Identifying PIDs playing FAIR
Abstract
This is an extended, revised version of Philipson (2017). Findability and interoperability of some PIDs, Persistent Identifers, and their compliance with the FAIR data principles are explored, where ARKs, Archival Reource Keys, were added in this version. It is suggested that the wide distribution and findability (e.g. by simple ‘googling’) on the internet may be as important for the usefulness of PIDs as the resolvability of PID URIs – Uniform Resource Identifiers. This version also includes new reasoning about why sometimes PIDs such as DOIs, Digital Object Identifiers, are not used in citations. The prevalence of phenomena such as link rot implies that URIs cannot always be trusted to be persistently resolvable. By contrast, the well distributed, but seldom directly resolvable ISBN, International Standard Book Number, has proved remarkably resilient, with far-reaching persistence, inherent structural meaning and good validatability, through fixed string-length, pattern-recognition, restricted character set and check digit. Examples of regular expressions used for validation of PIDs are supplied or referenced. The suggestion to add context and meaning to PIDs, making them “identify themselves”, through namespace prefixes and object types is more elaborate in this version. Meaning can also be inherent through structural elements, such as well defined, restricted string patterns, that at the same time make PIDs more “validatable”. Concluding this version is a generic, refined model for a PID with these properties, in which namespaces are instrumental as custodians, meaning-givers and validation schema providers. A draft example of a Schematron schema for validation of “new” PIDs in accordance with the proposed model is provided.
1.Introduction: Identifiers in science
Identifiers in science may refer to digital or physical objects, or concepts. PIDs such as ORCIDs (Open Researcher and Contributor IDs) [24] may refer to persons, or, like the recently launched ROR (Research Organization Registry) [39] identifiers, to research organizations. This paper will focus on PIDs for research outputs, ‘things’ such as articles, datasets, samples, concepts etc. But, as suggested in Section 7, ORCIDs or RORs may be an optional part of a modular, integrated identifier for research outputs. PIDs may be general or domain-specific. Among the more prevalent general PID-types are ARK, DOI, Handle and UUID (Universally Unique Identifier). There are also old, bibliographic identifiers like ISBN. Created in the 1960’s and 1970’s of the print era, how come they survived into this digital age? Some reasons might be: they are well distributed across the internet and widely used by stakeholders (libraries, publishers, readers). They have a semantic structure, identifying well-defined objects, and a fairly precise validation mechanism through fixed string-lengths, limited character-set and check digits. Some of these properties are shared by ARKs, DOIs, Handles and UUIDs, or other more domain specific identifiers used for scholarly data, but seldom all of them simultaneously. The focus here is on findability and ‘validatability’ of PIDs of different types.
2.Identifiers – why do we need them?
The general purpose of identifiers is to serve as references to the objects that they are supposed to identify. Preferably they should indicate, in and by themselves, what types of objects they are meant to identify. Far from all PIDs do that. It is often left to the names of things to provide context and meaning. Context may be added by means of location within an hierarchical system, e.g. as in Linnéan taxonomy, where scientific names situate a species within a genus, sometimes also containing the provenance of that name, serving to disambiguate between names of species belonging to widely different genera, e.g. Asterina gibbosa Gaillard 1897 – a fungus, and Asterina gibbosa (Pennant, 1777) – an echinoderm, a starfish. It also happens that ‘things’, objects are renamed later, as with the preceding fungus species now having the accepted scientific name Asterolibertia gibbosa (Gaillard) Hansf. 1949, or are assigned an identifier: urn:lsid:catalogueoflife.org:taxon:02af8238-ac8f-11e3-805d-020044200006:col20150401 [2]. However, even if a PID may well serve the need for disambiguation by uniquely identifying an object, it may still be no better – sometimes perhaps even worse – at giving access to said object, or at least to a page with metadata about it. The identifier assigned above is neither directly resolvable nor ‘googlable’, while the scientific name is at least easily findable via a search engine. The PID type here, a LSID (Life Science Identifier), represented as a uniform resource name (URN), has also been criticized for not being resolvable as a HTTP URI and violating the web architecture [46]. The initial objectives of LSIDs may be well worth pursuing, notably to specify a “method for discovering multiple locations for data-retrieval ... and ... to discover multiple independent sources of metadata for any identified thing” [46], but judging from individual instances these objectives seem not to be fully achieved yet.
While scientific names are often useful for describing objects, they have other drawbacks compared to PIDs, some of which were identified by [36]. For example, homonymy and disambiguation should be no problem for ‘globally unique identifiers’ [23]. And while concatenations or abbreviations may be problematic in the use of names for identification, string-length and pattern restrictions are useful for validation of identifiers. Missing or added characters, and some types of misspellings are easier to detect and validate in standardized identifiers of fixed string-length or well-defined character patterns. Inconsistent encoding should also not be a problem in PIDs with restricted character sets. However, these desired properties of some identifiers may conflict with the interest in having also transparent, meaningful PIDs that at least in part “speak for themselves”.
3.FAIR principles
The FAIR guiding principles aim “to make (meta)data Findable, Accessible, Interoperable, and Re-usable” [15]. As such they concern also PIDs, as is seen from some of the principles (Fig. 1).
The FAIR principles clearly need interpretation to become fully operational, and such work is also well in progress [9,11,48]. Further explications of some of the principles are also available in [16]. Figuring prominently in the explications of all these principles, particularly interoperability, is the requirement that metadata should be machine readable “a conditio sine qua non for FAIRness” [17]. Providing machine-readable metadata is also used by fairmetrics.org as a measure of Findability [13].
However, the FAIR principles do not say anything explicitly about validation. Here we argue by contrast, that particularly for Interoperability and Re-usability it is crucial that metadata can be properly validated as compliant with an accepted metadata standard. It has been remarked that this is already implied by the FAIR principle R1.3 above, but even so, only indirectly and in an ambiguous way. There are several cases where general data repositories, professing to be FAIR and to comply with accepted metadata standards both for their default output and export formats, nevertheless fail to validate against schemas of these same standards [37]. Fairmetrics.org [48] explicates R1.3, as measuring a “Certification from a recognized body, of the resource meeting community standards”, by means of a valid electronic signature, such as a verisign signature [12]. One might ask, then, whether general data repositories such as Harvard’s Dataverse,11 Figshare22 or Zenodo,33 qualify as “recognized bodies” in this respect, all being part of the test reported in “Evaluation_Of_Metrics/Supplementary Information_ FM Evaluation Results.pdf” [48], but none of which could be evaluated on R1.3. This comes as no surprise, since there is already a comment on R1.3 saying that “Such certification services may not exist, but this principle serves to encourage the community to create both the standard(s) and the verification services for those standards” [12]. True, in the rationale for FM_R1.3 there is mention of validation: “... As such, data should be (individually) certified as being compliant, likely through some automated process (e.g. submitting the data to the community’s online validation service)” [12]. But it remains unclear if the “community” here refers to a general metadata standard or a repository using its own standard and validation service? Some output metadata files from repositories even lack a schemaLocation reference, making it difficult to validate them, or, the schemaLocation given might be erroneous, as observed in one case [37]. “Community” independent validation is needed to test if repositories are keeping their promises of compliance with metadata standards. This concerns metadata in general, but naturally includes also identifiers. We must be sure that they are of the type or format they claim to be, even if they cannot be resolved to a dedicated landing page with metadata. Failed validation, caused by simple typos or wrong namespace, may help explain why an identifier or URI does not resolve as expected. Validation is also important for the possibility to export metadata to another format, thereby promoting the re-use of data, without exporting also potential errors. Resistance to transcription errors, e.g. by means of a restricted character set, using base32 for encoding, and fixed string-length (suffix has 2 times 4 characters, separated by a hyphen), has been promoted as an advantage of so-called “cool DOIs” [14]. These are precisely the kind of properties that make PIDs eminently “validatable”, and thereby machine-actionable, in the sense of making it possible for a machine to decide of what type a given PID is (cool DOI, ISBN, ISSN...), or – as is seldom the case in my experince – if it already comes “typed”, whether it is true to its given type. A real use-case at the National Library of Sweden proved this information to be crucial in order to export error-free metadata to a new environment, to make it searchable, findable and accessible through the library catalog, thus promoting a wider distribution and use of said PID. Although transformation or harvesting of metadata might be possible even without validation, trust in the results and quality as well as the eventual findability of the data (and so again the re-usability) might be seriously affected. The use of standardized, widely distributed PIDs are likely to enhance the chances of finding metadata for a resource, even when the PID-URI fails resolution.
4.Resolvable or findable?
The current FAIR principles of Accessibility, particularly A1 above, imply that identifiers should be resolvable, seemingly disregarding the general awareness of phenomena like ‘link rot’ and ‘reference rot’ [18,26,27,44]. A 2013 study in BMC Bioinformatics analyzed nearly 15,000 links in abstracts from Thomson Reuters’ Web of Science citation index and found that the median lifespan of web pages was 9.3 years, and just 62% were archived [25]. This happens although there is an understanding that “[u]nique identifiers, and metadata describing the data, and its disposition, should persist – even beyond the lifespan of the data they describe” [7]. A recent study of some 40 research data repositories found that only one of these (3%) was compliant with the FAIR principle of Accessibility requiring “a clear policy statement (or various examples of data this has actually happened to) indicating that metadata is still available even if the data is removed” [11]. The argument here is not that resolvable, persistent URIs should be avoided as identifiers, but they may not be sufficient to guarantee persistence. As has been eloquently remarked, “persistent URIs must be used to be persistent” [41] (my emphasis). Resolvable URIs as PIDs work by decoupling the location and the identification functions of URIs.
The custodian of a web resource maintains the correspondence between the identifying URI and the locating URI in the resolver’s look-up table as the resource’s location changes over time. ... The solution comes at a price because it requires operating a resolver infrastructure and maintaining the look-up table that powers it [41].
This is true of ARKs, DOIs, as well as Handles, PURLs (Persistent Uniform Resource Locators) and URNs. There are in fact numerous cases when the lookup-table is not maintained and updated as required. A case in point are two PURLs from the FAIR metrics found in [48], https://purl.org/fair-metrics/FM_F2 and https://purl.org/fair-metrics/FM_R1.3, both non-resolving currently (2019-07-30). That is why it may be wise not to rely on a single ‘custodian’ for the resolution of identifiers and access to associated metadata. Note that we are not talking here about simply having more than one proxy server acting as resolvers of the same PIDs. We already have that; provided the lookup-table is managed properly, these three different DOI-URIs from different proxy-servers all resolve to the same landing-page location: https://doi.org/10.1007/978-3-319-53637-8_11, https://hdl.handle.net/10.1007/978-3-319-53637-8_11 and https://identifiers.org/doi:10.1007/978-3-319-53637-8_11. ARKs (Archival Resource Keys) are resolved by identifiers.org and n2t.net, as well as by their “mother institutions”, e.g. n2t.net/ark:/67531/metapth346793/,44 identifiers.org/ark:/67531/metapth346793/55 and digital.library.unt.edu/ark:/67531/metapth346793/66 resolve the same content. It is rather the distribution and use of identifiers – whether resolvable or not – that is important here. It seems not even the authors of [41] are true to their own principles, since three of their references that actually have DOIs are cited without them: [10,28,50]. So, despite having DOIs or other PIDs assigned, documents are often not cited by those PIDs. One possible reason might be that the PID is not clearly displayed in the landing page with metadata, or in the document itself. In the case of [10] above it takes an extra click on a link ‘Cite as’ to actually have the DOI displayed. But that should hardly be the reason why it was not used for citation in [41], since the citation there is actually much more verbose and complex, than it would have been to just copy-paste from the ‘Cite as’ page above. Another, slightly ironic case concerns [47], the founding paper of the FAIR principles, where you either have to download the citation with the DOI from the landing page, where it is not displayed, or find it at the bottom of each page in the actual paper, but not prominently marked. This may partly explain why a recent paper on software sustainability and reproducibility, while arguing that one of the ways to make software more reproducible is to “use a persistent identifier such as a Digital Object Identifier (DOI) to help find and cite code” [6], failed itself to use the DOI when citing [47]. Another reason, gathered from one of the authors by personal communication, but which I believe could be generalized, is that inclusion of the DOI (or other PID) was not part of the citation style of the publisher. In fact, it sometimes happens that publishers impose their own citation formats or standards, excluding the use of PIDs. Again, PIDs must be used and cited to persist. Citations promote wide distribution of PIDs, provided these are validated as correct, so as not to export errors and ‘non-resolution’ as a result.
Again, going back to the question of resolvability, the relationship between identifiers such as DOIs and URIs is not always straightforward, and sometimes involves a chain of redirects (‘303s’), before reaching a destination holding also the appropriate metadata [42,43]. Faced with a non-resolving PID-URI an alternative might be to try the identifiers.org SPARQL endpoint [49]. But it only works if the potential corresponding URIs have been assigned the property owl:sameAs just as the submitted subject URI.
Assuming we have finally found a single seemingly reliable custodian for our PIDs and URIs, promising 24/7 resolution and top quality metadata, should we rest content with that? In law and journalism it is desirable not to judge by the testimony of only one witness or source. The evidence of at least two, mutually independent sources is generally preferred. Multiple resolution of any PID by several different proxy servers, as we already know, still means single custodianship of that lookup-table that has to be managed and updated in order for the PID to resolve as expected. Clark describes it as representing a stage in the evolution of PIDs, that will eventually be surpassed by a more mature age when we supply also data types to come with the PIDs, in order to make them more machine actionable [4]. But we want more than that. We want backup for custodians. We need trustworthy, independent witnesses from different loci in space-time to provide multiple access to, or identification (findability) of resources through PIDs. Thus, we accept “that an object may have multiple PIDs”. Ideally these multiple PIDs should get to “know about” each other as a way towards interoperability [4]. This can be achieved already, e.g. by means of Linked Open Data (LOD), sameAs-relationships and tools provided by n2t.net, unpaywall.org and the identifiers.org SPARQL endpoint referred to above. Multiple identifiers from different namespaces for the same object may even be desirable in order to ensure interoperability in different environments [35]. It is also in line with the principle of the semantic web known as the NUNA, Non-Unique Naming Assumption, implying that “things described in RDF data can have more than one name” and any object may be identified by more than one URI, serving in RDF as ‘names’ of things [5].
However, this does not imply that any identifier, any PID is as good as the other. In fact, there are significant differences in quality between identifiers, particularly in terms of ‘validatability’ and ‘meaningfulness’. We are getting there a bit later.
But first, having referred to linked data and sameAs-relationships as a possible solution to achieving interoperability, what about long-term sustainability? Are LOD, relying heavily on opaque URIs, fit for survival? Archival information packages for long-term preservation need to be independently understandable [3], carrying meaning within themselves, while external links may no longer be resolvable. Thus, opaque URI strings lacking an inherently meaningful structure will give little or no clue about content or provenance, unless they can import some meaning from outside, through resolution or sameAs links.
5.Which identifiers are FAIR enough?
Just how “persistent” are PIDs really? Even if not always resolvable, are they in general still ‘findable’, well distributed over the internet in time and space? Are they ‘validatable’ (e.g. through fixed string-length, pattern-recognition, restricted character set, built-in checkdigit, built-in type)? Are they FAIR?
Findability: Beginning with the F for findability, for comparison we go back in time to ‘old-fashioned’ ISBNs, International Standard Book Numbers. Publicly declaring what type of objects they are meant to identify, ISBNs are rarely directly resolvable. But they apparently fulfill all the FAIR requirements F1–F4, in particular F3, since they are “registered or indexed” most often in more than one “searchable resource”, e.g. in library catalogs, book-sellers online etc. Thus, a wide distribution gives them good findability also in terms of precision hits through a search-engine, as seen by simple ‘googling’, with good survival rate, longer than the median age of web-pages 9.3 years. For example, look at ISBN 0-14-029161-X: The Diversity of Life/Edward O. Wilson (2001). Simple googling of 014029161X, unprefixed and without hyphens results in 57/57 precision hits (date: 2017-01-30). A search by the query ‘014029161X’, with the same unprefixed ISBN without hyphens, in the probably single most comprehensive library union catalog Karlsruhe Virtual Catalog – KVK worldwide,77 yields 123/123 precision hits.
We try ‘googling’ an older, presumably less well-known example: ISBN:2130381030. L’Identité : séminaire interdisciplinaire dirigé par Claude Lévi-Strauss, 1974–1975 (Paris: PUF, 1983). Without prefix (2130381030) the precision is between 14/39 and 22/50; with prefix (ISBN2130381030) it reaches as high as 17/18 (date: 2017-01-30).
Accessibility: Data and (digital) objects are accessible only in so far as identifiers are findable or resolvable to landing pages with either direct availability of resources, or sufficient metadata to direct the user to such an access point. In this respect DOIs are often, but not always, as good as or sometimes better than ISBNs (for obvious reasons regarding print only material), while UUIDs retrieved as results of a search in the Global Names Architecture described below are all but useless. Regarding FAIR principle A2, however, ISBNs with their demonstrated survival rate and ability to provide metadata (while the object they identify may no longer be available) should compare well with ARKs, DOIs or UUIDs.
Interoperability and Re-usability are both intimately associated with correctness, which can be helped by ‘validatability’, as argued above. We will look more into detail at the performance of different PIDs regarding this below.
Archival Resource Key (ARK) Identifiers: ARKs have a well defined syntax [1]: [http://NMA/]ark:/NAAN/Name[Qualifier], where NMA is a (changeable) Name Mapping Authority, a “host” or proxy resolving agent. This is not part of an ARK’s core identity, as shown by the encompassing brackets and by the example ARKs below resolving from two or more different NMAs, with the NMAs spelled out in the URIs to make this clear. The NAAN is the Name Assigning Authority Number, corresponding to the prefix starting with ‘10.dddd’ in a DOI, and serving as a namespace for the following /Name. The NMA-supported [Qualifier] is not further defined in [1], but an example is given by the suffix s3/f8.05v.tiff, including also a file extension as we can see. As examples below, none of them having a qualifier, we find ARKs giving direct access to digital fulltext of Buffon’s Histoire naturelle at the BnF and a 20th Century Guide for mixing fancy drinks at the Internet Archive. The third case, shows a resource with a special feature of ARKs, their possible inflections, here represented by ‘??’ at the end of the URI, giving metadata for a photo of the Dallas Police Department from 1963, and the name and location of the collection holding it. This inflection property of ARKs could be a response to the FAIR principle of accessibility (A2), requiring that “metadata are accessible, even when the data are no longer available” [15], i.e. unavailable resources should at least leave a gravestone with metadata behind. Such direct retrieval of “metadata tombstones” would be an advantage of ARKs over DOIs, where one must first find out which of 10 different registrant agents produced the DOI, and then use their search-API to find the metadata of an unresolvable DOI. There is presently no direct link from the error message received to the responsible registrant agent. However, this inflection option for ARKs does not seem to be generally implemented yet. In the BnF-case below it appears to be simply ignored; adding /? or /?? to the ARK-URI in question does not change anything (irrespective of NMA used for resolution). In the second example, the response to “https://identifiers.org/ark:/13960/t6c25cm5g/?” is: “# inflections under construction # reference https://n2t.net/e/n2t_apidoc.html”. Another interesting case is that when the resolving agent, the NMA is changed from texashistory.unt.edu to either identifiers.org or n2t.net, the very same ARK does not resolve to the exact same landing page, the same location, as shown from the last three examples below.
What about the “validatability” of ARKs, then? As exemplified above, ARKs match the regular expression ^ark:\/[0-9]{5}\/\S+$. (The regex for ARKs registered in MIRIAM, MIR:00000592,1111 is more permissive for the NAAN part, but more restrictive for the following parts, than even the ARK specification [29], by excluding ‘#’ from the allowed character set.) However, following the NAAN/, there is no specific pattern or definite string-length of an ARK. The only restriction on the Name and Qualifier parts “as strings of visible ASCII characters” is that they “should be less than 128 bytes in length” using “letters, digits, or any of these six [sic!] characters: = # * + @ _ $”, allowing also four more characters with reserved meaning: % - . / [29]. This is not sufficient to discriminate properly between real and fake ARKs. While not required by the ARK specification, however, the first two examples above as well as most ARKs in the world (and some Handles), are generated using the NOID Check Digit Algorithm (NCDA) [31], which was created in conjunction with ARKs. According to the creators, despite variable string-length, NOIDs have stronger transcription error detection than ISBN- or ISSN strings.This may be true, but in order to apply the NCDA it takes some initial checking, first that the string-length of the NOID substring is less than R, the number of digits and characters in an ordered set of ‘xdigits’, i.e. the set of permitted characters. Secondly, the NOID substring must be checked to be “well-formed, that is, that all non-xdigit characters ... are exactly where expected; if not, the substring is not well-formed and the computation aborts” [31]. For these steps to be automated, it requires for each substring a machine-readable definition of the permitted set of xdigits, the number R and the structural location of allowed non-xdigit constant characters such as e.g. ‘/’. This is necessary already to detect possible non-xdigits that are nowhere allowed, which are otherwise assigned the same ordinal value 0, as ‘0’ or in our case ‘/’, and so risk go undetected in the rest of the computation, if they happen to replace one of those. But this means that at least “locally” some of the requisites for more efficient validation by means of a regular expression are already at hand: if not fixed string-length, at least a limited range, a restricted character set and a structural element, defining the allowed placement of certain characters.
Thus, ARKs have the potential to offer stricter constraints for validation locally, than those represented by the regular expression above. But for this, it would be desirable to have a kind of lookup service for the NAANS, a directory, which for each NAAN – a little like MIRIAM – informed about the permitted character sets of its substrings, string-length limits, possible structure and regular expressions for validation. This could balance out the lack of semantic content in ARKs, that might otherwise limit their use and, possibly, persistence.
The Findability by simple ‘googling’ and current Accessibility of the example ARKs above presently (July 2019) still seems quite good. At least the first of these examples seems to be well distributed, producing an impressive precision score of 27/27 by simple googling of “12148/bpt6k97497t” (each hit actually containing a reference to the same document by Buffon in the Gallica collection). The second example apparently has a narrower distribution, but the few items found still display good precision, 4/4. The third example, without inflection, has been used extensively as a paradigmatic case, so should perhaps be considered outside competition here, but anyway also shows good precision. The long-term sustainability and persistence of ARKs, that is, the future preservation of their connection with the objects they are supposed to identify may be difficult to predict, but given their present apparent “findability” and at least potential “validatability”, they might be able to compete with ISBNs in the future.
DOI: DOIs can look almost like anything. Here are some real cases, all at the time of writing resolvable and with multiple Findability also by simple googling, some of them identifying ‘old’ documents, although they got their DOIs assigned fairly recently. One from 1977 (doi: https://doi.org/10.1177/030631277700700112), produces an impressive precision score of 68/68 (date: 2018-11-07), mostly due to it quite high citation rate, yielding hits for all the citing sources.
Now, following are two DOIs from Wiley Online Library 1996 and Springer 2001 that still do not seem to resolve properly (tested 2017-01-31, 2018-11-11, 2019-07-30):
However, these DOIs, again from Wiley Online (1996, 1998) that were earlier unresolvable (at 2017-01-31), are proof that some PIDs might (re)gain resolvability later:
Obviously, all these DOIs, whether resolvable or not, vary substantially in string-length, from just 17 to over 60 characters, some involving abbreviations of journals or organisations, one an ISBN, and some containing characters in need of special XML-encoding, different from URI. Note that although the two last items in the first group are from the same journal, Scientometrics, they are quite different in structure. Anyway, all the above DOI examples are valid in accordance with the best we can offer as a regular expression restriction, with only partial pattern recognition: ^10\.[0-9]{4,}\/\S+$ meaning that any valid DOI must start by ‘10.’ followed by a minimum of 4 digits, before the slash ‘/’ and then a suffix of any length or characters, but no spaces in between.
But then, according to the same partial restriction, defined by the regex above, this entirely fake DOI is equally valid:
10.99999999/xxxxxxxx/x(y)x\:-{=?%%@@@@@
To be sure, there are other regular expression restrictions suggested for DOIs, those that are even more permissive (as DataCite 4.1, with the pattern value for doiType set to “10\..+/.+” [8], apart from not being PHP or JavaScript compliant, allowing also inline spaces, or the pattern registered for DOIs at identifiers.org as “^(doi\:)?\d{2}\.\d{4}.*$” MIR:00000019,2222 both of which also allow for the fake DOI above as valid, when tested in regex101.com2323). There are other patterns that are more restrictive, but then obviously not catching all the now prevalent and permitted DOIs by one singular regular expression [14 ,19]. Thus, unlike ISBNs, DOIs are difficult to validate properly. Or rather, it is hard to find sufficiently discriminatory criteria to distinguish proper DOIs from fake ones. They have no fixed string-length, and few character set restrictions. All we can have is a partial pattern recognition; the more restrictive the validation rule or regular expression, the more it is likely to leave out extant DOIs.
Handle: The Handle identifier system, of which DOIs are only a special case, seems fairly easy and handy at first glance. Handles come in two different flavors. One is the semantically opaque, which has the structure: Prefix/noid (10079/sqv9sf1), where the NOID-part (for Nice Opaque Identifier [30]) is a short alphanumeric string from the restricted character set “0123456789bcdfghjkmnpqrstvwxz”, with random minting order [22]. The other flavor is the semantically transparent, which could be of three different types: the URL handle: Prefix/local-PID (10079/bibid/123456),2424 the user handle: Prefix/netid/netid (10079/netid/guoxinji),2525 which as demonstrated here seems to be less persistent, as people tend to move, and the simpler group handle: Prefix/group (10079/ISPS).2626 While those of the second flavor might be more instantly “meaningful”, providing context, how are Handles faring regarding Findability and Accessibility? The Findability by googling will, as for other PIDs, largely depend on the use and citation rate of items, while the accessibility again rests largely on the maintenance of the lookup-table by the custodian. Even so, as just demonstrated, Handles may not always resolve to the page expected, especially when used as context dependent identifiers of individuals. In these cases, ORCID IDs should be preferred. What about the Interoperability and Re-usability of Handles then? Those of the NOID type, with a restricted character set, will in principle at least be effectively “validatable”, to the extent that the “namespace” or minting agent restricts the string-length, as e.g. 2077/366872727 – Gothenburg University: 4/5 characters, and 10079/31zcrtn2828 – Yale University: 5/7 characters. Those of the second, “semantic” flavor will apparently prove less “validatable” in the sense that there is no longer any fixed string-length or restricted character set.
UUID: UUIDs v5 were introduced to the field of biodiversity taxonomy in 2015 by the Global Names Architecture – GNA [20] to replace scientific name strings for certain functions, with the arguments that they save space as index keys in databases, and they have a fixed string length (36 characters, including the dashes) while scientific names are of variable length. UUIDs do not suffer, as names sometimes do, from encoding problems that are difficult to detect, and they are more easily distinguishable one from the other than name strings for closely related species variants. Specifically, it is argued that “UUIDs v5 ... can be generated independently by anybody and still be the same to the same name string... Same ID can be generated in any popular language following well-defined algorithm” [20]. By “popular language” here is meant code languages such as Go, Java, PHP, Python, Ruby, as seen from the GitHub link2929 in the source. Note, however, that it is actually the specific name string that is identified here, not the object – neither a specimen of an organism, i.e. the ‘thing itself’, – nor the concept, e.g., a species. Thus, the resulting UUID is completely dependent upon the particular name string (with its encoding), it cannot be used as a bridge between different name forms for the same organism, telling us that they are naming the same object. This is due to the fact that it is “generated by hashing a namespace identifier and name” [45]. As a result, UUIDs generated in this way by the GNA name resolver, are next to useless as instruments of Findability, often yielding 0 hits by simple googling, while a search on the scientific name alone will give plenty of precision hits for the sought after organism, providing rich metadata for the ‘thing itself’. Likewise, the same UUID is seldom or never Accessible, by being resolvable on its own. As an example, consider one of the most well studied organisms of all, the fruitfly Drosophila melanogaster. Using the Global Names Resolver [21] to get a UUID v. 5 for Drosophila melanogaster, <gni-uuid>1bc2f359-47e4-5da6-a748-74676b7c8c5d</gni-uuid>, googling it either unprefixed or prefixed gives a zero result (0 recall, 0 precision, date: 2017-01-30). Trying instead the same UUID in a general search of all databases of NCBI, the US National Center for Biotechnology Information), we get 0 hits (2018-11-15): 1bc2f359-47e4-5da6-a748-74676b7c8c5d.3030 Most notably, we get 0 hits in the NCBI Taxonomy database,3131 that on the face of it would seem to be the most relevant to our search.
By contrast, UUIDs v5 are eminently “validatable”, with a character set restricted to digits and lower case [a-f], and a fixed string length, 36 characters including hyphens, in a recognizable, precise pattern: “8-4-4-4-12”, allowing for validation by a regular expression such as ^[a-f\d]{8}-[a-f\d]{4}-[a-f\d]{4}-[a-f\d]{4}-[a-f\d]{12}?$, or by means of an online validator.3232 On the other hand, since these UUIDs are seldom or ever used for citation, and are not “fed back” to the source databases, it is doubtful whether this “validatability” is also sufficient to make them qualify for Interoperability and Re-usability. They might improve their findability and re-usability through “ping-back” and assign themselves to the records in the biodiversity database sources they were drawn from and further use schema.org markup to get incoming links and a better ranking by search engines.
6.Why context?
Generally speaking, although it is preferable that identifiers be findable and identifiable also in their unprefixed, pure form, typed identifiers give context by means of namespace prefixes of a metadata standard, a vocabulary or ontology. A typed identifier “introduces itself”, telling us what kind of identifier it is, and what type of objects it is used for. Most importantly the namespace tells us what schema(s) or which rules should be used for its validation.
Page [34] claimed that e.g. “dc:title” is adding “unnecessary complexity (why do we need to know that it’s a “dc” title?)” in the JSON expression:

A simple answer is that namespaces are important to retain meaning from context, serving as a key to interpretation for the future. Long-term preservation of archival information packages (AIP), in order to ensure that these will be “independently understandable” [3] for the future should mean in a case like this, that the dc specification and schemas valid at the time be archived together with the records [32], or at least that there is provenance metadata including timestamps and namespace of terms used. Metadatafiles in XML usually have a xsi:schemaLocation indicating which schema to validate against. This information, together with timestamped metadata elements such as ‘dateIssued’ should be sufficient to provide context. For JSON metadata there are name/value pairs such as { "protocol": "doi", ... "createTime": "2017-01-12T10:49:03Z", ...} that could fulfill the same function. And then, context is just as important for validation of records also in the present.
7.A “new” contextual, integrated, validatable PID?
As seen in the case of Handle above, validatability sometimes comes at a cost: transparency lost. Are we forced to make a choice between the two? Can we create identifiers that are both fully validatable and at the same time more meaningful, providing context? So, here we suggest a model for a “new” PID, with a limited character set, at least for the object id part, defined by namespace specifications and schemas.


It is a model of a contextual, validatable identifier, structured into modules (sub-strings) separated by a dot (.). To make it easier to implement, and more generalizable, there are no character set or string-length restrictions for the first two modules, except that they should not contain the dot (.), which is the module separator. Nevertheless, this means already existing namespaces and object types could already be used to create a PID in accordance with this model.
The third module, the objectId (local ID) has a limited character set, selected to escape ambiguous interpretations (excluding the letters ‘l’ and ‘o’, as possible to confuse with numbers) and, to avoid making local uniqueness case-dependent [33], restricted to lower case letter characters and digits. The full stop or dot (.) was chosen as module separator, since it works well in both xml- and http-environments, without encoding, and is not subject to confusion as sometimes hyphens and dashes (en-dash and em-dash) can be. It also works for tokenization of strings. The object type identified in the second module should belong to the initial namespace prefix. Every namespace can have as many object types as needed. Namespace schemas could also define valid data types for their different object types, thus supplying PIDs with data types, in order to make them even more machine actionable [4].
The scalability of this model will mainly depend on the 10-character objectId and the size of the permitted character set. An objectId limited to the proposed character set [a-kmnp-z0-9] will have 3410 permutations within each namespace (and possibly objectType), still better than e.g. a 7 character Handle with NOID.
The objectId module, thus, could be validated separately by a regular expression restricted to ^[a-kmnp-z0-9]{10}$. It may also be part of a more comprehensive validation schema, involving a random or pattern based minting algorithm, preferably including a check digit, with different rules invoked for different namespace contexts, checking also for example the correspondence between namespace (module 1) and objectType (module 2) as in this still crude Schematron schema:
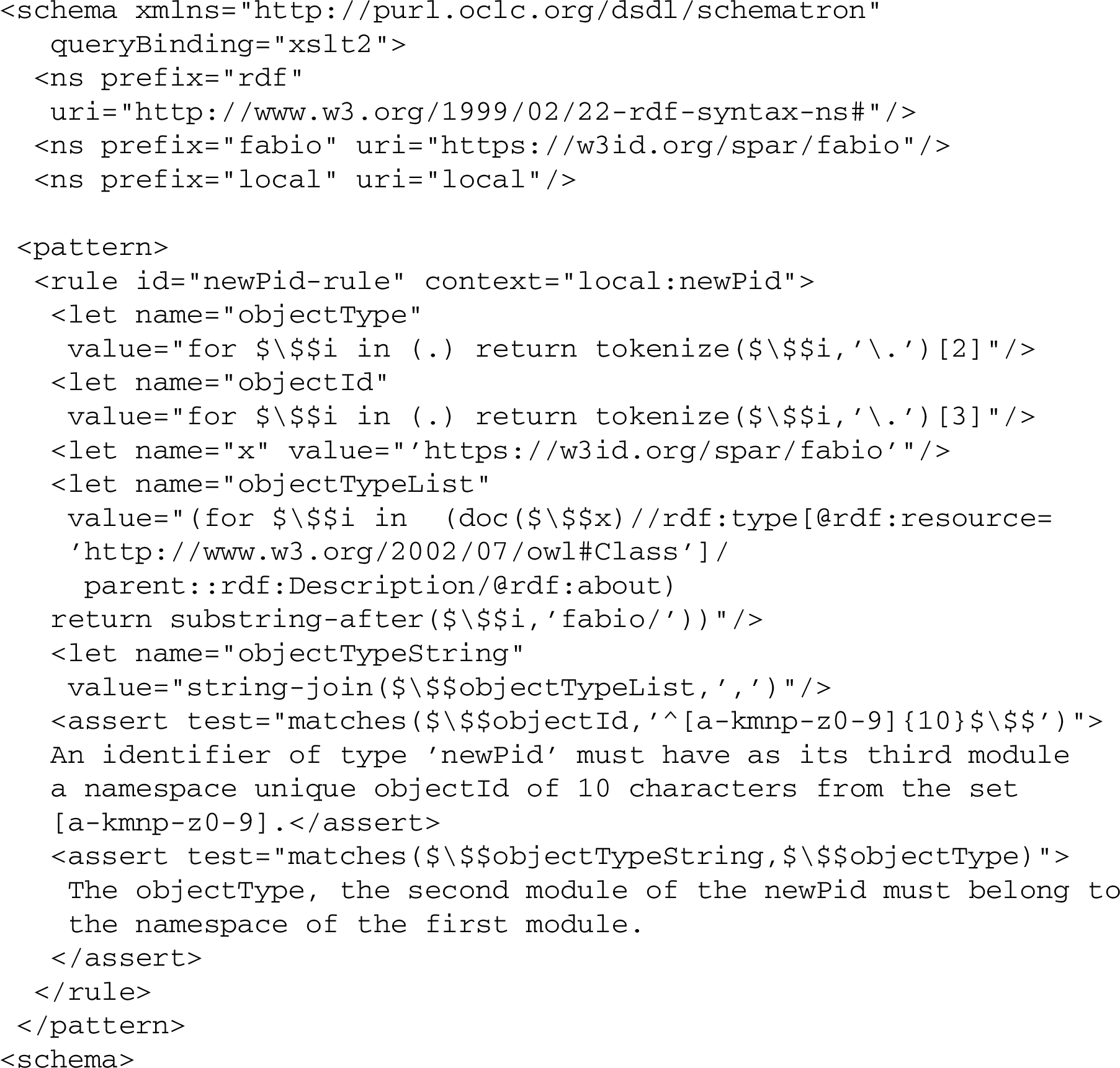
One might consider generalizing such a validation schema to the extent possible, so that the namespace URI in $objectTypeList, from which the $ objectType should be drawn, was automatically construed based on the namespacePrefix (module 1) of the newPid instance to be validated. This could be achieved by having the namespacePrefix expressed as a link with a namespace URI, e.g. such as fabio3333 in our case above. But that would also make the validation schema a bit more complicated, notably in the tokenization and separation of modules.
It is also conceivable, in order to allow for integration of already existing identifier schemes, that a namespace sets its own character set and string-length restrictions, to be declared by the validation rules of that namespace. For “narrow” namespaces, lacking defined diverse object types, possibly since they comprise basically only one type of object (as for ISBNs and ISSNs) we suggest as a default second module value ‘NOT’ = No Object Type. So we could have an IGSN, International Geo Sample Number [40], with string-length of objectID set to 9, expressed in this model:
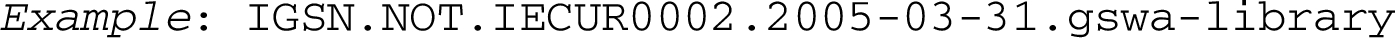
The identifier should be fully validatable, as a whole or in part (modules), in the corresponding namespace(s). The last two modules are optional, but they are meant to offer built in data provenance. For organisation identifiers, we hope that the recently launched ROR-IDs will become a global standard, like ORCIDs for persons. Then we could replace the last module in the IGSN-PID above with “05h2dda38”.
The resulting PIDs should be minted within the corresponding namespaces, which would also be the ‘custodians’ and resolving authorities of their respective PIDs, responsible for uniqueness within the namespace. Another task would be to monitor and assign sameAs-properties to PIDs that refer to the same ‘thing’ in other namespaces.
It has been suggested that in order “to build more connected, cross-linked and digitally accessible Internet content” it is necessary “to assign recognizable, persistent, globally unique, stable identifiers to ... data objects” [23]. The model proposed here aims to promote “new” PID strings that are universally unique and stable, recognizable through validation and enough inherent meaning to make them useful and understandable also in the future, thus, with a good potential for backup and persistence.
8.Conclusions
The purpose of this paper was to analyse some of the more prevalent general PIDs used in scholarly communication, identify some of their shortcomings and find out how PIDs could be made more FAIR. Real examples of PIDs were analysed to find out what additional requirements there might be to make them fully Findable, Accessible, Interoperable and Re-usable – FAIR. The “novelty” of the paper, if any, is the “widening” of the FAIR principles to have Findability include also rate of distribution or dissemination (e.g. as measured by means of ‘googling’) and Interoperability or Re-usability to include also ‘validatability’. Further, as against earlier insistence on the opaqueness of PIDs as a warrant for persistence, we argued for the importance of adding enough meaning to PIDs, through namespace prefixes and object types, so as to enhance their future use, distribution, findability and interpretability, and to safeguard against failed resolvability. The custodianship and minting of PIDs, we suggested to be the responsibility of the custodians of namespaces, as these are already assuming the administration of specifications, validation schemas, vocabularies or ontologies, and should be well qualified for the task. The minting algorithm, the patterns for PID-recognition, restriction in character set, string-length (with possible checkdigit) of objectId module should all be part of the validation schemas. These namespaces should then be able to register their schemes with n2t.net or identifiers.org, as already happens. And there might be several services such as the SPARQL endpoint of identifiers.org for registering sameAs-links. To create, maintain and make our PIDs truly persistent, widely used and FAIR should be a cooperative effort of the whole scholarly community.
Notes
References
[1] | California Digital Library, Archival Resource Key (ARK) Identifiers, 2018. http://n2t.net/e/ark_ids.html. |
[2] | Catalogue of Life: Annual Checklist, Asterolibertia gibbosa (Gaillard) Hansf. 1949, 2015. http://www.catalogueoflife.org/annual-checklist/2015/details/species/id/4f5bf9e96f36e1c530b147c7105e865b. |
[3] | CCSDS, Reference Model for an Open Archival Information System (OAIS): Recommended Practice. CCSDS 650.0-M-2. Magenta Book, Washington DC, 2012. https://public.ccsds.org/Pubs/650x0m2.pdf. |
[4] | J. Clark, PIDvasive:_What’s possible when everything has a persistent identifier? PIDapalooza, November 10, 2016. Retrieved Jan 16, 2017. doi:10.6084/m9.figshare.4233839.v1. |
[5] | K. Coyle et al., How Semantic Web differs from traditional data processing. RDF Validation in the Cultural Heritage Community. International Conference on Dublin Core and Metadata Applications, Austin, Oct. 2014. Date accessed: 24 Mar. 2017. http://dcevents.dublincore.org/IntConf/dc-2014/paper/view/311. |
[6] | M. Cruz, S. Kurapati and Y. Turkyilmaz-van der Velden, The Role of Data Stewardship in Software Sustainability and Reproducibility. Zenodo. 2018-09-14. doi:10.5281/zenodo.1419085. |
[7] | Data Citation Synthesis Group, Martone M. (ed.), Joint Declaration of Data Citation Principles, San Diego, CA: FORCE11, 2014. https://www.force11.org/group/joint-declaration-data-citation-principles-final. |
[8] | DataCite Metadata Working Group, DataCite Metadata Schema 4.1, 2017. doi:10.5438/0015. |
[9] | P. Doorn and I. Dillo, Assessing the FAIRness of Datasets in Trustworthy Digital Repositories: A Proposal. IDCC Edinburgh, 22 February 2017. http://www.dcc.ac.uk/webfm_send/2481. |
[10] | R.E. Duerr et al., On the utility of identification schemes for digital earth science data: An assessment and recommendations. Earth Science Informatics 4:139. ISSN: 1865-0473 (Print) 1865-0481 (Online), 2011. doi:10.1007/s12145-011-0083-6. |
[11] | A. Dunning, M. de Smaele and J. Böhmer, Are the FAIR Data Principles fair? Practice Paper, in: 12th International Digital Curation Conference (IDCC 2017), Edinburgh, Scotland, 20–23 February 2017. doi:10.5281/zenodo.321423. |
[12] | FAIRMetrics, FM_R1-3, 2018. https://github.com/FAIRMetrics/Metrics/blob/master/FM_R1.3. |
[13] | FAIRMetrics, FM-F2, 2019. https://github.com/FAIRMetrics/Metrics/blob/master/FM_F2. |
[14] | M. Fenner, Cool DOI’s, 2016. DataCite Blog. doi:10.5438/55e5-t5c0. |
[15] | Force11, The FAIR Data Principles, 2016. https://www.force11.org/group/fairgroup/fairprinciples. |
[16] | Force11, Guiding Principles for Findable, Accessible, Interoperable and Re-usable Data Publishing version B1.0, 2016. https://www.force11.org/fairprinciples. |
[17] | Force11, Guiding Principles for Findable, Accessible, Interoperable and Re-usable Data Publishing version b1.0 San Diego, CA: FORCE11, 2016. https://www.force11.org/node/6062/#Annex6-9. |
[18] | A. Gertler and J. Bullock, Reference Rot: An Emerging Threat to Transparency in Political Science. The Profession, 2017. doi:10.1017/S1049096516002353. |
[19] | A. Gilmartin, DOIs and matching regular expressions. Crossref Blog, 2015-08-11. https://www.crossref.org/blog/dois-and-matching-regular-expressions/. |
[20] | Global Names Architecture – GNA, New UUID v5 Generation Tool – gn_uuid v0.5.0, 2015. http://globalnames.org/news/2015/05/31/gn-uuid-0-5-0/. |
[21] | Global Names Architecture – GNA, Global Names Resolver, 2015. http://resolver.globalnames.org/. |
[22] | X. Guo, (2016). Yale Persistent Linking Service, PIDapalooza, November 10, 2016. Retrieved Jan 16, 2017. doi:10.6084/m9.figshare.4235822.v1. |
[23] | R. Guralnick et al., Community next steps for making globally unique identifiers work for biocollections data, ZooKeys 494: ((2015) ), 133–154. doi:10.3897/zookeys.494.9352. |
[24] | L. Haak et al., ORCID: A system to uniquely identify researchers, Learned Publishing 25: ((2012) ), 259–264. doi:10.1087/20120404. |
[25] | J. Hennessey and S. Xijin Ge, A cross disciplinary study of link decay and the effectiveness of mitigation techniques, Proceedings of the Tenth Annual MCBIOS Conference. BMC Bioinformatics 14: (Suppl 14) ((2013) ), S5. doi:10.1186/1471-2105-14-S14-S5. |
[26] | S.M. Jones, H. Van de Sompel, H. Shankar, M. Klein, R. Tobin and C. Grover, Scholarly context adrift: Three out of four URI references lead to changed content, PLoS ONE 11: (12) ((2016) ), e0167475. doi:10.1371/journal.pone.0167475. |
[27] | L.W. Kille, The growing problem of Internet “link rot” and best practices for media and online publishers, 2015. https://journalistsresource.org/studies/society/internet/website-linking-best-practices-media-online-publishers. |
[28] | M. Klein, H. Van de Sompel, R. Sanderson, H. Shankar, L. Balakireva, K. Zhou and R. Tobin, Scholarly context not found: One in five articles suffers from reference rot, PLoS ONE 9: (12) ((2014) ), e115253. doi:10.1371/journal.pone.0115253. |
[29] | J. Kunze and R. Rodgers, The ARK Identifier scheme, 2008. https://n2t.net/ark:/13030/c7cv4br18. |
[30] | J. Kunze and M. Russell, Noid – search.cpan.org, 2006. http://search.cpan.org/~jak/Noid/noid. |
[31] | J. Kunze and M. Russell, Noid Check Digit Algorithm, 2006. https://metacpan.org/pod/distribution/Noid/noid#NOID-CHECK-DIGIT-ALGORITHM. |
[32] | C. Li and S. Sugimoto, Provenance Description of Metadata using PROV with PREMIS for Long-term Use of Metadata, in: Proceedings of the International Conference on Dublin Core and Metadata Applications (Austin TX, 2014). http://dcpapers.dublincore.org/pubs/article/view/3709. |
[33] | J.A. McMurry et al., Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data, PLoS Biol 15: (6) ((2017) ), e2001414. doi:10.1371/journal.pbio.2001414. |
[34] | R. Page, Towards a biodiversity knowledge graph, Research Ideas and Outcomes 2: ((2016) ), e8767. doi:10.3897/rio.2.e8767. |
[35] | N. Paskin, Toward unique identiers, Proceedings of the IEEE 87: (7) ((1999) ), 1208–1227. doi:10.1109/5.771073. |
[36] | D. Patterson et al., Challenges with using names to link digital biodiversity information, Biodiversity Data Journal 4: ((2016) ), e8080. doi:10.3897/BDJ.4.e8080. |
[37] | J. Philipson, The Red Queen in the Repository: Metadata quality in an ever-changing environment, IDCC, In press, 2019. doi:10.5281/zenodo.2276777. |
[38] | J. Philipson, About a BUOI: Joint custody of persistent universally unique identifiers on the web, or, making PIDs more FAIR. SAVE-SD 2017. http://cs.unibo.it/save-sd/2017/papers/html/philipson-savesd2017.html. |
[39] | ROR, ROR, 2019. https://ror.org/about/. |
[40] | SESAR – System for Earth Sample Registration, What is the IGSN?, 2017. http://www.geosamples.org/aboutigsn. |
[41] | H. Van de Sompel, M. Klein and S.M. Jones, Persistent URIs Must Be Used To Be Persistent. WWW 2016. arXiv:1602.09102v1 [cs.DL] 29 Feb 2016. https://arxiv.org/abs/1602.09102v1. |
[42] | J. Wass, When PIDs aren’t there. Tales from Crossref Event Data. PIDapalooza, Reykjavik, November 2016. Retrieved: 11:57, Mar 20, 2017 (GMT). doi:10.6084/m9.figshare.4220580.v1. |
[43] | J. Wass, URLs and DOIs: A complicated relationship. CrossRef Blog, 2017 January 31. https://www.crossref.org/blog/urls-and-dois-a-complicated-relationship/. |
[44] | Wikipedia, Link rot. (Last modified on 13 March 2017, at 17:46. Retrieved 2017-03-14.) https://en.wikipedia.org/wiki/Link_rot. |
[45] | Wikipedia, Universally unique identifier. (Last modified on 29 January 2017, at 15:28. Retrieved 2017-01-30.) https://en.wikipedia.org/wiki/Universally_unique_identifier. |
[46] | Wikipedia, LSID. (Last edited on 20 February 2019, at 16:11 (UTC). Retrieved 2019-07-28.) https://en.wikipedia.org/wiki/LSID. |
[47] | M. Wilkinson et al., The FAIR guiding principles for scientific data management and stewardship, Scientific Data 3: ((2016) ), 160018. doi:10.1038/sdata.2016.18. |
[48] | M. Wilkinson, E. Schultes, L. Bonino, S. Sansone, P. Doorn and M. Dumontier, FAIRMetrics/Metrics: FAIR Metrics, Evaluation results, and initial release of automated evaluator code. Scientific Data. Zenodo, 2018. doi:10.5281/zenodo.321423. |
[49] | S. Wimalaratne et al., SPARQL-enabled identifier conversion with identifiers.org, Bioinformatics 31: (11) ((2015) ), 1875–1877. doi:10.1093/bioinformatics/btv064. |
[50] | K. Zhou et al., No more 404s: Predicting referenced link rot in scholarly articles for pro-active archiving, in: Proceedings of the 15th ACM/IEEE-CE on Joint Conference on Digital Libraries. JCDL’15, (2015) , pp. 233–236. doi:10.1145/2756406.2756940. |

![The FAIR data principles [15].](https://content.iospress.com:443/media/ds/2019/2-1-2/ds-2-1-2-ds190024/ds-2-ds190024-g001.jpg)



