A novel disulfidptosis-related prognostic gene signature and experimental validation identify ACTN4 as a novel therapeutic target in lung adenocarcinoma
Abstract
BACKGROUND:
Lung adenocarcinoma (LUAD) is a prevalent form of malignancy globally. Disulfidptosis is novel programmed cell death pathway based on disulfide proteins, may have a positive impact on the development of LUAD treatment strategies.
OBJECTIVE:
To investigate the impact of disulfidptosis-related genes (DRGs) on the prognosis of LUAD, developed a risk model to facilitate the diagnosis and prognostication of patients. We also explored ACTN4 (DRGs) as a new therapeutic biomarker for LUAD.
METHODS:
We investigated the expression patterns of DRGs in both LUAD and noncancerous tissues. To assess the prognostic value of the DRGs, we developed risk models through univariate Cox analysis and lasso regression. The expression and function of ACTN4 was evaluated by qRT-PCR, immunohistochemistry and in vitro experiments. The TIMER examined the association between ACTN4 expression and immune infiltration in LUAD.
RESULTS:
Ten differentially expressed DRGs were identified. And ACTN4 was identified as potential risk factors through univariate Cox regression analysis (
CONCLUSIONS:
Our study introduced a prognostic model based on DRGs, which could forecast the prognosis of patients with LUAD. The biomarker ACTN4 exhibits promise for the diagnosis and management of LUAD, given its correlation with tumor immune infiltration and m6A modification.
1.Introduction
According to research, lung cancer is the primary cause of cancer-related mortality on a global scale [1]. Specifically, non-small cell lung cancer comprises 85% of all cases, with lung adenocarcinoma (LUAD) being the most common subtype [2]. While driver mutations, including EGFR, KRAS, ALK, and TP53, play a critical role in LUAD [3, 4, 5], only about 30% of patients benefit from targeted treatment [6]. Despite the implementation of diverse therapeutic approaches, including surgery, radiotherapy, chemotherapy, and immunotherapy, certain patients continue to encounter postoperative recurrence and metastasis [7, 8]. Therefore, there exists a pressing necessity to devise dependable and efficacious prognostic biomarkers, construct risk prognostic models, and guide physicians in evaluating patients for individualized and optimal treatment. This is especially important given the rapid development of bioinformatics.
A recent study has identified a novel form of programmed cell death, termed disulfidptosis, which is reliant on disulfide proteins [9]. The current body of research indicates that the presence of disulfides is linked to changes in cellular redox state, which can induce the demise of neoplastic cells by modifying the conformation of cytoskeletal proteins. Consequently, this discovery may represent a significant advancement in the field of tumor therapy, and additional investigation and inquiry into the precise mechanism is warranted.
Disulfide metabolism pertains to the biochemical process through which disulfides are transformed into more stable compounds via diverse chemical reactions within the human body. Recent research has revealed that tumors exhibit anomalies in the expression and operation of enzymes responsible for disulfide metabolism, which could potentially be linked to the occurrence and treatment of cancer [10, 11, 12]. The aberrant expression and function of these disulfide-metabolizing enzymes may result in the accumulation and heightened toxicity of disulfides, thereby fostering the development and advancement of tumorigenesis. Moreover, the metabolism of disulfide bonds in cancer cells has been associated with various biological phenomena, including drug resistance, metastasis, and immune evasion, suggesting a potential correlation between disulfide-mediated apoptosis and tumor immune response [13, 14]. Notably, ACTN4, a cytoskeletal protein family member, is responsible for binding to actin filaments to uphold cytoskeletal architecture and cellular morphology [15, 16]. The migration of cancer cells within the extracellular matrix necessitates actin polymerization and rearrangement, thereby playing a pivotal role in cancer cell motility [17, 18]. Numerous studies have indicated that the upregulation of ACTN4 is commonly associated with an adverse prognosis, metastasis, and aggressive phenotype in various types of cancer [19, 20, 21]. High ACTN4 expression has been recognized as a prognostic indicator of platinum-based therapy outcome in LUAD. In the cohort of patients exhibiting high ACTN4 expression, the implementation of cisplatin-based adjuvant chemotherapy confers a significant clinical benefit in terms of overall survival [22, 23]. A growing body of literature suggests that tumor immunotherapy and N6-methyladenosine (m6A) are crucial factors in the development of LUAD [24, 25]. Nonetheless, the comprehension of ACTN4 in LUAD, particularly the correlation between ACTN4 and tumor immunotherapy and m6A modification, has received limited attention.
This research has developed a prognostic model utilizing DGRs to forecast the prognosis of LUAD patients. Additionally, the study has confirmed the role of ACTN4 in A549 and PC9 cells. A multidimensional analysis was performed to assess the gene and functional network associated with the expression of ACTN4 in LUAD, as well as to investigate the relationship between its expression and tumor immunity and m6A modification. The results of this investigation offer a theoretical foundation for identifying potential molecular mechanisms.
2.Materials and methods
2.1Data acquisitions
RNA-seq data and clinicopathological parameters sourced from The Cancer Genome Atlas and Genotype Tissue Expression Database, encompassing 574 LUAD samples and 288 normal lung samples. The DRGs set were derived from the latest research [11, 12].
2.2Patient tissue samples
From 2019 to 2022, fifty paired LUAD tissues and adjacent non-tumor tissues were obtained from patients who were diagnosed with LUAD at the Thoracic Surgery Department of the First Affiliated Hospital of Soochow University. The clinical information pertaining to these patients was retrieved from their medical records. The Ethics Committee of the First Affiliated Hospital of Soochow University granted approval for this study, and informed consent was obtained from all participants. And the study conforms with The Code of Ethics of the World Medical Association (Declaration of Helsinki), printed in the British Medical Journal (18 July 1964).
2.3Cell lines
The cell lines utilized in this study were procured from the Shanghai Institutes for Biological Sciences (China). Specifically, LUAD cells (NCI-H1975, PC9, and A549) were cultured in RPMI 1640 medium (KeyGene, Nanjing, China), and human bronchial epithelial cell (HBE) was cultured in DMEM medium (10% fetal bovine serum), respectively. All cells were maintained at 37∘C in a humidified incubator with 5% CO2.
2.4RNA extraction and qRT-PCR
The process of extracting Total RNA from tissue samples or cells was conducted through the utilization of TRIzol reagent (Invitrogen, Carlsbad, CA, USA) in adherence to the manufacturer’s guidelines. Subsequently, reverse transcription was executed using the Prime Script RT kit (Takara, Nanjing, China), and qRT-PCR was carried out using the SYBR Select Master Mix kit (KeyGEN, Nanjing, China). Each qRT-PCR reaction was performed in triplicate as follows: step 1: denaturation at 95∘C for 10 min; step 2: 40 cycles of 95∘C for 15 s and 60∘C for 1 min. The primers presented in the Supplementary Table 1.
2.5siRNA construction and cell transfection, cell proliferation assays
The siRNAs targeting ACTN4 was produced from RiboBio (Guangzhou, China), and the procedures for cell transfection and cell proliferation assays were executed in accordance with previously established protocols [26]. The sequences presented in the Supplementary Table 1.
2.6Immunohistochemistry (IHC)
IHC was conducted as described in refs. [26], and IHC was carried out using following antibodies: Anti-ACTN4 antibody (#15145; Cell Signaling Technology, USA).
2.7Identification of differentially expressed DRGs and prognostic genes
In this study, the “DESeq2” package was employed to detect DRGs that were differentially expressed (
2.8Construction of prognostic model based on DRGs
The standardized mRNA data from TCGA-LUAD was utilized to establish the risk score, which was determined through Cox regression analysis. The coefficient of DRGs was represented by X, while the expression level of DRGs was represented by Y. Subsequently, LUAD patients were classified into high-risk and low-risk groups based on the median risk score, and OS was analyzed.
The prognostic performance of the models was assessed using receiver operating characteristic (ROC) curves generated by the “timeROC” package.
2.9Construction of nomograms and calibration curves
Nomograms were generated utilizing the “RMS” package of R software to forecast individualized probabilities of survival, whereas calibration curves were constructed to prognosticate the survival rates of LUAD patients.
2.10TIMER analysis
TIMER2 (http://timer.cistrome.org/) is an interactive web-based tool that utilizes the deconvolution method to infer the gene expression spectrum of tumor-infiltrating immune cells across various cancer types from the TCGA dataset [27].
2.11Statistical analysis
The statistical analyses were performed using R software (version 4.1.1), and the experimental graphs were generated using GraphPad Prism software (version 8.0.). Unpaired t-tests were utilized for unpaired samples, while paired t-tests were employed for paired samples. The correlation of gene expression was evaluated using Spearman’s correlation. A statistical significance level of
3.Results
3.1Identification of differentially expressed DRGs in LUAD
Figure 1.
The differential expression of disulfidptosis-related genes (DRGs) in lung adenocarcinoma (LUAD) tissues compared to normal tissues. (A) Volcano plot indicated DRGs. (B) Protein-protein interaction network illustrated the interactions among DRGs. Heat map (C) and boxplots (D) of DRGs in LUAD compared the expression of DRGs in LUAD and normal tissues. (E) Mutation analysis of differentially expressed DRGs in TCGA-LUAD. *P < 0.05, and ***P< 0.001.
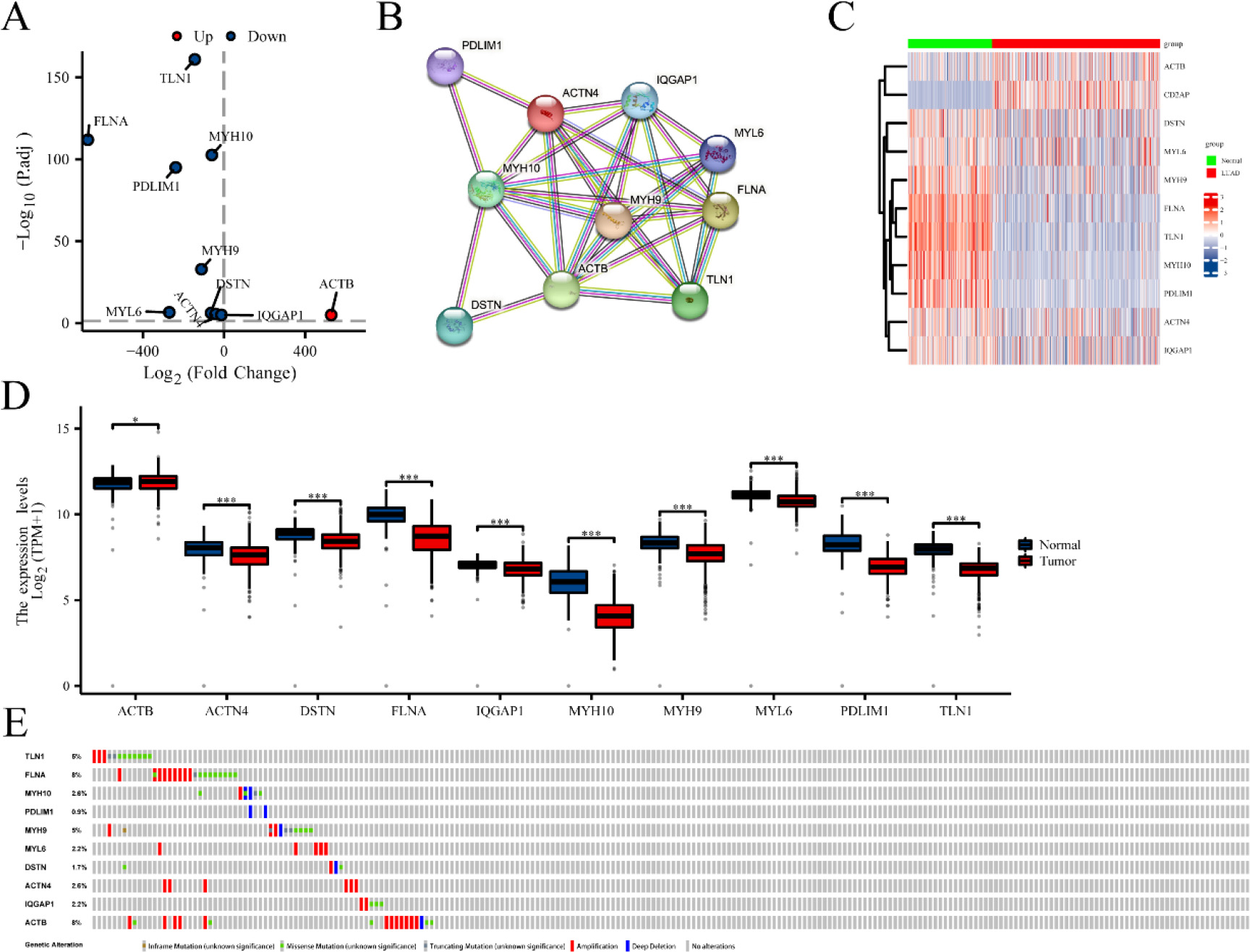
Fifteen genes (ACTB, ACTN4, CAPZB, CD2AP, DSTN, FLNA, FLNB, INF2, IQGAP1, MYH10, MYH9, MYL6, PDLIM1, SLC7A11 and TLN1) associated with disulfidptosis were cataloged [11, 12]. Differential expression of ten genes was identified from 15 DRGs using the R package “DESeq2” (
Figure 2.
Functional Annotation of disulfidptosis-related genes (DRGs) in TCGA-LUAD: Enrichment analyses in GO (A) and KEGG (B–C) pathways.
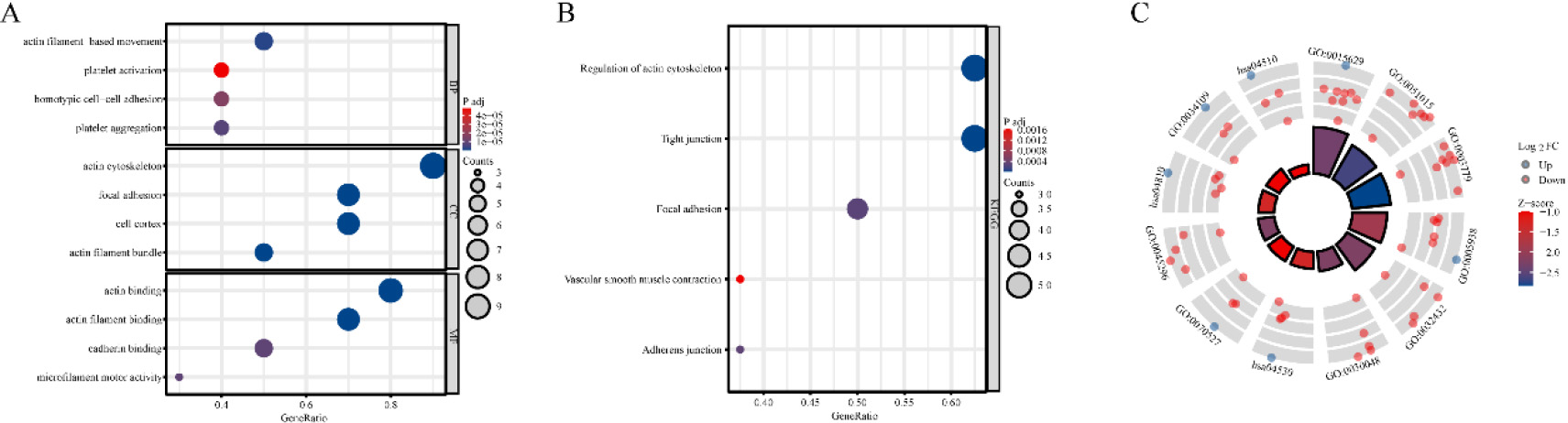
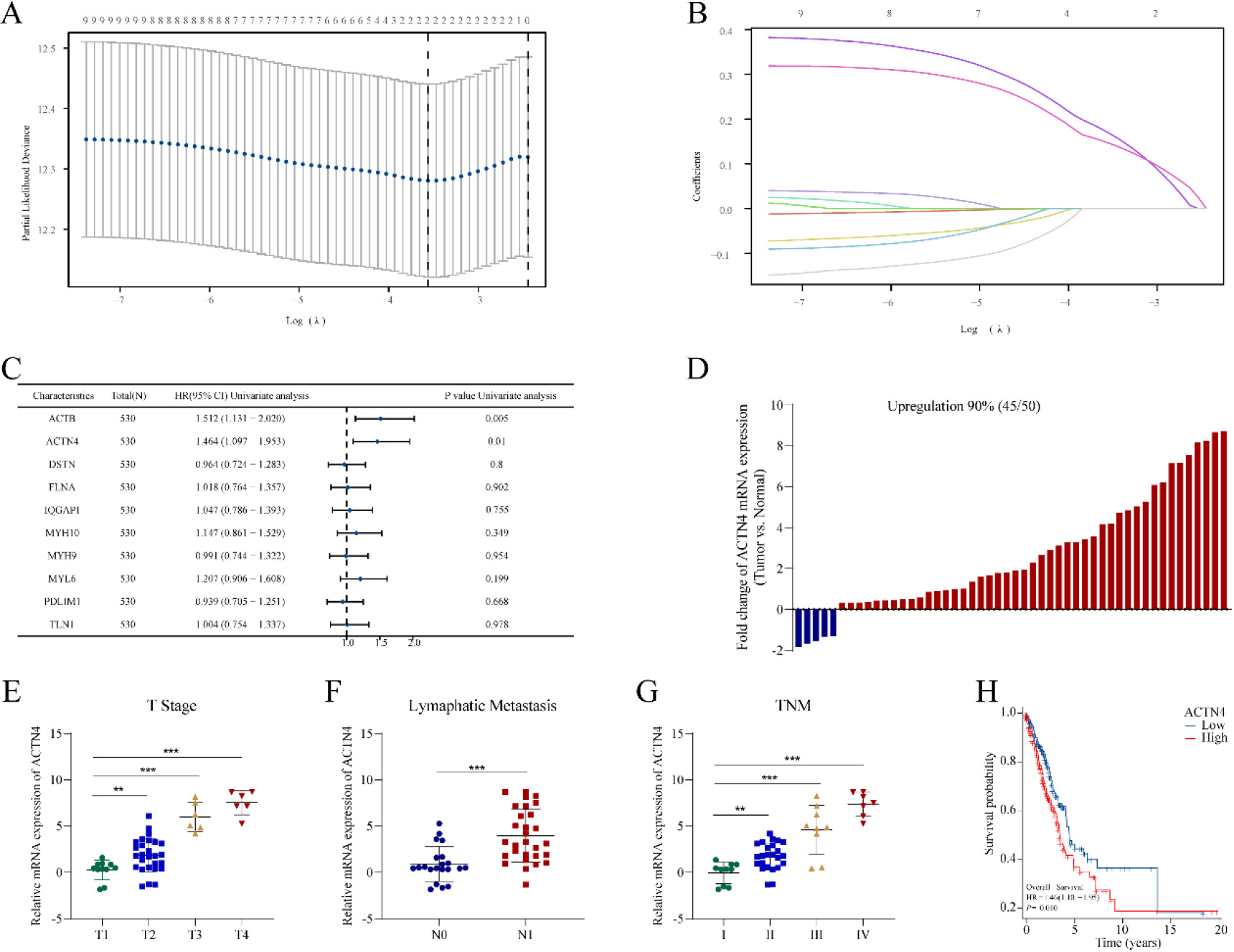
Figure 3.
Construction of a risk prognostic model based on disulfidptosis-related genes (DRGs) in TCGA-LUAD. (A) LASSO regression of DRGs. (B) Cross-validation for tuning the parameter selection in the LASSO regression. (C) Univariate Cox regression analysis of DRGs. (D) The mRNA expression of ACTN4 is upregulated in 90% of 50 lung adenocarcinoma (LUAD) tissues compared to normal tissues. The mRNA expression of ACTN4 was positively with (E) T stage (**P< 0.01), (F) lymphatic metastasis (**P< 0.001), and (G) TNM stage (**P < 0.01). (H) LUAD patients with high expression of ACTN4 have a lower percentage of overall survival. **P< 0.01, and ***P< 0.001.
To enhance the understanding of the functions of DRGs that exhibit differential expression, enrichment analyses were conducted on GO and KEGG pathways. The outcomes of the enrichment analyses indicated that these genes were predominantly associated with cytoskeletal components and cell-generated adhesion factors (Fig. 2A–C).
Table 1
Correlation between ACTN4 expression and clinicalpathological characteristics in TCGA-LUAD
| Characteristics | Low expression of ACTN4 | High expression of ACTN4 | |
|---|---|---|---|
|
| 269 | 270 | |
| Age, | 0.85995 | ||
| | 130 (25%) | 127 (24.4%) | |
| | 131 (25.2%) | 132 (25.4%) | |
| Gender, | 0.57664 | ||
| Female | 141 (26.2%) | 148 (27.5%) | |
| Male | 128 (23.7%) | 122 (22.6%) | |
| Smoker, | 0.18059 | ||
| No | 33 (6.3%) | 44 (8.4%) | |
| Yes | 229 (43.6%) | 219 (41.7%) | |
| T stage, | 0.04156 | ||
| T1 | 97 (18.1%) | 79 (14.7%) | |
| T2 | 147 (27.4%) | 145 (27.1%) | |
| T3 | 16 (3%) | 33 (6.2%) | |
| T4 | 8 (1.5%) | 11 (2.1%) | |
| N stage, | 0.05105 | ||
| N0 | 187 (35.8%) | 163 (31.2%) | |
| N1 | 42 (8%) | 55 (10.5%) | |
| N2&N3 | 31 (5.9%) | 45 (8.6%) | |
| M stage, | 0.30129 | ||
| M0 | 185 (47.4%) | 180 (46.2%) | |
| M1 | 10 (2.6%) | 15 (3.8%) | |
| TNM stage, | 0.01221 | ||
| Stage I | 166 (31.3%) | 130 (24.5%) | |
| Stage II | 53 (10%) | 72 (13.6%) | |
| Stage III | 34 (6.4%) | 50 (9.4%) | |
| Stage IV | 11 (2.1%) | 15 (2.8%) |
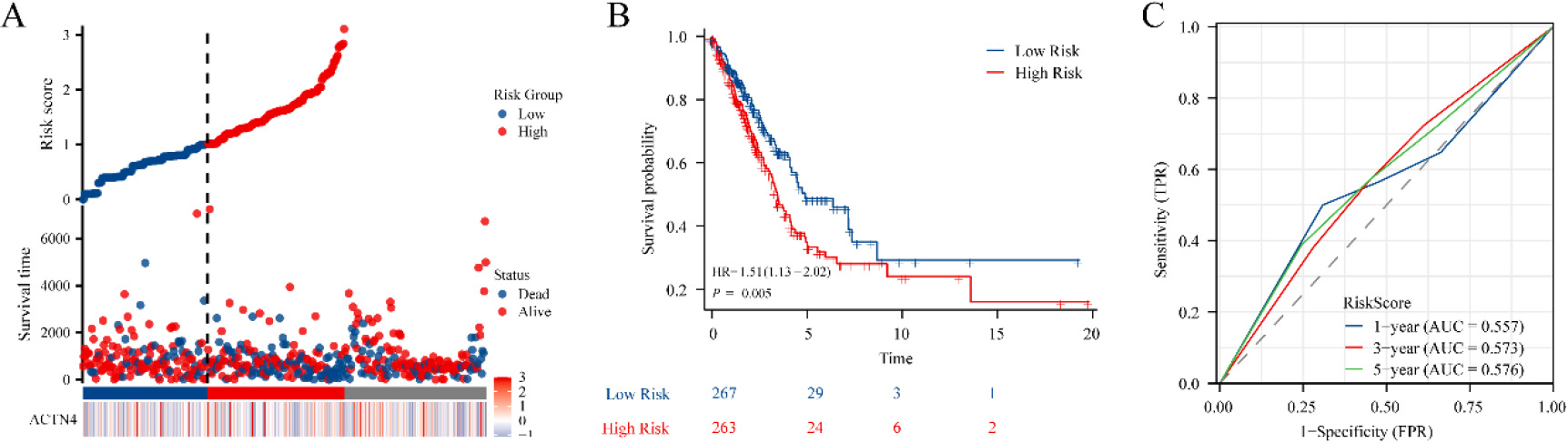
Figure 4.
Construction of the prognostic model based on ACTN4 and riskscore in TCGA-LUAD. (A) distribution of risk score, survival status and the expression of prognostic ACTN4. (B) Kaplan-Meier plot of the riskscore and overall survival. (C) ROCs for 1-year, 3-year and 5-year survival prediction.
3.2Construction of a prognostic model based on DRGs
A prognostic model was developed utilizing 10 DRGs through LASSO regression, as depicted in Fig. 3A–B. Subsequently, two genes (ACTB and ACTN4) (
Figure 5.
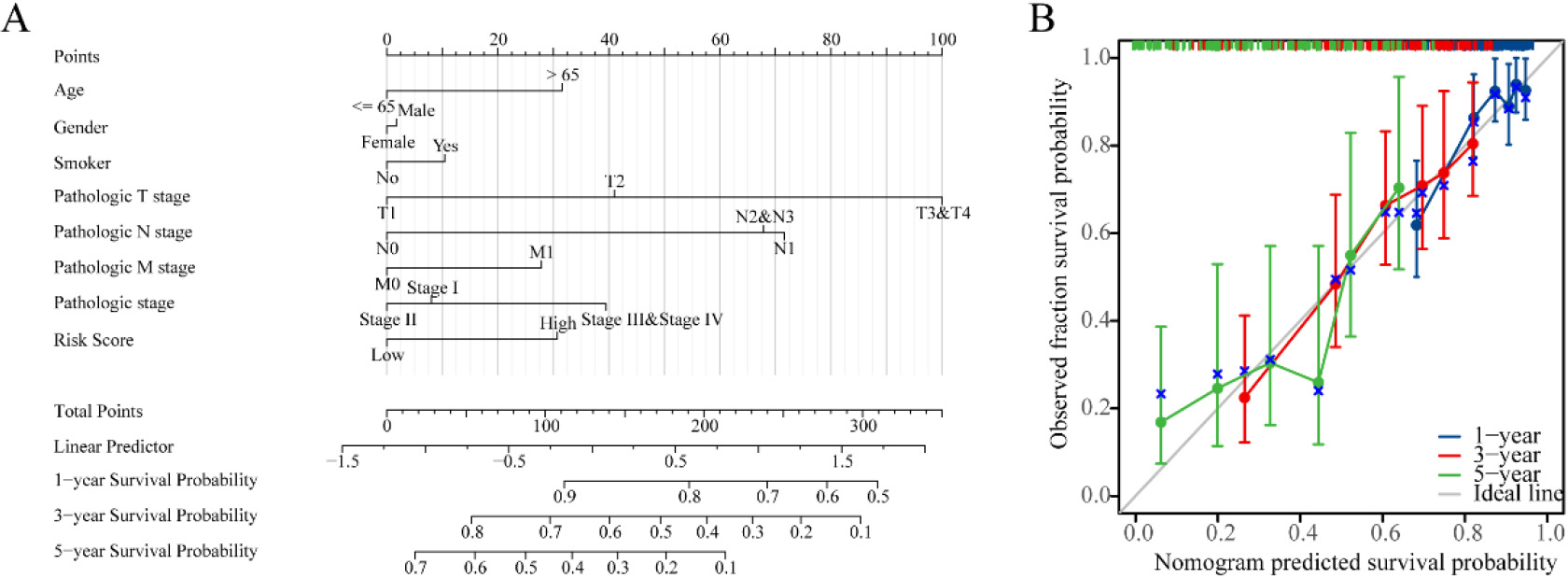
Nomogram development and validation. (A) Nomogram to predict the 1-year, 3-year and 5-year overall survival (OS) rate of lung adenocarcinoma (LUAD) patients. (B) Calibration curve for the OS nomogram model in LUAD.
3.3Construction of the prognostic model based on ACTN4 and Riskscore
To explore the potential of ACTN4 as a prognostic marker for LUAD, a prognostic model was constructed utilizing ACTN4 expression and riskscore. The findings revealed that the cohort identified as high-risk demonstrated a significantly higher mortality rate and a decreased duration of survival when compared to the low-risk cohort. Furthermore, elevated scores were indicative of an unfavorable prognosis in LUAD (Fig. 4A). The Kaplan Meier curve provided additional evidence that patients categorized in the high-risk group exhibited an unfavorable prognosis (
3.4Construction of nomogram and calibration curves
In order to improve the quantitative prognostication of LUAD, we developed a nomogram that integrates age, sex, smoking, T, N, M, stage and riskscore (Fig. 5A). Additionally, we generated a calibration curve that demonstrated a close alignment between the predicted and actual survival outcomes of LUAD patients (Fig. 5B). These results indicated that the inclusion of a risk score is a dependable method for forecasting the overall survival of individuals who have been diagnosed with LUAD.
3.5Knockdown of ACTN4 inhibited LUAD cell proliferation
Figure 6.
Knockdown of ACTN4 inhibited lung adenocarcinoma (LUAD) cell proliferation. (A) The ACTN4 staining score was up-regulated compared with that in adjacent normal tissues (
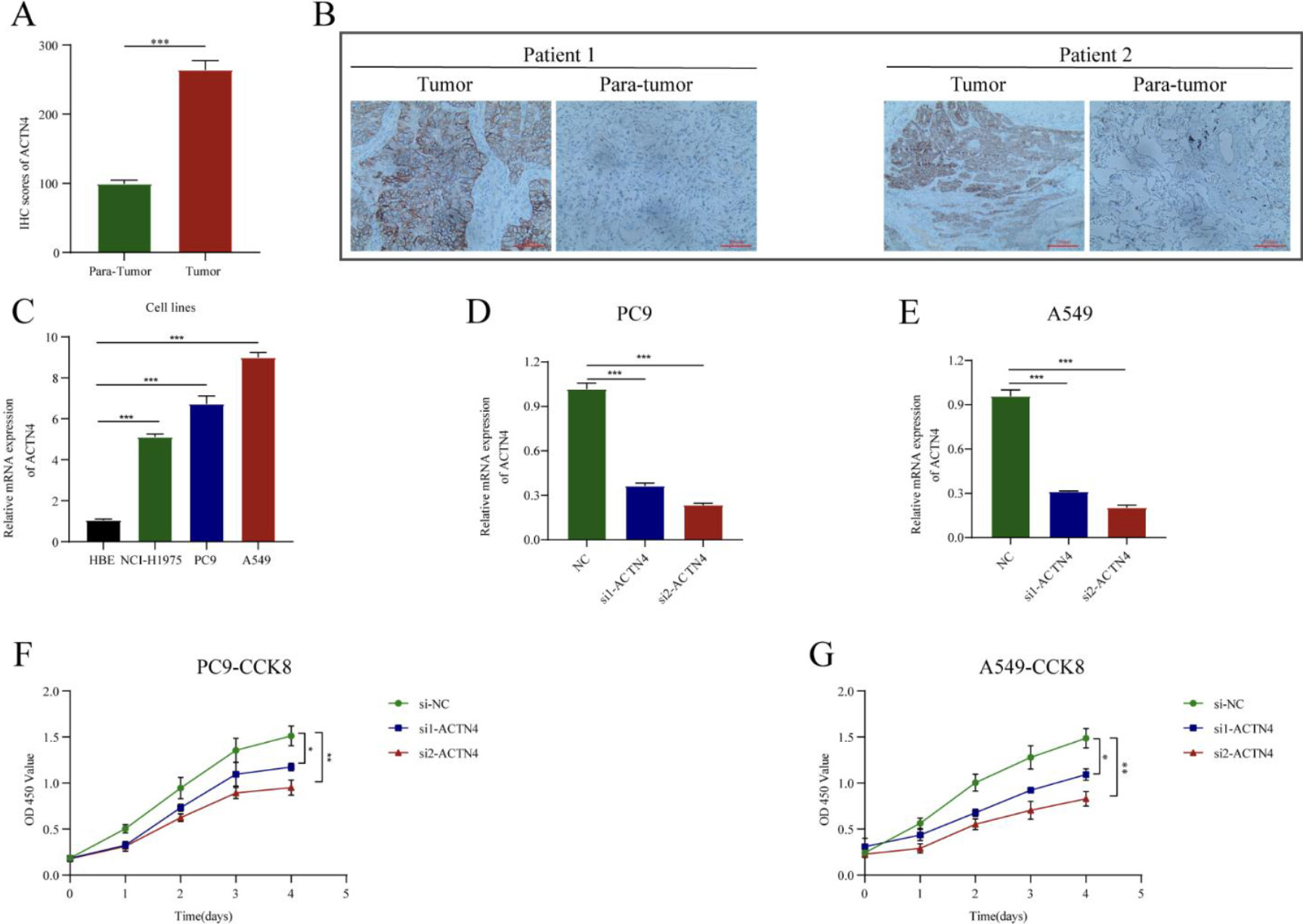
IHC staining showed that ACTN4 was significantly higher than in adjacent normal tissues compared to LUAD cancer tissues (
3.6ACTN4 expression is associated with immune signatures in LUAD
Figure 7.
Correlations of ACTN4 expression with immune infiltration level in lung adenocarcinoma (LUAD). (A) The expression of ACTN4 was significantly correlated with infiltrating levels of CD4
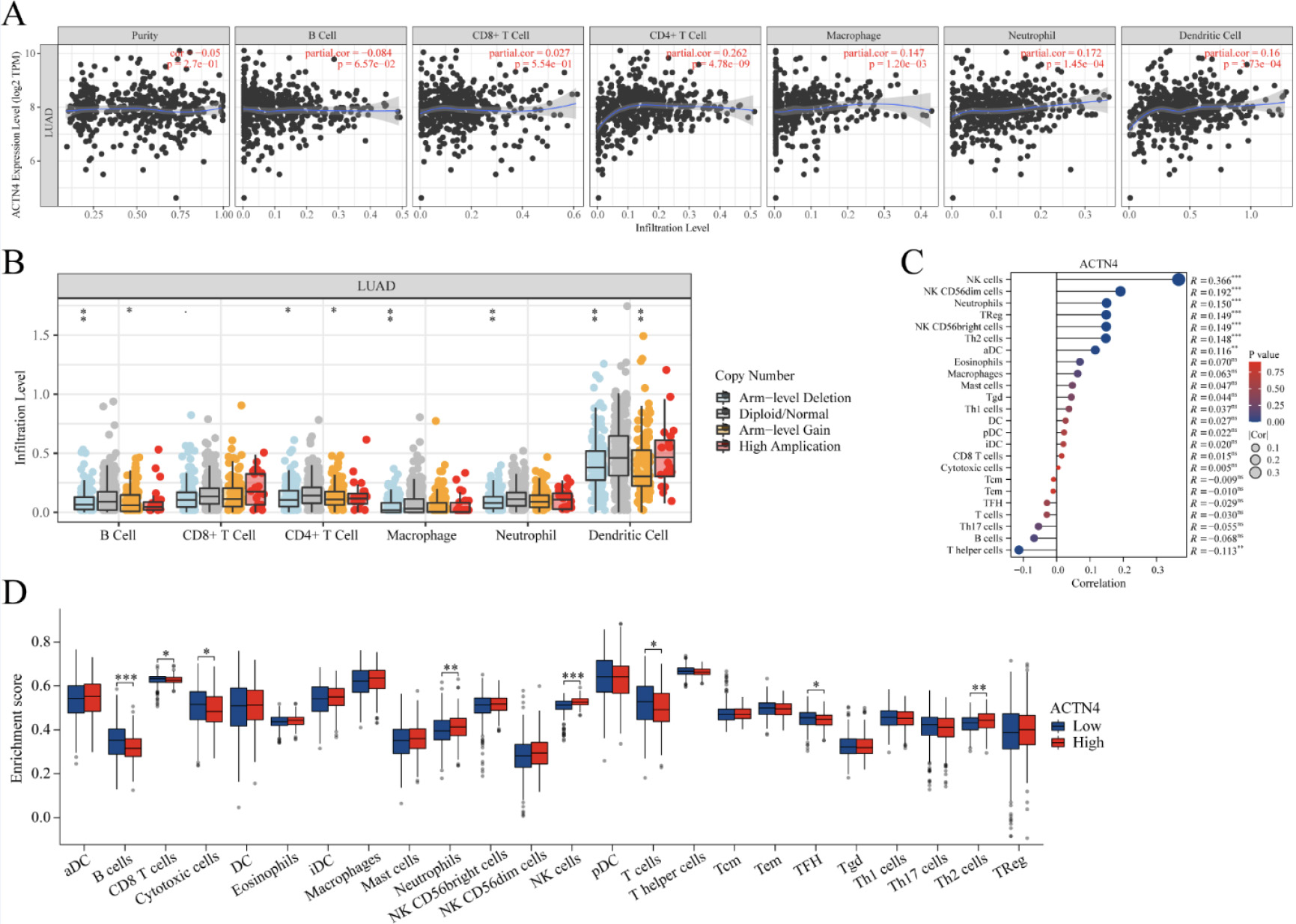
The existence of tumor-infiltrating lymphocytes is an autonomous predictor for both lymph node invasion and survival [28]. As depicted in Fig. 7A, TIMER2 website analysis indicated that ACTN4 expression was correlated with CD4
Table 2
Correlation analysis between ACTN4 and relate genes and markers of immune cells in TIMER
| Gene markers | Gene markers | partial.cor | |
|---|---|---|---|
| B cell | CD19 | 1.87E-02* | |
| CD20 | 7.44E-03** | ||
| CD70 | 0.082183 | 6.83E-02 | |
| CD8 | CD8A | 0.041848 | 3.54E-01 |
| CD8B | 0.000887 | 9.84E-01 | |
| CD25 | 0.040319 | 3.72E-01 | |
| Tfh | CD183 | 0.125097 | 5.41E-03** |
| CD185 | 0.013307 | 7.68E-01 | |
| CD278 | 2.71E-01 | ||
| Th1 | CD212 | 0.106543 | 1.80E-02* |
| CD191 | 0.081040 | 7.22E-02 | |
| CD195 | 0.129645 | 3.93E-03** | |
| Th2 | CD194 | 2.45E-01 | |
| CD198 | 0.043452 | 3.36E-01 | |
| CD365 | 0.031213 | 4.89E-01 | |
| Th17 | CD360 | 0.103248 | 2.19E-02* |
| IL23R | 4.34E-01 | ||
| CD196 | 1.76E-01 | ||
| Treg | FOXP3 | 0.141300 | 1.66E-03** |
| CD73 | 0.217631 | 1.07E-06*** | |
| CD127 | 0.025895 | 5.66E-01 | |
| T cell exhaustion | PD-1 | 0.120094 | 7.60E-03** |
| CTLA4 | 5.64E-01 | ||
| LAG3 | 0.149673 | 8.57E-04*** | |
| Macrophage | CD68 | 0.161359 | 3.22E-04*** |
| CD11b | 0.197407 | 1.01E-05*** | |
| M1 macrophage | NOS2 | 0.126003 | 5.08E-03** |
| IRF5 | 0.181242 | 5.18E-05*** | |
| M2 macrophage | CD163 | 0.150301 | 8.15E-04*** |
| CD206 | 0.042607 | 3.45E-01 | |
| TAM | CCL2 | 0.116461 | 9.65E-03** |
| CD86 | 0.161359 | 3.22E-04*** | |
| Monocyte | CD14 | 0.105238 | 1.94E-02* |
| CD33 | 0.013952 | 7.57E-01 | |
| Natural killer cell | CD57 | 0.081677 | 7.00E-02 |
| KIR3DL1 | 0.073553 | 1.03E-01 | |
| CD7 | 0.125883 | 5.12E-03** | |
| Neutrophl | CD16 | 0.114612 | 1.09E-02* |
| CD55 | 6.16E-03** | ||
| Dendritic cell | CD1C | 9.63E-03** | |
| CD141 | 0.165256 | 2.28E-04*** |
In order to examine the relationship between ACTN4 and a range of immune infiltrating cells in LUAD, the TIMER tool was employed to assess the correlation of ACTN4 with multiple immune cell markers in LUAD (Table 2). The findings indicated a noteworthy correlation between the expression of ACTN4 and the expression of B cell immune markers CD20 and CD19 (
Furthermore, 539 LUAD samples were classified into two distinct groups based on their ACTN4 expression levels (high
3.7ACTN4 Expression is Associated with m6A RNA Methylation Regulators in LUAD
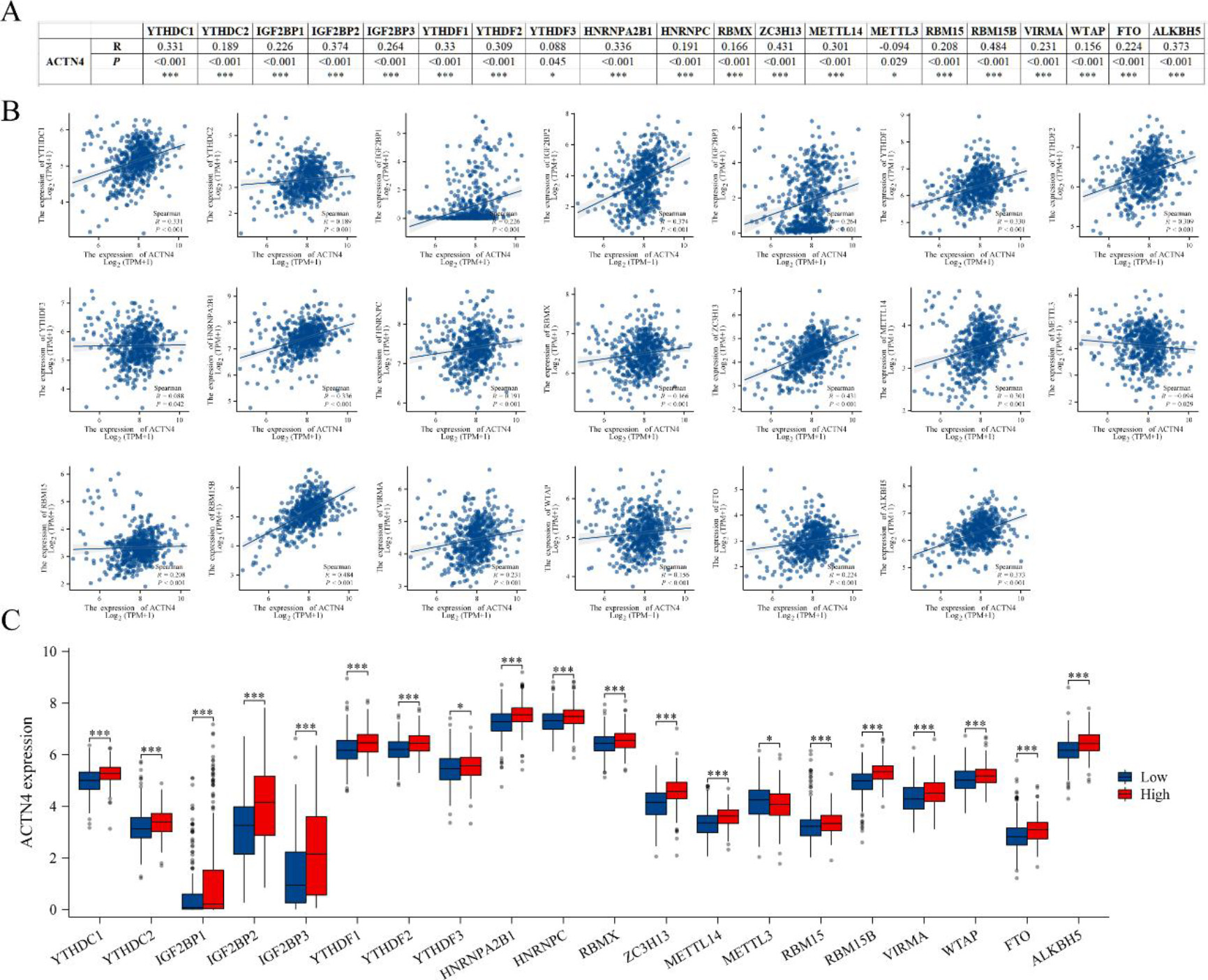
The acknowledged importance of m6A modification in the progression of LUAD prompted our analysis of the expression correlation between ACTN4 and 20 m6A-related genes in the TCGA LUAD dataset [25, 29]. Our findings indicate a significant correlation between ACTN4 expression and the 20 m6A-related genes in LUAD (
Figure 8.
Correlations of ACTN4 expression with m6A related genes in lung adenocarcinoma (LUAD). (A) TCGA-LUAD analyzed the correlation between the expression level of ACTN4 and m6A-related genes. (B) Draw a scatter plot to show the correlation between ACTN4 and m6A related genes. (C) The differential expression of m6A related genes in the high and low ACTN4 expression groups in LUAD. *P < 0.05, and ***P < 0.001.
4.Discussion
LUAD is a prevalent and perilous neoplasm characterized by a high incidence of metastasis and mortality, and represents a significant area of interest for cancer research [1]. To effectively address this disease and elucidate its underlying mechanisms, a more profound understanding is necessary. Recent investigations have uncovered a novel mode of cell death, known as disulfidptosis, which has been implicated in cancer-related pathways [9, 12]. Disulfide has been shown to mitigate cellular oxidative stress by activating the antioxidant enzyme system, thereby impeding tumorigenesis and progression [30]. For example, tumor cells have the ability to manipulate the intracellular redox environment by utilizing disulfides, which facilitates the growth and dissemination of tumors [31]. Antineoplastic agents, including cisplatin and paclitaxel, hinder tumor initiation and progression by interacting with intracellular disulfides [32, 33]. Consequently, the concept of disulfidptosis presents a promising opportunity for the advancement of therapeutic interventions for LUAD.
The aim of this study was to construct a prognostic model that employs DRGs to diagnose and forecast the prognosis of patients with LUAD. Initially, potential risk genes were identified through the use of univariate Cox and Lasso Cox regression analyses, with ACTN4 being singled out. Subsequently, a prognostic model was developed utilizing the expression of ACTN4 and riskscore, which exhibited effectiveness in prognosticating the outcome of LUAD. As a result, experimental investigations were conducted to elucidate the precise role of ACTN4. IHC analysis revealed that the expression of ACTN4 protein in LUAD was higher than in adjacent tissues, and its expression was correlated with clinicopathological features. In vitro cell experiments demonstrated that the inhibition of ACTN4 expression could suppress the proliferation of LUAD cells, which is in accordance with previous studies [34].
ACTN4 expression level was significantly correlated with a variety of immune cells and immune cell marker genes. Notably, high expression of ACTN4 was positively correlated with increased levels of neutrophils, NK cells, and Th2 cells, while the levels of B cells, CD8
The presence of M6A modification in eukaryotic RNA is evident in numerous biological processes, particularly in the promotion of tumor development within the tumor immune microenvironment [36]. The current investigation has demonstrated a noteworthy association between the expression of ACTN4 and 20 m6A-related genes. Furthermore, the heightened expression of ACTN4 has been linked to elevated levels of 19 m6A-related genes, except for METTL3 which displayed a reduction. These observations imply that ACTN4 may undergo m6A modification, thereby enhancing mRNA stability and ultimately influencing the tumor immune microenvironment, thereby facilitating tumor progression.
Disulfidptosis, a novel form of cell death, was identified by Liu et al. [9]. They observed that sugar-deficient cancer cells with high expression of SLC7A11 experienced disrupted disulfide binding between cytoskeletal proteins due to the accumulation of disulfide substances, leading to histone skeleton breakdown and cell death. SLC7A11, a key determinant of glucose deficiency-induced cell death in various cancer cell lines [37, 38], is frequently overexpressed in multiple cancers, often correlating with poor patient outcomes [39, 40, 41]. Studies have shown that SLC7A11 knockout mice exhibit no distinct phenotype in major organs, indicating its potential as a promising therapeutic target in cancer treatment [42]. Recent research has focused on exploiting SLC7A11-associated metabolic vulnerabilities (such as glucose or glutamine dependence) to induce disulfidptosis, offering new strategies for cancer therapy [43]. Liu et al. [44] demonstrated that compared to cancer cells with low SLC7A11 expression, inhibitors of glucose transporters (GLUT) were more effective in inducing disulfidptosis in cells with high SLC7A11 expression. The use of KL-11743, a potent inhibitor of GLUT1 and GLUT3, selectively suppressed the growth of SLC7A11-expressing tumors in cell line xenografts and lung cancer patient-derived xenografts [45]. Furthermore, high expression of SLC7A11 can serve as a biomarker for selecting cancer patients who may benefit from glutaminase or GLUT inhibitors for disulfidptosis treatment, offering a new avenue for targeting cancer metabolism weaknesses through metabolic therapy.
However, there are some limitations to our study. This was a retrospective study, with data from public databases lacking information such as treatment and relapse records. Our conclusions need to be validated in vivo or in vitro experiments and prospective clinical studies.
5.Conclusions
In brief, the current study has developed a novel prognostic model utilizing DRGs, which demonstrates a high degree of efficacy in predicting the prognosis of patients with LUAD. Furthermore, in vitro experiments have revealed a significant upregulation of ACTN4 in LUAD, which is closely associated with the clinicopathological features of patients and can facilitate the proliferation of LUAD cells. Additionally, a comprehensive analysis has been conducted to investigate the correlation between ACTN4 expression and tumor immune infiltration as well as m6A modification. The expression of ACTN4 displays a strong association with various immune cells, thereby potentially impeding the infiltration of memory B cells and influencing the immune response against tumors. The m6A-mediated modification of ACTN4 mRNA may augment its stability, consequently impacting the tumor immune microenvironment and facilitating the progression of LUAD.
Author contributions
Conception: Kai Xie, Haitao Ma, Yu Feng and Wei Jiang.
Interpretation or analysis of data: Bin Wang, Pei Pang and Guangbin Li.
Preparation of the manuscript: Kai Xie, Qianqian Yang and Chen Fang.
Revision for important intellectual content: Kai Xie, Haitao Ma, Yu Feng and Wei Jiang.
Supervision: Haitao Ma, Yu Feng and Wei Jiang.
Supplementary data
The supplementary files are available to download from http://dx.doi.org/10.3233/CBM-230276.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 82101215), Suzhou Ke Jiao Xing Wei Youth Science and Technology Projects (No. KJXW2022006, No. KJXW2022084), Suzhou Medical Innovation Applied Research Project (No. SKY2022142), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX23_3275).
References
[1] | R.L. Siegel, K.D. Miller, N.S. Wagle and A. Jemal, Cancer statistics, CA Cancer J Clin 73: ((2023) ), 17–48. |
[2] | R.S. Herbst, D. Morgensztern and C. Boshoff, The biology and management of non-small cell lung cancer, Nature 553: ((2018) ), 446–454. |
[3] | F. Skoulidis and J.V. Heymach, Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy, Nat Rev Cancer 19: ((2019) ), 495–509. |
[4] | A. Leonetti, S. Sharma, R. Minari, P. Perego, E. Giovannetti and M. Tiseo, Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer, Br J Cancer 121: ((2019) ), 725–737. |
[5] | H. Liu, B. Zhang and Z. Sun, Spectrum of EGFR aberrations and potential clinical implications: insights from integrative pan-cancer analysis, Cancer Commun (Lond) 40: ((2020) ), 43–59. |
[6] | Comprehensive molecular profiling of lung adenocarcinoma, Nature 511: ((2014) ), 543–50. |
[7] | H. Wang, Q. Deng, Z. Lv, Y. Ling, X. Hou, Z. Chen, X. Dinglin, S. Ma, D. Li, Y. Wu, Y. Peng, H. Huang and L. Chen, N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1, Mol Cancer 18: ((2019) ), 181. |
[8] | J. Zhan, P. Wang, S. Li, J. Song, H. He, Y. Wang, Z. Liu, F. Wang, H. Bai, W. Fang, Q. Du, M. Ye, Z. Chang, J. Wang and H. Zhang, HOXB13 networking with ABCG1/EZH2/Slug mediates metastasis and confers resistance to cisplatin in lung adenocarcinoma patients, Theranostics 9: ((2019) ), 2084–2099. |
[9] | X. Liu, L. Nie, Y. Zhang, Y. Yan, C. Wang, M. Colic, K. Olszewski, A. Horbath, X. Chen, G. Lei, C. Mao, S. Wu, L. Zhuang, M.V. Poyurovsky, M. James You, T. Hart, D.D. Billadeau, J. Chen and B. Gan, Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis, Nat Cell Biol 25: ((2023) ), 404–414. |
[10] | R.H. Wang, Y.H. Chu and K.T. Lin, The Hidden Role of Hydrogen Sulfide Metabolism in Cancer, Int J Mol Sci 22: ((2021) ). |
[11] | S. Zhao, L. Wang, W. Ding, B. Ye, C. Cheng, J. Shao, J. Liu and H. Zhou, Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework, Front Endocrinol (Lausanne) 14: ((2023) ), 1180404. |
[12] | H. Liu and T.J.b. Tang, Descriptive pan-cancer genetic analysis of disulfidptosis-related gene set, ((2023) ). |
[13] | Y. Wang, Y. Jiang, D. Wei, P. Singh, Y. Yu, T. Lee, L. Zhang, H.K. Mandl, A.S. Piotrowski-Daspit, X. Chen, F. Li, X. Li, Y. Cheng, A. Josowitz, F. Yang, Y. Zhao, F. Wang, Z. Zhao, A. Huttner, R.S. Bindra, H. Xiao and W. Mark Saltzman, Nanoparticle-mediated convection-enhanced delivery of a DNA intercalator to gliomas circumvents temozolomide resistance, Nat Biomed Eng 5: ((2021) ), 1048–1058. |
[14] | C. Chen, M. Shen, H. Liao, Q. Guo, H. Fu, J. Yu and Y. Duan, A paclitaxel and microRNA-124 coloaded stepped cleavable nanosystem against triple negative breast cancer, J Nanobiotechnology 19: ((2021) ), 55. |
[15] | K. Honda, T. Yamada, R. Endo, Y. Ino, M. Gotoh, H. Tsuda, Y. Yamada, H. Chiba and S. Hirohashi, Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion, J Cell Biol 140: ((1998) ), 1383–93. |
[16] | A. Virel and L. Backman, Molecular evolution and structure of alpha-actinin, Mol Biol Evol 21: ((2004) ), 1024–31. |
[17] | M.R. Mejillano, S. Kojima, D.A. Applewhite, F.B. Gertler, T.M. Svitkina and G.G. Borisy, Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end, Cell 118: ((2004) ), 363–73. |
[18] | S. Khurana and S.P. George, The role of actin bundling proteins in the assembly of filopodia in epithelial cells, Cell Adh Migr 5: ((2011) ), 409–20. |
[19] | K. Honda, The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer, Cell Biosci 5: ((2015) ), 41. |
[20] | D. Tentler, E. Lomert, K. Novitskaya and N.A. Barlev, Role of ACTN4 in Tumorigenesis, Metastasis, and EMT, Cells 8: ((2019) ). |
[21] | S. Park, M. Kang, S. Kim, H.T. An, J. Gettemans and J. Ko, α-Actinin-4 Promotes the Progression of Prostate Cancer Through the Akt/GSK-3β/β-Catenin Signaling Pathway, Front Cell Dev Biol 8: ((2020) ), 588544. |
[22] | H. Shiraishi, Y. Fujiwara, T. Kakuya, K. Tsuta, N. Motoi, N. Miura, Y. Watabe, S.I. Watanabe, R. Noro, K. Nagashima, W. Huang, T. Yamada, H. Asamura, Y. Ohe and K. Honda, Actinin-4 protein overexpression as a predictive biomarker in adjuvant chemotherapy for resected lung adenocarcinoma, Biomark Med 11: ((2017) ), 721–731. |
[23] | N. Miura, M. Kamita, T. Kakuya, Y. Fujiwara, K. Tsuta, H. Shiraishi, F. Takeshita, T. Ochiya, H. Shoji, W. Huang, Y. Ohe, T. Yamada and K. Honda, Efficacy of adjuvant chemotherapy for non-small cell lung cancer assessed by metastatic potential associated with ACTN4, Oncotarget 7: ((2016) ), 33165–78. |
[24] | C. Zhang, Q. Sun, X. Zhang, N. Qin, Z. Pu, Y. Gu, C. Yan, M. Zhu, J. Dai, C. Wang, N. Li, G. Jin, H. Ma, Z. Hu, E. Zhang, F. Tan and H. Shen, Gene amplification-driven RNA methyltransferase KIAA1429 promotes tumorigenesis by regulating BTG2 via m6A-YTHDF2-dependent in lung adenocarcinoma, Cancer Commun (Lond) 42: ((2022) ), 609–626. |
[25] | H. Song, D. Liu, L. Wang, K. Liu, C. Chen, L. Wang, Y. Ren, B. Ju, F. Zhong, X. Jiang, G. Wang, Z.S. Chen and C. Zou, Methyltransferase like 7B is a potential therapeutic target for reversing EGFR-TKIs resistance in lung adenocarcinoma, Mol Cancer 21: ((2022) ), 43. |
[26] | K. Xie, J. Feng, D. Fan, S. Wang, J. Luo, Z. Ren, C. Zheng, Y. Diao, R.A. De Mello, S. Tavolari, G. Brandi, A.C. Roden, B. Ren, Y. Shen and L. Xu, BARX2/FOXA1/HK2 axis promotes lung adenocarcinoma progression and energy metabolism reprogramming, Transl Lung Cancer Res 11: ((2022) ), 1405–1419. |
[27] | T. Li, J. Fu, Z. Zeng, D. Cohen, J. Li, Q. Chen, B. Li and X.S. Liu, TIMER2.0 for analysis of tumor-infiltrating immune cells, Nucleic Acids Res 48: ((2020) ), W509–w514. |
[28] | F. Azimi, R.A. Scolyer, P. Rumcheva, M. Moncrieff, R. Murali, S.W. McCarthy, R.P. Saw and J.F. Thompson, Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma, J Clin Oncol 30: ((2012) ), 2678–83. |
[29] | Y. Li, J. Gu, F. Xu, Q. Zhu, Y. Chen, D. Ge and C. Lu, Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma, Brief Bioinform 22: ((2021) ). |
[30] | H.Y. Min and H.Y. Lee, Oncogene-Driven Metabolic Alterations in Cancer, Biomol Ther (Seoul) 26: ((2018) ), 45–56. |
[31] | E.W. Iyamu, The redox state of the glutathione/glutathione disulfide couple mediates intracellular arginase activation in HCT-116 colon cancer cells, Dig Dis Sci 55: ((2010) ), 2520–8. |
[32] | S. Sideris, F. Aoun, M. Zanaty, N.C. Martinez, S. Latifyan, A. Awada and T. Gil, Efficacy of weekly paclitaxel treatment as a single agent chemotherapy following first-line cisplatin treatment in urothelial bladder cancer, Mol Clin Oncol 4: ((2016) ), 1063–1067. |
[33] | T. Mitin, D. Hunt, W.U. Shipley, D.S. Kaufman, R. Uzzo, C.L. Wu, M.K. Buyyounouski, H. Sandler and A.L. Zietman, Transurethral surgery and twice-daily radiation plus paclitaxel-cisplatin or fluorouracil-cisplatin with selective bladder preservation and adjuvant chemotherapy for patients with muscle invasive bladder cancer (RTOG 0233): a randomised multicentre phase 2 trial, Lancet Oncol 14: ((2013) ), 863–72. |
[34] | M.C. Wang, Y.H. Chang, C.C. Wu, Y.C. Tyan, H.C. Chang, Y.G. Goan, W.W. Lai, P.N. Cheng and P.C. Liao, Alpha-actinin 4 is associated with cancer cell motility and is a potential biomarker in non-small cell lung cancer, J Thorac Oncol 10: ((2015) ), 286–301. |
[35] | B.A. Helmink, S.M. Reddy, J. Gao, S. Zhang, R. Basar, R. Thakur, K. Yizhak, M. Sade-Feldman, J. Blando, G. Han, V. Gopalakrishnan, Y. Xi, H. Zhao, R.N. Amaria, H.A. Tawbi, A.P. Cogdill, W. Liu, V.S. LeBleu, F.G. Kugeratski, S. Patel, M.A. Davies, P. Hwu, J.E. Lee, J.E. Gershenwald, A. Lucci, R. Arora, S. Woodman, E.Z. Keung, P.O. Gaudreau, A. Reuben, C.N. Spencer, E.M. Burton, L.E. Haydu, A.J. Lazar, R. Zapassodi, C.W. Hudgens, D.A. Ledesma, S. Ong, M. Bailey, S. Warren, D. Rao, O. Krijgsman, E.A. Rozeman, D. Peeper, C.U. Blank, T.N. Schumacher, L.H. Butterfield, M.A. Zelazowska, K.M. McBride, R. Kalluri, J. Allison, F. Petitprez, W.H. Fridman, C. Sautès-Fridman, N. Hacohen, K. Rezvani, P. Sharma, M.T. Tetzlaff, L. Wang and J.A. Wargo, B cells and tertiary lymphoid structures promote immunotherapy response, Nature 577: ((2020) ), 549–555. |
[36] | H. Zhou, M. Zheng, M. Shi, J. Wang, Z. Huang, H. Zhang, Y. Zhou and J. Shi, Characteristic of molecular subtypes in lung adenocarcinoma based on m6A RNA methylation modification and immune microenvironment, BMC Cancer 21: ((2021) ), 938. |
[37] | C.S. Shin, P. Mishra, J.D. Watrous, V. Carelli, M. D’Aurelio, M. Jain and D.C. Chan, The glutamate/cystine xCT antiporter antagonizes glutamine metabolism and reduces nutrient flexibility, Nat Commun 8: ((2017) ), 15074. |
[38] | N.N. Pavlova and C.B. Thompson, The Emerging Hallmarks of Cancer Metabolism, Cell Metab 23: ((2016) ), 27–47. |
[39] | X. Ji, J. Qian, S.M.J. Rahman, P.J. Siska, Y. Zou, B.K. Harris, M.D. Hoeksema, I.A. Trenary, C. Heidi, R. Eisenberg, J.C. Rathmell, J.D. Young and P.P. Massion, xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression, Oncogene 37: ((2018) ), 5007–5019. |
[40] | M.A. Badgley, D.M. Kremer, H.C. Maurer, K.E. DelGiorno, H.J. Lee, V. Purohit, I.R. Sagalovskiy, A. Ma, J. Kapilian, C.E.M. Firl, A.R. Decker, S.A. Sastra, C.F. Palermo, L.R. Andrade, P. Sajjakulnukit, L. Zhang, Z.P. Tolstyka, T. Hirschhorn, C. Lamb, T. Liu, W. Gu, E.S. Seeley, E. Stone, G. Georgiou, U. Manor, A. Iuga, G.M. Wahl, B.R. Stockwell, C.A. Lyssiotis and K.P. Olive, Cysteine depletion induces pancreatic tumor ferroptosis in mice, Science 368: ((2020) ), 85–89. |
[41] | T. Goji, K. Takahara, M. Negishi and H. Katoh, Cystine uptake through the cystine/glutamate antiporter xCT triggers glioblastoma cell death under glucose deprivation, J Biol Chem 292: ((2017) ), 19721–19732. |
[42] | H. Sato, A. Shiiya, M. Kimata, K. Maebara, M. Tamba, Y. Sakakura, N. Makino, F. Sugiyama, K. Yagami, T. Moriguchi, S. Takahashi and S. Bannai, Redox imbalance in cystine/glutamate transporter-deficient mice, J Biol Chem 280: ((2005) ), 37423–9. |
[43] | P. Koppula, L. Zhuang and B. Gan, Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy, Protein Cell 12: ((2021) ), 599–620. |
[44] | X. Liu, K. Olszewski, Y. Zhang, E.W. Lim, J. Shi, X. Zhang, J. Zhang, H. Lee, P. Koppula, G. Lei, L. Zhuang, M.J. You, B. Fang, W. Li, C.M. Metallo, M.V. Poyurovsky and B. Gan, Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer, Nat Cell Biol 22: ((2020) ), 476–486. |
[45] | L. Jiang, N. Kon, T. Li, S.J. Wang, T. Su, H. Hibshoosh, R. Baer and W. Gu, Ferroptosis as a p53-mediated activity during tumour suppression, Nature 520: ((2015) ), 57–62. |




