FAIR scientific information with the Open Research Knowledge Graph
Abstract
The Open Research Knowledge Graph is an infrastructure for the production, curation, publication and use of FAIR scientific information. Its mission is to shape a future scholarly publishing and communication where the contents of scholarly articles are FAIR research data.
Despite improved digital access to scientific information in recent decades, scholarly communication continues to be document-based, and the scholarly record managed by document repositories. Scientific information is therefore locked into Expressions (see FRBR model [4]) that are inadequate for machine processing. Given the enormous value of the scholarly record for knowledge societies and the time pressure of many societal challenges, transitioning to a scholarly communication infrastructure that manages FAIR scientific information is of utmost importance.
The rapidly growing and inefficient to process document-based scholarly record is a source of substantial friction in the research lifecycle. Systematic review teams, for instance, routinely manually extract [3] inaccurate information from scholarly articles in order to build databases capable of supporting their research. Answering relatively straightforward questions, such as best-performing algorithms for given tasks, relies on enormous manual efforts.
Various communities have identified the problem and have created databases that manage curated content extracted from the literature and developed services that support content use. Examples can be found in invasion biology (hi-knowledge.org), social sciences (cooperationdatabank.org), computer science (paperswithcode.com), cognitive science (langcog.github.io/metalab), biodiversity (plazi.org), environmental science (covid-aqs.fz-juelich.de), life science (geneontology.cloud), to name a few. While these FAIR Supporting Resources are technologically rather diverse, they share disciplinary focus and tailored services as common traits. Beyond such local initiatives, scientific information remains buried in narrative text documents [1,6].
As a FAIR Supporting Service, the Open Research Knowledge Graph [2] (ORKG, https://orkg.org) addresses the challenge at hand as-a-Service by providing research communities with a readily usable and sustainably governed infrastructure that implements best practices (e.g., FAIR principles) and provides services to support the production, curation, publication and use of FAIR scientific information.
Methodologically, these activities are supported in a semi-automated manner. FAIR scientific information production and curation is implemented using techniques that ensure FAIRification either pre-publication while information is produced (e.g., in data analysis) or post-publication by means of automated (Natural Language Processing) or manual (Crowdsourcing) information extraction from articles. Various features support these activities. Specifically, the Web-based user interface supports the structured description of contributions published in articles, in particular the research problem addressed by the contribution with the results and utilized materials and methods. Such structured descriptions may be guided by templates, i.e. graph patterns that specify properties and values. Leveraging automated techniques, the user interface includes features such as the abstract annotator and suggestions to guide users in identifying relevant information. To complement these post-publication techniques, ORKG also features pre-publication approaches to FAIR scientific information production. Specifically, the ORKG Python library [8] and R package [9] integrate with ORKG templating to support the production of FAIR scientific information as an activity of data analysis.
FAIR scientific information is published and can be accessed openly via user and application programming interfaces. The modern, interactive, and dynamic Web-based user interface is complemented with REST and SPARQL programmatic interfaces [7].
To support the efficient use of scientific information, e.g., in downstream data science, ORKG provides libraries for Python and R that enable loading or producing ORKG content in computational environments. Additionally, ORKG features numerous generic services that make use of FAIR scientific information. For example, the ORKG Comparison service automatically compares research contributions of selected articles. Given a research problem, e.g. open access download advantage [5], the service can automatically compare contributions across the relevant literature. Figure 1 provides a graphical overview of ORKG and some key services, such as comparison data visualization, production of thematic reviews, and observatories as virtual spaces for knowledge organization.
Fig. 1.
Overview of the primary ORKG services: Tabular comparisons of scientific information, visualizations of quantitative comparison data, thematic reviews including ORKG contents, and expert-based scientific knowledge organization in observatories.
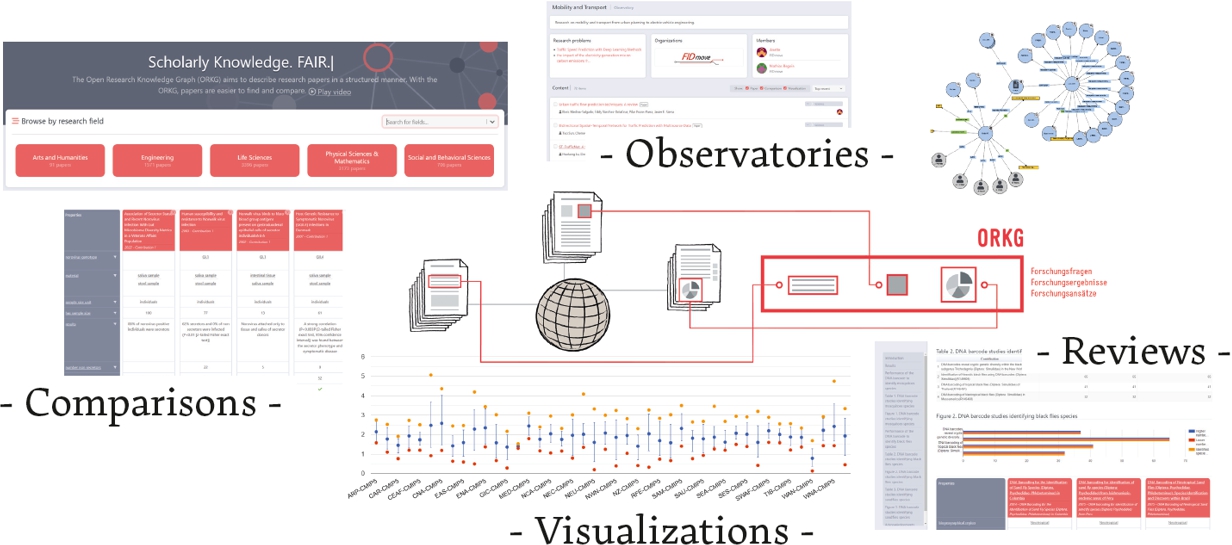
The ORKG initiative is actively engaging research communities in numerous ways. As an important instrument, ORKG Observatories are virtual communities in which experts in a domain collaborate on relevant content production and curation, and can thus create community-specific entry points. Activities include the identification and ORKG-Templates-based specification of scientific information essential to their domain; taxonomic organization of research problems in their field; and the production and quality control of content relevant to the observatory. Furthermore, the initiative engages with publishers and conferences to integrate the ORKG into manuscript submission processes. Finally, the initiative engages with research groups as well as individual researchers to test and deploy all aspects of the infrastructure in order to mature the services and ensure their integration in the research lifecycle.
We are at the beginning of a journey aiming at frictionless scientific information use with advanced machine processing. Various initiatives in disciplines ranging from invasion biology to social science have shown what conducting science with FAIR scientific information can look like. The ORKG initiative is driving the transformations seen in these communities in research more fundamentally by increasing productivity through reusable infrastructure and services, organizing needed activities, delivering training and support, and building capacity.
Acknowledgements
The authors like to thank colleagues who have contributed to this work, in particular Irina Sens, Oliver Koepler, Philipp Dillschneider, and Anette Ganske, as well as numerous students.
Funding
Parts of the work described in this article have been co-funded by the European Research Council for the project ScienceGRAPH (GA: 819536).
Disclosures and conflict of interest
All authors report no conflict of interest.
References
[1] | T.K. Attwood, D.B. Kell, P. McDermott, J. Marsh, S.R. Pettifer and D. Thorne, Calling international rescue: Knowledge lost in literature and data landslide! Biochemical Journal 424: (3) ((2009) ), 317–333. doi:10.1042/BJ20091474. |
[2] | S. Auer, A. Oelen, M. Haris, M. Stocker, J. D’Souza, K.E. Farfar, L. Vogt, M. Prinz, V. Wiens and M.Y. Jaradeh, Improving access to scientific literature with knowledge graphs, Bibliothek Forschung und Praxis 44: (3) ((2020) ), 516–529. doi:10.1515/bfp-2020-2042. |
[3] | Cochrane handbook for systematic reviews of interventions: Extracting data from reports. https://training.cochrane.org/handbook/current/chapter-05#section-5-5 (Accessed: 29.11.2022). |
[4] | Functional requirements for bibliographic records: Final report. https://repository.ifla.org/handle/123456789/811 (Accessed: 29.11.2022). |
[5] | D. Hopf, ORKG Comparison – Open access download advantage, 2022. https://orkg.org/comparison/R202380/ (Accessed: 29.11.2022). |
[6] | B. Mons, Which gene did you mean?, BMC Bioinformatics 6: ((2005) ), 142. doi:10.1186/1471-2105-6-142. |
[7] | ORKG Help Center. https://orkg.org/help-center (Accessed: 29.11.2022). |
[8] | ORKG Python library documentation. https://orkg.readthedocs.io/ (Accessed: 29.11.2022). |
[9] | ORKG R package. https://gitlab.com/TIBHannover/orkg/orkg-r/ (Accessed: 29.11.2022). |




