Realizing health data interoperability in low connectivity settings: The case of VODAN-Africa
Abstract
VODAN Africa has produced FAIR data in low resource settings. Federated machine actionable data is available in a triple store for visiting. Interoperability facets (I1–I3) were followed to achieve semantic interoperability. Vertical interoperability was also realized with DHIS2.
1.Context
A challenge in digital health is interoperability as uncoordinated and fragmented digital health initiatives lead to lack of sustainability and re-usability of digital health solutions. Solutions that create data of unknown provenance and merit prevent interoperability and reuse. An example is data generated from past Ebola epidemics in Liberia, which is difficult to re-utilize since data is no longer findable, accessible, interoperable and reusable (FAIR) [12] which has made the data poorly available to the population that was affected by the disaster [11].
Interoperability is the ability to interact with and exchange data, services, and processes with clearly defined semantics and compliance to regulatory frameworks at the place where data is produced [1] Lack of interoperability is a bottleneck to the digital transformation of healthcare [8].VODAN-Africa has tested the data-interoperability in low/no-connectivity settings and across different jurisdictions. This article discusses the realization of horizontal and vertical interoperability across different settings.
2.Description
2.1.VODAN-Africa
VODAN-Africa (VODAN-A) curates patient data at point of care in clinics according to FAIR Guiding Principles. The community produces federated, machine readable data in residence. Data ownership, Localisation and Regulatory (OLR) [9] compliance elevates federated interoperability for analytical purposes. The VODAN-A Minimum Viable Product (MVP) tried to achieve semantic interoperability. The diversity of formats with which medical information is registered includes: ICD-10, SNOMED, LOINC, RxNorm, and NDC. Those diversities of standards and formats in a medical information registry pose semantic challenges. It is challenging to achieve semantic level interoperability for data exchange from multiple sources. Different terminologies can be used for the same concept, which impedes conducting data analytics in heterogeneous systems. This is a problem because community-defined data exchange formats serve the community itself, but not beyond [3,5].
Fig. 1.
VODAN Africa architecture for clinical data.img.
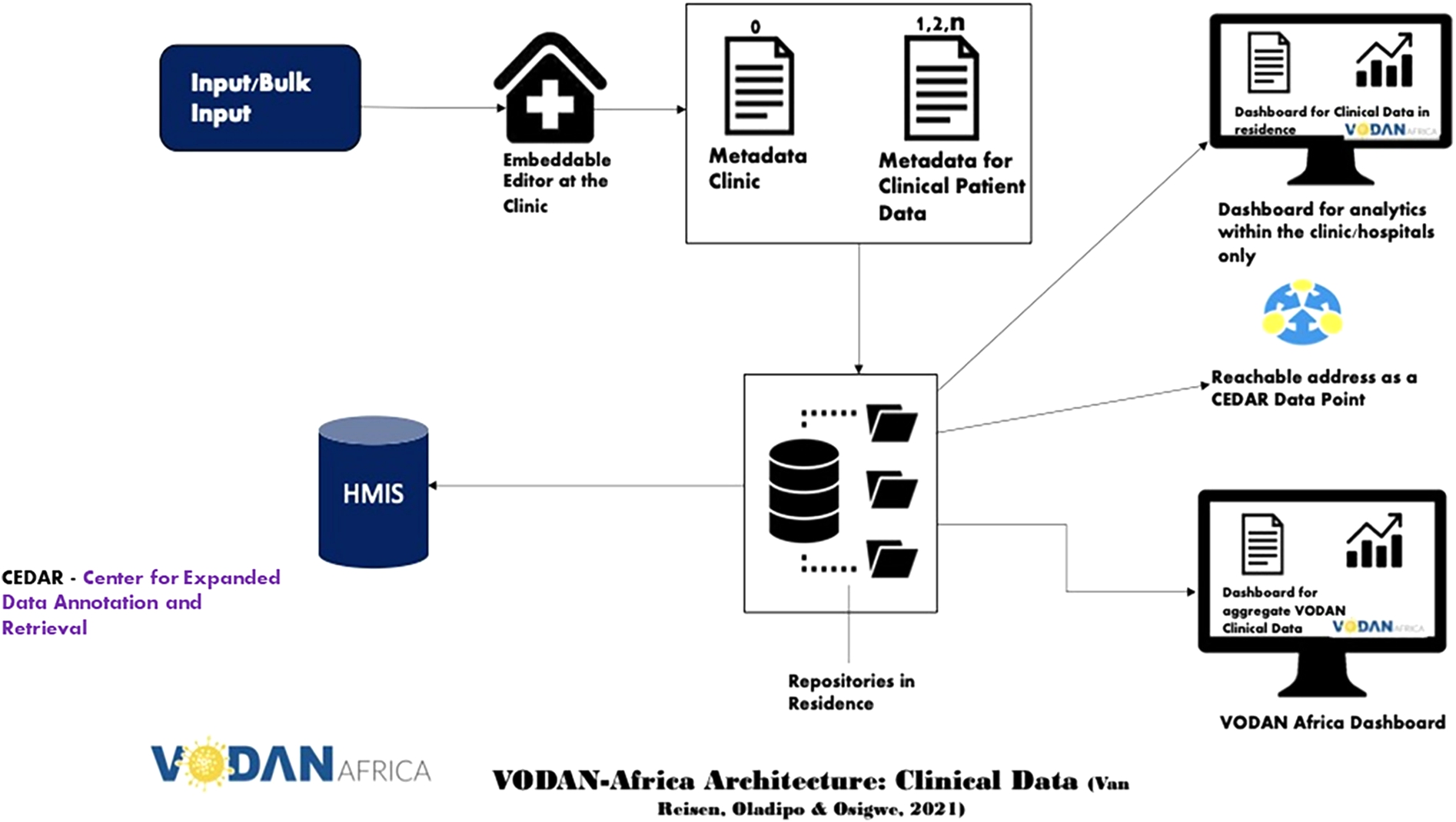
The architecture of VODAN-Africa (Fig. 1) is a blueprint for the MVP deployed in 88 health facilities in 9 African countries [10]. It enables us to FAIRify data and visualize it through internal and external dashboards, to store machine readable data in residence and to make data accessible through a data visiting tool under well-defined conditions. For data FAIRification, two approaches were followed. First was engineering of a tool that makes bulk upload possible to FAIRify existing data. It converts existing comma-separated data into RDF triples. It reverse-models the vocabularies designed in the template and stores the resulting JSON-LD instance simultaneously in MongoDB and RDF triples in AllegroGraph [2]. Second was collection of data at point of service from registries in clinics, which are stored in JSON-LD format in MongoDB and in the form of RDF triples in AllegroGraph. The FAIR-ready data in a triple store can be exposed to data visiting algorithms. Different use-cases can then be envisaged, such as the export of the data to Health Management Information Systems (HMIS), e.g. District Health Information System (DHIS2). To this end, the MVP has used AllegroGraph as a triple store for querying using SPARQL. VODAN-Africa has implemented two different interoperability methodologies: vertical and horizontal interoperability [7].
2.2.Vertical interoperability
The MVP provides the ability to compute and send aggregated monthly reports to the HMIS systems, which replaces manual computation of indicators. VODAN-A provides a solution to enable interoperability between a point of service application, the MVP and health management information systems. The MVP prepares the statistics based on the data element report skeleton templates prepared in JSON format and posts the aggregated indicators into DHIS2. DHIS2 provides a web API to accept data element values from the MVP, see Fig. 2.
Fig. 2.
Interoperability between VODAN – A MVP and DHIS2.
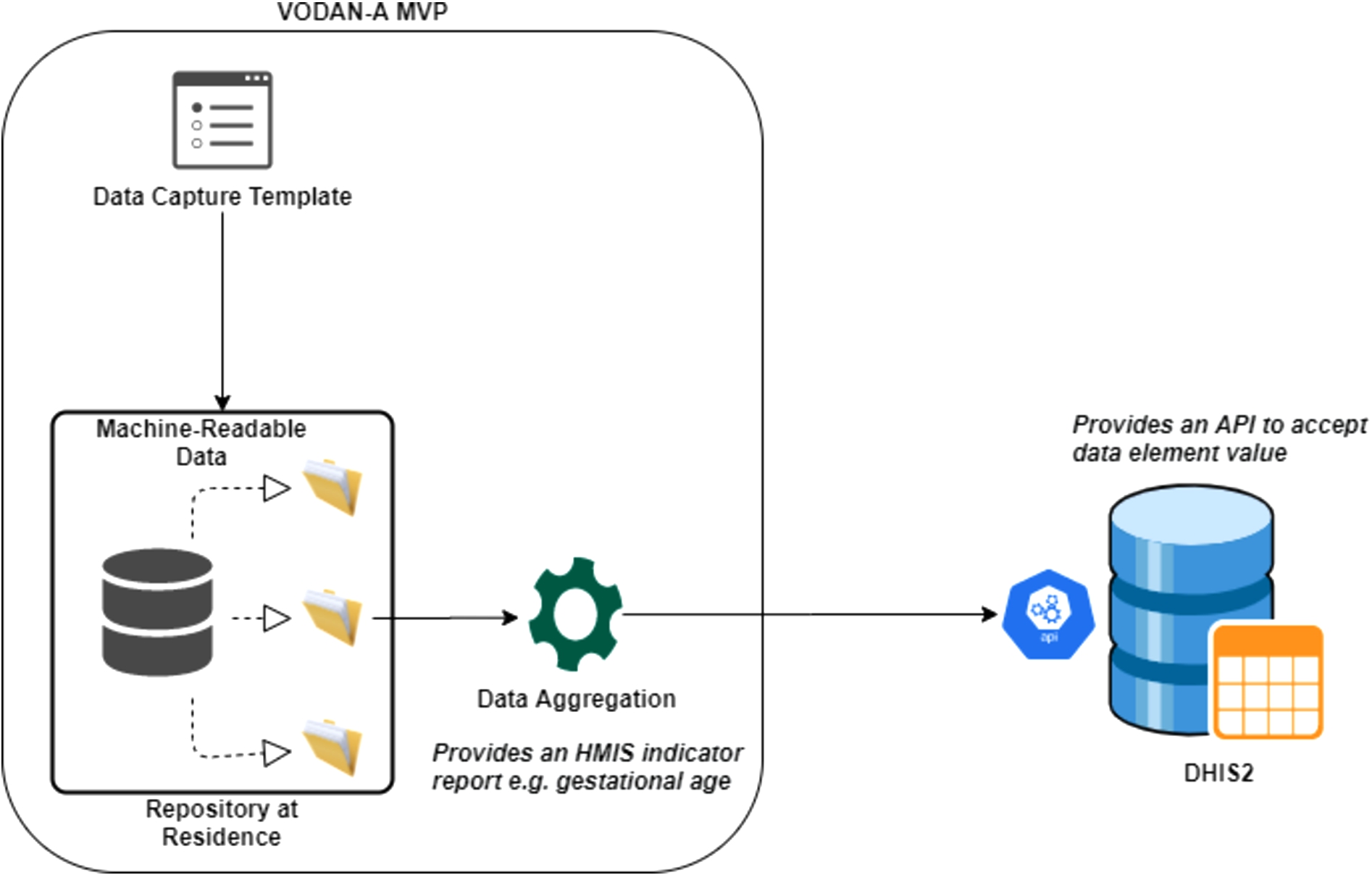
To create semantic interoperability, terminology services are required to perform the mapping of terms used in the MVP as well as DHIS2 to the terms stored in the ontology server. VODAN-A interoperates with DHIS2, using JSON standard reporting templates generated from DHIS2, which increases syntactic interoperability with point-to-point integration. To add flexibility and ensure security of communication during interoperability, having a separate enterprise service bus is envisaged as a micro-service. It provides authentication, mediation, logging and management of interoperability services. Having such a layer can encourage and promote interoperability as other systems will delegate the task of interoperability to the dedicated service.
2.3.Horizontal interoperability
Health clinics in the VODAN-Africa implementation network sign data sharing agreements to enable generation of knowledge at different levels to facilitate data and service interoperation. The community has created shared vocabularies, metadata templates and other digital objects. As the system is interoperable by design, it creates horizontal interoperability. To realize seamless interoperability, VODAN Africa followed guidance of FAIR principal interoperability facets (I1–I3). Thus, different health facilities are connected to create and use common terminologies which are displayed in a shared VODAN-Africa dashboard Fig. 3.
Principle I1: (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation:
Fig. 3.
Interoperability across health facilities in VODAN Africa.
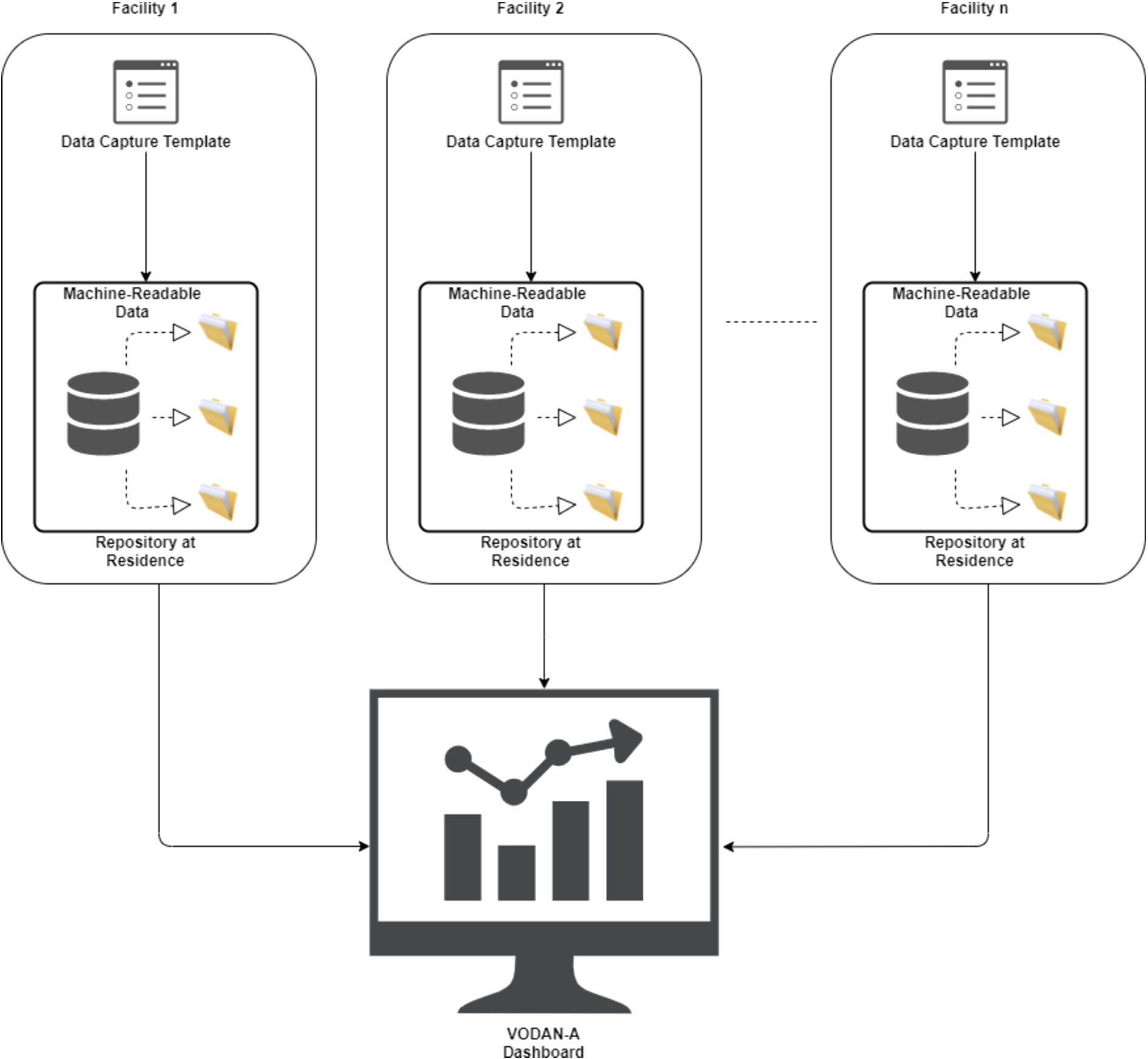
To create a common understanding of digital resources, we used a globally understood language, Simple Knowledge Organization System (SKOS), to produce vocabularies. VODAN-A used CEDAR template110, using SKOS vocabularies loaded into BioPortal. The templates are presented in JSON format, which can be reused and are interoperable for other systems. Data produced using the templates is represented in JSON-LD as well as RDF triples.
Principle I2: (meta)data use vocabularies that follow FAIR principles:
Reusability of data by generic agent, rather than a community-specific agent is the key consideration in this principle [6]. This was made possible through the use of controlled vocabularies. To this end, we created templates using CEDAR, which utilize vocabularies we had uploaded into BioPortal. VODAN-A has attempted to deploy OntoPortal locally, named as VodanaPortal, to allow for storing data in the residence where the data is produced. As the terminology service in CEDAR is developed to work with BioPortal, adjustments were needed in the code-base to localize the data storing. Despite attempts to do so, the result was not satisfactory for technical reasons.
Fig. 4.
VODAN Africa architecture for low resource and offline setting.
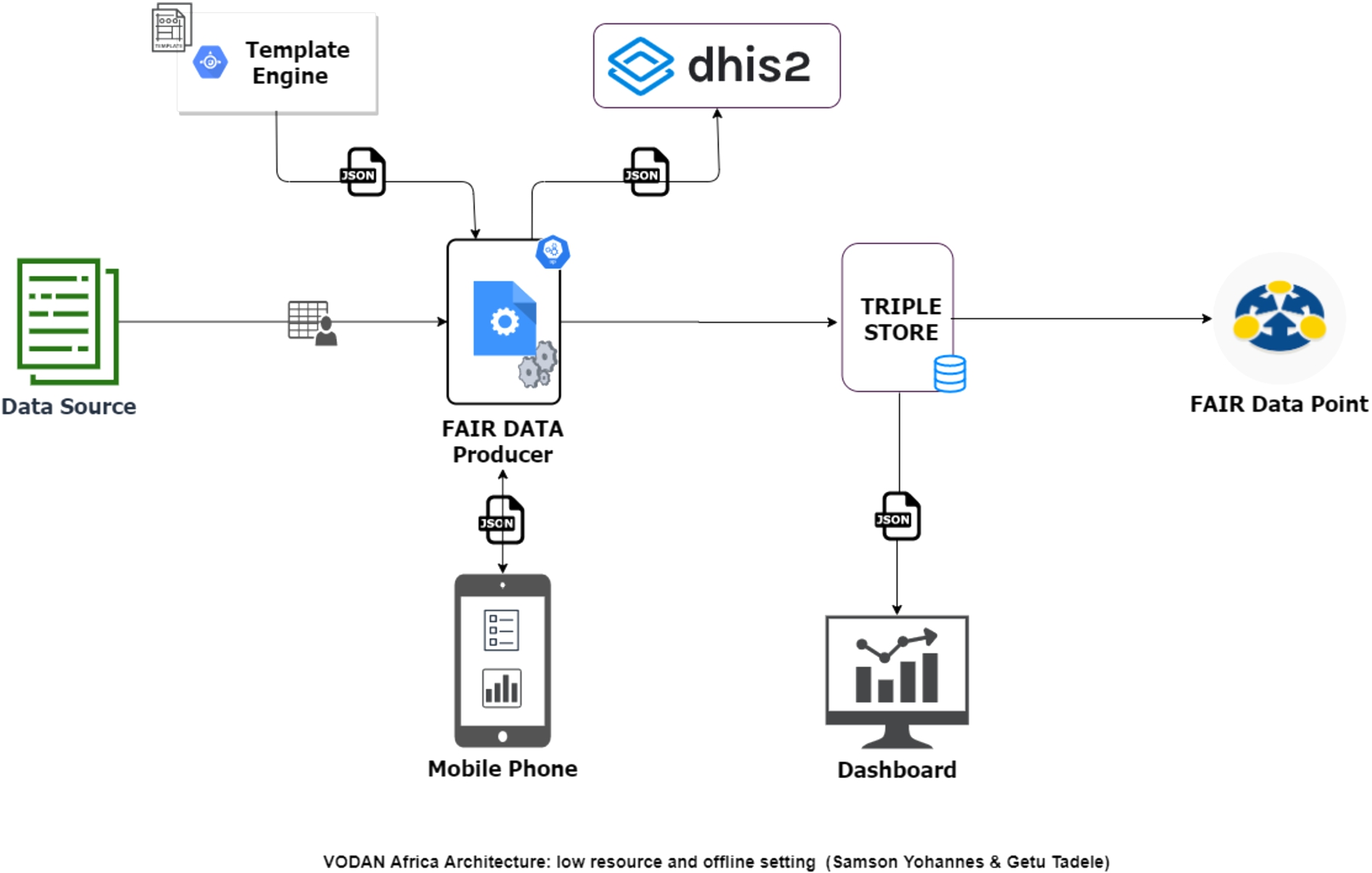
Data production has been challenging in health facilities with low or no connectivity settings because the ontology portals are remote services that prompt Internet connectivity. This has affected the performance of the system in data production for low connectivity and made it impossible when there is no connectivity. Some of the sites changed the templates removing the controlled vocabularies and replacing them with local controlled lists that users can select. Through these solutions, the spirit of the principle is adapted to what is possible within the context of deployment. The issue was then addressed in a new architecture, see Fig. 4, by making controlled vocabularies accessible from a local repository.
Principle I3: (meta)data include qualified references to other (meta)data:
The semantic modelling was conducted by reviewing terminologies from the registry forms. VODANA-GENERAL terms were identified and stored on BioPortal. Each country has also defined its own templates, vocabularies, and types of (meta)data to be collected. The metadata templates which are in JSON format have as fully qualified reference a resolvable URI to the vocabulary classes and acronyms used. The metadata instances created by filling the templates are represented in JSON-LD. Even metadata information for the terms used is stored using a unique IRI that can help in interpretation, query and visualization. This facilitates both human and machine agents to interpret and understand concepts in the same way.
To address issues of performance for low/no connectivity settings, a lightweight application was developed, using a new architecture Fig. 4, which can produce FAIR data using vocabularies that is accessible offline. Currently, it is being tested in health clinics in Tigray, Ethiopia, where there is a communication blackout [4]. To ensure principle I2, the templates are designed in CEDAR using vocabularies from BioPortal. The template JSON and the vocabularies in CSV format are downloaded from CEDAR and Bioportal, respectively, then loaded to the light MVP. The light MVP renders the templates parsing the JSON and loading the vocabularies offline. During data entry, the system gives an autocomplete feature utilizing the controlled vocabularies and keeps the IRIs of the terms. It produces and displays the resulting JSON-LD and an RDF and offline first manner. The system also stores the RDF triples in AllegroGraph. Statistics are then calculated and loaded into the internal dashboard and the external dashboard. This creates potential for the development of further use-cases through data-visiting that includes low/no connectivity settings [12].
3.Conclusion
VODAN-Africa is a FAIR-implementation realizing data-interoperability by starting FAIRification and exposing FAIR-ready data for visiting algorithms. The FAIR Guiding Principles are technology agnostic and provide freedom to operate. An MVP was created to generate FAIR-compliant medical data, stored in a federated manner on AllegroGraph, which made the RDF triples available for visiting, using SPARQL-queries. Use of controlled vocabularies for low/no connectivity settings was tested and found to be feasible. Vertical interoperability has also been tested with another platform, DHIS2, and horizontal interoperability was tested with two community dashboards. The test was successful; it included different settings, with low/no connectivity settings. Ensuring that such data is curated as FAIR and Federated, AI-Ready data would accelerate the availability of inclusive, quality data-pipelines that enhance the application of machine learning and future AI to discover meaningful patterns in epidemic outbreaks.
Acknowledgements
We gratefully acknowledge Invest International and Philips Foundation for the support to VODAN-Africa, and Go FAIR Foundation for the partnership.
Funding
The Invest International and Philips Foundation.
Conflict of interest
The authors have no conflict of interest to report.
References
[1] | A. Aerts and D. Bogdan-Martin, Leveraging data and AI to deliver on the promise of digital health, International Journal of Medical Informatics 150: ((2021) ), 104456. doi:10.1016/j.ijmedinf.2021.104456. |
[2] | Allelograph, Available from: https://allegrograph.com/ (accessed: 04.11.2022). |
[3] | FIP Wizard, Available from: https://fip-wizard.ds-wizard.org/ (accessed: 03.12.2022). |
[4] | H. Gesesew, K. Berhane, E.S. Siraj, D. Siraj, M. Gebregziabher, Y.G. Gebre, S.A. Gebreslassie, F. Amdes, A.G. Tesema, A. Siraj and M. Aregawi, The impact of war on the health system of the Tigray region in Ethiopia: An assessment, BMJ Global Health 6: (11) ((2021) ), e007328. |
[5] | Interoperability standards in digital health – A white paper from the medical technology industry, Available from: https://www.medtecheurope.org/resource-library/interoperability-standards-in-digital-health-a-white-paper-from-the-medical-technology-industry/ (accessed: 08.10.2022). |
[6] | A. Jacobsen, R. de Miranda Azevedo, N. Juty, D. Batista, S. Coles, R. Cornet et al., FAIR principles: Interpretations and implementation considerations, Data Intelligence 2: ((2020) ), 10–29. doi:10.1162/dint_r_00024. |
[7] | W. Kerber and H. Schweitzer, Interoperability in the Digital Economy, JIPITEC 8: ((2017) ), 39 para 1. |
[8] | G. Mehl, Ö. Tunçalp, N. Ratanaprayul, T. Tamrat, M. Barreix, D. Lowrance, K. Bartolomeos, L. Say, N. Kostanjsek, R. Jakob and J. Grove, WHO SMART guidelines: Optimising country-level use of guideline recommendations in the digital age, The Lancet Digital Health 3: (4) ((2021) ), e213-6. doi:10.1016/S2589-7500(21)00038-8. |
[9] | M. Van Reisen et al., Connecting health data across jurisdictions through FAIR data-visiting: Ownership, localisation and regulatory compliance (OLR), FAIR Connect (forthcoming). |
[10] | M. Van Reisen, F. Oladipo, M. Mpezamihigo, R. Plug, M. Basajja, A. Aktau, P.H. Purnama Jati, R. Nalugala, S. Folorunso, Y.S. Amare, I. Abdulahi, O.O. Afolabi, E. Mwesigwa, G.T. Taye, A. Kawu, M. Ghardallou, Y. Liang, O. Osigwe, A.A. Medhanyie and M. Mawere, Incomplete COVID-19 data: The curation of medical health data by the virus outbreak data network-Africa, Data Intelligence 4: (4) ((2022) ). doi:10.1162/dint_a_00166. |
[11] | M. Van Reisen, F. Oladipo, M. Stokmans, M. Mpezamihgo, S. Folorunso, E. Schultes, M. Basajja, A. Aktau, S.Y. Amare, G.T. Taye, P.H. Purnama Jati, K. Chindoza, M. Wirtz, M. Ghardallou, G. Stam, W. Ayele, R. Nalugala, I. Abdullahi, O. Osigwe et al., Design of a FAIR digital data health infrastructure in Africa for COVID-19 reporting and research, Advanced Genetics 2: (2) ((2021) ), e10050. doi:10.1002/ggn2.10050. |
[12] | M. Wilkinson, M. Dumontier, I. Aalbersberg, G. Appleton, M. Axton, A. Baak et al., The FAIR guiding principles for scientific data management and stewardship, Sci. Data 3: (1) ((2016) ), 1–9. doi:10.1038/sdata.2016.18. |



