Automating the pre-review of research
Abstract
This paper gives an overview of Ripeta, an organization whose mission is to assess, design, and disseminate practices and measures to improve the reproducibility of science with minimal burden on scientists, starting with the biomedical sciences. Ripeta focuses on assessing the quality of the reporting and robustness of the scientific method rather than on the quality of the science. Their long-term goal includes developing a suite of tools across the broader spectrum of sciences to understand and measure the key standards and limitations for scientific reproducibility across the research lifecycle and enable an automated approach to their assessment and dissemination. For consumers of scientific research who want an easier way to evaluate the science, Ripeta offers research reports – similar to credit reports - to quickly assess the rigor of publications. Unlike the current burden of extensive due diligence and sifting through lengthy scientific publications, Ripeta extracts the key components for reproducible research from the manuscripts and intuitively present them to you.
1.Introduction
An estimated fifty to eighty-five percent research resources are wasted due to the lack of reproducibility [1,2]. High-profile research integrity, data-quality, and replication disputes have plagued many scientific disciplines including psychology, climate science, and clinical oncology. Each year over $114.9 billion (see [1]) is spent by federal funders (e.g., National Institutes of Health (NIH), National Science Foundation (NSF)) to support the scholarly endeavors of academics nationwide; yet, within the NIH, estimates suggest 25% of this funded research is not replicable [3]. In 2015 alone, an estimated $28 billion (see [1]) of publicly and privately funded preclinical biomedical research failed to reproduce. This not only wastes resources, but also reduces the confidence in science as a whole.
There are many reasons as to why a research project may fail to reproduce, including unclear protocols and methods, an inability to recreate exact experimental conditions, not making code and data available [4], and not robustly reporting research practices in a journal article or supplementary information, to name a few [5]. Many facets moved us away from transparently reporting reproducible scientific practices in a published article. While software development and scientific automation have excelled over the past decades, processes for capturing the full, complex scientific processes have lagged [6]. For example, computational researchers have a plethora of software systems, yet they do not have a systematic way of detailing their complex experiments and are not routinely taught how to capture every facet of their computational processes. Journals have also limited word counts. Thus, when researchers prepare a publication, many inputs are either omitted or incomplete due to space constraints, posing significant challenges in presenting sufficient information to rerun the original experiment (i.e., lacking empirical reproducibility). To improve this plight, researchers must individually determine and coalesce research reproducibility elements (e.g., datasets, software versions) because globally- accepted, aggregated reporting standards do not currently exist. Once a paper is published, stakeholders - researchers, funders, institutions, publishers - do not have a streamlined method of assessing the quality and completeness of the reported scientific research; stakeholders must manually review lengthy manuscripts to assess scientific rigor.
Surmounting these research reproducibility challenges includes having the availability, verifiability, and understanding of not only the experimental research, but also the methods, processes, and tools used to conduct the experiments. Thus, assessing and reporting the transparency of reporting practices (TRP) using a robust reproducibility framework paired with automated information extraction would be of high value to scientific publishers, funders, reviewers, and scientists to rapidly assess the 2.5 million scholarly papers produced yearly across twenty-eight thousand journals.
2.Rationale for Ripeta
People look at scientific publications to make decisions and further science, yet while technological innovations have accelerated scientific discoveries, they have complicated scientific reporting. Most research publications often focus on reporting the impact of results; yet little attention is given to the methods that drive these impacts. Moreover, the structure, pieces, and operation of this scientific methodology is often opaque, ambiguous, or non-existent in reporting of research methods, leading to limited research transparency, accessibility, and reproducibility.
We need a faster and more scalable means to assess and improve scientific reporting practices to reduce the barriers in reusing scientific work, supporting scientific outcomes, and assessing scientific quality.
As the crisis of non-reproducible research continues to emerge and the field explores the diverse causes across the research landscape, the available remedies and strategies grow in nuance for detecting, evaluating, and bolstering limitations to research responsibility and reproducibility. In addition, understanding what pieces of information and related materials ought to be included in a scientific publication is challenging given the complexity of both modern computational workflows and a growing number of guidelines from multiple sources.
Part of the solution is moving from a sole focus of evaluating the potential reproducibility of published research to assessing the overall responsibility of the science - including how researchers describe their research hypothesis, methods, computational workflows, underlying limitations, and the availability of related research materials such as data and code.
3.Responsible research reporting
At Ripeta (https://www.ripeta.com/), we believe research should be transparent and our mission of ensuring responsible science ought to be shared among multiple stakeholders. We are developing automation tools to make research methods and supporting research materials transparent, enabling diverse stakeholders across the research landscape to evaluate the verifiability, falsifiability, and reproducibility of the entire research pipeline.
There are hundreds of check-lists and guidelines available providing best practices for transparently reporting research [7]. We have reviewed these guidelines, distilled and prioritized them, and currently focus on extracting a handful of key variables that will improve the reporting of data sharing, analysis, and code availability. In addition to being highly-ambiguous, assessing the reproducibility of a publication using existing frameworks of reproducibility best practices and policies can only be done knowing which domain or methodological specific checklist to use, then through manually comparing the publication to the human-readable checklists. Automating this process increases efficiency and ease of assessing highly-granular elements crucial to reproducing the entirety of the research lifecycle.
4.Automating information extraction
The ripetaSuite facilitates comparisons of research transparency and accessibility across domains and institutions. Ripeta’s unique approach generates usable findings in an audience-agnostic and easily- intelligible format providing an intuitive and evidence-supported method for multiple stakeholders to evaluate the potential for reproducibility within published literature. Using natural language processing (NLP) and other machine learning (ML) methods, the software extracts salient parts of the manuscript to highlight in a report. The algorithms are compared against manually- curated texts and Ripeta reports to both build and validate the algorithms. During the training-test-validation pipeline of NLP, we compare the machine-generated output with the gold-standard of manually-curated publications.
For example, to develop the automated detection to find a 'study purpose or research hypothesis' in a publication, we use machine learning processing and human curation to develop a statistically accurate algorithm. Once the algorithm is developed, the code integrated in our software and exposed through the Ripeta report (see next section). When a research publication is run through the software, if the study purpose/hypothesis is present, the supporting text is presented in the report, otherwise 'None Found' will be shown. This advanced manuscript parser completes in a few seconds, thus greatly reducing the time to assess the scientific rigor and reproducibility of the work.
5.Reports
To support the evaluation of research by multiple stakeholder groups, we have generated two report styles providing a summary of the responsibility of individual publications or multiple publications.
5.1.Individual publication reports
Our report (Fig. 1a and b) depicts extractions from a manuscript and summarises the performance of the publication’s reported science against a framework of best practices categorized into five sections: study overview, data collection, analytics, software and code, and supporting material. Each of these sections has sub-elements (Fig. 1b) describing granular best practices, the importance of the stated practice, if the extracted text meets the best practice, and manners in which the text can be meaningfully enhanced to satisfy the best practice.
Fig. 1.
(a): A sample Ripeta report manuscript overview with synthetic data. (b): A sample Ripeta report with sections and descriptive variables.
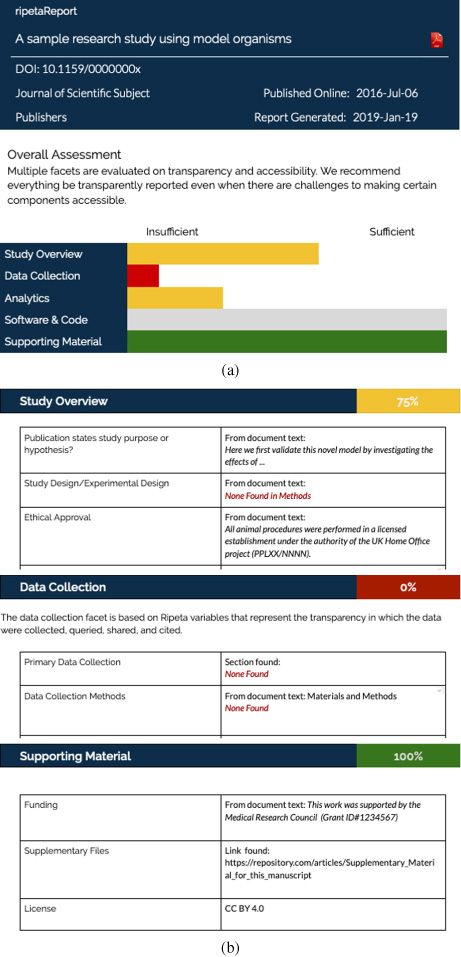
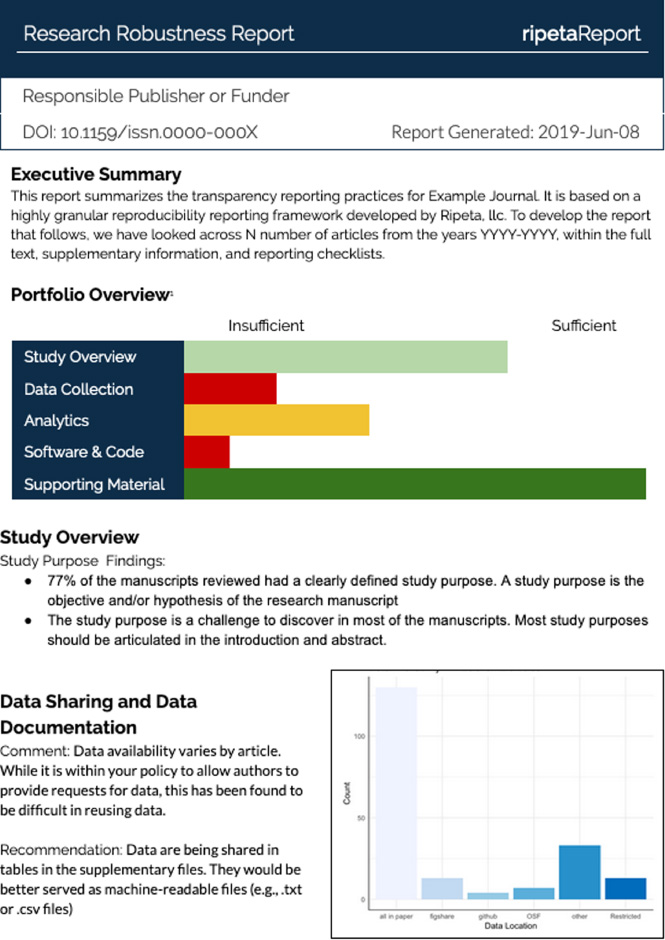
Fig. 2.
A sample Ripeta research robustness report across a portfolio of journal articles.
In addition, the individual report packages key publication metadata and a brief quantitative summary of the publication’s performance into a one-page executive summary. These individual summaries can: (i) Provide an additional check and balance for peer-reviewers throughout the publication process; (ii) Support individual researchers throughout multiple stages of publication or pre-publication preparation; and, (iii) Evaluate the responsibility of other research evidence, which researchers may integrate within their own research or practice.
5.2.Aggregated portfolio reports
To support the evaluation of overall trends across a portfolio of publications (e.g., one journal, publications from a grant), we report quantitative and qualitative summaries evaluating the performance of multiple publications against elements of responsible science. Such aggregate reports could provide a tool for publishers to evaluate the responsibility of the published research across domains or years, granting agencies to evaluate the responsibility and reproducibility of their funded projects, and academic or research institutions to evaluate the reproducibility of research throughout their department or student cohort. See Fig. 2 for a sample portfolio report.
6.Better science made easier
Ripeta was created to solve a few challenges related to responsible research reporting and supporting scientific rigor and reproducibility. In the future, we must not only showcase which areas need improving, but also suggestions for how to robustly report research. The ultimate goal is and will be to support researchers in making science better by making robust science easier.
About the First Author
Dr. Leslie McIntosh, PhD, MPH is a Co-Founder and the CEO of Ripeta. She is an internationally-known consultant, speaker, and researcher passionate about mentoring the next generation of data scientists. She is active in the Research Data Alliance, grew the St. Louis Machine Learning and Data Science Meetup to over fifteen hundred participants, and was a fellow with a San Francisco based VC firm. She recently concluded as the Director of Center for Biomedical Informatics (CBMI) at Washington University in St. Louis, MO where she led a dynamic team of twenty-five individuals facilitating biomedical informatics services. Dr. McIntosh has a focus of assessing and improving the full research cycle (data collection, management, sharing, and discoverability) and making the research process reproducible. She holds a Masters and PhD in Public Health with concentrations in Biostatistics and Epidemiology from Saint Louis University. Telephone: +1(314) 479-3313; Email Address: [email protected].
References
[1] | L.P. Freedman, I.M. Cockburn and T.S. Simcoe, The economics of reproducibility in preclinical research, PLoS Biol. 13: (6) ((2015) ). doi:10.1371/journal.pbio.1002165. |
[2] | M.R. Macleod, Biomedical research: increasing value, reducing waste, Lancet 383: (9912) ((2014) ), 101–104. doi:10.1016/S0140-6736(13)62329-6. |
[3] | Historical Trends in Federal R\&D, American Association for the Advancement of Science (AAAS), Available at: https://www.aaas.org/programs/r-d-budget-and-policy/historical-trends-federal-rd, accessed June 25, 2019. |
[4] | P.D. Schloss, Identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research, MBio 9. Jg.: (3) ((2018) ), S. e00525-18. doi:10.1128/mBio.00525-18, https://mbio.asm.org/ content/9/3/e00525-18, accessed June 25, 2019. |
[5] | J.J. Van Bavel, P. Mende-Siedlecki, W.J. Brady and D.A. Reinero, Context sensitivity in scientific reproducibility, Proceedings of the National Academy of Sciences 113: (23) ((2016) ), 6454–6459. doi:10.1073/pnas.1521897113. |
[6] | V. Stodden, Enhancing reproducibility for computational methods, Science 354: (6317) ((2016) ), 1240–1241. doi:10.1126/science.aah6168. |
[7] | Research Reporting Guidelines and Initiatives: By Organization, U.S. National Library of Medicine, 2019 [update 2019 April 16; cited 2019 June 23] Available at: https://www.nlm.nih.gov/services/research_report_guide.html, accessed June 25, 2019. |




