Long-chain noncoding RNA LINC01569 upregulates filamin A-interacting protein 1-like to prevent metastasis of triple-negative breast cancer via sponging miR-300
Abstract
BACKGROUND:
Long-chain noncoding RNA (lncRNA), LINC01569, is important for regulating the extracellular matrix, which affects cell migration. However, its involvement in the occurrence and development of triple-negative breast cancer (TNBC) remains unclear.
OBJECTIVE:
This study is aimed to investigate the role of LINC01569 on TNBC.
METHODS:
Online database was used for clinical data analysis. Cell viability and migration capability were monitored using cell counting kit-8 and transwell assays, respectively. Luciferase reporter assay and RNA pull-down were used to confirm the binding capability between noncoding RNAs and filamin A-interacting protein 1-like (FILIP1L). Western blotting was used to determine the protein content.
RESULTS:
Compared with normal breast tissue, LINC01569 was significantly reduced in patients with TNBC subtype, and LINC01569 expression gradually decreased with the progression of tumor stage. Patients with TNBC with high lncRNA LINC01569 levels had a better prognosis than did patients with low LINC01569 levels. LINC01569 overexpression inhibited the migration capability, whereas siRNA-mediated LINC01569 downregulation promoted the migration capability in TNBC cells. Using ENCORI and lncRNA SNP online databases, miR-300 was screened as the potential sponge of LINC01569. The binding of LINC01569 to miR-300 was confirmed using the dual-luciferase reporter and RNA pull-down assays. miR-300 was negatively correlated with LINC01569, and miR-300 mimics eliminated the anti-proliferation and anti-migration effects of LINC01569 on TNBC cells. Additionally, FILIP1L was further verified as the downstream target of miR-300. miR-300 mimics blocked LINC01569 upregulation-mediated elevation of FILIP1L. Importantly, the anti-tumor effects mediated by LINC01569 overexpression were abolished by miR-300 mimics and further restored by FILIP1L upregulation.
CONCLUSIONS:
LINC01569 was expressed at a low level in TNBC and could sponge miR-300 to promote FILIP1L expression, reducing the proliferation and metastasis capability of TNBC. Thus, LINC01569 might be a useful biomarker in the diagnosis and prognosis of metastatic TNBC.
1.Introduction
Breast cancer (BC) is the most common cancer in women and has become the leading cause of cancer-related death worldwide. BC accounts for 24.5% of female malignant tumor cases and 15.5% of malignant tumor-related deaths [1, 2]. BC mainly consists of the following four subtypes: luminal A, luminal B, human epidermal growth factor (HER2)-enrich, and triple-negative breast cancer (TNBC). Compared with the other BC subtypes, TNBC mostly occurs in young women with a late mortality rate of 40% in the first 5 years of diagnosis [3]. TNBC is usually more aggressive with a worse prognosis than that of the other BC subtypes [4]. Both conventional targeted and endocrine therapies are ineffective in TNBC [5]. In addition, patients with TNBC have a stable and persistent response to immunotherapy (e.g., PD-1/PD-L1 inhibitors), and TNBC is less sensitive to immunosuppression, with only a 5% response [6]. Owing to its highly heterogeneous nature and lack of specific molecular targets, the targeted therapy for migrating TNBC has stagnated. Therefore, there are still great challenges in treating migratory TNBC tumors.
Long-chain noncoding RNA (lncRNA) is closely related to biological processes such as development, differentiation, apoptosis, autophagy, inflammation, and cancer. Recently, an increasing number of lncRNAs have been found to play an important role in TNBC tumorigenesis and tumor metastasis [7, 8, 9]. For example, lncRNA HOTAIR is overexpressed in BC tissues, thus participating in the occurrence and metastasis of BC [10, 11]. NAMPT (NAMPT-AS), a long-chain noncoding anti-sense transcript, is highly upregulated in TNBC tumors. Moreover, it was positively correlated with prognosis, lymph node metastasis, distant migration, and pathologic grade of TNBC, suggesting that lncRNA NAMPT-AS participates in TNBC development and distant migration [12]. In addition, some lncRNAs with a tumor suppressor role were found in TNBC. Upregulation of lncRNA GAS5 regulates the invasion and migration of TNBC cells through FoxO1/PI3K/Akt signal transduction and may serve as a biomarker to evaluate TNBC migration and prognosis [13]. Therefore, lncRNAs are important regulators in monitoring tumor cell metastasis. LINC01569 expression is upregulated in colorectal cancer and promotes the proliferation and migration of colorectal cancer cells through the miR-381-3p/RAP2A signal axis [14]. LINC01569 has a significant effect on the migration of cancer cells, but its involvement in TNBC tumor migration remains unknown.
lncRNAs always function through a competing endogenous RNA (ceRNA) mechanism. Studies have demonstrated that miR-300 plays a key role in the migration of tumors by targeting or binding lncRNA. For example, miR-300 can target pituitary tumor-transforming gene 1 to inhibit tumorigenesis of pituitary tumor cells. lncRNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder cancer [15]. miR-300 affects tumor proliferation and metastasis by inhibiting lymphoid enhancer binding factor 1 in hepatocellular carcinoma [16]. Moreover, lncRNA SDHAP1 affects the proliferation and invasion of non-small cell lung cancer via sponging miR-300 [17]. The downregulation of LINC00472 promotes the occurrence of osteosarcoma by reducing FoxO1 expression through miR-300 [18]. These results suggest that miR-300 plays a key role in the regulation of the migration of cancer cells via a ceRNA mechanism. However, whether miR-300 can affect TNBC migration through binding LINC01569 remains unknown.
This study clarifies the role and regulatory mechanism of lncRNA LINC01569 in the progression of TNBC through in vitro and in vivo experiments. These results lay a foundation for mining lncRNA LINC01569 that mediate TNBC tumor cell migration and provide a new strategy for targeting lncRNA LINC01569 to treat migratory TNBC.
2.Materials and methods
2.1Data collection and analysis
Differential gene expression in human breast carcinoma (BRCA) in different molecular subtypes was analyzed using Breast Cancer Gene-Expression Miner v4.4 [19] and Hu’s subtypes. The Breast Cancer Gene-Expression Miner v4.4 and the online Kaplan–Meier Plotter database were used to determine the overall survival rates in patients with BC. The expression of LINC01569 in different clinical pathological stages was obtained using GEPIA 2.0 [20]. The association between LINC01569 and miR-300 expression or miR-300 and FILIP1L was assessed using SPSS software 22.0. The binding of miR-300 to LINC01569 was monitored using ENCORI and lncRNASNP online databases, and the interaction between FILIP1L and miR-300 was determined through the databases TargetScan, miRDB, ENCORI, and miRWalk.
2.2Patient samples
Fresh TNBC tissues (
2.3Cell culture and transfection
Four human TNBC cell lines, including Hs578t, MDA-MB-468, BT549, and MDA-MB-231, and a normal mammary epithelial cell line MCF-10A were purchased from the Cell Line Bank of the Chinese Academy of Sciences. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) plus 100 U/mL penicillin and 0.1 mg/mL streptomycin in an atmosphere containing 5% CO2 at 37∘C. All cells were passaged every 3 days. For cell transfection, MDA-MB-231 and MDA-MB-468 cells were cultured in 6-well plates at 80% confluence and transfected with LINC01569 overexpressed plasmid or vector plasmid, LINC01569 siRNA or NC siRNA, NC mimic or miR-300 mimic and FILIP1L overexpressed or FILIP1L vector plasmids. PcDNA3.1 and pcDNA3.1-LINC01569 plasmids were obtained from GenePharma (Shanghai, CN). The short hairpin RNA (shRNA) targeting LINC01569 (sence: caccGG ACAGGACACTTCTTTATctcgagATAAAGAAGTGTCCTGTCC; anti-sence: aaacGGACAGGACACTTCTTTATctcgagATAAAGAAGTGTCCTGTCC) and negative control shRNA (GTCTCGCTTGGGCGAGAGTAAGTAGTGAAGCCACAGATGTACTTACTCTCGCCCAAGCGAGAC) were provided by Liaoning Biology (Wuhan, Hubei, China). miR-300 mimic, sense 5′-UAUACAAGGGCAGACUCUCUCU-3′ and anti-sense 5′-AGAGAGAGUCU GCCCUUGUAUA-3′; miR negative control, sense 5′-UUCUCCGAACGUGUCACGU TT-3′ and anti-sense 5′-ACGUGACACGUUCGGAGA ATT-3′; miR-300 inhibitor, sense 5′-GAGAGAGUCU GCCCUUGUAU-3′; miR-300-inhibitor NC, sense 5′-CAGUACUUUUGUGUAGUACAA-3′ were synthesized by RiboBio Corporation (Guangzhou, China). FILIP1L cDNA fragments were sub-cloned into the pcDNA3.1 vector, and the pcDNA3.1 vector served as the mock-vehicle. Before transfection, the cells were cultured for 24 h and then transiently transfected with the corresponding vector using Lipofectamine 3000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. After 48 h, cells transfected with the corresponding vector were harvested for subsequent experiments. Experiments were performed in triplicate.
2.4Real-time quantitative PCR (RT-PCR)
Total RNA was extracted from TNBC tissue and cultured cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and RNA concentration was measured using the NanoDrop system. Additionally, miRNeasy Serum/Plasma Kit (QIAGEN, Germany) was used to extract total microRNAs from tissues and cells. The cDNA was synthesized from total RNA (1000 ng), respectively, using RevertAid first strand cDNA (Fermentas; Thermo Fisher Scientific, Inc.) or a TaqMan microRNA reverse transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, the expression levels of LINC01569 and others were measured using RT-PCR using 10
Table 1
Primer sequences of RT-PCR
| Gene | Forward | Reverse |
|---|---|---|
| LINC01569 | 5′-CAGTGCCACCTTCTCTACCTGCT-3′ | 5′-GGCATGACCTCACACTCACGC-3′ |
| GAPDH | 5′-ACCACAGTCCATGCCATCAC-3′ | 5′-TCCACCACCCTGTTGCTGTA-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACATATACT-3′ | 5′-ACGCTTCACGAATTTGCGTGTC-3′ |
| FILIP1L | 5′-ACAGGCTGTACATAAGAAGGCA-3′ | 5′-TGGTACGGGTCCCTCTTCTT-3′ |
| MiR-300 | 5′-TATACAAGGGCAGACTCTCTCT-3′ | 5′-CGCAAGGATGACACGCAAATTCGT-3′ |
2.5Cell counting kit-8 (CCK-8)
CCK-8 (HYK0301, MCE, US) was used to detect cell proliferation according to the manufacturer’s instructions. After attaining 80% confluence, the cells were washed twice with phosphate-buffered saline and separated using 0.25% trypsin to form a single-cell suspension. Cells were seeded into 96-well plates, 3000 cells/well, and cultured. After transfection for 24, 48, and 72 h, 10
2.6Transwell and invasion assay
To detect cell migration capability, transwell chambers coated with and without Matrigel (Becton Dickinson, CA) were employed and utilized a 24-well transwell chamber (8
2.7Western blot
Total protein was extracted from tissues/cells using RIPA lysis buffer (Beyotime, Shanghai, CN), containing protease and phosphatase inhibitors cocktails. The BCA Protein Assay Kit (Beyotime) was used to measure protein concentrations according to the manufacturer’s protocol. An identical protein amount (20
2.8Luciferase reporter assay
The predicted binding sites of miR-300 with LINC01569-3′UTR and FILIP1L-3′UTR were obtained from RNA hybrid. The binding and mutant sequences were cloned into pmirGLO Dual-luciferase vectors (Gene Pharma, Shanghai, China). MDA-MB-231 and MDA-MB-468 cells were cultured in 96-well plates and subsequently co-transfected with the wild-type pmirGLO-LINC01569 reporter plasmid (LINC01569-WT, gccuguucuagaagCUUGUAUc) or the mutated type (LINC01569-MUT, gccuguucuagaagGAACAUAc) and miR-300 mimics or NC using Lipofectamine 3000. To analyze the binding between miR-300 and FILIP1L, cells were co-transfected with the FILIP1L-3′UTR reporter plasmid (FILIP1L-WT, auauuuAGUCUGCACUUGUAUa) or the mutated type (FILIP1L-MUT, auauuuUCAGACGAGAACAUAa) and the above-listed mimics. The pRL-CMV plasmid (Renilla luciferase, Promega) were transfected together into cells with the above plasmids. Dual-Luciferase Reporter Assay System (Promega, Madison, WI USA) was used to analyze luciferase activity, which was recorded as the ratio of firefly luciferase activity to Renilla luciferase activity. Experiments were performed at least three times.
2.9RNA pull-down assay
RNeasy Mini Kit (QIAGEN, 74104) was used for the RNA pull-down assay. Briefly, 1
Figure 1.
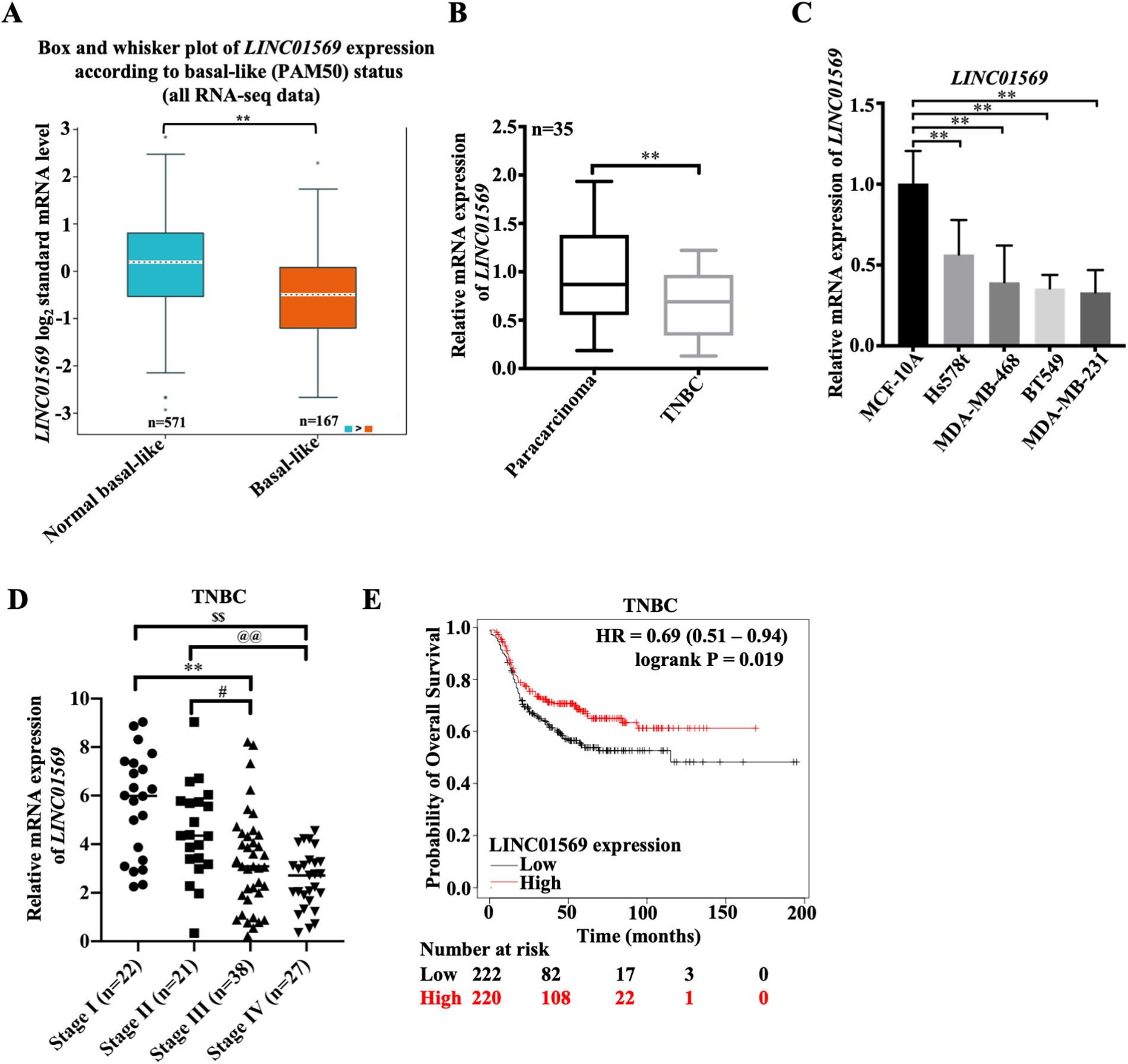
Expression of IncRNA LINC01569 in TNBC and its clinical significance. (A) The differential expression of LINC01569 in TNBC (
2.10Statistical analysis
All the tests were conducted at least three times. All methods were carried out in accordance with relevant guidelines and regulations in the Ethics declaration section including the Informed consent statement. The Kaplan–Meier approach was used to estimate the overall survival. All statistical analyses were performed with GraphPad 9.0 software (LaJolla, CA, USA). Different groups were compared using unpaired Student’s two-tailed
3.Results
3.1LINC01569 is low expressed in TNBC cells
To investigate the effect of LINC01569 on the development of TNBC, an online database was used to determine the expression pattern of LINC01569. According to GEPIA, there was no significant difference in LINC01569 expression in healthy subjects (
3.2LINC01569 inhibits the growth and migration ability of TNBC cells
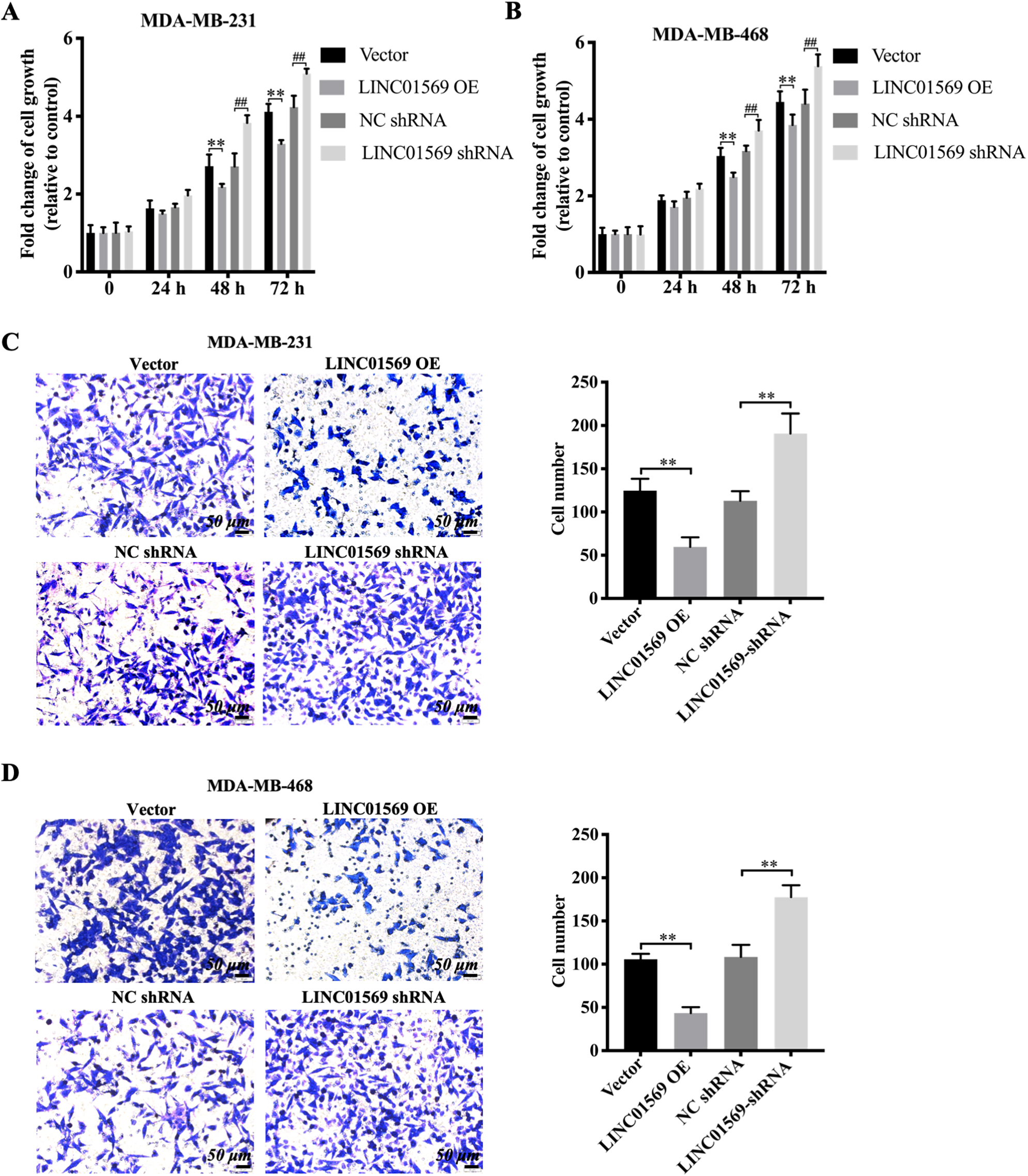
To determine whether LINC01569 affects the progression of TNBC, LINC01569-overexpressed TNBC or LINC01569-knocked TNBC cells were produced. In MDA-MB-231 and MDA-MB-468 cells, qRT-PCR was used to identify the transfection efficiency of LINC01569 overexpressing plasmid and the siRNA targeting LINC01569 (Figs. S2A and S2B). As shown in Fig. 2, it was found that cell viability was inhibited 48 and 72 h after the transfection of LINC01569 overexpression plasmid into MDA-MB-231 and MDA-MB-468 cells. However, siRNA-mediated LINC01569 knockdown significantly induced cell viability after 48 h in both cell lines (Fig. 2A and B). Additionally, the upregulation of LINC01569 markedly inhibited cell migration. The number of TNBC cells migrating to the lower chamber decreased drastically, whereas LINC01569 knockdown significantly stimulated cell migration in MDA-MB-231 and MDA-MB-468 cells (Figs. 2C and D). Therefore, LINC01569 can lessen the growth and migration capabilities of TNBC cells.
Figure 2.
The Influence of LINC01569 on the viability and migration of TNBC cells. Transfection of MDA-MB-231 and MDA-MB-468 cells with overexpression plasmid (LINC01569 OE), siRNA targeting LINC01569 (LINC01569 siRNA), and the corresponding control vector/negative siRNA. (A–B) MDA-MB-231 (A) and MDA-MB-468 (B) cells were cultured for 0, 24, 48, and 72 h. Cell growth was measured using CCK-8. ** Indicated vector vs. LINC01569 OE. ## Indicated NC siRNA vs. LINC01569 siRNA. (C–D) Migration capability of MDA-MB-231 (C) and MDA-MB-468 cells (D) were transferred using transwell assay. Relative quantitation of migration cells, as illustrated in the right panel. ** Indicated LINC01569-overexpressed or LINC01569 siRNA vs. vector or NC siRNA. NC, negative control. Scale bar: 50
3.3LINC01569 directly targets miR-300 in TNBC cells
LincRNAs play a significant role in tumor migration via ceRNA. Therefore, the potential binding partner of LINC01569 was subsequently screened to better study the relevant mechanism of LINC01569 in regulating TNBC progression. Six potential target genes including hsa-miR-381-3p, hsa-miR-1323, hsa-miR-548o-3p, hsa-miR-103a-3p, hsa-miR-193a-5p, and hsa-miR-300 were screened combining the ENCORI and lncRNASNP online databases (Fig. 3A). Owing to the negative association between miR-300 and LINC01569 in the TNBC tissues, miR-300 was selected for the subsequent experiments (Fig. 3B). Moreover, miR-300 expression was remarkably increased in TNBC compared with the corresponding adjacent cancer tissues (Fig. 3C). The luciferase activity of the LINC01569 WT reporter was almost completely abrogated by the transfection of miR-300 mimics, whereas miR-300 mimics did not affect the luciferase activity of cells transfected with a LINC01569 reporter with site-directed mutation (Fig. 3D). In addition, the pull-down test further demonstrated that miR-300 is directly associated with LINC01569 (Fig. 3E). Moreover, in response to the challenge of exogenous LINC01569, miR-300 was robustly reduced, and increased significantly as LINC01569 was depleted in MDA-MB-468 cells (Fig. 3F). Thus, these data provide evidence that LINC01569 acts as a sponge of miR-300.
Figure 3.
Association of LINC01569 and miR-300. (A) Potential downstream target microRNAs were analyzed by combining ENCORI and LncRNASNP online databases. (B) Relative expression of LINC01569 and miR-300 in TNBC tissues (
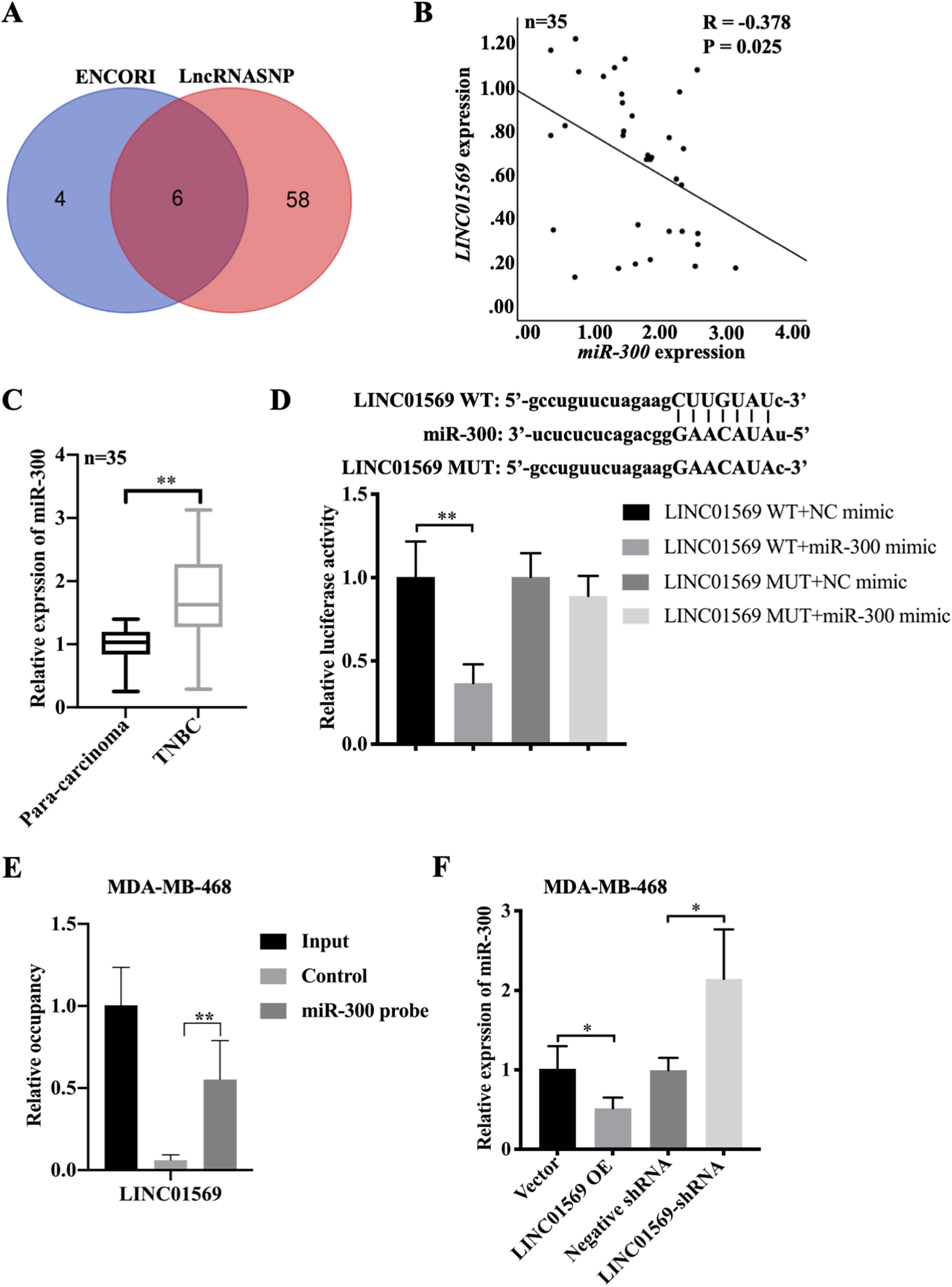
Figure 4.
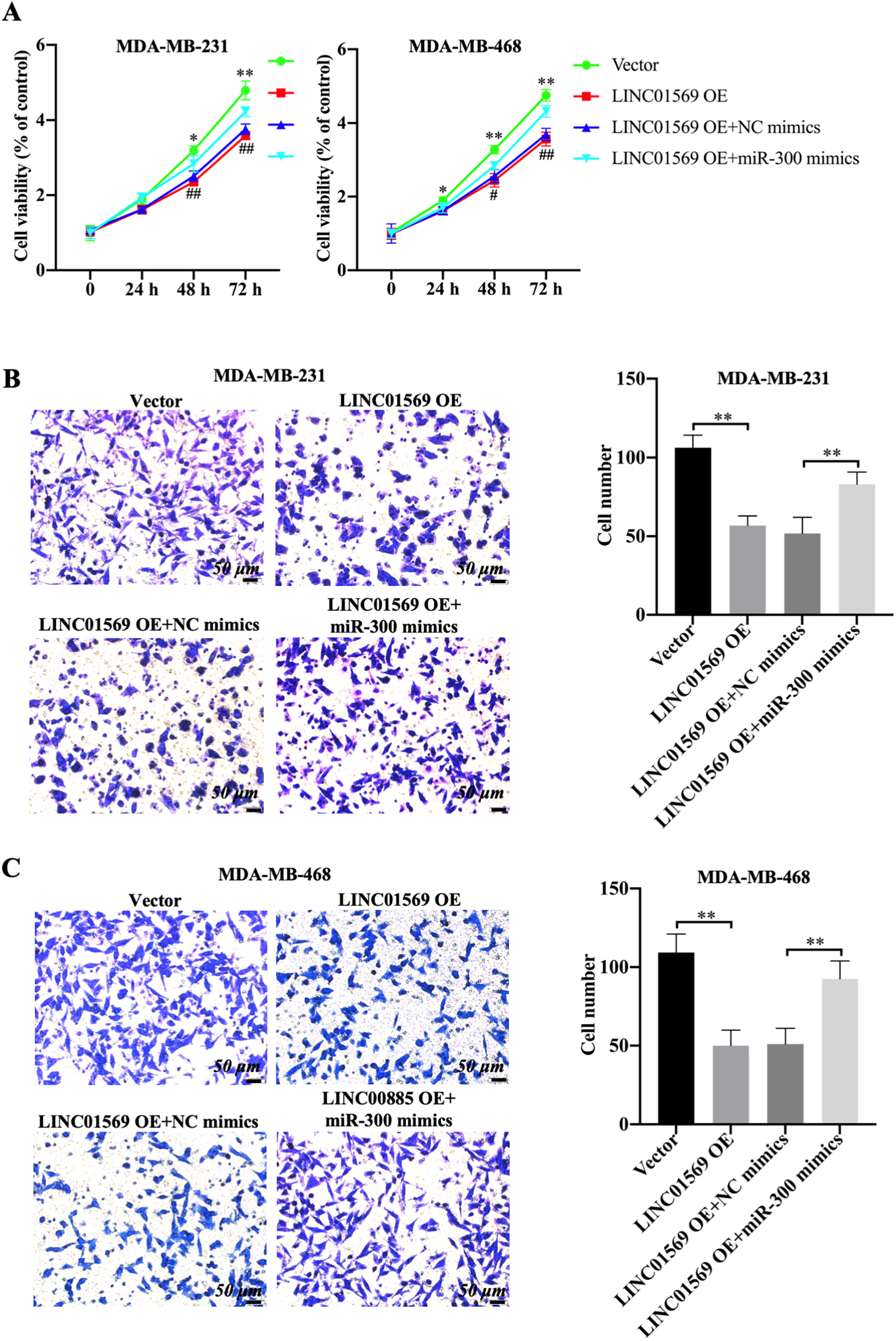
Effect of miR-300 on the migration of TNBC cells mediated by LINC01569 overexpression. MDA-MB-231 and MDA-MB-468 cells were transfected with vector, LINC01569 OE or LINC01569 OE plus NC mimics, LINC01569 OE plus miR-300 mimics. (A) After transfection, cell viability was determined using CCK8 assay at 0, 24, 48, and 72 h. (B–C) Transwell assay was used to determine the migration ability of MDA-MB-231 (B) and MDA-MB-468 (C) cells. Relative quantitative analysis of migrating cells, as illustrated in the right panel. Scale: 50
3.4miR-300 inhibits the tumor suppressive effect of LINC01569 in TNBC
Rescue experiments were conducted to identify whether miR-300 influences the biological role of LINC01569 in TNBC. MDA-MB-231 and MDA-MB-468 cells were transfected with vector, LINC01569-OE plasmid, the combination of LINC01569-OE and NC mimics or the combination of LINC01569-OE and miR-300 mimics, followed by the measurement of tumor cell behaviors. As expected, the introduction of miR-300 mimics significantly reversed the decrease in cell viability caused by LINC01569 overexpression in MDA-MB-231 and MDA-MB-468 cells (Fig. 4A). Consistently, transwell experiments showed that LINC01569 overexpression significantly inhibited cell migration capability in MDA-MB-231 and MDA-MB-468 cells, whereas miR-300 mimics notably abolished the anti-migration effects of LINC01569 overexpression in TNBC cells, increasing the number of migratory cells (Fig. 4B and C). These results indicate that miR-300 upregulation can inhibit the tumor suppressive effect induced by LINC01569 overexpression in TNBC cells.
3.5Filamin A–interacting protein 1-like (FILIP1L) is a downstream target gene of miR-300
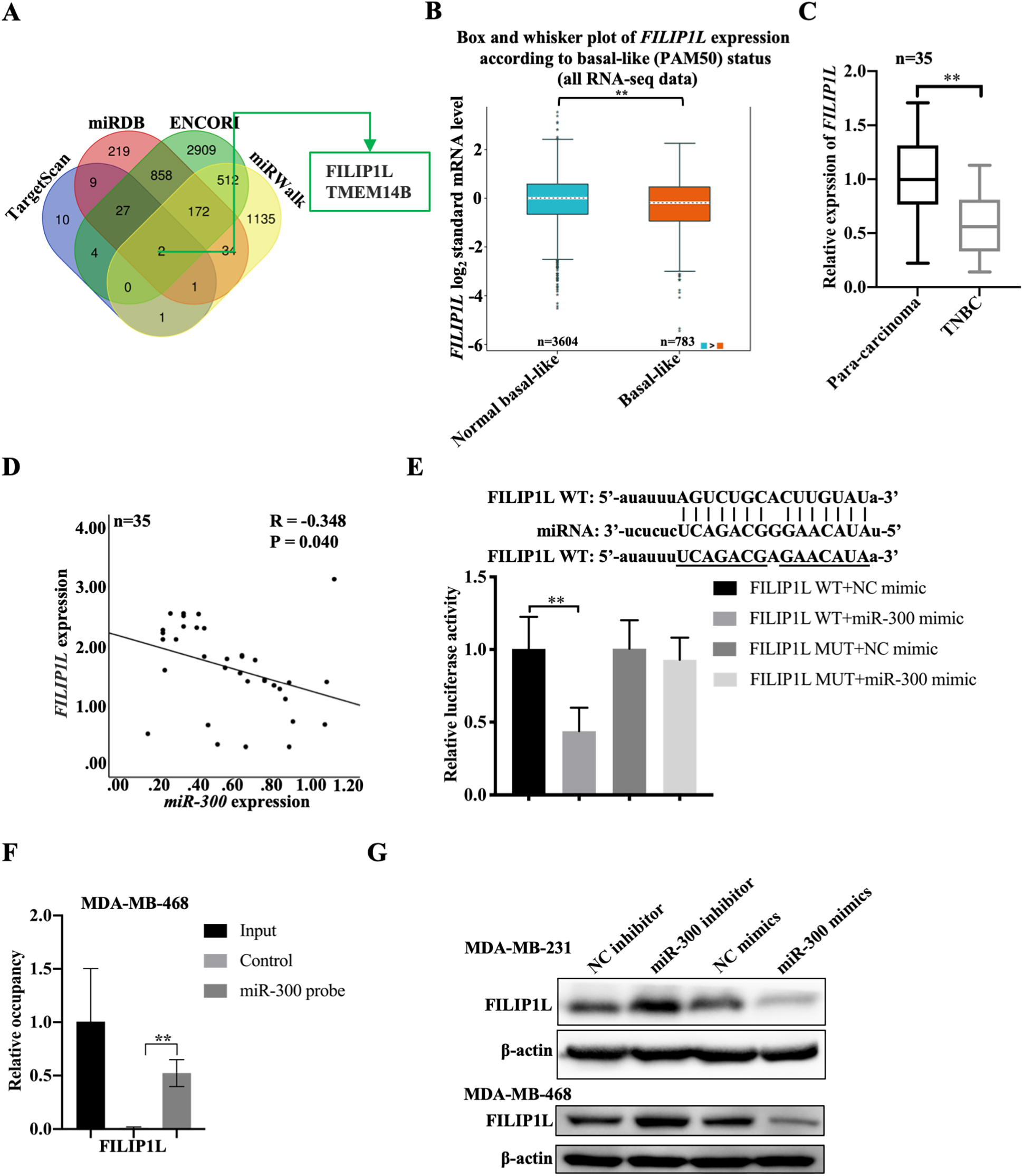
We explored the underlying mechanism of LINC01569/miR-300 in regulating the development of TNBC. Combined with online databases such as TargetScan, miRDB, ENCORI, and miRWalk, two potential downstream targets were screened, including FILIP1L and transmembrane protein 14B (TMEM14B) (Fig. 5A). Actually, TMEM14B is at a high level in TNBC tissues compared to non-TNBC tissues (the below image) which was in contradiction with the previous findings in this study (Fig. S3). FILIP1L was selected as a downstream target gene of miR-300 for the subsequent experiments owing to the significant differential expression of FILIP1L in TNBC and non-TNBC. Based on the analysis of All RNA SEQ data, it was found that the expression level of FILIP1L in TNBC was lower in normal subjects (
Figure 5.
Relationship between FILIP1L and miR-300. (A) The downstream targets of miR-300 as analyzed through TargetScan, miRDB, ENCORI, and miRWalk online databases. (B) The differential expression of FILIP1L in normal basal-like (
3.6LINC01569/miR-300/FILIP1L signaling axis is involved in the proliferation and migration of TNBC cells
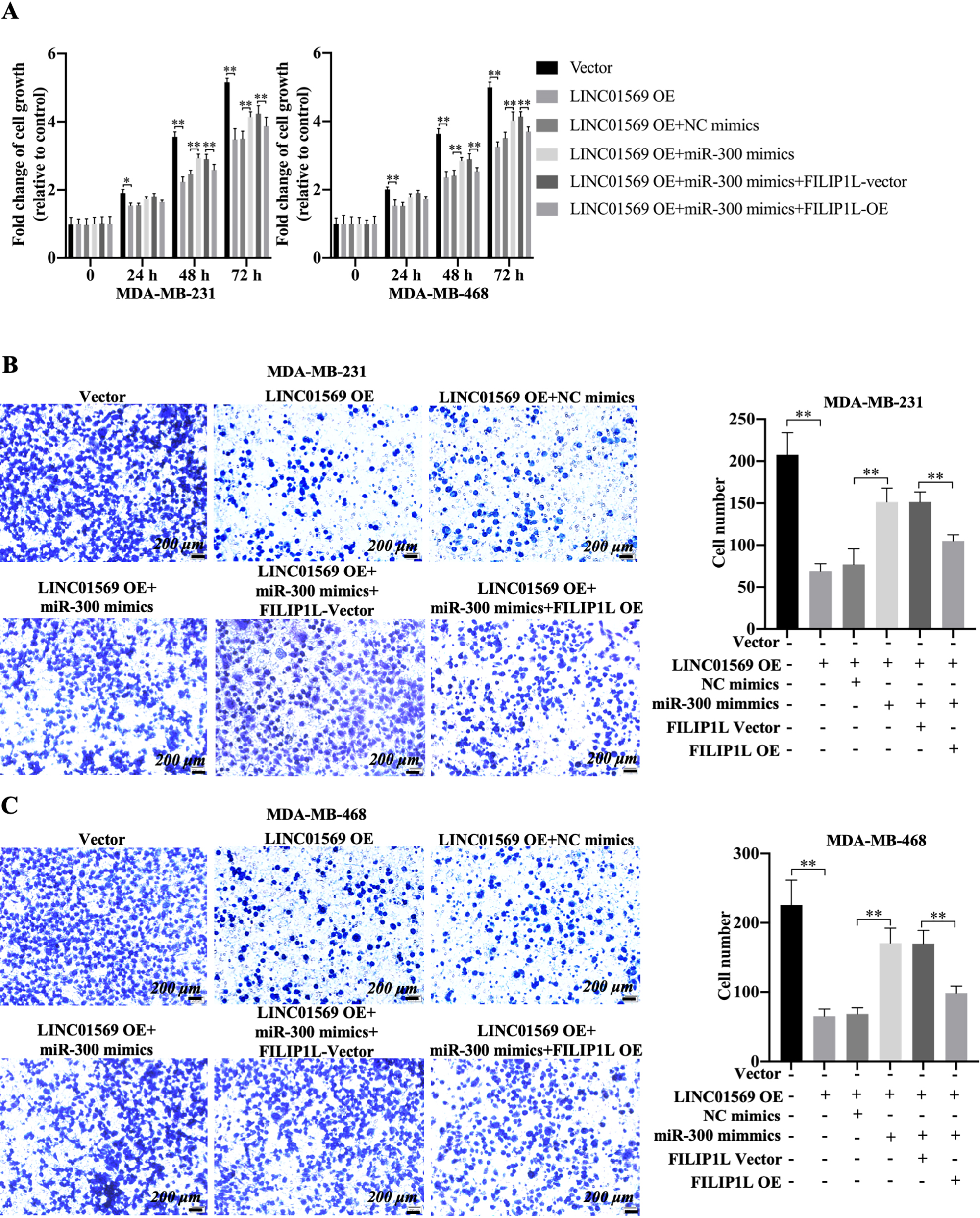
To determine the association among LINC01569, miR-300, and FILIP1L in TNBC cells, FILIP1L protein levels were monitored in MDA-MB-231 and MDA-MB-468 cells after the transfection of LINC01569 overexpression plasmid alone, or LINC01569 overexpression plasmid plus miR-300 mimics. LINC01569 overexpression (OE) promoted the expression of FILIP1L, whereas miR-300 mimics could reverse the increased expression of FILIP1L caused by LINC01569 OE (Fig. S5). Consistently, miR-300 mimics reversed the anti-proliferation effects mediated by LINC01569 OE for 48 h using CCK8 assay in TNBC cells (Fig. 6A). However, FILIP1L overexpression reversed the increased cell viability of TNBC cells under the joint action of LINC01569 overexpression and miR-300 mimics compared with the corresponding control group for 48 h (Fig. 6A). Using a transwell experiment, FILIP1L overexpression further suppressed the upregulated migration and invasive capabilities of MDA-MB-231 cells and also increased the migration and invasive capabilities of MDA-MB-468 cells compared with cells transfected with the combination of LINC01569-OE plasmid, miR-300 mimics, and vector (Fig. 6B–C and S6A). Importantly, the LINC01569 overexpression-mediated decline of MMP9/MMP2 was reversed by the transfection of miR-300 mimics, which was partly restored by the transfection of exogenous FILIP1L (Fig. S6B). These data indicate that LINC01569/miR-300/FILIP1L pathway regulates the progression of TNBC, especially cell proliferation and migration.
Figure 6.
Effects of LINC01569/miR-300/FILIP1L axis in cell viability and migration of TNBC cells. MDA-MB-231 and MDA-MB-468 cells were transfected with vector, LINC01569 OE, LINC01569 OE plus NC mimics, LINC01569 OE plus miR-300 mimics, the combination of LINC01569 OE, miR-300 mimics plus FILIP1L vector, and the combination of LINC01569 OE, miR-300 mimics plus FILIP1L-OE. (A) CCK-8 assays were performed to evaluate the cell viability for 0, 24, 48, and 72 h. ** Indicated LINC01569-overexpression or LINC01569-overexpression with miR-300 mimics or LINC01569-overexpression and miR-300 mimics with FILIP1L-overexperssion vs. Vector or LINC01569-overexpression with NC mimics or LINC01569-overexpression and miR-300 mimics with NC-overexpression. Transwell assay was used to determine the migration ability of MDA-MB-231 (B) and MDA-MB-468 cells (C). The relative quantitation of the migrating cells is shown in the right panel. Scale: 200
4.Discussion
TNBC tends to be more aggressive with a worse prognosis than that of the other types of BC, and has a higher recurrence rate than other subtypes [4]. Less than 30% of patients with migrating TNBC live beyond 5 years after diagnosis [21]. Studies have shown that migratory TNBC is easily resistant to chemotherapeutic drugs, leading to a mean survival time of 8–13 months [22, 23]. Increased extracellular matrix (ECM) deposition is the main cause of poor prognosis in patients with TNBC [24]. Moreover, therapeutic methods can inhibit the development of TNBC by acting on ECM. A novel ECM-targeted nano-therapy epirubicin-loaded anti-LOX liposomes can be used as a viable therapeutic strategy for TNBC [25]. Therefore, it is reasonable to suspect that ECM degradation may be one of the reasons for the high migration characteristics of TNBC cells. Studies have shown that FILIP1L can inhibit invasion and metastasis of ovarian cancer cells in situ by inhibiting the Wnt signaling pathway and MMP expression, suggesting that FILIP1L may regulate and affect cell migration [26]. In this study, we concluded that lncRNA LINC01569 was low expressed in TNBC cells and could downregulate the expression of FILIP1L through miR-300 to promote the metastasis of TNBC. Therefore, understanding the underlying regulation mechanism of TNBC metastasis is very important.
Studies have shown that the expression of LINC01569 was significantly increased in colorectal cancer, hypopharyngeal carcinoma and endometriosis [14, 27, 28]. These data hinted that LINC01569 may be served as a tumor oncogene. lncRNA LINC01569 was at a higher level in human BC tissue and breast cancer cell lines than that in normal control, but LINC01569 was not associated with tumor stage and overall survival of patients with BC. LINC01569 level was lower in TNBC tissues than that in other BC subtypes, indicating that LINC01569 might be a cancer suppressor gene in TNBC. Several studies have suggested that lncRNAs can be an important prognostic indicator in TNBC [29, 30]. In this study, patients with TNBC with high expression of lncRNA LINC01569 had a better prognosis. LINC01569 expression was gradually decreased along with the increased tumor stage in TNBC. Therefore, LINC01569 had a negative association with TNBC progression and might also be defined as a diagnostic and prognostic indicator for TNBC. lncRNA LINC01569 upregulation promotes cancer proliferation and migration in colorectal cancer [14]. However, the increased expression of lncRNA LINC01569 inhibited the proliferation and migration of TNBC cells in this study. Therefore, the poor prognosis caused by the abnormal low expression of LINC01569 in TNBC may be related to the high mobility of TNBC cells.
lncRNAs play an important role in reducing their regulation of the target mRNA by means of ceRNA by the “miRNA sponge adsorption” mechanism [31, 32]. lncRNA LINC01569 exerts pro-proliferation and pro-migration effects in colorectal cancer cells through regulating miR-381-3p/Ras-related protein Rap-2 (RAP2A signal axis) [14]. Therefore, LINC01569 may also affect tumor migration and progression in TNBC through the ceRNA mechanism. Downregulation of miR-300 promotes the expression of fatty acid 2-hydroxylase and inhibits the proliferation and apoptosis of gastric cancer cells [33]. miR-300 overexpression promotes the invasion, metastasis, and proliferation of osteosarcoma cells by downregulating pituitary tumor-transforming gene 1 [34]. This indicates that miR-300 is closely related to tumor invasion and migration. Moreover, lncRNA SDHAP1 can sponge miR-300 to affect the proliferation and invasion of non-small cell lung cancer [17], and lncRNA FTX promotes tumorigenesis of lung adenocarcinoma by targeting miR-300 [35]. Therefore, miR-300 can influence tumor cell migration by ceRNA integration of lncRNAs. Herein, LINC01569 negatively correlates with miR-300 in TNBC, and miR-300 mimics can abrogate the anti-tumor effects mediated by LINC01569 overexpression in TNBC cells. Therefore, LINC01569 exerts anti-proliferation and anti-metastasis functions in TNBC by sponging miR-300.
Simultaneously, it has also been shown that lncRNAs play a key role in the migration of breast, colon, lung, and pancreatic cancers [36]. FILIP1L is an important inhibitor of ovarian cancer cell migration and invasion [26]. Knockdown of trophoblast cell surface antigen 2 can significantly enhance the proliferation and migration of hepatobiliary hepatoma cell lines, and alter the expression of the myristoylated alanine-rich c-kinase substrate, epithelial membrane protein genes 1 and FILIP1L [37]. Therefore, FILIP1L is key in monitoring tumor cell metastasis in several tumor types. This study found that miRNA-300 has multiple binding sites to FILIP1L, and LINC01569 can compete against FILIP1L for miR-300. Additionally, there is a negative correlation between FILIP1L and miR-300, and miR-300 mimics can reduce FILIP1L expression, indicating that FILIP1L was a potential downstream target gene of miR-300. Studies have shown that LINC00472 can promote osteogenic differentiation and alleviate osteoporosis by upregulating fibroblast growth factor receptor 2 expression through sponging miR-300 [38]. lncRNA HAND2-AS1 upregulation improves the viability of hepatocellular carcinoma cells by targeting the miRNA-300/suppressor of cytokine signaling 5 axis [39]. In this study, overexpression of FILIP1L reversed the effect of miR-300 mimics. Therefore, the LINC01569/miR-300/FILIP1L signaling axis plays an important role in TNBC cell migration. In the present study, LINC01569 induced the upregulation of FILIP1L expression in TNBC cells, which were abolished by miR-300 mimics. In addition, LINC01569 overexpression-mediated anti-tumor effects, including the reduced cell viability and cell migration, were reversed by miR-300 mimics, whereas the transfection of exogenous FILIP1L restored the anti-tumor role of LINC01569 in TNBC cells. Possibly, LINC01569 ameliorated cell viability and metastasis of TNBC cells by regulating the miR-300/FILIP1L signaling axis. It was worth noting that the LINC01569 overexpression-mediated a decline of MMP9/MMP2 that were reversed by the miR-300 mimics/FILIP1L signaling pathway. Actually, the knockdown of FILIP1L could enhance EMT and ECM synthesis and promote migration of residual lens cells and FILIP1L overexpression was able to inhibit cell migration possibly through ECM degradation [40]. Therefore, LINC01569/miR-300/FILIP1L signaling pathway may balance the processes of ECM degradation or synthesis to affect cell migration and invasion in TNBC.
Although many studies have been performed in this regard, many problems remain to be solved. The underlying mechanism still needs to be confirmed in a tumor migratory animal model in vivo. However, the LINC01569-overexpressed TNBC cell line was not suitable for the in vivo animal experiment. Another cell lines should be constructed to prove the findings in animal model in the future research. Additionally, whether FILIP1L influences tumor cell migration through the regulation of ECM degradation remains unclear. Moreover, a large-scale clinical study is required to confirm the results and improve the clinical research value and application in TNBC.
5.Conclusion
lncRNA LINC01569 was low expressed and participated in TNBC cell migration. Mechanistically, LINC01569 competitively inhibited miR-300 through the ceRNA mechanism, subsequently inducing the expression of FILIP1L, resulting in the metastasis inhibition of TNBC cells. The results will help understand the pathogenesis and progression of metastasis of TNBC and improve the therapy of metastatic TNBC.
Author contributions
Interpretation or analysis of data: Xinyu Jiang and Juli Lin.
Preparation of the manuscript: Zhanlin Zhu.;Revision for important intellectual content: Xinyu Jiang, Juli Lin and Zhanlin Zhu.
Supervision: Zhanlin Zhu.
All authors edited and approved the manuscript.
Ethics approval and Consent to participate
All experiments were approved by the ethics committee of Women and Children’s Hospital of Xiamen University (2021-XMFYBI0-073).
Ethics declaration
All methods were carried out in accordance with relevant guidelines and regulations. All subjects have signed the informed consent form.
Patients consent for publication
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Funding
The present study was supported by Lin Qiaozhi scientific research project of Women and Children’s Hospital of Xiamen University (FYLQZ2020005).
List of abbreviations
FILIP1L, Filamin A–interacting protein 1-like; TNBC, triple-negative breast cancer; BC, breast cancer; HER2, human epidermal growth factor; LncRNA, Long-chain noncoding RNA; ECM, extracellular matrix; ceRNA, competitive endogenous RNA; MMP9, matrix metalloproteinase-9; MMP2, matrix metallopro-teinase-2.
Supplementary data
The supplementary files are available to download from http://dx.doi.org/10.3233/CBM-230261.
Acknowledgments
We thank Gene Expression Profiling Interactive Analysis 2.0 (GEPIA2).
Conflict of interest
The authors declare that they have no competing interests.
References
[1] | U. Mehraj, I.A. Mir, M.U. Hussain, M. Alkhanani, N.A. Wani and M.A. Mir, Adapalene and Doxorubicin Synergistically Promote Apoptosis of TNBC Cells by Hyperactivation of the ERK1/2 Pathway Through ROS Induction, Front Oncol 12: ((2022) ), 938052. |
[2] | C. Song, V.J. Lowe and S. Lee, Inhibition of Cdc20 suppresses the metastasis in triple negative breast cancer (TNBC), Breast Cancer 28: ((2021) ), 1073–1086. |
[3] | M. Bou Zerdan, T. Ghorayeb, F. Saliba, S. Allam, M. Bou Zerdan, M. Yaghi, N. Bilani, R. Jaafar and Z. Nahleh, Triple Negative Breast Cancer: Updates on Classification and Treatment in 2021, Cancers (Basel) 14: ((2022) ), 1253. |
[4] | Y. Hayashi, H. Satake, S. Ishigaki, R. Ito, M. Kawamura, H. Kawai, S. Iwano and S. Naganawa, Kinetic volume analysis on dynamic Contrast-enhanced MRI of Triple-Negative breast cancer: Associations with survival outcomes, Br J Radiol ((2019) ), 20190712. |
[5] | S. Zheng, Y. Zou, Y. Tang, A. Yang, J.Y. Liang, L. Wu, W. Tian, W. Xiao, X. Xie, L. Yang, J. Xie, W. Wei and X. Xie, Landscape of cancer-associated fibroblasts identifies the secreted biglycan as a protumor and immunosuppressive factor in triple-negative breast cancer, Oncoimmunology 1: ((2022) ), 2020984. |
[6] | L.Y. Dirix, I. Takacs, G. Jerusalem, P. Nikolinakos, H.T. Arkenau, A. Forero-Torres, R. Boccia, M.E. Lippman, R. Somer, M. Smakal, L.A. Emens, B. Hrinczenko, W. Edenfield, J. Gurtler, A. von Heydebreck, H.J. Grote, K. Chin and E.P. Hamilton, Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study, Breast Cancer Res Tr 167: ((2018) ), 671–686. |
[7] | W. Zhang, X. Guan and J. Tang, The long non-coding RNA landscape in triple-negative breast cancer, Cell Prolif 54: ((2021) ), e12966. |
[8] | E. Mou and H. Wang, LncRNA LUCAT1 facilitates tumorigenesis and metastasis of triple-negative breast cancer through modulating miR-5702, Biosci Rep 39: ((2019) ), BSR20190489. |
[9] | D. Jiang, C. Wang and J. He, Long non-coding RNA DGCR5 incudes tumorigenesis of triple-negative breast cancer by affecting Wnt/beta-catenin signaling pathway, J BUON 25: ((2020) ), 702–708. |
[10] | H. Xiong, Z. Ni, J. He, S. Jiang, X. Li, J. He, W. Gong, L. Zheng, S. Chen, B. Li, N. Zhang, X. Lyu, G. Huang, B. Chen, Y. Zhang and F. He, LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells, Oncogene 36: ((2017) ), 3528–3540. |
[11] | F. Ferre, A. Colantoni and M. Helmer-Citterich, Revealing protein-lncRNA interaction, Brief Bioinform 17: ((2016) ), 106–116. |
[12] | H.W. Zhang, N. Zhang, Y. Liu, P. Su, Y.R. Liang, Y.M. Li, X.L. Wang, T. Chen, X.J. Song, Y.T. Sang, Y. Duan, J.S. Zhang, L.J. Wang, B. Chen, W.J. Zhao, H.Y. Guo, Z.J. Liu, G.H. Hu and Q.F. Yang, Epigenetic Regulation of NAMPT by NAMPT-AS Drives Metastatic Progression in Triple-Negative Breast Cancer, Cancer Res 79: ((2019) ), 3347–3359. |
[13] | S. Li, J. Zhou, Z. Wang, P. Wang, X. Gao and Y. Wang, Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p, Biomed Pharmacother 104: ((2018) ), 451–457. |
[14] | G.Y. Ye, Z.Z. Zhang, C.C. Zhu, Z.J. Cong, Z. Cui, L. Chen and G. Zhao, Long Non-Coding RNA LINC01569 Promotes Proliferation and Metastasis in Colorectal Cancer by miR-381-3p/RAP2A Axis, Front Oncol 11: ((2021) ), 727698. |
[15] | F. Ma, S.H. Wang, Q. Cai, L.Y. Jin, D. Zhou, J. Ding and Z.W. Quan, Long non-coding RNA TUG1 promotes cell proliferation and metastasis by negatively regulating miR-300 in gallbladder carcinoma, Biomed Pharmacother 88: ((2017) ), 863–869. |
[16] | Y. Chen, Y. Guo, Y. Li, J. Yang, J. Liu, Q. Wu and R. Wang, miR300 regulates tumor proliferation and metastasis by targeting lymphoid enhancerbinding factor 1 in hepatocellular carcinoma, Int J Oncol 54: ((2019) ), 1282–1294. |
[17] | Z. Lu, X. Xu, H. Peng, R. Li and J. Zeng, Long chain noncoding RNA SDHAP1 targeting miR-300 affects the proliferation and invasion of non-small cell lung cancer, Minerva Surg ((2021) ). |
[18] | J. Zhang, J. Zhang, D. Zhang, W. Ni, H. Xiao and B. Zhao, Down-regulation of LINC00472 promotes osteosarcoma tumorigenesis by reducing FOXO1 expressions via miR-300, Cancer Cell Int 20: ((2020) ), 100. |
[19] | J.H. Li, S. Liu, H. Zhou, L.H. Qu and J.H. Yang, starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data, Nucleic Acids Res 42: ((2014) ), D92–D97. |
[20] | Z.F. Tang, C.W. Li, B.X. Kang, G. Gao, C. Li and Z.M. Zhang, GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses, Nucleic Acids Res 45: ((2017) ), W98–W102. |
[21] | R. Dent, M. Trudeau, K.I. Pritchard, W.M. Hanna, H.K. Kahn, C.A. Sawka, L.A. Lickley, E. Rawlinson, P. Sun and S.A. Narod, Triple-negative breast cancer: Clinical features and patterns of recurrence, Clin Cancer Res 13: ((2007) ), 4429–4434. |
[22] | W.D. den Brok, C.H. Speers, L. Gondara, E. Baxter, S.K. Tyldesley and C.A. Lohrisch, Survival with metastatic breast cancer based on initial presentation, de novo versus relapsed, Breast Cancer Res Tr 161: ((2017) ), 549–556. |
[23] | F. Kassam, K. Enright, R. Dent, G. Dranitsaris, J. Myers, C. Flynn, M. Fralick, R. Kumar and M. Clemons, Survival Outcomes for Patients with Metastatic Triple-Negative Breast Cancer: Implications for Clinical Practice and Trial Design, Clin Breast Cancer 9: ((2009) ), 29–33. |
[24] | S.A. Fertal, J.E. Poterala, S.M. Ponik and K.B. Wisinski, Stromal Characteristics and Impact on New Therapies for Metastatic Triple-Negative Breast Cancer, Cancers (Basel) 14: ((2022) ), 1238. |
[25] | A. De Vita, C. Liverani, R. Molinaro, J.O. Martinez, K.A. Hartman, C. Spadazzi, G. Miserocchi, F. Taraballi, M. Evangelopoulos, F. Pieri, A. Bongiovanni, L. Mercatali, E. Tasciotti and T. Ibrahim, Lysyl oxidase engineered lipid nanovesicles for the treatment of triple negative breast cancer, Sci Rep 11: ((2021) ), 5107. |
[26] | M. Kwon, S.J. Lee, Y. Wang, Y. Rybak, A. Luna, S. Reddy, A. Adem, B.T. Beaty, J.S. Condeelis and S.K. Libutti, Filamin A interacting protein 1-like inhibits WNT signaling and MMP expression to suppress cancer cell invasion and metastasis, Int J Cancer 135: ((2014) ), 48–60. |
[27] | S. Shan, Y. Yang, J. Jiang, B. Yang, Y. Yang, F. Sun, J. Zhang, Y. Lin and H. Xu, Extracellular vesicle-derived long non-coding RNA as circulating biomarkers for endometriosis, Reprod Biomed Online 44: ((2022) ), 923–933. |
[28] | Q. Gong, H. Li, J. Song and C. Lin, LncRNA LINC01569 promotes M2 macrophage polarization to accelerate hypopharyngeal carcinoma progression through the miR-193a-5p/FADS1 signaling axis, J Cancer 14: ((2023) ), 1673–1688. |
[29] | Y. Ma, L. Liang, Y. Zhang, M. Wang, W. Jiang, F. Zhao and S. Zhang, Upregulated IncRNA WTAPP1 in Triple Negative Breast Cancer Predicts Survival, Crit Rev Eukaryot Gene Expr 32: ((2022) ), 67–78. |
[30] | Z. Xing, M. Zhang, J. Liu, G. Liu, K. Feng and X. Wang, Overexpression of IncRNA SAMMSON Promotes Triple-Negative Breast Cancer Cell Proliferation by Interacting with p53, Crit Rev Eukaryot Gene Expr 31: ((2021) ), 1–8. |
[31] | J.J. Quinn and H.Y. Chang, Unique features of long non-coding RNA biogenesis and function, Nat Rev Genet 17: ((2016) ), 47–62. |
[32] | E.A. Braga, M.V. Fridman, A.A. Moscovtsev, E.A. Filippova, A.A. Dmitriev and N.E. Kushlinskii, LncRNAs in Ovarian Cancer Progression, Metastasis, and Main Pathways: ceRNA and Alternative Mechanisms, Int J Mol Sci 21: ((2020) ), 8855. |
[33] | B. Hong, J. Li, C. Huang, T. Huang, M. Zhang and L. Huang, miR-300/FA2H affects gastric cancer cell proliferation and apoptosis, Open Med (Wars) 15: ((2020) ), 882–889. |
[34] | D. Liang, X. Wu, J. Bai, L. Zhang, C. Yin and W. Zhong, [MiR-300 inhibits invasion and metastasis of osteosarcoma cell MG63 by negatively regulating PTTG1], Nan Fang Yi Ke Da Xue Xue Bao 41: ((2021) ), 285–291. |
[35] | W. Jiang, B. Zhang, J. Sun, Y. Liu, Y. Bi and H. Wei, LncRNA FTX promotes the tumorigenesis of lung adenocarcinoma by targeting miR-300, Panminerva Med ((2020) ), 116–117. |
[36] | A. Bhan, M. Soleimani and S.S. Mandal, Long Noncoding RNA and Cancer: A New Paradigm, Cancer Res 77: ((2017) ), 3965–3981. |
[37] | K. Sawanyawisuth, N. Tantapotinan, C. Wongkham, G.J. Riggins, R. Kraiklang, S. Wongkham and A. Puapairoj, Suppression of trophoblast cell surface antigen 2 enhances proliferation and migration in liver fluke-associated cholangiocarcinoma, Ann Hepatol 15: ((2016) ), 71–81. |
[38] | H.L. Guo, X. Wang, G.Y. Yang, Y.Y. Wu, Y.C. Chen, H.S. Zhan and Y.F. Zhao, LINC00472 promotes osteogenic differentiation and alleviates osteoporosis by sponging miR-300 to upregulate the expression of FGFR2, Eur Rev Med Pharmacol Sci 24: ((2020) ), 4652–4664. |
[39] | C.M. Roake and S.E. Artandi, Regulation of human telomerase in homeostasis and disease, Nat Rev Mol Cell Biol 21: ((2020) ), 384–397. |
[40] | R. Jing, C. Hu, T. Qi, J. Yue, G. Wang, M. Zhang, C. Wen, C. Pei and B. Ma, FILIP1L-mediated cell apoptosis, epithelial-mesenchymal transition and extracellular matrix synthesis aggravate posterior capsular opacification, Life Sci 286: ((2021) ), 120061. |




