Protocols.io: Reducing the knowledge that perishes because we do not publish it
The way life scientists communicate has hardly changed since the days of Gregor Mendel. Even though mobile and Internet technology has transformed a wide array of communication channels, academic publishing has largely resisted change. As a consequence, it is impossible to stay up-to-date with the rapidly evolving methodology in the life sciences. Most corrections and optimizations of previously-published methods are never properly communicated beyond the confines of a given laboratory. Researchers are constantly re-discovering knowledge simply due to the lack of an efficient system to communicate such findings. Protocols.io is a central platform that leverages modern web and mobile technology to facilitate easy discovery and sharing of this scientific knowledge.
1.Introduction
Our publishing system turned 350 years old just two months ago, and Wiley publishers celebrated the birthday with “Why has the scholarly journal endured?” – a post extolling the values of the academic journal [8]. While the celebrated features of the academic journal are debatable, Wiley is correct that the way academics publish and communicate has hardly changed. This is despite the advent of the Internet, web and mobile technology. This is despite the creation of the web by Tim Berners-Lee in 1989, with the express purpose of facilitating communication and exchange between academics [2,9]. In the past 25 years, the web has transformed seemingly everything with the exception of science communication.
The outdated publishing system is particularly problematic in light of the high incidence of wrong and non-reproducible published results. In pre-clinical research, Bayer and Amgen reported failure to reproduce published results in 70–90% of the cases [1]. John Ioannidis has argued that most published research in general is wrong [5]. Methodology reporting is poor across a wide-range of research domains, with more than half of the publications not identifying resources in enough detail for reproducibility [12].
The key issue is not the lack of reproducibility, which may just be inherent to the scientific process [4,10]. Rather, the problem is that our publishing system provides no effective means for correcting and improving the published science. For example, the discovery of a critical mistake in a single-step of a published technique is typically not published. The correction of a single step of a previously-published method is not a new method, but just a modification, and there exists no good infrastructure for communicating such knowledge. There is also no proper way to get credit for such work.
Fig. 1.
Checklist functionality for following a protocols.io method. Panel (A) demonstrates the website interface and panel (B) shows the iOS mobile view. (Colors are visible in the online version of the article; http://dx.doi.org/10.3233/ISU-150769.)
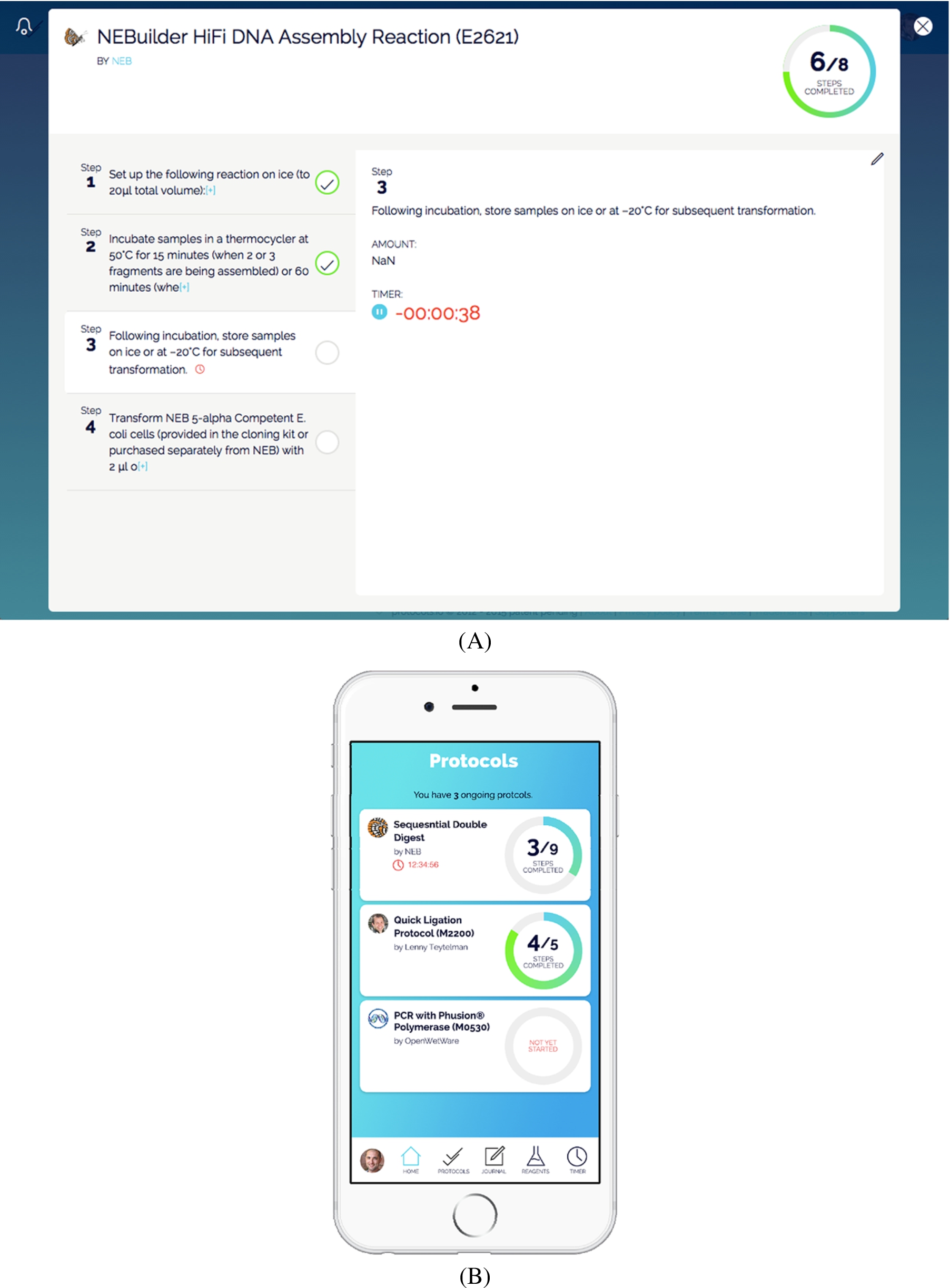
The mission of Protocols.io is to give researchers credit for their corrections to and optimizations of research methods and to allow them to easily share and discover this knowledge. We focus on the protocols specifically because the frequent and tiny tweaks of research methods have a tremendous impact on the reproducibility of experiments [3,7].
2.Protocols.io platform functionality
Available on the web at protocols.io and as native iOS and Android applications, the platform offers three main benefits: (a) step-by-step following of protocols during labwork; (b) versioning and editing of protocols; and (c) easy sharing and discovery of corrections and optimizations. All user-contributed public content is open access under the CC-BY license and the web and mobile tools are free for the scientists.
2.1.Working with digital protocols
Researchers can create protocols and enter methods that they commonly use. By default, all new protocols are private, but can be shared with collaborators of choice or made public for everyone. Protocol instructions may be followed step-by-step in real time during the experimental work. That is, a checklist interface provides the means for tracking the progress of the experiments, with the ability to record any changes during the particular execution (Fig. 1). A protocol-centered digital notebook records all of the edits during the protocol execution. All protocols and notebook records are easily exportable in a variety of formats, including text, PDF and JSON.
2.2.Forking (cloning) and versioning protocols
The central problem protocols.io is designed to solve is the inability to keep published methods up-to-date. To that end, any method in protocols.io can be “forked” or “cloned” with a single click. A private copy of the method is instantly created, allowing flexible editing. In addition to forking, a protocol’s original author can also create a new version that will supersede the previous record, with the history available on demand. All public protocols, versions and forks can be assigned a unique DOI to facilitate citations in the methods sections of research manuscripts.
Fig. 2.
Annotations. The purple arrow points to the step-level annotation and the orange arrow indicates the protocol-level comments. (Colors are visible in the online version of the article; http://dx.doi.org/10.3233/ISU-150769.)
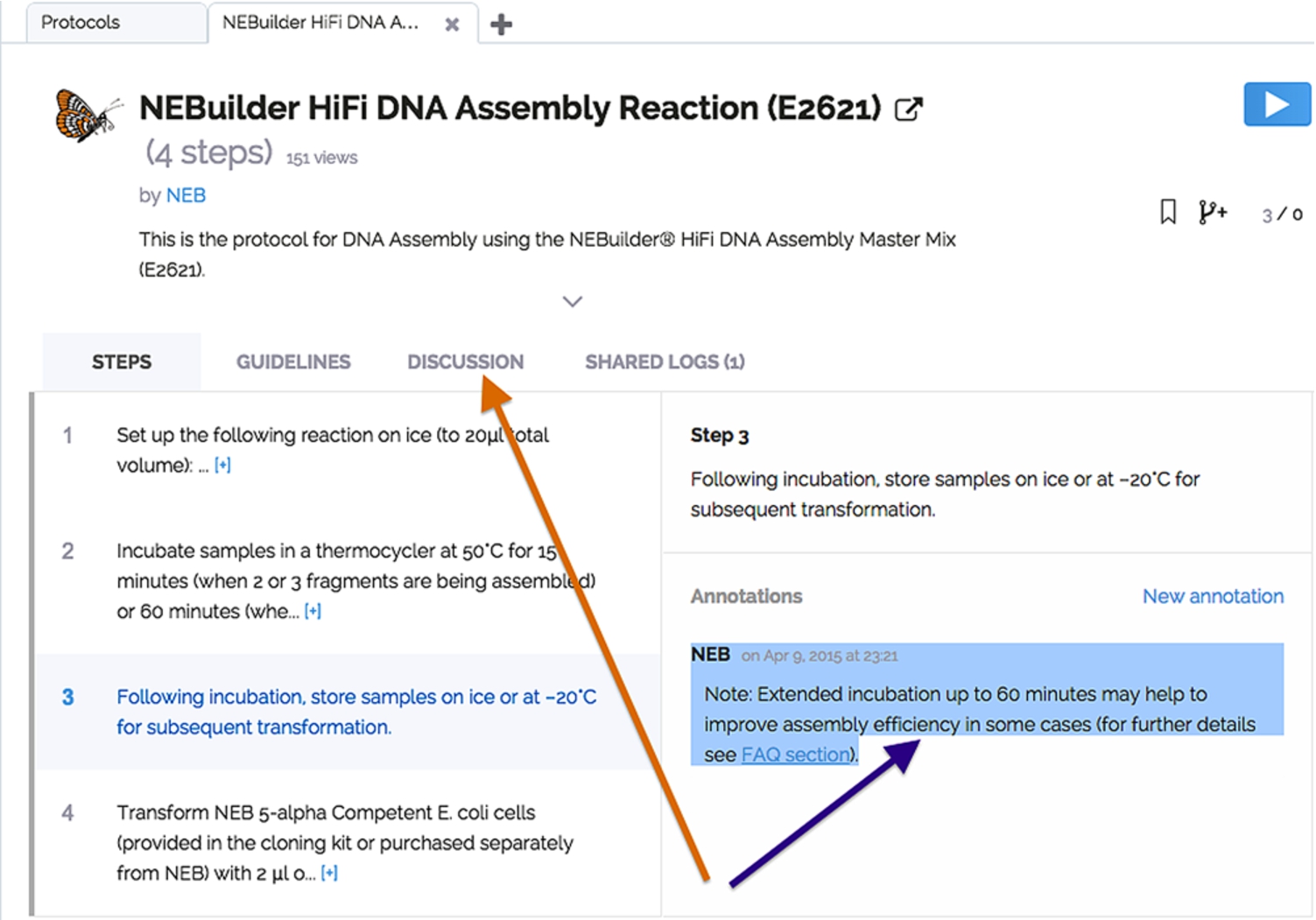
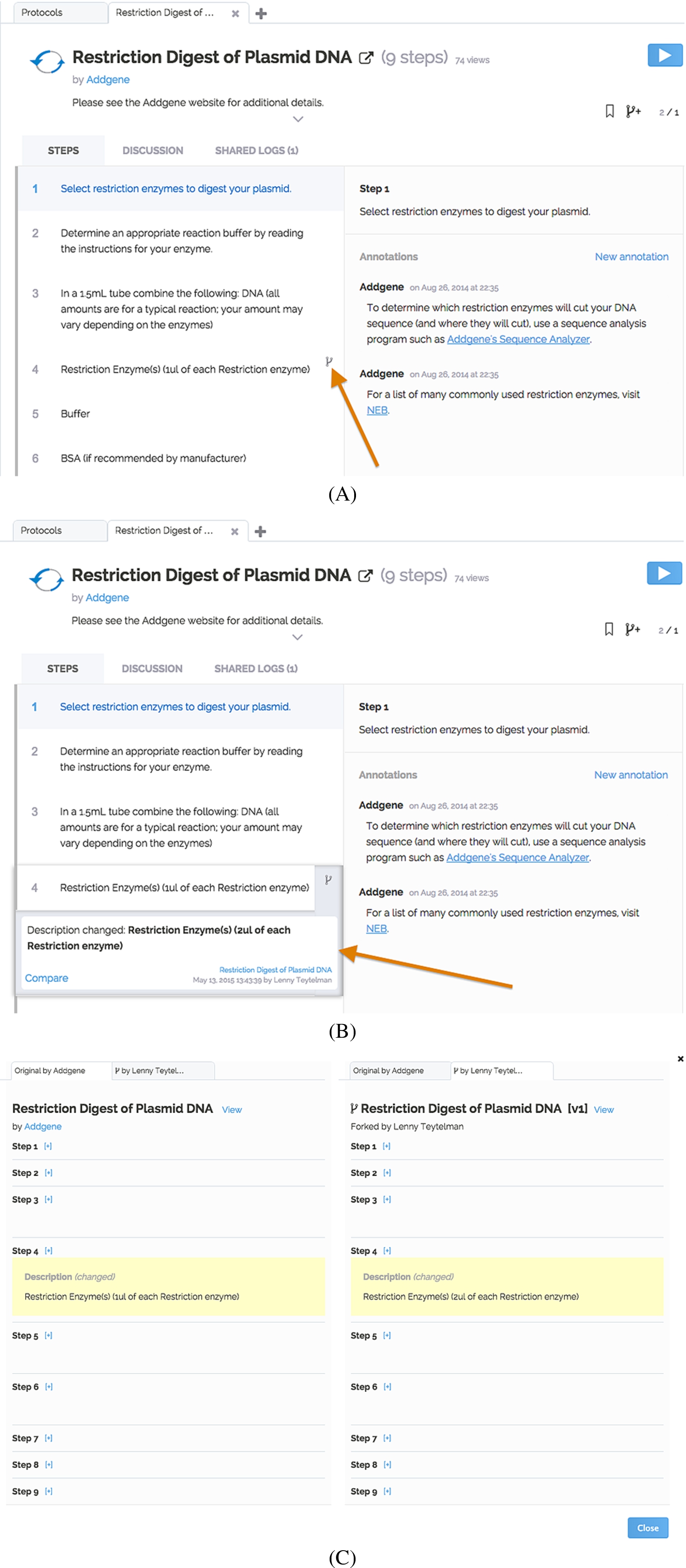
Fig. 3.
Forked changes. Panel (A) shows that a modification exists on step 4. Panel (B) provides the drop-down detail on the edit and panel (C) displays the side-by-side comparison of the original and forked protocols. (The colors are visible in the online version of the article; http://dx.doi.org/10.3233/ISU-150769.)
2.3.Communicating and discovering protocol changes
Every protocol may be annotated on the protocol- or step-level (Fig. 2). Moreover, all modifications in a published fork are discoverable on the original master protocol (Fig. 3(A), (B)). It is possible to compare in detail the master protocol and any of its branches (Fig. 3(C)). To aid discoverability of the changes, users of the protocol are instantly notified by e-mail or upon logging into protocols.io if there are shared annotations, forks or new versions. Usage and adoption statistics on each protocol and fork provide an additional way to discover important improvements.
Though the focus of the platform is on communicating corrections and improvements, many non-functional tweaks of methods are sampled every day. The protocols.io platform also encourages the sharing of changes that do not work by allowing researchers to “share” protocol runs from their journal. These shared logs are visible on the master protocol.
3.Discussion
Razib Khan and colleagues recently published, “Dragging scientific publishing into the 21st century” [6], highlighting the many deficiencies of the traditional science publishing. We agree that innovation is desperately needed in science communication, but we believe that startups and non-profits are in a better position to deliver this innovation [11].
We hope that protocols.io will save researchers countless hours by providing a central place to share and discover new method knowledge. We hope that the platform will significantly reduce the needless re-discovery that is currently plaguing biomedical research. However, realizing this vision depends on the crowdsourcing of the knowledge from the research community. We invite scientists to experiment with protocols.io, to share their knowledge and to give us input and feedback on how we can improve the platform.
Acknowledgements
We thank Anjuli Manche for thoughtful comments on this manuscript. We also thank all of the employees and consultants who have worked incredibly hard to develop protocols.io. The support of our advisors and investors has been invaluable. Most of all, we thank the countless scientists who have supported our vision; we are deeply grateful for their use of the platform, their sharing, and their invaluable feedback on our software and ideas.
About the authors
Lenny Teytelman has over a decade of computational and experimental biology experience. He did his graduate studies at UC Berkeley and finished his postdoctoral research at MIT. For three years, prior to switching from math and computer science to molecular biology, he was a database developer. Lenny brings to protocols.io a strong passion for sharing science and improving research efficiency through technology.
Alexei Stoliartchouk has spent a major part of his career focusing on building web-based systems and analyzing the large data sets behind them. He was director at CNET, engineering manager at Yahoo, director at thisMoment and CTO of rewardStyle.com. He has a Master’s degree in Computer Science and a Bachelor’s in Applied Math.
References
1 | [[1]] C.G. Begley and L.M. Ellis, Drug development: Raise standards for preclinical cancer research, Nature 483: ((2012) ), 531–533. doi:10.1038/483531a. |
2 | [[2]] D. Butler, Science in the web age: Joint efforts, Nature 438: ((2005) ), 548–549. doi:10.1038/438548a. |
3 | [[3]] D. Crotty, Nevermind the data, where are the protocols?, The Scholarly Kitchen on WordPress.com, The Scholarly Kitchen [Internet], 2014 [cited 14 May 2015], available at: http://scholarlykitchen.sspnet.org/2014/11/18/nevermind-the-data-where-are-the-protocols/. |
4 | [[4]] S. Heard, Our literature isn’t a big pile of facts, Scientist Sees Squirrel on WordPress.com [Internet], 2015 [cited 14 May 2015], available at: https://scientistseessquirrel.wordpress.com/2015/04/07/our-literature-isnt-a-big-pile-of-facts/. |
5 | [[5]] J.P.A. Ioannidis, Why most published research findings are false, PLoS Med. 2: (8) ((2005) ), e124. doi:10.1371/journal.pmed.0020124. |
6 | [[6]] R. Khan, L. Goodman and D. Mittelman, Dragging scientific publishing into the 21st century, Genome Biology 15: ((2014) ), 556. doi:10.1186/s13059-014-0556-2. |
7 | [[7]] M. Loukides, Beyond lab folklore and mythology, O’Reilly Radar [Internet], 2015 [cited 14 May 2015], available at: http://radar.oreilly.com/2015/01/beyond-lab-folklore-and-mythology.html#comment-1798673964. |
8 | [[8]] A. Meadows, Why has the scholarly journal endured?, Exchanges, Wiley Exchanges [Internet], 2015 [cited 13 May 2015], available at: http://exchanges.wiley.com/blog/2015/03/06/why-has-the-scholarly-journal-endured/. |
9 | [[9]] C. O’Luanaigh, Twenty years of a free, open web, CERN [Internet], 2013 [cited 14 May 2015], available at: http://home.web.cern.ch/about/updates/2013/04/twenty-years-free-open-web. |
10 | [[10]] L. Teytelman, We can and should improve reproducibility. But it’s not a crisis, so can do it without panic, The Spectroscope [Internet], 2015 [cited 13 May 2015], available at: http://www.thespectroscope.com/read/we-can-and-should-improve-reproducibility-but-its-not-a-by-lenny-teytelman-309. |
11 | [[11]] L. Teytelman, Subscription publishing is the hotel industry. Science needs Airbnb, The Spectroscope [Internet], 2015, [cited 13 May 2015], available at: http://www.thespectroscope.com/read/subscription-publishing-is-the-hotel-industry-science-needs-airbnb-by-lenny-teytelman-285. |
12 | [[12]] N.A. Vasilevsky, M.H. Brush, H. Paddock, L. Ponting, S.J. Tripathy, G.M. LaRocca et al., On the reproducibility of science: Unique identification of research resources in the biomedical literature, PeerJ 1: ((2013) ), e148. doi:10.7717/peerj.148. |




