HNRNPA2B1 induces cell proliferation and acts as biomarker in breast cancer
Abstract
BACKGROUND:
Numerous studies have shown that m6A plays an important regulatory role in the development of tumors. HNRNPA2B1, one of the m6A RNA methylation reading proteins, has been proven to be elevated in human cancers.
OBJECTIVE:
In this study, we aimed to identify the role of HNRNPA2B1 in breast cancer.
METHODS:
HNRNPA2B1 expression was investigated via RT-qPCR and TCGA database in breast cancer. Then, the function of HNRNPA2B1 on cancer cell was measured by CCK8 assays, colony formation and scratch assays. In addition, HNRNPA2B1 expression in BRCA was explored via the Wilcoxon signed-rank test, KruskalWallis test and logistic regression. The association with HNRNPA2B1 expression and survival were considered by KaplanMeier and Cox regression analyses. The biological function of HNRNPA2B1 was analyzed via gene set enrichment analysis (GSEA) and the cluster Profiler R software package.
RESULTS:
We found that HNRNPA2B1 was highly expressed and induced cell proliferation and migration in breast cancer. Moreover, we observed HNRNPA2B1 induced tumor growth in vivo. In addition, we also found HNRNPA2B1 expression was associated with characteristics and prognosis in breast cancer patients.
CONCLUSION:
Our findings suggested that HNRNPA2B1 promoted tumor growth and could function as a new potential molecular marker in breast cancer.
1.Introduction
Breast cancer is the most common malignant tumor in women [1]. The majority of breast cancer is invasive carcinoma (BRCA), which accounts for approximately 80% [2]. During the development of cancer, the cancer shows the heterogeneity of metastasis, with obvious metastasis advantages in different organs, leading to differences in the prognosis and treatment response of cancer patients [3]. Approximately 30% of breast cancers eventually develop into advanced cancer, and 90% of cancer deaths are attributed to primary metastases and the 5-year overall survival rate of patients without distant metastasis is more than 80% [4].
Significant heterogeneity differences in breast cancer have been recognized via advances in cancer diagnosis and treatment (Yeo and Guan, 2017). Recent study showed that precision medicinewas playing an increasingly important role in the clinical diagnosis and treatment of cancer patients [5]. The molecular mechanisms in the development of breast cancer was helpful to identify potential molecular therapeutic targets and improve the survival rate and quality life of patients [6].
N6-Methyladenosine (m6A) is one of the most abundant and representative modification in messenger RNA (mRNA) including mRNA transcription, translation, degradation and stability [7]. During this process, m6A methylation is transferred by m6A methyltransferase (Writers), removed by m6A demethylases (Erasers), and recognized by methylated reading proteins (Readers) [8].
Heterogeneous nuclear ribonucleoprotein A2B1(HNRNPA2B1), one member of m6A Readers, is widely expressed in human tissues and affects transcriptional processes by binding to RNA-specific nucleic acid sequences. Previous studies showed that HNRNPA2B1 induced cancer cell migration and invasion by regulating the processes of aerobic glycolysis [9], telomere maintenance [10], and cell metabolism [11] and promoted tumor progression [12, 13]. These findings suggested that HNRNPA2B1 played an extremely important role in tumor progression, and the research on the role of HNRNPA2B1 should be further strengthened in breast cancer.
In the present study, we investigated the role of HNRNPA2B1 in the development of breast cancer. The results suggested that HNRNPA2B1 was highly expressed and induced cell proliferation and migration in breast cancer. In addition, the expression of HNRNPA2B1 was associated with the characteristics of breast cancer patients.
2.Materials and methods
2.1Cell culture
The human breast cancer cell lines MCF-7 and MDA-MB-231 were purchased from Zhong Qiao Xin Zhou Biotechnology (Shanghai, China). Cell lines were cultured in Roswell Park Memorial Institute 1640 (Thermo Fisher Scientific, Waltham, MA, USA) in a 25-cm cell culture flask containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific, Waltham, MA, USA) and were grown in humidified 5% CO2 incubator at 37∘C. Cell transfection was performed with Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA).
2.2RNA extraction and RT-qPCR
Total RNA from breast cancer cells was isolated by using RNAeasyTM Animal RNA Isolation Kit with Spin Column (Beyotime Biotechnology, Shanghai, China). Total RNA from the serum of patients was isolated via Blood RNA Kit (Yeasen Biotechnology, Shanghai, China). The primer sequences were shown in Table S1 of the Supplementary materials. RNA extraction and RT-qPCR were performed using the HiScript II One Step RT-qPCR SYBR Green Kit-V22.1 (Vazyme Biotech, Nanjing, China). The relative expression was standardized as the internal control, and the 2- ΔΔCT method was used for calculation.
2.3Western blotting
Total protein from approximately 5
2.4CCK8 assay
Cell proliferation was measured by cell counting kit-8 assay (CCK-8, Beyotime Biotechnology, China) according to the manufacturer’s instructions. Briefly, cancer cells were seeded into 96-well plate. The absorbance was detected by Benchmark PlusTM microplate spectrometer (Bio-Rad Laboratories Inc., USA).
2.5Colony formation assay
Breast cancer cells were seeded in 6-well plates and cultured for 10 days. Cell colonies were fixed in 10% ethanol for 30 minutes and stained with crystal violet (Beyotime Biotechnology, Shanghai, China) for 5 minutes. Cell colonies were imaged, counted and analyzed by Image J.
2.6Scratch migration assay
Breast cancer cells were cultured in 6-well plates. The wound healing assay was performed using serum-free medium after transfection for 48 hours. The cells were cultured at 37∘C in 5% CO2 incubators, and photographs were taken at 0, 24, and 48 hours.
2.7Establishment tumor model in nude mice
Nude mice were purchased from Cavenslasales (SCXK-SU: 2021-0013, Changzhou, China). All 15 male mice were approximately 6 weeks. Food, drinking water and bedding were sterilized by high pressure and refreshed every four days. The cells were divided into three groups, including MCF-7 cells (blank control group), MCF-7 cells transfected with nonspecific siRNA for 24 hours (nonspecific siRNA transfection group) and MCF7 cells transfected with HNRNPA2B1 shRNA for 24 hours (HNRNPA2B1 shRNA transfection group). Each mouse was injected with 0.1 ml of cell suspension (5
2.8Patient information
Tissue samples (35 pairs of breast cancer tissues and adjacent tissues) and serum samples (from 14 healthy control subjects, 15 benign breast tumor patients and 23 breast cancer patients) were obtained from the affiliated Changzhou Second People’s Hospital of Nanjing Medical University. The ethical committees of the Affiliated Changzhou Second People’s Hospital of Nanjing Medical University approved this study (Ethics Committee Approval Number, 2019KY028-1) and conformed to the guidelines of the Helsinki Declaration (2014).
RNA-sequencing data and patient clinical information were collected from the TCGA data repository (https://portal.gdc.cancer.gov/repository, accessed on 26 May 2022), involving 1091 BRCA samples and 113 normal samples (workflow type: HTSeq-TPM). Features such as age, TNM classification, pathologic stage, histological type, PR status, ER status, HER2 status, Prediction Analysis of Microarray 50 (PAM50), menopause status and radiation therapy were included, and other missing or uncertain features were removed. For further verification, we downloaded the gene expression profiles of GSE10810 and GSE42568 from the Gene Expression Omnibus (GEO) database and analyzed the expression of HNRNPA2B1 in BRCA tissues and normal tissues.
2.9Enrichment and correlation analyses
Pearson correlation analysis of HNRNPA2B1 and other genes was performed via TCGA BRCA data. Enrichment analysis was performed from the MSigDB database in cluster Profiler R software package 3.6.3 with the following parameters: nPerm
2.10Immune cell infiltration
We used ssGSEA (single-sample GSEA) and Spearman rank correlation analysis to estimate immune cell infiltration scores. Briefly, BRCA samples were divided into two groups according to the median HNRNPA2B1 expression (high vs. low level), and immune cell infiltration levels were compared. Chord pictures using the circlize R software package were also tested by Spearman rank correlation analysis.
2.11Construction of the PPI network
The HNRNPA2B1-related PPI network was established via the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) to explore the association between HNRNPA2B1 and other genes at the protein level. A minimum needed interaction score
2.12Statistical analyses
Data are expressed as the means
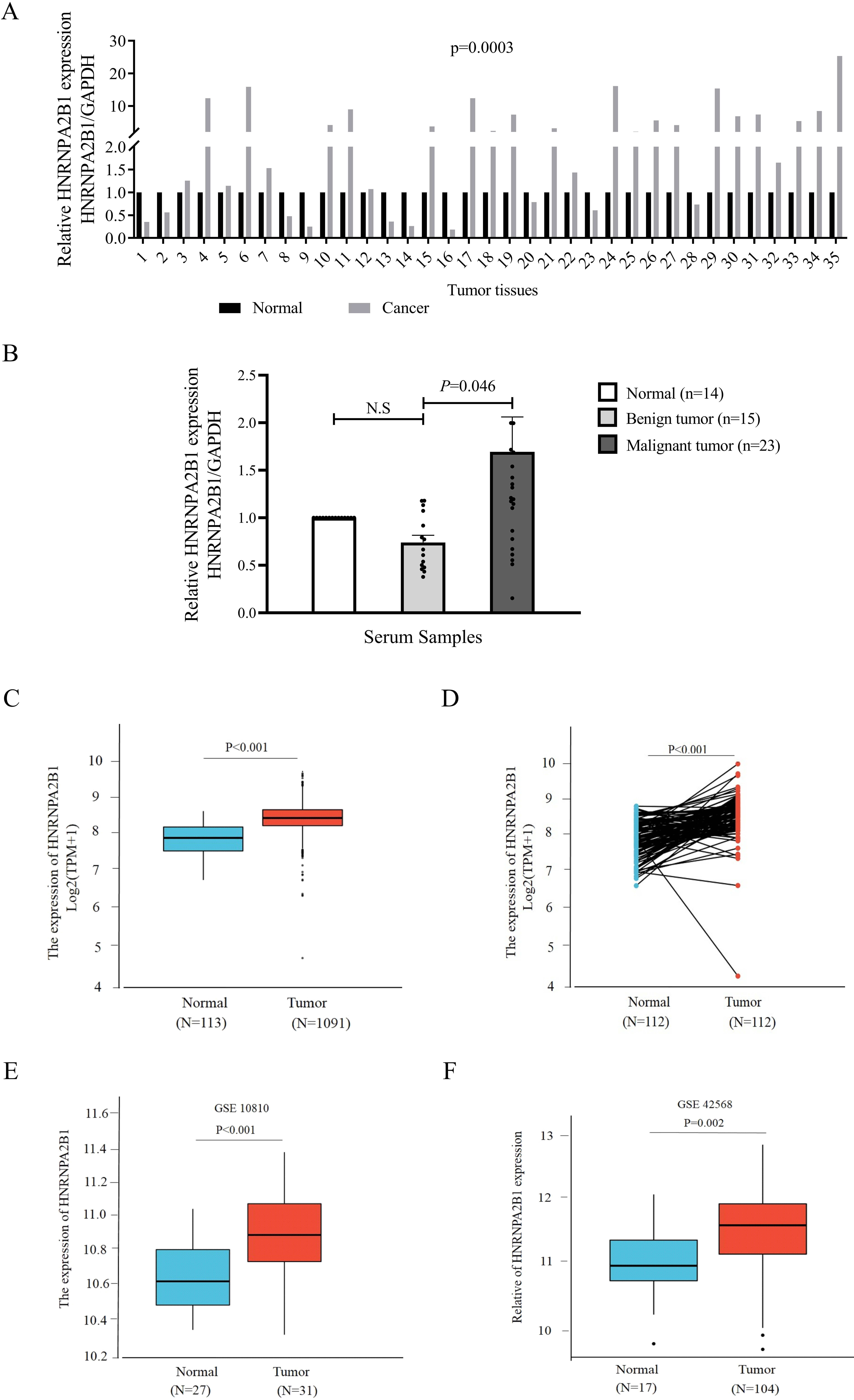
Figure 1.
Expression of HNRNPA2B1 in breast cancer. (A) HNRNPA2B1 expression in 35 primary breast cancer tissues and adjacent normal tissues was analyzed through RTqPCR. (B) HNRNPA2B1 expression in the serum of 14 healthy volunteers, 15 benign breast tumor patients and 23 breast cancer patients. (C) The expression of HNRNPA2B1 in BRCA compared by the Wilcoxon rank sum test (unpaired,
3.Results
3.1HNRNPA2B1 was highly expressed in breast cancer
To investigate the role of HNRNPA2B1 in breast cancer, we collected 35 breast cancer specimens and adjacent normal tissues. We performed RT-qPCR to measure expression and the data showed that compared with adjacent normal tissues, the expression of HNRNPA2B1 in breast cancer tissue was significantly increased (Fig. 1A). And then, HNRNPA2B1 expression in peripheral blood serum of 23 patients with breast cancer were significantly increased compared with in peripheral blood serum of 14 serum healthy volunteers, 15 benign breast tumor patients (Fig. 1B). Furthermore, HNRNPA2B1 expression was significantly elevated in BRCA tissues compared with normal tissues via Wilcoxon rank sum test (Fig. 1C). In addition, the expression of HNRNPA2B1 was significantly increased in BRCA tissues compared with paired normal tissues (Fig. 1D). Consistently, the expression of HNRNPA2B1 was higher than that in normal tissues in GSE10810 and GSE42568 via the GEO database (Fig. 1E, 1F). The results showed that HNRNPA2B1 was highly expressed and played an important role in breast cancer progression.
Figure 2.
HNRNPA2B1 induced cell proliferation and migration in breast cancer cells. (A) CCK-8 assays in MDA-MB-231 cells. (B) CCK8 assays in MCF-7 cells. (C) Colony formation assay in MDA-MB-231 and MCF-7 cells. (D) Bar chart of the colony formation assay. (E) Scratch assay in MDA-MB-231 and MCF-7 cells. (F) A bar chart of the scratch assay in MDA-MB-231 cells. (G) A bar chart of the scratch assay in MCF-7 cells.
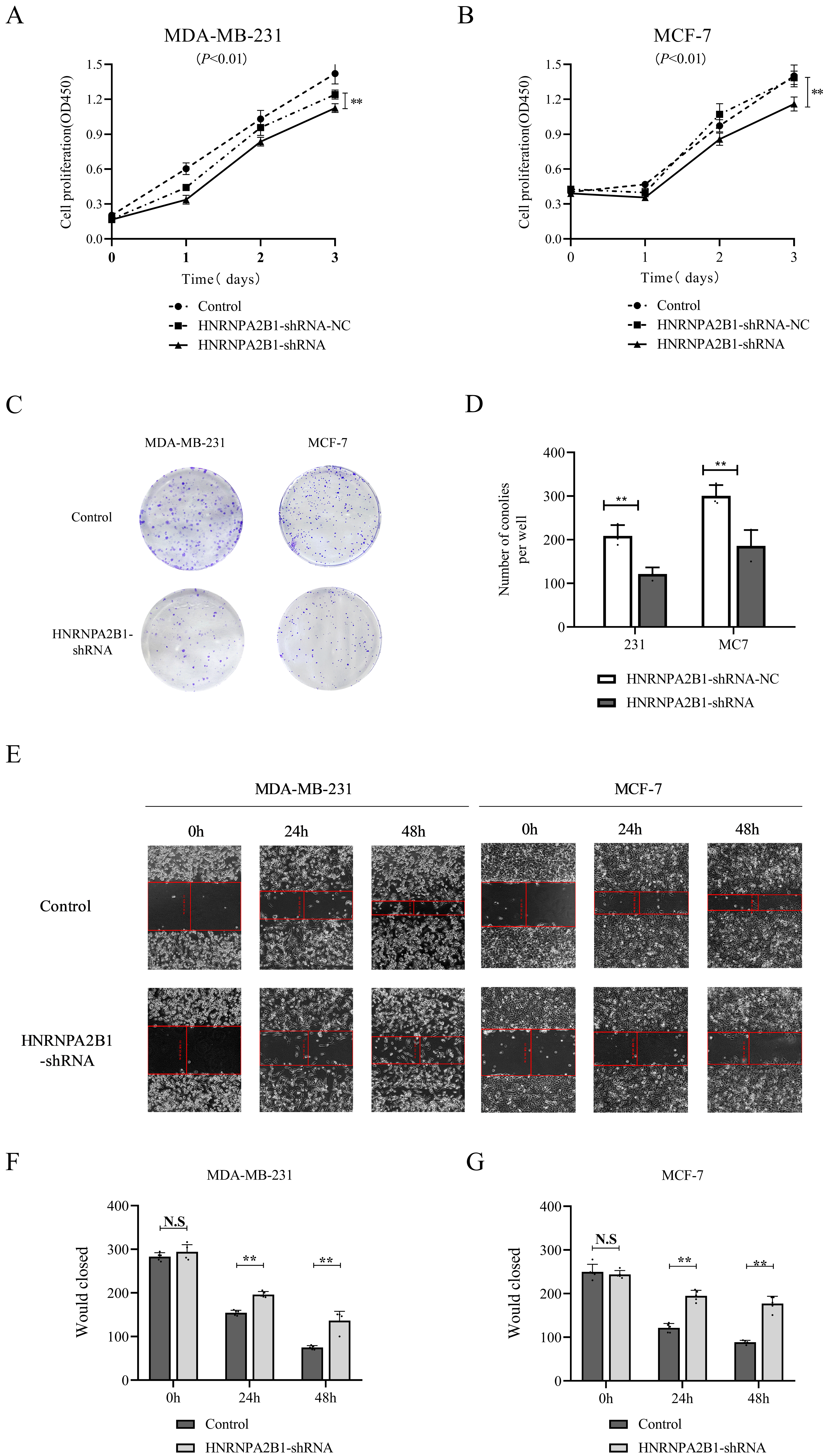
3.2Silencing HNRNPA2B1 expression inhibited cell proliferation and migration in breast cancer
To explore the function of HNRNPA2B1 in breast cancer, we silenced HNRNPA2B1 expression by interfering with HNRNPA2B1 shRNA in breast cancer cells. We observed that interfering with HNRNPA2B1 expression significantly inhibited cell proliferation in MDA-MB-231 (Fig. 2A) and MCF-7 cells (Fig. 2B) via CCK-8 assays and colony forming assays in breast cancer (Fig. 2C–2D). Then, we found that silencing HNRNPA2B1 expression inhibited cell migration via scratch assays in breast cancer (Fig. 2E–2G). The data suggested that HNRNPA2B1 induced cell proliferation and migration in breast cancer.
Figure 3.
HNRNPA2B1 induced breast tumor growth in vivo. (A) Images of breast tumors cultivated in nude mice. (B) Tumor growth curves of nude mice. (C) Tumor weight statistics. (D) Relative HNRNPA2B1 mRNA expression in breast cancer tissues detected by RTqPCR. (E) HNRNPA2B1 protein expression in breast cancer tissues detected by western blot.
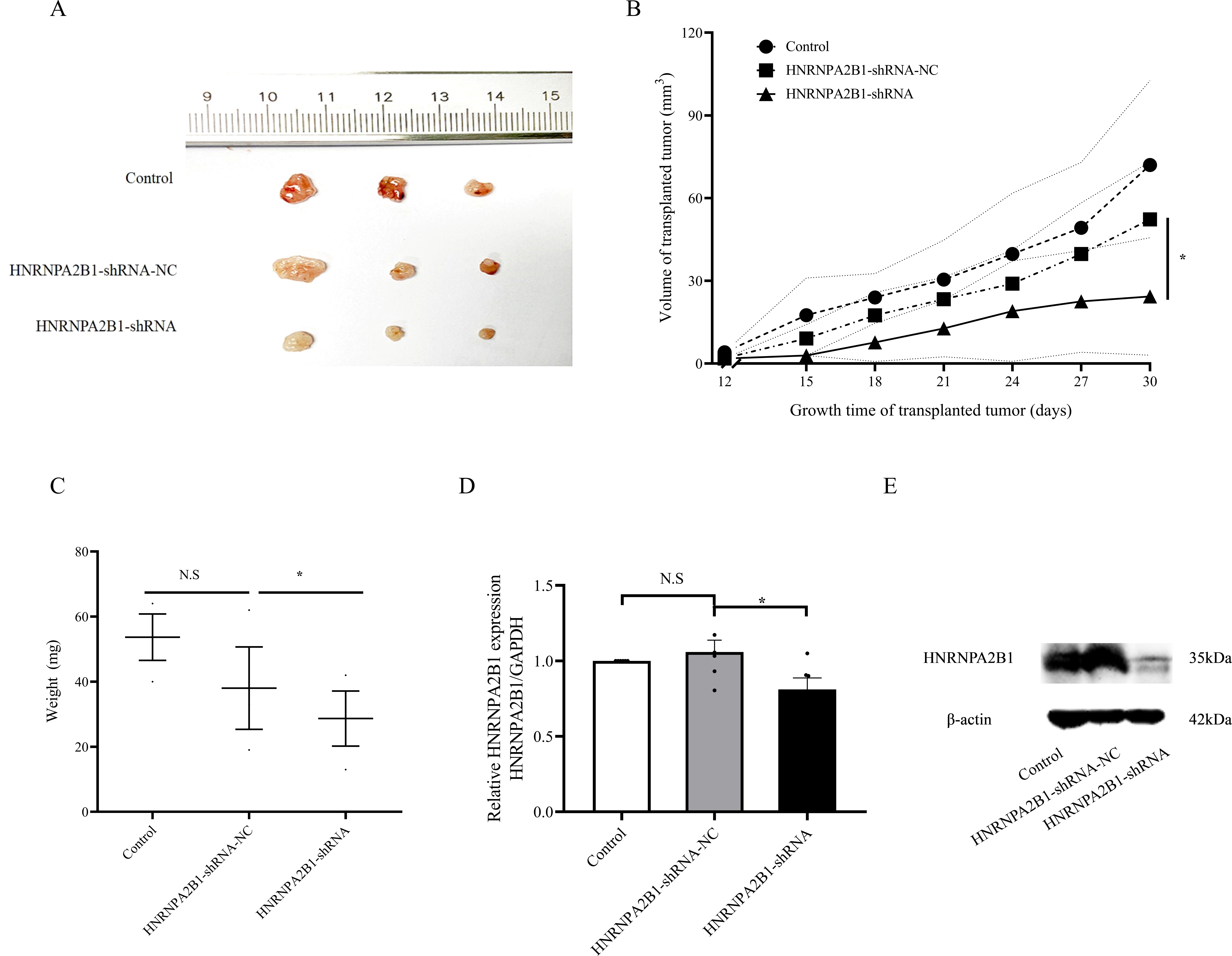
3.3HNRNPA2B1 induced breast tumor growth in vivo
To further investigate the function of HNRNPA2B1 during breast cancer progression, HNRNPA2B1-silenced MCF-7 cells treated with HNRNPA2B1 shRNA were collected and subcutaneously injected into nude mice. We observed that compared with the control group, the tumor mass was significantly smaller in the HNRNPA2B1 shRNA group (Fig. 3A), and the trend of tumor growth in the HNRNPA2B1 shRNA group was slow, but the difference was not statistically significant (Fig. 3B). In addition, the data showed that the average tumor weight was significantly lower in the HNRNPA2B1 shRNA group than in the control group (Fig. 3C). Moreover, we found that the expression of HNRNPA2B1 decreased significantly in the HNRNPA2B1 shRNA group compared with the control group (Fig. 3D–E). The results demonstrated that HNRNPA2B1 induced tumor growth in breast cancer.
Figure 4.
HNRNPA2B1 was associated with characteristics in breast cancer. (A) The expression of HNRNPA2B1 in breast cancer patients was associated with (A) age, (B) menopause status, (C) Her2 status, (D) T stage, (E) pathologic stage, (F) PAM50, (G) histological type, and (H) relationship between HNRNPA2B1 and characteristics using logistic regression.
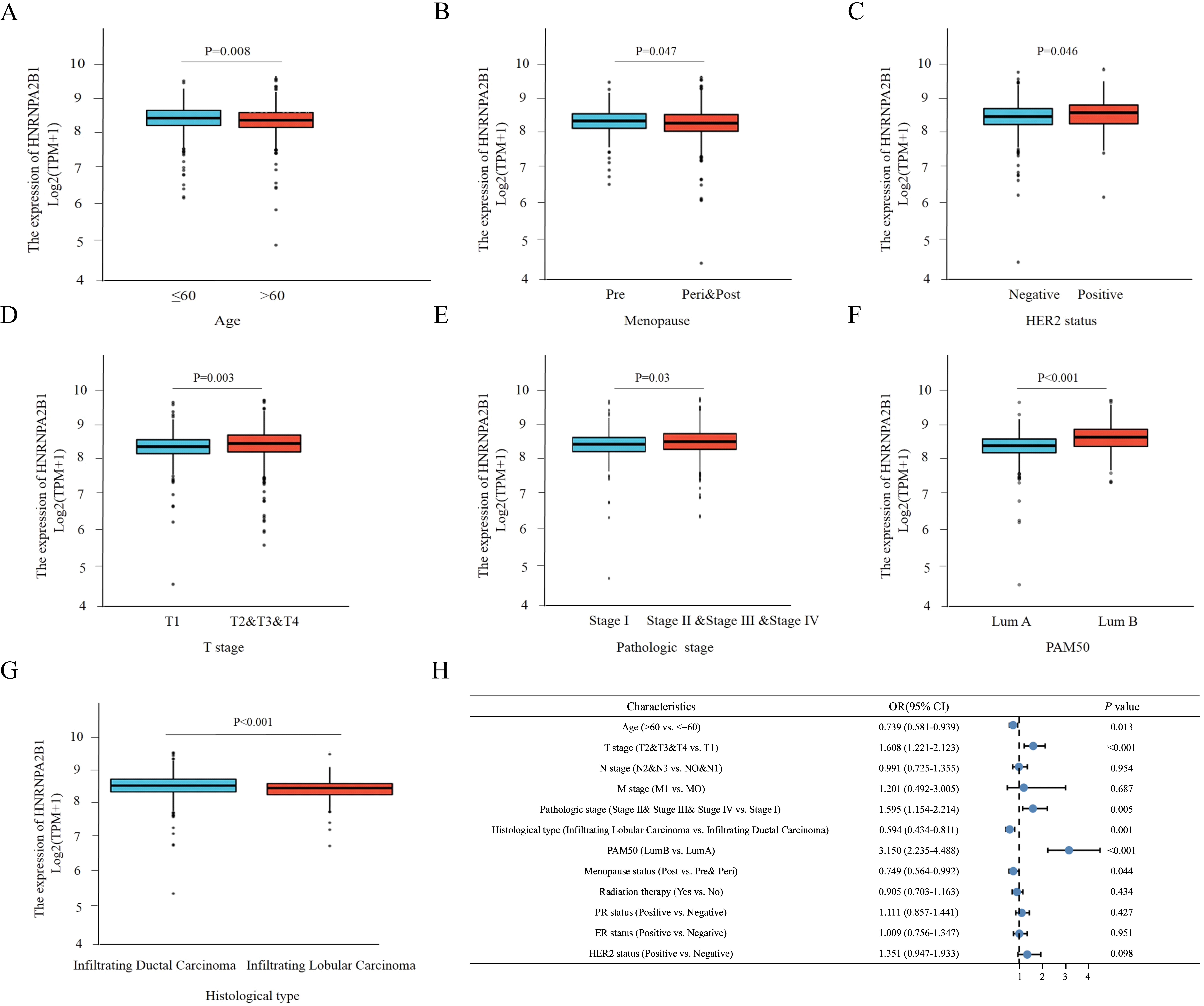
3.4HNRNPA2B1 was associated with clinicopathological characteristics in breast cancer
To evaluate the characteristics of HNRNPA2B1 in breast cancer patients, we used the TCGA database to analyze the association of HNRNPA2B1 and characteristics in breast cancer patients. We found that the expression of HNRNPA2B1 in breast cancer patients aged less than or equal to 60 years old was significantly higher than that in breast cancer patients aged over 60 years old (Fig. 4A). In addition, the data showed that the expression of HNRNPA2B1 was associated with menopause status (Pre vs Peri&Post, Fig. 4B), Her2 status (negative vs. Positive, Fig. 4C), T stage (T1 vs. T2/T3/T4, Fig. 4D), pathologic stage (Stage I vs. Stage II/Stage III/Stage IV, Fig. 4E), PAM50 (LumA vs. LumB, Fig. 4F) and histological type (infiltrating ductal carcinoma vs. infiltrating lobular carcinoma, Fig. 4G). Moreover, we performed logistic regression to analyze HNRNPA2B1 expression and clinicopathological characteristics for risk factors in BRCA patients. The data showed that HNRNPA2B1 expression with age (OR
Figure 5.
Association between HNRNPA2B1 expression level and survival. Relationship between HNRNPA2B1 expression level and patient survival in TCGA tumors using KaplanMeier Plotter, including (A) overall survival (OS), (B) relapse-free survival (RFS), (C) distant metastasis-free survival (DMFS), and (D) postprogression survival (PPS).
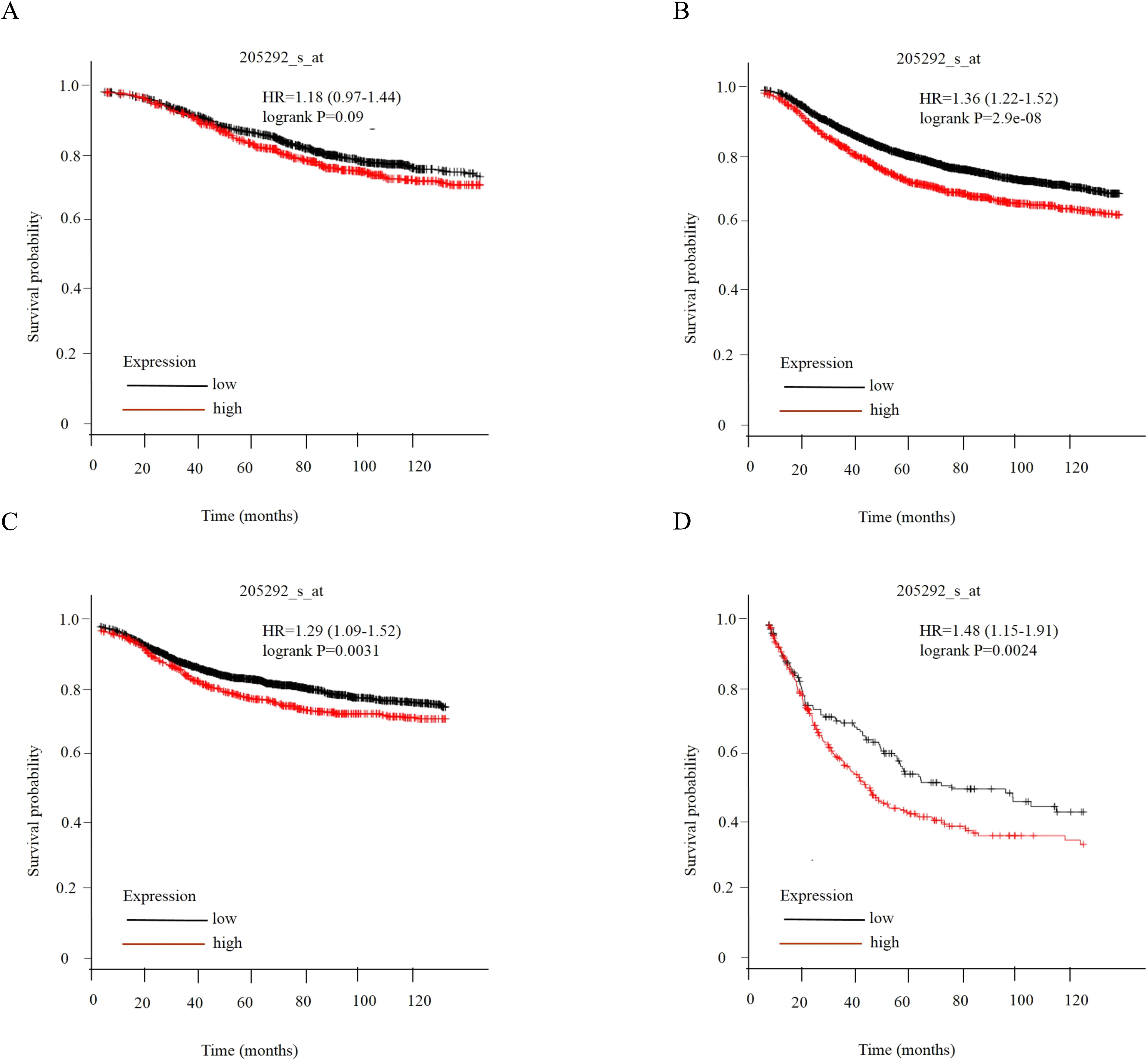
3.5HNRNPA2B1 was associated with the prognosis of breast cancer patients
To demonstrate the effect of HNRNPA2B1 on the prognosis of breast cancer patients, KaplanMeier analysis (http://kmplot.com/analysis/) was performed. The data showed that the overall survival (OS) of patients with high expression of HRNRPA2B1 was worse than that of patients with low expression of HRNRPA2B1, and the difference was not statistically significant (Fig. 5A). The relapse-free survival (RFS) of patients with high expression of HRNRPA2B1 was worse than that of patients with low expression of HRNRPA2B1 (Fig. 5B). In addition, the distant metastasis-free survival (DMFS) of patients with high expression of HRNRPA2B1 was worse than that of patients with low expression of HRNRPA2B1 (Fig. 5C). Moreover, the post-progression survival (PPS) of patients with high expression of HRNRPA2B1 was worse than that of patients with low expression of HRNRPA2B1 (Fig. 5D). The results suggested that HNRNPA2B1 was associated with prognosis and acted as potential molecular marker of breast cancer patients.
Figure 6.
Correlation and enrichment analysis of HNRNPA2B1 in BRCA. (A) Heatmap of the top 50 genes most positively associated with HNRNPA2B1. The upper part shows the expression of HNRNPA2B1, and the lower part shows the expression of the top 50 genes after Z score transformation (∗∗∗:
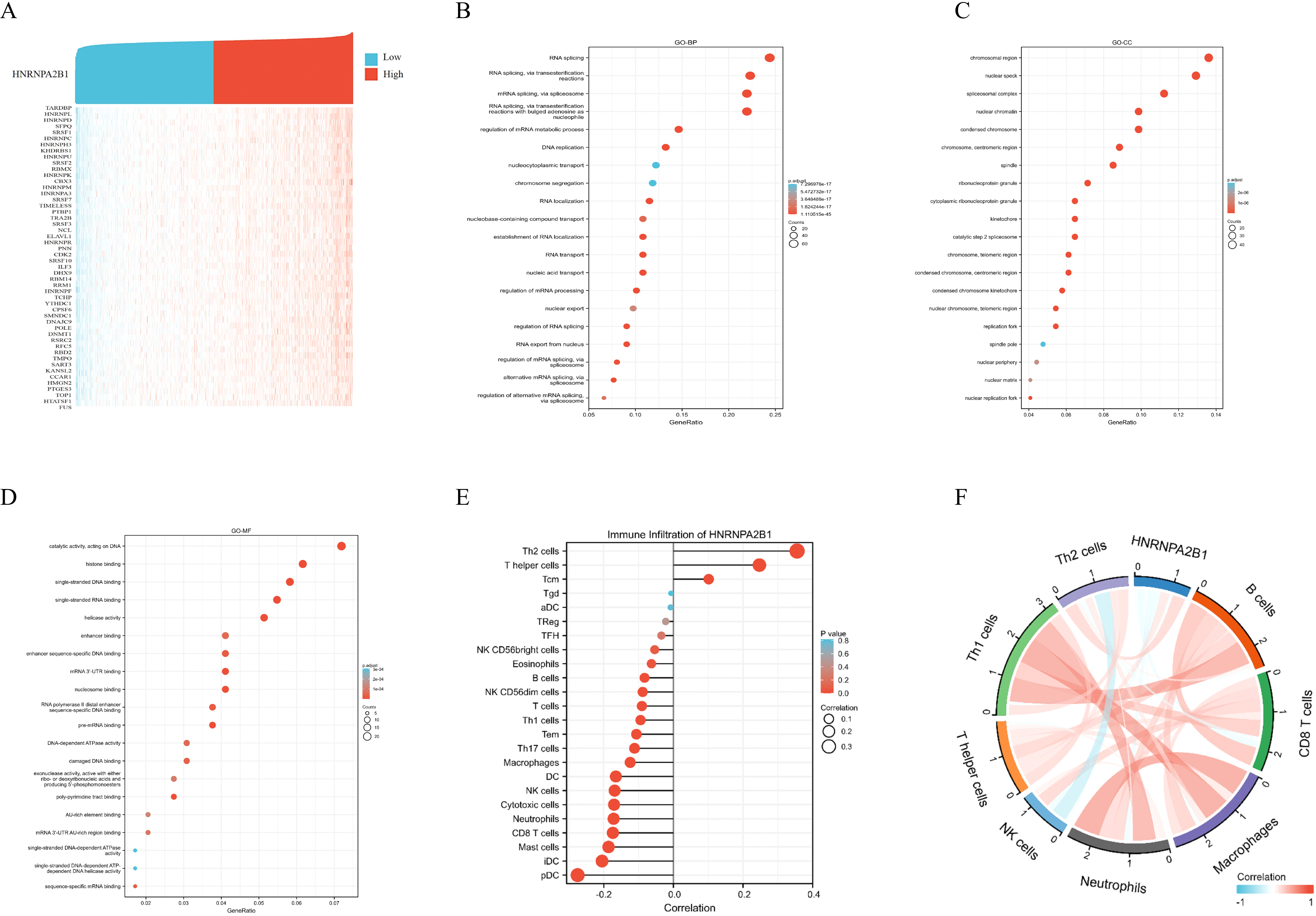
3.6HNRNPA2B1 accelerated immune cell infiltration in BRCA
To investigate the potential roles of HNRNPA2B1 in BRCA carcinogenesis, we selected the positively associated with the expression of HNRNPA2B1 for enrichment analysis via TCGA data (Fig. 6A). Then, we mapped the HNRNPA2B1 co-expression pattern by calculating the Pearson correlation coefficients of the expression value with the target mRNA and chosed significantly different target mRNAs to conduct GO analyses. GO analysis was performed from three functional domains: biological process (Fig. 6B), cellular component (Fig. 6C), and molecular function (Fig. 6D), which revealed that HNRNPA2B1 was associated mainly with RNA processing and modification pathways, including RNA splicing and mRNA splicing. In addition, we analyzed immune cell infiltration in BRCA patients via the TCGA database. The infiltration scores of Th2 cells and T helper cells were higher in the high HNRNPA2B1 expression cohort than in the low HNRNPA2B1 expression cohort (Fig. 6E). The expression of HNRNPA2B1 was significantly positively correlated with Th2 cell and T helper cell infiltration (Fig. 6F), suggesting that high HNRNPA2B1 expression promoted Th2 cell accumulation in BRCA. Consistent with the Th1/Th2 equilibrium drift theory, the Th2-type immune response was dominant and led to the suppression of Th1 cell-mediated cellular immunity, which may be one of the mechanisms of immune escape of tumor cells promoting the occurrence or metastasis in breast cancer tissues.
4.Discussion
Breast cancer is one of the most common malignant tumors and the leading cause of cancer-related deaths among women worldwide. While there has been no significant progress in breast cancer surgical treatment in recent years, the number of new cases of breast cancer surpassed that of lung cancer in 2020 [1]. Considering the drug resistance and side effects of chemotherapy drugs, the application of endocrine and molecular targeted therapy in early diagnosis makes sense. It is essential to customize individualized, precise and integrated treatments to improve the survival rate of breast cancer patients [14].
In this study, we found that the expression of HNRNPA2B1 was significantly elevated in breast cancer using the TCGA database and clinical specimens. Kaplan-Meier survival curve analysis revealed that BRCA patients with high expression of HNRNPA2B1 tend to have worse prognosis. The data showed that the expression of HNRNPA2B1 was higher in situ than in adjacent tissues via breast cancer clinical specimens. Similarly, the expression of HNRNPA2B1 was elevated in the serum of patients with breast cancer. In addition, interference with HNRNPA2B1 expression significantly inhibited cell proliferation and migration in breast cancer. In summary, HNRNPA2B1 may play a pathogenic role in breast cancer progression, acting on tumor cells, oncogene signaling pathways and tumor-infiltrating immune cells comprehensively.
HNRNPA2B1 has been reported to play regulatory roles in various kinds of human cancers in recent years [15]. In multiple myeloma (MM), for example, HNRNPA2B1 was elevated and negatively correlated with favorable prognosis. The depletion of HNRNPA2B1 in MM cells inhibited cell proliferation and induced apoptosis, while the overexpression of HNRNPA2B1 promoted cell proliferation in vitro and in vivo [16]. Knockdown of hnRNPA2B1 attenuated H19-induced migration and invasion in colorectal cancer (CRC) cells [17]. HNRNPA2B1 also functions as an oncogenic factor in promoting esophageal cancer (ESCA) progression via upregulation of the fatty acid synthesis enzymes ACLY and ACC1 [18]. A high expression level of HNRNPA2B1 was observed in 16 types of tumor tissues compared with normal tissues by data mining using TCGA datasets, suggesting that HNRNPA2B1 was closely related to the tumorigenicity of cancer cells and may be a promising prognostic biomarker and therapeutic target for human cancers.
RNA-binding proteins (RBPs) mainly participate in transcriptional and posttranscriptional events in tumor progression [19]. HNRNPA2B1, an important member of BNPs, regulates mRNA processing of specific gene sets [20]. Different downstream targets (SATA3, ERK1/2, PKM2/PKM1, BIRC5, COX-2, AXL, KRAS, PI3K/AKT, ACLY/ACC1, Lin28B, for instance) are recruited by hnRNPA2B1 in biological processes involving proliferation, migration, and apoptosis [20, 21, 22, 23, 24]. The association between downstream genes of hnRNPA2B1 in different tumor microenvironments, such as SATA3 and ERK1/2, in breast cancer [25].
In our study, the results showed that HNRNPA2B1 induced cell proliferation in breast cancer via RNA processing and modification pathways. Furthermore, stromal cells in the tumor microenvironment, especially immune cells, were essential elements that profoundly impacted the regulation of the malignant behaviors of tumor cells. Studies have proven that immune cells have clinical significance in predicting the prognosis and treatment outcome of cancer patients. Furthermore, the expression of immune infiltration was increased, which mainly induced angiogenesis genes, the high proliferation rate and Th2 cell deviation. It has been reported that Th1/Th2 balance bias contributes to breast cancer progression [26]. The expression of HNRNPA2B1 was positively associated with Th2 cells in prostate cancer [27].
5.Conclusion
In summary, our present study indicated that the expression of HNRNPA2B1 was significantly higher in human breast cancer tissues than in paracancerous tissues and that silenced HNRNPA2B1 expression inhibited cell proliferation and tumor growth in breast cancer. Moreover, the expression of HNRNPA2B1 was associated with characteristics and prognosis in breast cancer patients and immune cell infiltration in BRCA. Our findings indicated that HNRNPA2B1 could be a molecular targeting site and biomarker in breast cancer.
Funding
This work was supported by the Changzhou Sci & Tech Program (CE20235061), the Changzhou Medical Center Project (CMCB202226), and the Changzhou High-Level Medical Talents Training Project (2022CZBJ077).
Availability of data and materials
The data used to support the findings of this study were included with in the article and Additional files.The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request
Author contributions
Conception: Tongbao Feng, Chunfu Zhu.
Interpretation or analysis of data: Yi Yang, Yi Zhang.
Preparation of the manuscript: Yi Yang, Tongbao Feng.
Revision for important intellectual content: Tongbao Feng.
Supervision: Chunfu Zhu.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the ethical committees of the Affiliated Changzhou Second People’s Hospital of Nanjing Medical University approved this study (Ethics Committee Approval Number, 2019KY028-1). All experiments were performed in accordance with relevant guidelines and regulations
Consent for publication
Not applicable.
Supplementary data
The supplementary files are available to download from http://dx.doi.org/10.3233/CBM-230576.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
[1] | H. Sung, J. Ferlay, R.L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA Cancer J Clin 71: ((2021) ), 209–249. |
[2] | T. Feng, P. Zhang, Y. Sun, Y. Wang, J. Tong, H. Dai and Z. Hua, High throughput sequencing identifies breast cancer-secreted exosomal LncRNAs initiating pulmonary pre-metastatic niche formation, Gene 710: ((2019) ), 258–264. |
[3] | Y. Liang, H. Zhang, X. Song and Q. Yang, Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets, Semin Cancer Biol 60: ((2020) ), 14–27. |
[4] | C. Allemani, T. Matsuda, V. Di Carlo, R. Harewood, M. Matz, M. Niksic, A. Bonaventure, M. Valkov, C.J. Johnson, J. Esteve, O.J. Ogunbiyi, E.S.G. Azevedo, W.Q. Chen, S. Eser, G. Engholm, C.A. Stiller, A. Monnereau, R.R. Woods, O. Visser, G.H. Lim, J. Aitken, H.K. Weir, M.P. Coleman and C.W. Group, Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries, Lancet 391: ((2018) ), 1023–1075. |
[5] | E.S. McDonald, A.S. Clark, J. Tchou, P. Zhang and G.M. Freedman, Clinical Diagnosis and Management of Breast Cancer, J Nucl Med 57: (Suppl 1) ((2016) ), 9S–16S. |
[6] | U. Veronesi, P. Boyle, A. Goldhirsch, R. Orecchia and G. Viale, Breast cancer, Lancet 365: ((2005) ), 1727–41. |
[7] | Y. An and H. Duan, The role of m6A RNA methylation in cancer metabolism, Mol Cancer 21: ((2022) ), 14. |
[8] | L. He, H. Li, A. Wu, Y. Peng, G. Shu and G. Yin, Functions of N6-methyladenosine and its role in cancer, Mol Cancer 18: ((2019) ), 176. |
[9] | H. Yang, R. Zhu, X. Zhao, L. Liu, Z. Zhou, L. Zhao, B. Liang, W. Ma, J. Zhao, J. Liu and G. Huang, Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma, Oncogene 38: ((2019) ), 4915–4931. |
[10] | C. Patry, L. Bouchard, P. Labrecque, D. Gendron, B. Lemieux, J. Toutant, E. Lapointe, R. Wellinger and B. Chabot, Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines, Cancer Res 63: ((2003) ), 7679–88. |
[11] | C.V. Clower, D. Chatterjee, Z. Wang, L.C. Cantley, M.G. Vander Heiden and A.R. Krainer, The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism, Proc Natl Acad Sci U S A 107: ((2010) ), 1894–9. |
[12] | K. Moran-Jones, J. Grindlay, M. Jones, R. Smith and J.C. Norman, hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration, Cancer Res 69: ((2009) ), 9219–27. |
[13] | Y. Yan-Sanders, G.J. Hammons and B.D. Lyn-Cook, Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells, Cancer Lett 183: ((2002) ), 215–20. |
[14] | H.J. Burstein, G. Curigliano, B. Thurlimann, W.P. Weber, P. Poortmans, M.M. Regan, H.J. Senn, E.P. Winer, M. Gnant and C. Panelists of the St Gallen Consensus, Customizing local and systemic therapies for women with early breast cancer: the St. Gallen International Consensus Guidelines for treatment of early breast cancer 2021, Ann Oncol 32: ((2021) ), 1216–1235. |
[15] | G. Dreyfuss, V.N. Kim and N. Kataoka, Messenger-RNA-binding proteins and the messages they carry, Nat Rev Mol Cell Biol 3: ((2002) ), 195–205. |
[16] | F. Jiang, X. Tang, C. Tang, Z. Hua, M. Ke, C. Wang, J. Zhao, S. Gao, A. Jurczyszyn, S. Janz, M. Beksac, F. Zhan, C. Gu and Y. Yang, HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA, J Hematol Oncol 14: ((2021) ), 54. |
[17] | Y. Zhang, W. Huang, Y. Yuan, J. Li, J. Wu, J. Yu, Y. He, Z. Wei and C. Zhang, Long non-coding RNA H19 promotes colorectal cancer metastasis via binding to hnRNPA2B1, J Exp Clin Cancer Res 39: ((2020) ), 141. |
[18] | H. Guo, B. Wang, K. Xu, L. Nie, Y. Fu, Z. Wang, Q. Wang, S. Wang and X. Zou, m(6)A Reader HNRNPA2B1 Promotes Esophageal Cancer Progression via Up-Regulation of ACLY and ACC1, Front Oncol 10: ((2020) ), 553045. |
[19] | B. Pereira, M. Billaud and R. Almeida, RNA-Binding Proteins in Cancer: Old Players and New Actors, Trends Cancer 3: ((2017) ), 506–528. |
[20] | A.E. Brinegar and T.A. Cooper, Roles for RNA-binding proteins in development and disease, Brain Res 1647: ((2016) ), 1–8. |
[21] | C.J. David, M. Chen, M. Assanah, P. Canoll and J.L. Manley, HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer, Nature 463: ((2010) ), 364–8. |
[22] | X.H. Qu, J.L. Liu, X.W. Zhong, X.I. Li and Q.G. Zhang, Insights into the roles of hnRNP A2/B1 and AXL in non-small cell lung cancer, Oncol Lett 10: ((2015) ), 1677–1685. |
[23] | Y. Xuan, J. Wang, L. Ban, J.J. Lu, C. Yi, Z. Li, W. Yu, M. Li, T. Xu, W. Yang, Z. Tang, R. Tang, X. Xiao, S. Meng, Y. Chen, Q. Liu, W. Huang, W. Guo, X. Cui and W. Deng, hnRNPA2/B1 activates cyclooxygenase-2 and promotes tumor growth in human lung cancers, Mol Oncol 10: ((2016) ), 610–24. |
[24] | W.Z. Peng, J. Zhao, X. Liu, C.F. Li, S. Si and R. Ma, hnRNPA2B1 regulates the alternative splicing of BIRC5 to promote gastric cancer progression, Cancer Cell Int 21: ((2021) ), 281. |
[25] | Y. Hu, Z. Sun, J. Deng, B. Hu, W. Yan, H. Wei and J. Jiang, Splicing factor hnRNPA2B1 contributes to tumorigenic potential of breast cancer cells through STAT3 and ERK1/2 signaling pathway, Tumour Biol 39: ((2017) ), 1010428317694318. |
[26] | X. Zhao, J. Liu, S. Ge, C. Chen, S. Li, X. Wu, X. Feng, Y. Wang and D. Cai, Saikosaponin A Inhibits Breast Cancer by Regulating Th1/Th2 Balance, Front Pharmacol 10: ((2019) ), 624. |
[27] | Z. Liu, J. Zhong, J. Zeng, X. Duan, J. Lu, X. Sun, Q. Liu, Y. Liang, Z. Lin, W. Zhong, W. Wu, C. Cai and G. Zeng, Characterization of the m6A-Associated Tumor Immune Microenvironment in Prostate Cancer to Aid Immunotherapy, Front Immunol 12: ((2021) ), 735170. |




