A Reversible Data Hiding Based on Histogram Shifting of Prediction Errors for Two-Tier Medical Images
Abstract
Clinics and hospitals have already adopted more technological resources to provide a faster and more precise diagnostic for patients, health care providers, and institutes of medicine. Security issues get more and more important in medical services via communication resources such as Wireless-Fidelity (Wi-Fi), third generation of mobile telecommunications technology (3G), and other mobile devices to connect medical systems from anywhere. Furthermore, cloud-based medical systems allow users to access archived medical images from anywhere. In order to protect medical images, lossless data hiding methods are efficient and easy techniques. In this paper, we present a data hiding of two-tier medical images based on histogram shifting of prediction errors. The median histogram shifting technique and prediction error schemes as the two-tier hiding have high capacity and PSNR in 16-bit medical images.
1Introduction
Mobile and digital media are getting more and more popular on Internet and cloud services. In social networks, many multi-media such as images, videos, and audios are produced by different aspects of the human activities. Moreover, telemedicine applies telecommunication and information technology to offer clinical health care from a distance. In critical care and emergency situations, health informatics, medical, and imaging data are transmitted from doctors and healthcare professionals to discuss patient issues. Therefore, E-Healthcare data management plays a crucial role in modern hospitals. Over Internet and cloud services, digitized healthcare systems have provided easy access, viewing and sharing of digitized medical images to patients, doctors, medical professionals, health care providers, and institutes of medicine. Medical images may provide for teleconferences among clinicians, interdisciplinary exchanges between radiologists for consultative purposes, and distant learning of medical personnel. For illegal purposes used for insurance, for example, digital medical malignant nodule images may be modified on lung parenchyma in chest CT scan images. Therefore, data hiding schemes (Wang et al., 2017; Huang et al., 2017; Li et al., 2018; Wang et al., 2018) are much simpler and cheaper than an intrusion detection system (Huang and Hwang, 2012) to protect security and privacy of images by image with an unique digital identity.
Reversible data hiding schemes (Li et al., 2016; Huang et al., 2013a; Chen and Guo, 2020; Kim et al., 2009) (RDH) are applied to embed secrets inside an image as a stego-image with minimal distortion. After the secrets are retrieved from a stego-image at the extraction stage, the original images can be reconstructed exactly. The major applications of RDH are authentication, diagnostic image, military imagery, astronomical images, satellite, and artwork preservation. Shi et al. (2016) classified RDH techniques into six categories: histogram shifting, image compressing (e.g., JPEG), semi-fragile authentication, image contrast enhancement, encrypted images, and RDH based on audio and video. In general, there are two types of popular RDH techniques: histogram shifting (HS) (Thodi and Rodriguez, 2007; Hong et al., 2008) and difference expansion (DE). In 2006, Ni et al. (2006) proposed the first HS-based RDH by modifying the generated histogram. Some extensions of Ni et al.’s HS-based RDH methods proposed such as block-based HS (Fallahpour and Sedaaghi, 2007), difference-histogram (Lee et al., 2006), high-frequency IWT coefficients (Xuan et al., 2007; Huang et al., 2017). In 2002, Tian (2002) proposed a high capacity DE method. Furthermore, DE has been developed in three species: integer-to-integer transformation (Qiu et al., 2016), prediction-error expansion (PEE) (Dragoi and Coltuc, 2015), and adaptive embedding (Hong et al., 2015). There are three approaches in image compressing: RDH with quantized DCT coefficients modification (Huang et al., 2016), RDH with quantization table modification (Wang et al., 2013), RDH with Huffman table modification (Wu and Deng, 2011). In 2003, Vleeschouwer et al. (2003) presented the first robust RDH based on the correlations among the neighbouring pixels. In general, the approaches of contrast enhancement RDH applied some functionalities such as histogram bin expansion operations to improve the visual quality. There are four RDH approaches with contrast enhancement: method by histogram bin expansion (Wu et al., 2015a), method with contrast enhancement for medical images (Wu et al., 2015b), method with the controlled contrast enhancement (Gao and Shi, 2015), and automatic contrast enhancement method (Kim et al., 2015). Due to protection of the privacy of data and enabling the cloud server to easily manage the data, more and more researchers study reversible data hiding in encrypted images. The techniques of encrypted images are divided into three types: vacating room before encryption (Cao et al., 2016), vacating room after encryption (Zhang, 2011), and reversible image transformation (Zhang et al., 2016).
Fig. 1
The image I is partioned into a set of

Considering resource sharing data in Cloud computing environments, stego-images may be disturbed by various techniques such as a chaotic Hénon function to obtain encrypted images (Huang et al., 2013b). Abbasy and Shanmugam (2011) presented biological aspects of the DNA to increase the level of data confidentiality among clients. Some other authors (Zhang, 2012; Surekha and Swamy, 2013) converted stego-images into encryption data transferred to public Cloud environments. Based on prediction error expansion, a predictive value is calculated by predictors. Then the secret bit-stream will be embedded into the cover image according to the expansion of the difference between a pixel and its predictive values.
Most of image data hiding techniques (Huang et al., 2012; Wu et al., 2009; Zhang S. et al., 2016; Jana et al., 2016) are developed in 2D images. However, many medical images are produced and processed (Tseng and Huang, 2010) as stacks of slices such as CT, MRI, and PET. These slices can be used to generate 3D image volumetric information. Thus, it is important to apply data hiding in 3D images efficiently. Recently, for digital media it is getting more and more important to apply data hiding to the quality of compressed video. Shanableh (2012) applied data hiding schemes to embed secrets into a compressed video bit stream for copyright protection.
In this paper, two-tier structures, histogram-shifting of median and prediction error, are utilized to embed secret messages. The rest of this paper is presented as follows. The second section demonstrates our method. The fourth section shows the outcome of the experiments. Conclusions are presented in the last section.
2Proposed Method
In this paper, we apply a block median preservation technique (Luo et al., 2011), a prediction-based reversible hiding method (Chang et al., 2012), and a histogram-based overflow/underflow process (Huang et al., 2013b) to embed n bits of secret messages for each block in gray level images. Adopting overlapping blocks partition and a histogram-based overflow/underflow process, the capacity can be improved. Two-tier data hiding will be adopted to embed messages. The detail of a two-tier data hiding scheme is described as follows.
2.1Data Embedding
At first, we partition image I into a set of
2.1.1Tier-1 Data Hiding
In the tier-1 data hiding, we adopt the histogram shifting method (Luo et al., 2011) by adjusting the peak point to zero points of the image histogram. The tier-1 data hiding consists of four steps: constructing difference, classifying block type, emptying peak points, and embedding data.
Step 1: Construct difference
Fig. 2
Diagram for the framework of two-tier medical image data hiding based on histogram shifting of prediction errors at the sender side.
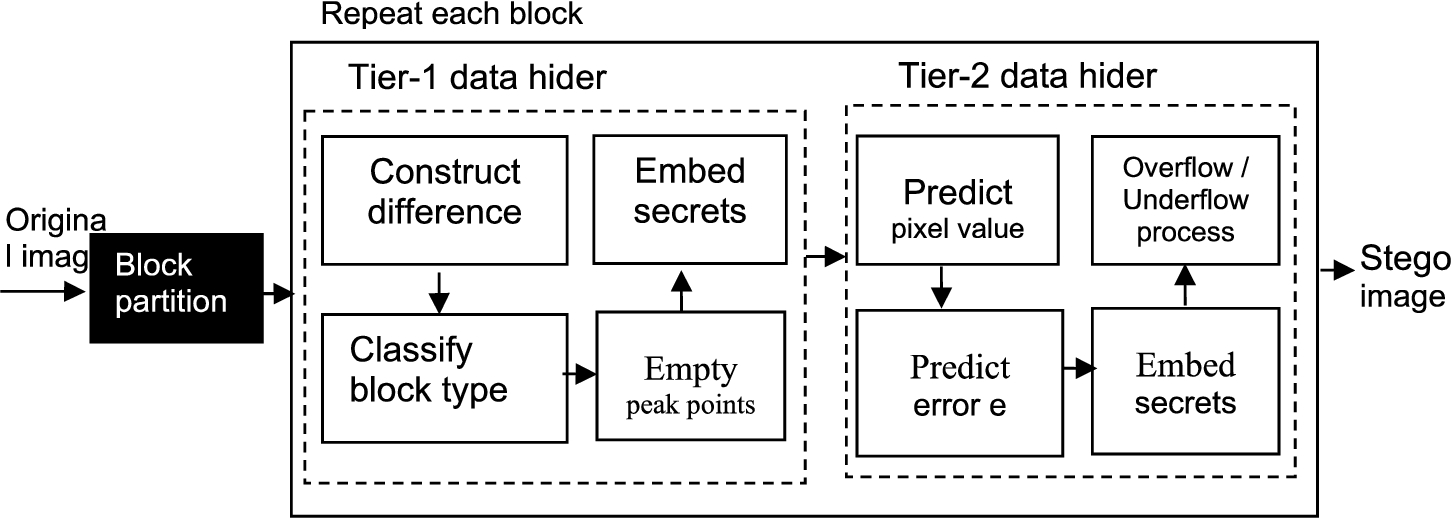
At this step, we find median values of each block. Let four pixel values
(1)
Step 2: Classify block type
In this step, all blocks will be classified into two types, Type I and Type II. Let
Step 3: Empty peak points
Embedding level (EL) is a parameter to adjusting capacity by histogram shifting. Let EL = i, the bins of
(2)
(3)
Step 4: Embed data
In this step, we apply histogram shifting to differentiate types of blocks. The strategies of the secret embedding are demonstrated as follows:
(1) Type I.
The definition of Type I is
(4)
The secrets will be embedded as Eq. (4) repeatedly.
(5)
(2) Type II-1.
The definition of block Type II-1 is presented as follows:
(6)
We will apply Eq. (6) to embed secrets w repeatedly.
(7)
(3) Type II-2.
In a Type II-2 block, the definition is demonstrated as
(8)
(9)
(4) Type II-3.
In a Type II-3 block, the definition is demonstrated as
(10)
(11)
2.1.2Tier-2 Data Hiding
In order to adjust the quality of stego-image, we apply threshold in the embedding process. Let a set of
Step 1: Compute the prediction pixel value
(12)
Step 2: Compute the prediction error e
Let predict pixel p be an element a
(13)
Step 3: Data embedding
(14)
Step 4: Overflow/Underflow process
In order to restore the cover image correctly, we process the overflow/underflow by the shifting histogram from the embedding stage without a location map or compression (Kim et al., 2009). At first, we compute the utilization rate of the medical image which consists of intensity rate and underflow/overflow rate. In order to get the intensity rate, we divide the range of intensity by 2 to the power of bit depth. The range of intensity is the difference in the intensity of the cover image between max value and min value. Bit depth is the image quality expressing how many unique shades are available. Images with higher bit depths are able to encode more intensities because there are combinations of 0’s and 1’s available.
(15)
(16)
(17)
Further, we adjust image histograms to prevent overflow/underflow problems. On low underflow/overflow rates of high bit-depth medical images, unsinged bit images will produce underflow problems.
Because modern medical devices provide high quality images, medical images can range from 0 to 65535 for a 16-bit image. Therefore, the utilization rate of 16-bit depth is lower than 8-bit depth shown in equations (15), (16). In other words, we can utilize empty histogram bin to process overflow/underflow problem by shifting histogram of the whole image.
In respect of preserving boundary pixels, our method can restore the original-image from the stego-image successfully. Thus, the boundary pixels will be preserved. After the process of embedding, our method also can preserve the boundary pixels shown in Fig. 5. The 16-bit depth unsigned images are tested with threshold 21. In Fig. 5(b) and (c) the preserved boundary of the medical image when EL are changed from 5 to 20 is shown. In the same way, another test of the preserved boundary is shown in Fig. 5(e), (f).
2.2Data Extraction
Fig. 3
Diagram for the framework of two-tier medical image data extraction based on histogram shifting of prediction errors at receiver side.
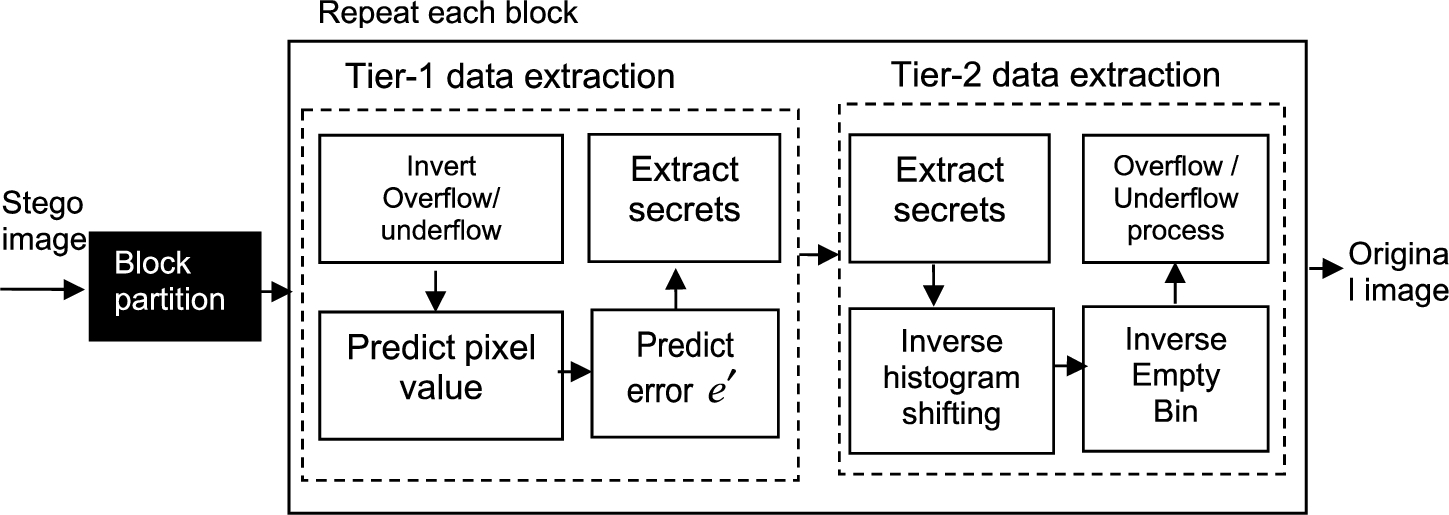
In order to extract data, we need the embedding level and overflow/underflow distance as parameters. The data extraction will extract secret messages correctly and restore the stego-image back to the original without any distortions, described in detail below as shown in Fig. 3 Similar to the data embedding process, we partition the stego-image into overlaying blocks with
2.2.1Tier-2 Data Extraction
In tier-2 data extraction, we perform the extraction of prediction-based reversible data hiding method. There are four stages: inverting overflow/underflow process, predicting pixel value, predicting error e, and extracting secrets. At the first stage, we adopt histogram shifting with distance overflow/underflow to invert overflow/underflow process. At the predict pixel value stage, we construct a difference block like in the Tier-1 data hiding, step 2. Then we apply the formula (11) to obtain the predicted pixel value
2.2.2Tier-1 Data Extraction
There are three stages: extracting secrets, inversing histogram shifting, and inversing empty bin in tier-2 data extraction demonstrated as follows.
a. Extracting secrets
In order to extract secrets and restore the original image, we apply histogram shifting to the intermediate image block from tier-1 data extraction at this stage. Let the elements of the intermediate block be
(18)
Next step, repeat n times when EL is equal to n. In the case EL = 1,
b. Inversing histogram shifting
According to Embedding level (EL), there are different strategies to inverse histogram described as follows.
If EL = 0,
(19)
If EL > 0,
(20)
c. Inversing empty bin
If EL is greater than 0, we perform the inversion of empty bin according to the following formula:
(21)
After inversing empty bin, we add a median value of a block to elements of the block. Finally, we construct each block as the original image.
3Experimental Results
In this paper, we demonstrate two-tier medical images data hiding based on histogram shifting of prediction errors to experiment on medical images with DICOM. The peak signal-to-noise ratio (PSNR) is applied to measure the quality.
(22)
(23)
Fig. 4
(a) CT image with signed
Table 1
The results for a signed 16-bit grayscale medical image.
| Image name from the national cancer imaging archive | Image size | Embedding level | Threshold | Capacity | bpp | PSNR |
| 1.3.6.1.4.1.9328.50.14.2.dcm | 350 × 512 | 1 | 1 | 99,633 | 0.555987723 | 81.92740097 |
| 6 | 120,508 | 0.672477679 | 79.74795677 | |||
| 11 | 150,878 | 0.841953125 | 77.48244098 | |||
| 16 | 179,047 | 0.999146205 | 75.77926976 | |||
| 21 | 197,696 | 1.103214286 | 74.64371168 |
Table 2
The results for an unsigned 16-bit grayscale medical image with different embedding levels.
| Image name from the national cancer imaging archive | Image size | Embedding level | Threshold | Capacity | bpp | PSNR |
| 1.3.6.1.4.1.9328.50.14.2.dcm | 350 × 512 | 1 | 21 | 197,696 | 1.103214286 | 74.64371168 |
| 6 | 304,972 | 1.701852679 | 68.91926164 | |||
| 11 | 365,123 | 2.037516741 | 66.1103813 | |||
| 16 | 406,108 | 2.266227679 | 64.16268337 | |||
| 21 | 434,770 | 2.426171875 | 62.72562295 |
Table 3
The results for a signed 16-bit grayscale medical image.
| Image name from the national cancer imaging archive | Image size | Embedding level | Threshold | Capacity | bpp | PSNR |
| 1.3.6.1.4.1.9328.50.14.159.dcm | 395 × 512 | 21 | 21 | 501,905 | 2.481729628 | 62.81406295 |
| 1.3.6.1.4.1.9328.50.14.1207.dcm | 429 × 512 | 574,925 | 2.617483428 | 63.33996413 | ||
| 1.3.6.1.4.1.9328.50.14.1210.dcm | 429 × 512 | 515,352 | 2.346263112 | 62.08233045 | ||
| 1.3.6.1.4.1.9328.50.14.1278.dcm | 444 × 512 | 526,871 | 2.317668743 | 62.05008477 | ||
| 1.3.6.1.4.1.9328.50.14.1280.dcm | 444 × 512 | 589,797 | 2.594475823 | 63.22757547 | ||
| 1.3.6.1.4.1.9328.50.14.157.dcm | 395 × 512 | 455,072 | 2.250158228 | 61.92850025 | ||
| 1.3.6.1.4.1.9328.50.14.1141.dcm | 458 × 512 | 555,916 | 2.370684361 | 62.12956713 | ||
| 1.3.6.1.4.1.9328.50.14.1908.dcm | 547 × 512 | 739,699 | 2.641178445 | 63.24405029 |
In this paper, we also used Lena and airplane with 8-bit depth
Fig. 5
The result for boundary preserving with EL 5 and 20.
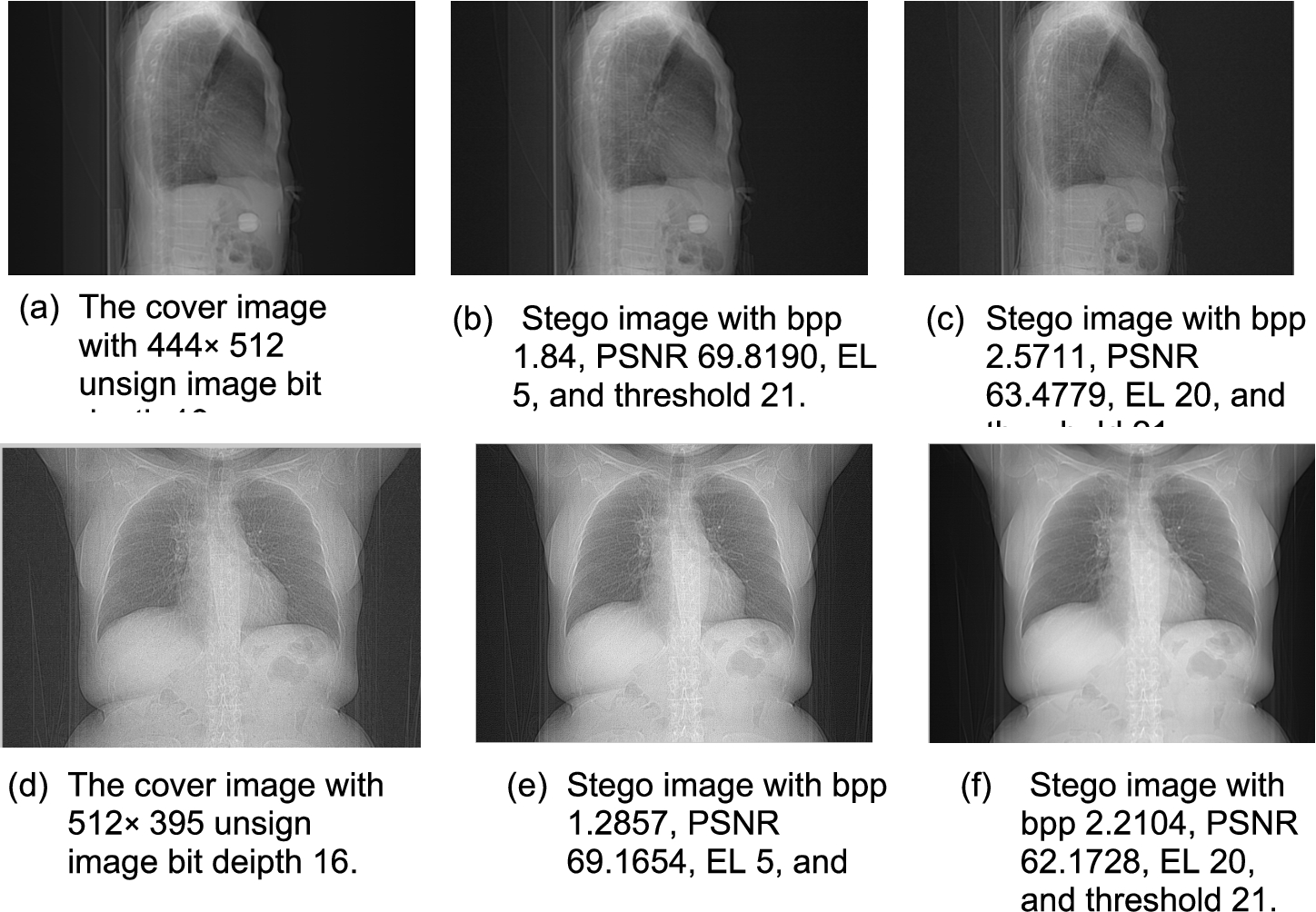
Fig. 6
Two cover images with stego-images: (a), (b) Lena (c), (d) F-16.

Table 4
The results for Lena and airplane with 8-bit depth.
| Image name from the national cancer imaging archive | Image size | Embedding level | Threshold | Capacity | bpp | PSNR |
| Lena | 512 × 512 | 1 | 1 | 158,702 | 0.605400085 | 33.63540908 |
| 2 | 230,835 | 0.880565643 | 30.32126065 | |||
| 3 | 290,965 | 1.109943390 | 28.06630698 | |||
| 4 | 340,771 | 1.299938202 | 26.39720695 | |||
| Airplane | 512 × 512 | 1 | 1 | 241,027 | 0.919445038 | 34.05170498 |
| 2 | 324,556 | 1.238082886 | 31.01245276 |
Fig. 7
The results for a 16-bit grayscale medical Image and a 8-bit image.
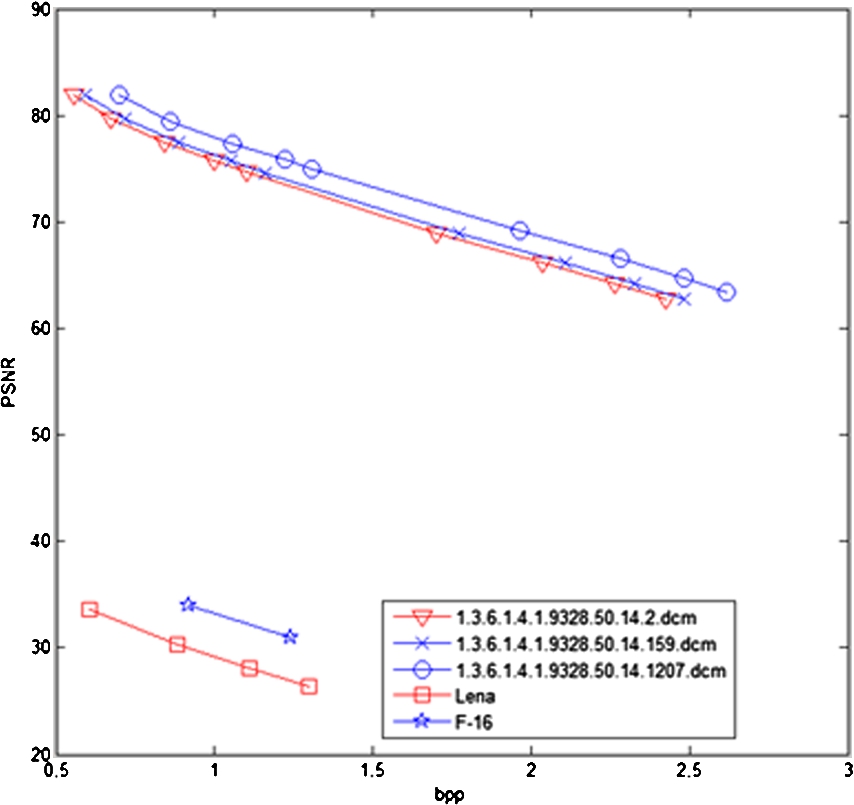
Table 5
The results for the comparison of median and prediction methods.
| Image name from the national cancer imaging archive | Image size | Median | Prediction | Our method | |||
| Capacity | PSNR | Capacity | PSNR | Capacity | PSNR | ||
| 1.3.6.1.4.1.9328.50.14.159 | 395 × 512 | 0.71 | 69.9889 | 0.99 | 69.1440 | 1.68 | 69.6712 |
| 1.3.6.1.4.1.9328.50.14.1207 | 429 × 512 | 0.71 | 69.6336 | 0.99 | 69.2379 | 1.88 | 69.9551 |
| 1.3.6.1.4.1.9328.50.14.1280 | 444 × 512 | 0.71 | 69 | 0.98 | 69.747 | 1.84 | 69.8189 |
| 1.3.6.1.4.1.9328.50.14.1210 | 429 × 512 | 0.72 | 73.6699 | 0.96 | 73.396 | 1.02 | 73.3560 |
| 1.3.6.1.4.1.9328.50.14.1278 | 444 × 512 | 0.73 | 73.3000 | 0.952 | 73.063 | 1.01 | 73.1843 |
| 1.3.6.1.4.1.9328.50.14.157 | 395 × 512 | 0.73 | 73.3590 | 0.96 | 73.077 | 0.96 | 73.073 |
| 1.3.6.1.4.1.9328.50.14.1141 | 458 × 512 | 0.73 | 73.7324 | 0.96 | 73.727 | 1.04 | 73.459 |
4Security Aspects
The embedding algorithm is used to embed the secret data into the cover image as a stego-image. Stego-images are transmitted over the Internet. If stego-images are altered or destroyed due to certain attacks, our method has the basic characteristics of data hiding to protect secret data. Because the secret data is hidden in the cover image, it is difficult for the human visual system to detect any unperceivable secret data.
In our method, there are two layers of block median retention technology and a predictive reversible hiding method to hide secret data. Therefore, our method is a technique that resists the attack of this technique without parameters such as threshold, EL and overflow/underflow methods. We also applied 2 × 2 overlapping blocks to partition the image. If a hacker changes the secure covert image over the network, the secure covert image cannot be extracted correctly.
5Conclusion
We have proposed the data hiding of two-tier medical images based on histogram shifting of prediction error. The median histogram shifting technique and prediction error schemes as the two-tier hiding have high capacity and PSNR in 16-bit medical images. The overlapping block partitions have also been adopted in this scheme, and it has performed well. At the Tier-1 data hider, the median histogram shifting technique is based on a multi-level histogram shifting mechanism. Due to adopting two types of embedding strategies, the block medians are able to be preserved. Therefore, the stego-image utilizes these block medians to convert the stego image to the original image successfully. At the Tier-2 data hider, the prediction error scheme applies a pre-determined threshold and the absolute difference of the neighbouring pixels to decide whether the current pixel is embeddable.
Acknowledgements
In this paper, we adopt medical images from the cancer imaging archive (NBIA, 2018) and thank National Cancer Institute for providing the CT image data for this study for free. Most of the medical devices provide 16 or more bits of image information at each pixel (Culjat et al., 2012). Therefore, a very large number of histogram is empty. Those histograms can be utilized to process overflow/underflow problems.
References
1 | Abbasy, M.R., Shanmugam, B. (2011). Enabling data hiding for resource sharing in cloud computing environments based on DNA sequences. In: IEEE World Congress on Services, pp. 385–390. |
2 | Cao, X., Du, L., Wei, X., Meng, D., Guo, X. ((2016) ). High capacity reversible data hiding in encrypted images by patch-level sparse representation. IEEE Transactions on Cybernetics, 46: , 1132–1143. |
3 | Chang, C.C., Huang, Y.H., Tsai, H.Y., Qin, C. ((2012) ). Prediction-based reversible data hiding using the difference of neighboring pixels. International Journal of Electronics and Communications, 66: , 758–766. |
4 | Chen, X., Guo, W. ((2020) ). Reversible data hiding scheme based on fully exploiting the orientation combinations of dual stego-images. International Journal of Network Security, 22: , 126–135. |
5 | Culjat, M., Singh, R., Lee, H. ((2012) ). Medical Devices: Surgical and Image-Guided Technologies. Wiley. |
6 | Dragoi, I.C., Coltuc, D. ((2015) ). On local prediction based reversible watermarking. IEEE Transactions on Image Processing, 24: , 1244–1246. |
7 | Fallahpour, M., Sedaaghi, M.H. ((2007) ). High capacity lossless data hiding based on histogram modification. IEICE Electronics Express, 4: , 205–210. |
8 | Gao, G., Shi, Y.Q. ((2015) ). Reversible data hiding using controlled contrast enhancement and integer wavelet transform. IEEE Signal Processing Letters, 22: , 2078–2082. |
9 | Hong, W., Chen, T.S., Shiu, C.W. ((2008) ). Reversible data hiding based on histogram shifting of predict errors. International Workshop on Geoscience and Remote Sensing, 2: , 578–581. |
10 | Hong, W., Chen, T.S., Chen, J. ((2015) ). Reversible data hiding using Delaunay triangulation and selective embedment. Information Sciences, 308: , 140–154. |
11 | Huang, L.C., Hwang, M.S. ((2012) ). Study of an intrusion detection system. Journal of Electronic Science and Technology, 10: , 77–83. |
12 | Huang, L.C., Tseng, L.Y., Hwang, M.S. ((2012) ). The study of data hiding in medical image. International Journal of Network Security, 14: , 243–251. |
13 | Huang, L.C., Hwang, M.S., Tseng, L.Y. ((2013) a). Reversible and high-capacity data hiding in high quality medical images. KSII Transactions on Internet and Information Systems, 7: , 132–148. |
14 | Huang, L.C., Hwang, M.S., Tseng, L.Y. ((2013) b). Reversible data hiding for medical images in cloud computing environments based on Chaotic Hénon map. Journal of Electronic Science and Technology, 11: , 230–236. |
15 | Huang, F., Qu, X., Kim, H.J., Huang, J. ((2016) ). Reversible data hiding in JPEG images. IEEE Transactions on Circuits and Systems for Video Technology, 26: , 1610–1621. |
16 | Huang, L.C., Feng, T.H., Hwang, M.S. ((2017) ). A new lossless embedding techniques based on HDWT. IETE Technical Review, 34: , 40–47. |
17 | Jana, B., Giri, D., Mondal, S.K. ((2016) ). Dual-Image based reversible data hiding scheme using pixel value difference expansion. International Journal of Network Security, 18: , 633–643. |
18 | Kim, K.S., Lee, M.J., Lee, H.Y., Lee, H.K. ((2009) ). Reversible data hiding exploiting spatial correlation between sub-sampled images. Pattern Recognit, 42: , 3083–3096. |
19 | Kim, S., Lussi, R., Qu, X., Kim, H.J. (2015). Automatic contrast enhancement using reversible data hiding. In: Proc. IEEE International Workshop on Information Forensics and Security, 1–5. |
20 | Lee, S.K., Suh, Y.H., Ho, Y.S. ((2006) ). Reversible image authentication based on watermarking. In: Proceedings of the IEEE International Conference on Multimedia and Expo, pp. 1321–1324. |
21 | Li, F., Mao, Q., Chang, C.C. ((2016) ). A reversible data hiding scheme based on IWT and the Sudoku method. International Journal of Network Security, 18: , 410–419. |
22 | Li, J.J., Wu, Y.H., Lee, C.F., Chang, C.C. ((2018) ). Generalized PVO-K embedding technique for reversible data hiding. International Journal of Network Security, 20: , 65–77. |
23 | Luo, H., Yu, F.X., Chen, H., Huang, Z.L., Li, H., Wang, P.H. ((2011) ). Reversible data hiding based on block median preservation. Information Sciences, 181: , 308–328. |
24 | National Cancer Institute, National Biomedical Imaging Archive (NBIA) (2018). Institute, National Biomedical Imaging Archive (NBIA). https://imaging.nci.nih.gov/ (accessed March 2 2018). |
25 | Ni, Z., Shi, Y.Q., Ansari, N., Su, W. ((2006) ). Reversible data hiding. IEEE Transactions on Circuits and Systems for Video Technology, 16: , 354–362. |
26 | Qiu, Y., Qian, Z., Yu, L. ((2016) ). Adaptive reversible data hiding by extending the generalized integer transformation. IEEE Signal Processing Letters, 23: , 130–134. |
27 | Shanableh, T. ((2012) ). Data hiding in MPEG video files using multivariate regression and flexible macroblock ordering. IEEE Transactions on Information Forensics and Security, 7: , 455–464. |
28 | Shi, Y.Q., Li, X., Zhang, X., Wu, H.T., Ma, B. ((2016) ). Reversible data hiding: Advances in the past two decades. IEEE Access, 4: , 3210–3237. |
29 | Surekha, B., Swamy, G.N. ((2013) ). Sensitive digital image watermarking for copyright protection. International Journal of Network Security, 15: , 113–121. |
30 | Thodi, D.M., Rodriguez, J.J. ((2007) ). Expansion embedding techniques for reversible watermarking. IEEE Transactions on Image Processing, 6: , 721–730. |
31 | Tian, J. ((2002) ). Wavelet-based reversible watermarking for authentication. In: Proceedings SPIE 4675, Security and Watermarking of Multimedia Contents, IV: , 679–690. |
32 | Tseng, L.Y., Huang, L.C. ((2010) ). Automatic fissure detection in CT images based on the genetic algorithm. In: Proceedings of the International Conference on Machine Learning and Cybernetics, Vol. 5: , pp. 2583–2588. |
33 | Vleeschouwer, C.D., Delaigle, J.F., Macq, B. ((2003) ). Circular interpretation of bijective transformations in lossless watermarking for media asset management. IEEE Transactions on Multimedia, 5: , 97–105. |
34 | Wang, K., Lu, Z.M., Hu, Y.J. ((2013) ). A high capacity lossless data hiding scheme for JPEG images. Journal of Systems and Software, 86: , 1965–1975. |
35 | Wang, Y.L., Shen, J.J., Hwang, M.S. ((2017) ). An improved dual image-based reversible hiding technique using LSB matching. International Journal of Network Security, 19: , 858–862. |
36 | Wang, Y.L., Shen, J.J., Hwang, M.S. ((2018) ). A survey of reversible data hiding for VQ-compressed images. International Journal of Network Security, 20: , 1–8. |
37 | Wu, Y., Deng, R.H. ((2011) ). Zero-error watermarking on JPEG images by shuffling Huffman tree nodes. In: Proceedings of the IEEE Visual Communications and Image Processing, pp. 1–4. |
38 | Wu, C.C., Hwang, M.S., Kao, S.J. ((2009) ). A new approach to the secret image sharing with steganography and authentication. Imaging Science Journal, 57: , 140–151. |
39 | Wu, H.T., Dugelay, J.L., Shi, Y.Q. ((2015) a). Reversible image data hiding with contrast enhancement. IEEE Transactions on Signal Processing, 22: , 81–85. |
40 | Wu, H.T., Huang, J., Shi, Y.Q. ((2015) b). A reversible data hiding method with contrast enhancement for medical images. Journal of Visual Communication and Image Representation, 31: , 146–153. |
41 | Xuan, G., Shi, Y.Q., Chai, P., Cui, X., Ni, Z., Tong, X. (2007). Optimum histogram pair based image lossless data embedding. International Workshop on Digital Watermarking, 264–278. |
42 | Zhang, X. ((2011) ). Reversible data hiding in encrypted image. IEEE Signal Processing Letters, 18: , 255–258. |
43 | Zhang, X. ((2012) ). Separable reversible data hiding in encrypted image. IEEE Transactions on Information Forensics and Security, 7: , 826–832. |
44 | Zhang, W., Wang, H., Hou, D., Yu, N. ((2016) ). Reversible data hiding in encrypted image by reversible image transforming. IEEE Transactions on Multimedia, 18: , 1469–1479. |
45 | Zhang, S., Gao, T., Yang, L. ((2016) ). A reversible data hiding scheme based on histogram modification in integer DWT domain for BTC compressed images. International Journal of Network Security, 18: , 718–727. |




