data as well as their optimized use, sharing and recognition for data producers. Input from the scientific information community would be highly appreciated at this stage.
Much biomedical/epidemiological research is based on using bioresources (sample collections and databases) [2]. Sharing such resources is essential for optimizing knowledge production as their access to all relevant researchers is crucial. It enables research that builds on existing resources. It helps construct communities, optimizes fund investment and it fulfills ethical imperatives. Promoting bioresource sharing is crucial, but does not mean “just” putting files on the web! It requires work and this work is poorly recognized. De facto, although institutional incentives to sharing are there, bioresources are until now poorly shared. Since 2011, the BRIF initiative has initiated a work towards the development of bioresource standard citation and rewarding tools.
1.Sharing resources in research
1.1.The ethical foundation of data sharing
Following the Open Science movement, sharing data11 is now an institutional request (see Table 1 for key milestones). A commitment to share the information content of bioresources within the research community is paramount to advancing translational research. In 2004, the Ministries of science and technology asked the Organisation for Economic Co-operation and Development (OECD) to define guidelines based on agreed principles in order to facilitate access to digitalized data issued from publicly funded research. OECD principles and guidelines, published in 2007 [16], promoted and stimulated exchange of good practices in data access and sharing. In 2011 the joint statement of 17 major national health research funders sent a powerful signal that health research resources must be shared to maximize the potential of publicly funded resources [6,11,12,22]. The vision is to ‘
Now that the supra-national institutional incentives do exist, every scientific community needs to be empowered and organized. Many efforts have been and are currently put in place to improve the quality and interoperability of the publicly released data. The OECD (2011) Quality Framework and Guidelines for OECD Statistical Activities identified key aspects of data quality [17]. The ODE project (Opportunity for Data Exchange, a European Commission-funded research project) developed key models for standardizing data curation. In some scientific communities standards for data quality still lack and are being developed.
Meanwhile, the FAIR Guiding Principles for scientific data management and stewardship were established and formally published in 2016 [8]. These principles provide a precise and measurable set of qualities a good data publication should exhibit – that is Findable, Accessible, Interoperable, and Reusable (FAIR) – in order to ensure proper reuse for further discoveries. “The principles support a wide range of new international initiatives, such as the European Open Science Cloud and the NIH Big Data to Knowledge, by providing clear guidelines that help to ensure that all data and associated services in the emergent ‘Internet of Data’ will be Findable, Accessible, Interoperable and Reusable, not only by people, but notably also by machines.” The management plan for data generated through research from the start and long after a project is completed becomes a required practice in research planning. Education to processes regarding management and reuse of health data starts to be proposed to professionals in research, e.g. Inserm Workshop, France in 2017 (http://english.inserm.fr/inserm-workshops/future-inserm-workshops-schedule) and the different training initiatives (webinars, workshops etc.) developed within the EU funded project OpenAIRE22 in different European countries.
1.2.Specific case of bioresources
In addition to data, bioresources include biological samples scientifically documented that may come from patients (hospital collections) or from epidemiological sources (cohorts, population based collections). These bioresources should organize themselves to allow the sharing of samples. Beyond the value of sharing bioresources for knowledge advancement, there is also an ethical imperative to respect the will of bioresource participants as expressed in their consent. They often want to support research largely, while of course being respected in all their rights – “
1.3.The obstacles to sharing bioresources
Despite good will from the research community, the shared use of bioresources usually blocks at different levels: (i) technical ones (quality etc.); lack of skills to perform data management and data-sharing tasks, (ii) institutional ones: no exchange, sharing or access policy; no practical help; cost; (iii) intellectual property ones; information ones, as readily available information on content is often not available; restricted use to pre-defined professional circles, while nowadays metabolic pathways are transversal and shared among diseases of different categories; (iv) ethical/legal restrictions (data protection) that also evolve and put possible new restrictions (v) no obvious positive spin-off as the immediate benefit of the effort to share resources is often invisible for quite a long time; (vi) lack of rewarding mechanisms – see also ‘The nine challenges identified by the OECD Global Science Forum’ (Table 2). As a result, collections or databases are under-exploited or duplicated uselessly generating a lot of waste. Furthermore, abusive authorship is observed as some researchers may need their work to be recognized and do not find alternative ways to do so. They may end up being co-author of scientific works completely outside their own competencies just because the only way to report the use of the bioresource they established is to be a co-author.
1.4.Need for an appropriate set of tools
Although there is a continuous movement that aims coherently at fostering scientific resource sharing (Table 1), statements indicating practical tools or instruments to reach the objectives are scarce or non-existent in some communities. In the biomedical community, some tools currently exist (for example some access policies; quality tool kits) [5,20], but globally an insufficient level of coordination and systematic implementation makes it difficult to appreciate their positive impact on the overall organization of health research activities [3,16].
As regards bioresource access policies, there is a real need for an explicit sample/data access policy that is part of a transparent value based (not opportunistic) governance model, adapted to the context and aims of the resource, although no “one size fits all” policies are possible. Any such policy should take into account all stakeholders’ views. Incentives are required to implement the policy and assess the use of the resource, in accordance with the sharing policy.
Different models of data access policy currently exist often opposing strong restriction of access to open access. Some examples of data sharing policies are shown in the links below:
The situation is different in the fields of genomics and astronomy: they cover large scale research that pushes to collaboration and to sharing more easily. The resources are more efficiently used because of visibility, accessibility and sharing. What makes them being visible are the infrastructures, catalogues, description (metadata), publications. What makes them being shared are their scientific interest, recognition and sustainability. What makes them being cited/acknowledged are the agreements (MTA, DTA,
As described in 2015 OECD Making open science a reality report, an important alternative way to foster sharing could be through data citation: the possibility for researchers to be acknowledged for their work of data/samples collection and curation through mechanisms similar to the one already in place for citations of academic articles [4,14,18]. This would lead to reward mechanisms that are currently under discussion and that include widespread use of dataset citation and/or proper acknowledgment of open science and data, sharing efforts as criteria to be actually taken into account in career advancement mechanisms or in grant attribution to research teams.
In the meantime, as a beginning of a response to the lack of standard tools, the BRIF initiative has been working over the last years on a frame for developing tools to share data and samples.
2.The BRIF initiative: An integrative approach
Although the BRIF33 initiative [2] was initiated much before the FAIR principles were formulated, it yet underpinned the same ideas of reusability. BRIF constitutes an enabling condition to open science in developing direct and indirect means to share data and samples by recognizing and quantifying the specific contribution of bioresources to the production of novel scientific knowledge.
The basis of the BRIF concept is that making feasible to trace the use of a bioresource and to calculate a corresponding impact factor should encourage institutions, researchers, bioresource managers and other actors involved in bioresource work, to share them. Sharing would then be seen as a gain rather than a loss of control or than an additional non recognized work. Many researchers tend not to be willing to share the resource because they see it as a possible loss rather than valuing their own effort; or they want to share it in principle but the number of practical obstacles requiring efforts to do so and the complete lack of recognition of these efforts discourage them. These issues are now a concern in many biological and biomedical communities. Although the concept could be used in many areas (for example for primary resources in humanities and for ecological collections) we focus on human biological and biomedical resources because their very existence is depending on the willingness of patients and participants to give their samples and to allow the use of their data and there is an ethical imperative of making their contribution useful and recognized.
BRIF is an ongoing initiative that encompasses considerations and actions from various stakeholders (researchers, funders, industrials, editors) within dedicated working groups towards (i) standardized identification schemes and reporting for better visibility and tracing of bioresources on the web; (ii) incentive policies from hosting institutions; (iii) creation of tools allowing follow-up of their use.
Tracing the use of bioresource is the first step in this process and new tools have been or are being developed to make it feasible: the CoBRA guideline (Citation of BioResources in journal Articles), the Open Journal of Bioresources (OJB) and the BRIF metrics.
3.Citing bioresources: The CoBRA guideline
At present, bioresources are either cited in a confusing, heterogeneous way or they are not cited at all. The use of a bioresource in a research article is not retrievable systematically via PubMed or other bibliographic databases [13]. Traceability and visibility of bioresources in scientific literature or in other (online) sources would highlight their use. By being properly cited, bioresource use would be valued and their sharing thus encouraged. The CoBRA guideline [1] was hence developed to standardize citation of bioresources in scientific articles in order to trace their use on the web. This was achieved through close collaboration between the BRIF journal editors’ subgroup, the European Association of Science Editors (EASE44), the EQUATOR55 (Enhancing the QUAlity and Transparency Of health Research) network and the research community managing and/or using bioresources. CoBRA recommends mainly that each individual bioresource used to perform a research work should be mentioned in the Methods section and should be cited as an individual “reference [BIORESOURCE]” according to a given format, using a unique identifier when possible. The detailed recommendation is also given by the CoBRA checklist reported on the EQUATOR’s website66 which gathers the major reporting guidelines internationally recognized and referred to by journal editors (see also Table 3).
CoBRA needs now to be implemented [15,19]. The first way is through dissemination, notably in national and international conferences and workshops. While doing so, one important issue was raised by researchers regarding the use of reporting guidelines in the process of writing a manuscript: the lack of time necessary to go through all of them and learn how to apply them. Aiming to help the researcher to overtake this problem and facilitate the use of the CoBRA guideline, an e-learning tool entitled “How to use the CoBRA guideline” was conceived and presented at the ISBER 2016 annual meeting (Biobank Education Tools, BET-137, ISBER 2016 Annual Meeting & Exhibits – Berlin) in the section dedicated to the Biobank Education Tools. The video and audio tracks were recorded in digital multimedia format by using First. This e-learning video is freely available at the website: https://zenodo.org/record/55785.
Implementing the CoBRA guideline further points to the necessity of integrating scientific editorial policies in the loop using several strategies. One way to enforce CoBRA use in articles is to include it in instructions to reviewers and editorial assistants as part of the checklist used to process manuscripts. A second way is to add CoBRA in the list of reporting guidelines that is usually part of the instructions to authors. We also aim to obtain recommendations by the International Committee of Medical Journal Editors (ICMJE). In any such case though, compliance to the guideline is not guaranteed unless it is strictly verified by either reviewers or editorial staff (or made mandatory).
Associations of editors such as the EASE4 are of great help in reaching and empowering journal editors and authors of scientific publications. EASE Guidelines for authors and translators of articles to be published in English already include the necessity to mention the origin and identity of experimental materials used in the methods section, and refer to the CoBRA guideline. The more key associations or committees of scientific journals editors will be aware of CoBRA, the more it will be applied. There is a need to go beyond the European dimension. Worldwide associations such as WAME (World Association of Medical Editors), AMERBAC (Asociación Mexicana de Editores de Revistas Biomédicas, A.C.), Canadian Editors Association and CSE (Council of Science Editors) must be informed about the existence of CoBRA and should promote it.
Other stakeholders are also key players in developing good practices and could contribute to the implementation of CoBRA. Institutions hosting bioresources as well as funding agencies can guide researchers in good reporting of bioresource use. In France, the National Institute of Science and Techniques Information (INIST77 – CNRS) has been a great support in disseminating and promoting the guideline. The European Research Infrastructure of biobanking and biomolecular resources (BBMRI-ERIC88) has actively supported the BRIF initiative. BBMRI-ERIC has so far 20 member countries, observer countries and one observer international institution (Austria, Belgium, Cyprus, Czech Republic, Estonia, Finland, France, Germany, Greece, Italy, Latvia, Malta, Netherlands, Norway, Poland, Sweden, Switzerland, Turkey, United Kingdom and The International Agency for Research on Cancer – IARC – of the World Health Organization). BBMRI-ERIC included the BRIF initiative in its 2015–2016 workplan to facilitate notably the implementation of CoBRA among its members. It will be added to the MTA/DTA and specified in publication policies. Other infrastructures could be interested in helping implementing CoBRA as one of the tools of their own strategy. As a matter of fact, “Research infrastructures in the biological and medical thematic area of the European Strategy Forum on Research Infrastructures (ESFRI99) roadmap are committed to provide access to the most advanced, unique, and large-scale biological resources, instruments and expertise in Europe to support research and development in all life sciences.” In order to better organize the collaboration and mutualization of resources between life sciences and European infrastructures, a project “Corbel” is ongoing as part of the H2020 framework. Tools like BRIF and CoBRA are part of those developed and promoted in this context. On a global scale, consortia or scientific societies such as the Public Population Project in genomics and society (P3G1010), the International Society for Biological and Environmental Repositories (ISBER1111) and the European, Middle Eastern & African Society for Biopreservation & Biobanking (ESBB1212) would help in extending these actions. Patient’s associations could have a role in this too. Contributors to bioresources give importance to the fact that they are used and do not stay as sleeping resources. Thus accessing data on the use of such resources would also be valuable for them.
Over the last years, other initiatives throughout the world have flourished within the open access and sharing move to better identify and trace different types of resources (OpenAire, DataCite, CODATA, Force 11, ORCID and others). Among them, the Research Data Alliance (RDA) Working Group on Dynamic Data Citation1313 has provided recommendations about making subsets of data citable. Connecting to these groups would certainly facilitate CoBRA implementation and foster a better granularity by using suitable identifiers. Such identifiers of subsets or combination of subsets of bioresources must first be worked out with the idea of keeping traceable their “genealogy” (origin of parental resources). In general, coordination of all these actions has become an urge if one wishes to develop standard citation tools and improve good reporting practices. The participation of the BRIF initiative in two RDA workshops in Paris, the first one on September 2015 – “Persistent Identifiers: Enabling Services for Data Intensive Research” co-organized with DataCite – and the second one on April 2016 the 3rd RDA Europe Science Workshop, is a first move to link these initiatives.
4.Publishing a bioresource: A new type of journal
The Open Journal of Bioresources1414 is one journal in a series of so-called ‘metajournals’ published by Ubiquity Press. These journals are dedicated to opening up and aiding the discoverability of all research elements involved in the research lifecycle, such as data, software, bioresources and hardware (forthcoming). The idea behind the metajournals is that researchers need to be able to discover and cite these research elements, but they also want credit for sharing them and the ability to track their impact. Given this, the metajournals offer credit – in the form of citation and altmetric data – for researchers to make their resources permanently available and discoverable in accordance with community norms.
In the case of bioresources, the idea behind this journal is to provide a permanent marker paper so that users can definitively cite a bioresource they have accessed or referred to. The best way to do this is by integrating the bioresource into the traditional process for obtaining scholarly credit: the peer-reviewed journal article (Fig. 1). In this way, users simply cite the bioresource as they would do for any other journal article – and this is facilitated by the application of a digital object identifier (DOI) to all articles. This means that each article acts as a permanent marker for a bioresource and conforms to the standard processes for citing research.
OJB publishes bioresource papers, which are structured summaries of bioresources that are peer-reviewed to ensure they are accurately described. Papers are published in accordance with a structured template that describes the bioresource, outlines how it is preserved, the methods used in its creation, and how it can be accessed in the biobank.
These papers are not lengthy descriptions of bioresources but more akin to a short online form. Contents are therefore structured not by paragraphs, but by individual sentences and one-word answers. The result is a highly structured, objective description of a bioresource.
Because the bioresource paper is an objective description, so too is the peer review process. Importantly, OJB papers are not peer reviewed for their significance, but rather that the information is accurately filled out and presented in accordance with the standards set by the CoBRA guidelines (see above). Because of this, the peer review process is relatively quick and articles can be published within a matter of weeks from submission. Articles are published open-access under the CC BY license, ensuring anyone can access the final contents. For this, the journal charges a small APC of £100 – which is completely waivable if an author does not have access to funding for publication fees.
The published article then becomes a permanent marker paper for the described bioresource. Users cite the paper directly when they have accessed, used or simply referenced a bioresource. Citations are tracked and displayed on the article page alongside numbers of article views, tweets and Facebook likes. In this way, the bioresource paper allows authors to understand the true impact of their bioresource, which would not have been possible previously.
Articles are also sent to various scholarly indexes to aid discoverability, ensuring they become part of the permanent scholarly record. We have also been in discussion with PubMed about indexing articles there – which we are confident will happen in the future.
4.1.Bioresource papers in the scientific literature
As a new publication type, bioresource papers are designed to connect research articles, data repositories and biobanks. Research articles tend to focus on the results or application of a particular bioresource, whereas bioresource papers focus on the resource itself. In acting as a marker paper for the resource, bioresource papers introduce a level of permanence and consistency in citing the resource. This can only be achieved by working within the existing norms of publishing standards, particularly around citation credit.
As a formal, peer-reviewed publication, the bioresource paper can be cited like any other article and will appear in article reference lists along with its digital object identifier. Traditionally, bioresources would be referenced by mentioning the name of the biobank and sometimes the biobank’s website. However, many scientific journals prevent authors from citing websites (as they are not peer-reviewed publications) and so web resources often take the form of footnotes with hyperlinks. Not only is this a non-standardized way of referring to a bioresource, as journals have different policies for referring to websites, but web addresses also change or expire. Citing a bioresource paper is the easiest way to ensure that the bioresource enters the scientific record and will remain there in perpetuity. Because the bioresource paper is associated with a digital object identifier (DOI), citations can be easily tracked in the same way as research articles. The number of citations a paper receives is an indication of how well accessed and utilized the biobank/resource is, which helps building a picture of its impact and use within the scientific community. This could not be achieved without a standard and consistent way of citing the bioresource, which the paper itself provides. Furthermore, each paper is accompanied by a list of so-called altmetrics, highlighting article views and downloads and Tweets, Facebook posts and blogposts referring to the paper. This allows researchers to understand the impact of their bioresource with more granularity than mere citations.
Finally, articles are indexed in common scientific databases so that readers can discover the bioresource in the same way as any other item in the scientific literature. Article indexes can search across titles, abstracts, keywords and full contents, meaning that bioresource papers can be easily discovered alongside other articles of interest. Bioresource papers therefore ensure that bioresources can participate in the complex networks of web-based science, harnessing the power of social media, scientific databases and the traditional scientific literature for discoverability.
5.Towards a new metrics: The BRIFs
To measure the impact of a bioresource on the web, it is first necessary to have a persistently identifiable and traceable bioresource. The aim of BRIF is not to create a new identifier scheme specifically for bioresources, notably for biobanks. Rather, its aim is to identify frameworks which are already established (or well on their way to becoming so), such as registries for clinical trials and other more general ID schemes [10], and to subsequently assess them and recommend their use as appropriate for bioresources.
This major step is presently being discussed between BBMRI-ERIC and DataCite. A summary of the present state was presented at a workshop held in Toulouse on December 4, 2015 entitled ‘BRIF: From identifiers, parameters and sharing policies, towards metrics to measure bioresources impact’.
Once the bioresource is fully traceable and indexed, the impact of its use could be measured using the metrics tools offered on the net or by various stakeholders or providers. Those tools are mainly based on citation indexes and assume that citation reflects the ‘success’ of the enterprise. But in the case of bioresources this is not sufficient. They do not reflect the full range of utility of a bioresource. For example, a clinical and biological collection of rare diseases will be used by a restricted community, whereas the resource has a high value, requiring a worldwide coordination effort and the contribution of different stakeholders. Other metrics are needed that take relevant parameters into consideration.
As part of the BRIF initiative, a dedicated working subgroup worked out this issue and provided a first list of relevant parameters to take into account in the impact measure. An online survey was sent to selected biobanks in order to assess those parameters in the evaluation of the impact of a bioresource. The answers from 28 biobanks (mainly from Italy and France) were used to classify parameters of scientific impact for bioresources. Several groups of parameters were defined according to their availability and to the feasibility of their retrieving for calculating the impact using one or several specifically designed algorithm(s). The main parameters relate to indicators of research productivity and sustainability; indicators of sample/data value; indicators of workflow and efficiency and indicators of collaboration and visibility (Table 4). An extended study on various types of bioresources in more countries will allow refining the list and characteristics of such parameters.
On the basis of the selected parameters, algorithms will be proposed for measuring the use and impact of bioresources. They will be tested in the wider context of European biobanks covered by the National Nodes of BBMRI-ERIC. As part of its perspective BBMRI-ERIC indicates that the collective adoption of an identification scheme for bioresources and the creation of a necessary bioresource ID database/registry will become an integral part of the BBMRI-ERIC Directory. The involvement of the Common Service IT in the creation of a tool for calculating the research impact of bioresources is part of the H2020 ADOPT BBMRI-ERIC project.
6.Conclusion
The tools proposed here foster easier access to samples and associated data, their optimized use and sharing as well as the recognition of data producers. Input from the scientific information management and technology community would be highly relevant at this stage. This work could benefit from initiatives in other domains, in particular the long standing work performed in earth and physical sciences [21] and could serve as a reference for other communities, beyond human biological and medical bioresources.
Table 1
The landscape of Data Sharing (key milestones)
| ∙ | 1996: Principles for rapid release of genome sequence data from the Human Genome Project formulated at a meeting in Bermuda [http://hdl.handle.net/10161/7715] |
| ∙ | 1997 and 1998: Further Bermuda meetings |
| ∙ | 2003: Fort Lauderdale meeting, rapid prepublication release (resource for scientific community): responsibilities of the resource producers, resource users, and the funding agencies [https://wellcome.ac.uk/sites/default/files/wtd003207_0.pdf] |
| ∙ | 2008: Amsterdam meeting extended and adapted the same principles to proteomics |
| ∙ | 2009: Toronto statement on pre-publication data release |
| ∙ | 2010: Oxford International Conference on data sharing |
| ∙ | 2011: Sharing research data to improve public health: joint statement of purpose [http://www.wellcome.ac.uk/About-us/Policy/Spotlight-issues/Data-sharing/Public-health-and-epidemiology/WTDV030690.htm] |
| ∙ | 2011: A data sharing Code of Conduct for international genomic research [http://genomemedicine.com/content/3/7/46] |
| ∙ | 2011: Quality Framework and Guidelines for OECD Statistical Activities |
| ∙ | 2012: The tension between data sharing and the protection of privacy in genomics research [doi:10.1146/annurev-genom-082410-101454] |
| ∙ | 2013: Global alliance for Genomics and Health; Geneticists push for global data-sharing; this international organization aims to promote exchange and linking of DNA sequences and clinical information [http://www.nature.com/news/geneticists-push-for-global-data-sharing-1.13133] |
| ∙ | 2014: within Global alliance, Framework for Responsible Sharing of Genomic and Health-Related Data [http://genomicsandhealth.org/] |
| ∙ | 2015: EU launches Open Research data Pilot in Horizon 2020 in selected areas |
| ∙ | 2016: EU announces that in 2017 work programme the Open Research Data pilot has been extended to cover all the thematic areas of Horizon 2020 |
Table 2
The nine challenges identified by the OECD Global Science Forum
| The OECD Global Science Forum has recently identified a number of challenges related to data-driven and evidence-based research. |
| Challenge 1 – Massive amounts of digital data are being generated at an unprecedented scale, thanks partly to the advent of ICTs. The reliability, statistical validity and generalisability of new forms of data are not yet fully understood. |
| Challenge 2 – While administrative, survey and census data are widely collected by national statistical agencies and government departments, micro-data records are available to a much lesser extent. |
| Challenge 3 – New forms of personal data, such as social networking data, are increasingly created and collected. The use of those data may generate risks to individual privacy. |
| Challenge 4 – Legal, cultural, language and proprietary rights of access barriers hinder cross-national collaboration and international data exploitation, especially in the social sciences. |
| Challenge 5 – Global research agendas require increasingly interdisciplinary and international co-ordination. |
| Challenge 6 – Collaboration and experience sharing across countries in the development of comparable data resources is necessary to fully exploit the potential of data sets. |
| Challenge 7 – Researchers often lack the resources or the skills to make sure that the data they use, gather and produce are available for reuse. |
| Challenge 8 – National investments in skills and infrastructure related to data creation and curation are essential to avoid risk of data loss or degradation. |
| Challenge 9 – Researchers need to have the right set of incentives to ensure effective data sharing. |
Source: Adapted from OECD (2013), “New data for understanding the human condition – International perspectives”, OECD Global Science Forum Report on Data and Research Infrastructure for the Social Sciences.
Table 3
| Article text section | Item # | Guidance | Additional information |
| Abstract | 1 | Indicate whether the work has used one or more bioresources, and specify the number of bioresources if relevant. | Adapt according to the number of words allowed. |
| Introduction | 2 | Indicate that the work has used one or more bioresources. Specify the type. | The types of bioresources include: data, samples and data, database, registry. |
| Methods | 3 | Report each individual bioresource used to perform the study: By their name and other ID, if existant, and By a single bibliographic reference. | The format of the reference is detailed in item 6 in the section “Reference”. The bioresource name should be the original name as reported in official documents such as MTA(a)and DTA(b). The name should be reported in the original language without translation. Specify any relevant characteristics of the bioresource, such as sample number and type of biospecimens, if this information is not available from the bioresource reference. Number of accesses can be also reported here, for instance as the MTA(a)/DTA(b) registration number associated with each access. If the dates of actual bioresource availability for the user (e.g. reception of samples) are distant from those in the MTA(a) signature, this can be reported here. |
| Results | 4 | Indicate the relevance of the bioresource(s) used for the study (Optional). | |
| Discussion | 5 | Standard rules should apply. | |
| Reference | 6 | Cite each bioresource as a reference as follows: ID/Bioresource Name (acronym if available)/organization or network partnership, membership (optional)/Number of access(es), Date of last access; [BIORESOURCE] Specifications for ID: Unique ID can be DOI, catalogue number, or the name only. If the only ID is the name then add Town and Country. | Each citation includes three fields: Identification/Institution/Access. The “use” of the bioresource is distinguished within the citation by adding “[BIORESOURCE]” at the end of the reference. ID: citing the ID, rather than, or in addition to, the name is essential in order to avoid any confusion and facilitate retrieval. DOI: if the detailed description of the bioresource is available in a marker paper, it should be cited here, this being one way of providing a DOI. Name: the name should be the original name as reported in official documents such as MTA(a)and DTA(b). The name should be reported in the original language of the residence country without translation. Place of residence (town) and country should be translated in the article language. |
Table 3
(Continued)
| Article text section | Item # | Guidance | Additional information |
| Acronym: when available, stable and consolidated, it is recommended to add the acronym to the reference. If the bioresource requires mentioning membership or partnership in consortia, networks or organizations, a dedicated field should be included. When the bioresource is a physical resource such as a biobank or collection, the number of accesses should be specified, in addition to the date of last access. These data will generally correspond to the data signature of the MTA(a)/DTA(b). When the bioresource is a digital resource such as a database, dataset, or registry, only the last access should be reported. | |||
| Authorship | 7 | Standard rules should apply. Providing samples or data is not sufficient to justify authorship. | |
| Acknowledgements | 8 | Standard rules should apply. |
* In the case of bioresources not used as a source of material for the study, but only referred to, follow the citation format: ID/Bioresource Name (acronym if available)/organization or network partnership, membership (optional).
(a) MTA: Material Transfer Agreement.
(b) DTA: Data Transfer Agreement.
Fig. 1.
OJB publication process.
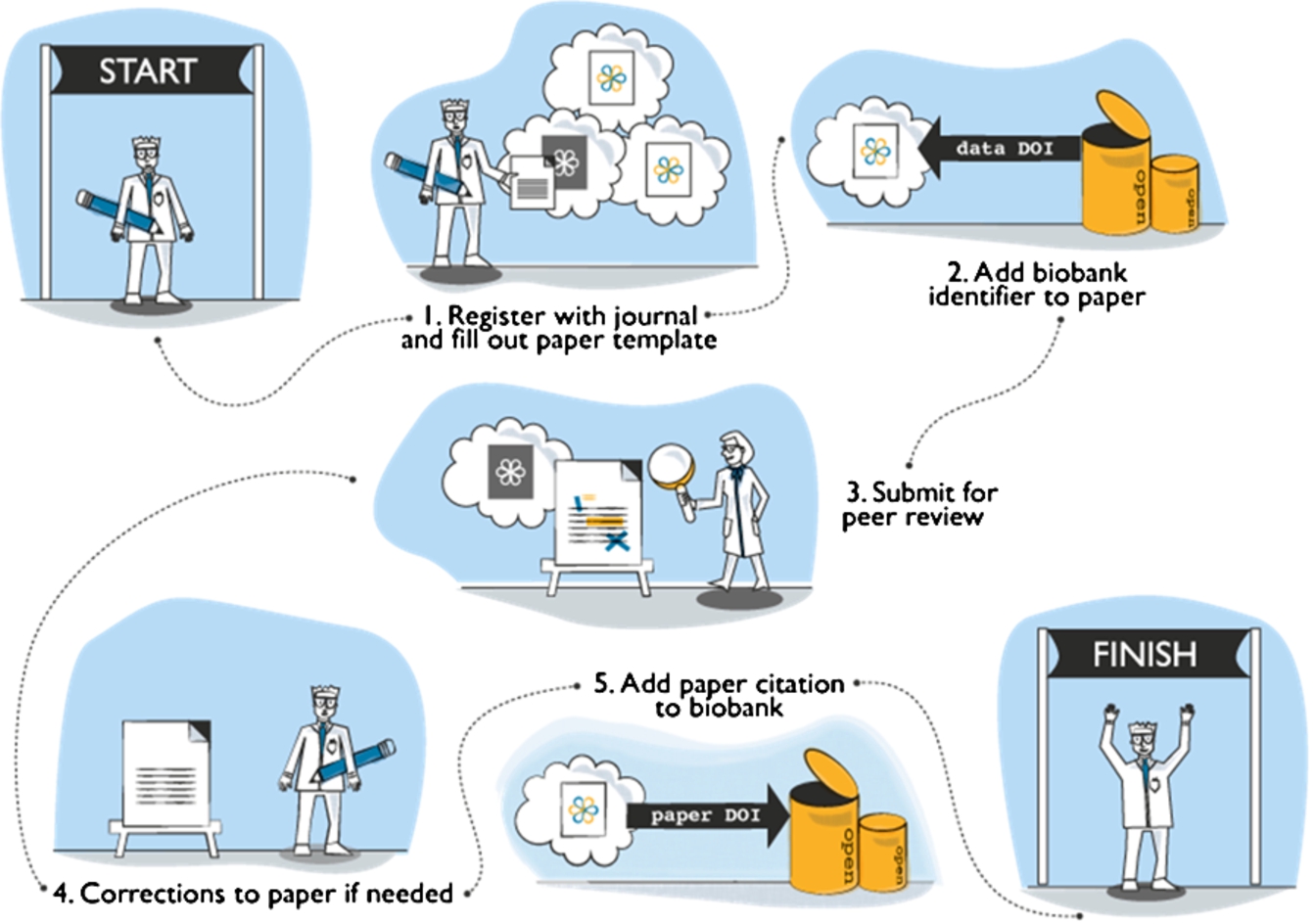
Table 4
List of parameters to consider when measuring the re-use of a bioresource from the web as a function of their accessibility
| Indicator | Type of indicator | Measurable | Often documented | Easily traceable | BBMRI-ERIC Directory |
| Turnaround time for requests | efficiency | yes | yes | no | no |
| Website | interoperability | yes | yes | yes | yes |
| On-line catalogue | interoperability | yes | yes | yes | yes |
| Participation in geographic networks | interoperability | yes | yes | yes | yes |
| Participation in thematic networks | interoperability | yes | yes | yes | yes |
| Access and prioritization policies | interoperability | yes | yes | no | yes |
| Measures to encourage resource sharing | interoperability | no | no | no | no |
| Journal articles (authorship) | research productivity | yes | yes | yes | no |
| Journal articles (acknowledgement) | research productivity | yes | yes | yes | no |
| Journal articles with impact factor | research productivity | yes | yes | yes | no |
| Citations of the marker paper | research productivity | yes | yes | yes | no |
| Citations in BioDBcore (for databases) | research productivity | yes | yes | yes | no |
| National grants obtained | research productivity | yes | yes | no | no |
| European grants obtained | research productivity | yes | yes | no | no |
| International grants obtained | research productivity | yes | yes | no | no |
| Patents for which the BioResource is a patent holder | research productivity | yes | yes | no | no |
| Patents obtained by using the BioResource | research productivity | yes | yes | no | no |
| Citation and authorship policies | research productivity | yes | yes | no | no |
| Policies to obtain return of research results | research productivity | yes | yes | no | no |
| Long term funding | research productivity | no | no | no | no |
| Variety of biospecimens | sample/data value | yes | yes | yes | yes |
| Rare disease samples | sample/data value | yes | yes | yes | yes |
| Long term clinical follow up data | sample/data value | yes | yes | no | no |
| Frequency of data updating | sample/data value | yes | yes | no | no |
| Capacity to re-identifying samples | sample/data value | yes | yes | no | yes |
| Quality control of samples and data | sample/data value | yes | yes | no | yes |
| Format of information linked to the samples | sample/data value | yes | yes | no | no |
| Number of different diseases | sample/data value | yes | yes | yes | yes |
| Size in terms of operators | structural factors | yes | yes | no | yes |
| Size in terms of samples | structural factors | yes | yes | no | yes |
| Age of the BioResource | structural factors | yes | yes | no | yes |
| Number of requests filled per year | workflow | yes | yes | no | no |
| Number of samples received per year | workflow | yes | yes | no | no |
| Number of samples distributed per year | workflow | yes | yes | no | no |
| Material Transfer Agreements number per year | workflow | yes | yes | no | no |
| Number of times accessed per year | workflow | yes | yes | no | no |
| Data Transfer Agreements number per year | workflow | yes | yes | no | no |
Notes
1 Research data are defined as ‘factual records (numerical scores, textual records, images and sounds) used as primary sources for scientific research, and that are commonly accepted in the scientific community as necessary to validate research findings. A research data set constitutes a systematic, partial representation of the subject being investigated.’
Acknowledgements
We wish to thank Federica Napolitani, Anna Maria Rossi, Alessia Calzolari from Istituto Superiore di Sanità, Rome, Italy, as well as Petr Holub, Jan Eric Litton, Outi Törnwall and Michaela Mayrhofer from BBMRI-ERIC, Graz, Austria for very fruitful discussions and constructive interactions.
The BRIF initiative was supported by funds from BBMRI-LPC project, grant agreement number 313010, theme FP7 – INFRA-2012-1 and is part of the actions supported by BBMRI-ADOPT project. The workshops held in Toulouse the 9th October 2015 and 4th December 2015 benefited from the support of DIST-INIST CNRS and from the help of the Genotoul Societal platform funded by ITMO “Santé Publique”.
References
[1] | E. Bravo, A. Calzolari, P. De Castro, L. Mabile, F. Napolitani, A.M. Rossi and A. Cambon-Thomsen, Developing a guideline to standardize the citation of bioresources in journal articles (CoBRA), BMC Med. 13: ((2015) ), 33. doi:10.1186/s12916-015-0266-y. |
[2] | A. Cambon-Thomsen, Assessing the impact of biobanks, Nat. Genet. 34: (5) ((2003) ), 25–26. doi:10.1038/ng0503-25b. |
[3] | A. Cambon-Thomsen, The social and ethical issues of post-genomic human biobanks, Nat. Rev. Genet. 5: (11) ((2004) ), 866–873. doi:10.1038/nrg1473. |
[4] | CODATA-ICSTI Task Group on Data Citation Standards and Practices, Out of cite, out of mind: The current state of practice, policy, and technology for the citation of data, Data Science Journal 12: ((2013) ), CIDCR1–CIDCR7. doi:10.2481/dsj.OSOM13-043. |
[5] | A.L. Delbecq, A.H. VandeVen and D.H. Gustafson, Group Techniques for Program Planners, Scott Foresman and Company, Glenview, IL, (1975) . |
[6] | Editorial, The scientific social network, Nat. Med. 17: (2) ((2011) ), 137. doi:10.1038/nm0211-137. |
[7] | European Commission Directorate – General for Research & Innovation, H2020 programme. Guidelines on open access to scientific publications and research data in Horizon 2020, available from: http://ec.europa.eu/research/participants/data/ref/h2020/grants_manual/hi/oa_pilot/h2020-hi-oa-pilot-guide_en.pdf. |
[8] | European Commission Directorate – General for Research & Innovation, H2020 programme. Guidelines on FAIR data management in Horizon 2020, available from: http://ec.europa.eu/research/participants/data/ref/h2020/grants_manual/hi/oa_pilot/h2020-hi-oa-data-mgt_en.pdf. |
[9] | European Society of Human Genetics, Data storage and DNA banking for biomedical research: Technical, social and ethical issues. Recommendations of the European Society of Human Genetics, European Journal of Human Genetics 11: ((2003) ), S8–S10. doi:10.1038/sj.ejhg.5201115. |
[10] | F. Kauffmann and A. Cambon-Thomsen, Tracing biological collections – Between books and clinical trials, JAMA 299: (19) ((2008) ), 2316–2318. doi:10.1001/jama.299.19.2316. |
[11] | J. Kaye, C. Heeney, N. Hawkins, J. de Vries and P. Boddington, Data sharing in genomics – Re-shaping scientific practice, Nat. Rev. Genet. 10: (5) ((2009) ), 331–335. doi:10.1038/nrg2573. |
[12] | B.M. Knoppers, J.R. Harris, A.M. Tasse, I. Budin-Ljosne, J. Kaye, M. Deschenes and M.H. Zawati, Towards a data sharing Code of Conduct for international genomic research, Genome Med. 3: ((2011) ), 46. doi:10.1186/gm262. |
[13] | L. Mabile, R. Dalgleish, G.A. Thorisson, M. Deschênes, R. Hewitt, J. Carpenter, E. Bravo, M. Filocamo, P.A. Gourraud, J.R. Harris, P. Hofman, F. Kauffmann, M.A. Muñoz-Fernàndez, M. Pasterk, A. Cambon-Thomsen and BRIF working group, Quantifying the use of bioresources for promoting their sharing in scientific research, Gigascience 2: (1) ((2013) ), 7. doi:10.1186/2047-217X-2-7. |
[14] | H. Mooney and M.P. Newton, The anatomy of a data citation: Discovery, reuse, and credit, Journal of Librarianship and Scholarly Communication 1: (1) ((2012) ), eP1035. doi:10.7710/2162-3309.1035. |
[15] | F. Napolitani, A. Calzolari, A. Cambon-Thomsen, L. Mabile, A.M. Rossi, P. De Castro and E. Bravo, Biobankers: Treat the poison of invisibility with CoBRA, a systematic way of citing bioresources in journal articles, Biopreservation and Biobanking 14: (4) ((2016) ), 350–352. doi:10.1089/bio.2015.0105. |
[16] | Organisation for Economic Co-operation and Development, OECD principles and guidelines for access to research data from public funding, 2007, available from: http://www.oecd.org/sti/sci-tech/38500813.pdf. |
[17] | Organisation for Economic Co-operation and Development, Quality framework and guidelines for OECD statistical activities, 2011, available from: http://www.oecd.org/std/qualityframeworkforoecdstatisticalactivities.htm [cited 2016 Oct 27]. |
[18] | Organisation for Economic Co-operation and Development, Making open science a reality, OECD Science, Technology and Industry Policy Papers, No. 25, 2015. doi:10.1787/5jrs2f963zs1-en. |
[19] | A.M. Rossi, P. De Castro, E. Bravo, A. Calzolari, F. Napolitani, A. Cambon-Thomsen and L. Mabile, Editors as promoters of good practices in bioresource research, European Science Editing 42: (1) ((2016) ), 18–19. |
[20] | N.R. Tague, The Quality Toolbox, 2nd edn, ASQ Quality Press, Milwaukee, WI (2004) , pp. 364–365. |
[21] | P.F. Uhlir, For Attribution – Developing Data Attribution and Citation Practices and Standards: Summary of an International Workshop, The National Academies Press, Washington, DC, (2012) . |
[22] | Wellcome Trust, Sharing research data to improve public health: Full joint statement by funders of health research, available from: https://wellcome.ac.uk/what-we-do/our-work/sharing-research-data-improve-public-health-full-joint-statement-funders-health [cited 2016 Oct 27]. |



