KIAA1429-mediated RXFP1 attenuates non-small cell lung cancer tumorigenesis via N6-methyladenosine modification
Abstract
BACKGROUND:
N6-methyladenosine (m6A) modification has been associated with non-small cell lung cancer (NSCLC) tumorigenesis.
OBJECTIVES:
This study aimed to determine the functions of Vir-like m6A methyltransferase-associated (KIAA1429) and relaxin family peptide receptor 1 (RXFP1) in NSCLC.
METHODS:
A quantitative real-time polymerase chain reaction was used to analyze the mRNA levels of KIAA1429 and RXFP1 in NSCLC. After silencing KIAA1429 or RXFP1 in NSCLC cells, changes in the malignant phenotypes of NSCLC cells were assessed using cell counting kit-8, colony formation, and transwell assays. Finally, the m6A modification of RXFP1 mediated by KIAA1429 was confirmed using luciferase, methylated RNA immunoprecipitation, and western blot assays.
RESULTS:
KIAA1429 and RXFP1 were upregulated and downregulated in NSCLC, respectively. Silencing of KIAA1429 attenuated the viability, migration, and invasion of NSCLC cells, whereas silencing of RXFP1 showed the opposite function in NSCLC cells. Moreover, RXFP1 expression was inhibited by KIAA1429 via m6A-modification. Therefore, silencing RXFP1 reversed the inhibitory effect of KIAA1429 knockdown in NSCLC cells.
CONCLUSION:
Our findings confirmed that the KIAA1429/RXFP1 axis promotes NSCLC tumorigenesis. This is the first study to reveal the inhibitory function of RXFP1 in NSCLC via KIAA1429-mediated m6A-modification. These findings may help identify new biomarkers for targeted NSCLC therapy.
1.Introduction
Lung cancer is a common malignancy with two subtypes: non-small cell lung cancer (NSCLC) and small cell lung cancer [1]. Among all lung cancers, NSCLC accounts for 85% [2]. Metastasis and chemoresistance are the two major obstacles to NSCLC therapy [3, 4]. Targeted therapy is an emerging therapy that can prolong the overall survival of patients with NSCLC to a certain extent [5, 6]. Therefore, investigating the key regulators of NSCLC development is crucial for improving NSCLC therapies.
N6-methyladenosine (m6A) is a post-transcriptional modification of RNAs that regulates RNA homeostasis [7]. Recently, increasing evidence has shown that abnormal m6A levels are correlated with tumorigenesis in NSCLC [8, 9, 10]. Vir-like m6A methyltransferase-associated (KIAA1429), a member of the m6A methyltransferase family, plays a key role in multiple human cancers [11, 12, 13]. For example, KIAA1429 upregulation in hepatocellular carcinoma can mediate the m6A methylation of GATA3 to promote the malignant phenotypes of hepatocellular carcinoma [14]. In colorectal cancer, high expression of KIAA1429 is associated with a poor prognosis and aggravates tumor growth [15]. In breast cancer, KIAA1429 is reported to be an oncogenic factor that regulates SMC1A mRNA stability [16] and CDK1 m6A modification [17]. Tang et al. confirmed that high expression of KIAA1429 in gefitinib-resistant NSCLC cells could enhance gefitinib resistance in NSCLC [18]. Here, this study aimed to deeply explore the regulatory mechanism of KIAA1429 in NSCLC.
Relaxin Family Peptide Receptor 1 (RXFP1), located on chromosome 4, is detected in multiple human tissues, including the lungs, heart, and ovaries [19]. The aberrant expression of RXFP1 is associated with cancer progression. For example, RXFP1, upregulated by the long noncoding RNA UCA1, promotes tumor growth in endometrial cancer [20]. Downregulation of RXFP1 in prostate cancer decreases metastasis and tumor growth [21]. The activation of RXFP1 by CTRP8 leads to the migration of glioblastoma cells [22]. However, the role of RXFP1 in NSCLC has not been elucidated.
This study aimed to verify the mechanism of action of KIAA1429 and RXFP1 in NSCLC progression and confirm their regulatory relationship between KIAA1429 and RXFP1 in NSCLC. The findings of our study may help identify new biomarkers for targeted NSCLC therapy.
2.Material and methods
2.1Bioinformatic analysis
The online platform UALCAN (https://ualcan.path.uab.edu/index.html) [23] was used to analyze the levels of KIAA1429 and RXFP1 in TCGA-lung adenocarcinoma samples. GEPIA (http://gepia.cancer-pku.cn/index.html) [24] was used to analyze the correlation between KIAA1429 expression and the prognosis in lung adenocarcinoma. In addition, the correlation between KIAA1429 and RXFP1 expression in lung adenocarcinoma was analyzed using Pearson’s correlation analysis according to data from GEPIA.
2.2Clinical samples
Table 1
Clinical characteristics of the NSCLC patients
| Characteristics | No. (%) | |
| Age, years | 17 (42.5) | |
| 23 (57.5) | ||
| Gender | Female | 11 (27.5) |
| Male | 29 (72.5) | |
| Tumor size, cm |
| 26 (65.0) |
| 14 (35.0) | ||
| Tumor differentiation | High | 7 (17.5) |
| Moderate | 9 (22.5) | |
| Low | 24 (60.0) | |
| Lymph node metastasis | No | 12 (30.0) |
| Yes | 28 (70.0) | |
| TNM stage | I | 15 (37.5) |
| III | 25 (62.5) | |
NSCLC samples and paired adjacent normal samples were collected from 40 patients (average age, 60 years; 72.5% male) diagnosed with NSCLC at the Wuhan Third Hospital between July 1, 2021 and December 1, 2022. All patients enrolled in this study were initially diagnosed with NSCLC and did not receive any antitumor treatment. The ethics committee of Wuhan Third Hospital approved this study, and informed consent was obtained from all 40 patients. Table 1 shows patient characteristics.
Table 2
List of primers used in this study
| Primer names | Sequences |
|---|---|
| KIAA1429 forward | 5’-AGTGCCCCTGTTTTCGATAGG-3’ |
| KIAA1429 reverse | 5’-TACCAGCCTCTTAGCACCAG-3’ |
| RXFP1 forward | 5’-AAAAGAGATGATCCTTGCCAAACG-3’ |
| RXFP1 reverse | 5’-CCACCCAGATGAATGATGGAGC-3’ |
| GAPDH forward | 5’-GCACCGTCAAGGCTGAGAAC-3’ |
| GAPDH reverse | 5’-TGGTGAAGACGCCAGTGGA-3’ |
2.3Cell culture
ProCell (China) provided all cell lines used in this study, including the human normal lung epithelial cell line BEAS-2B and two NSCLC cell lines (HCC827 and PC-9). All NSCLC cells were cultured in RPMI-1640 medium (ProCell) supplemented with 10% FBS (ProCell), whereas BEAS-2B cells were cultured in DMEM (ProCell) supplemented with 10% FBS. An incubator was used to incubate all cells at 37
2.4Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using a Total RNA Extraction Kit (R1200, Solarbio, China). Thereafter, 1
2.5Cell transfection
Two small interfering RNAs (siRNAs) targeting KIAA1429 (si-KIAA1429#1 and si-KIAA1429#2) and RXFP1 (si-RXFP1), negative control (si-NC), KIAA1429 overexpression vector (OE-KIAA1429), RXFP1 overexpression vector (OE-RXFP1), and negative control empty vector (OE-NC) were purchased from RiboBio (China). NSCLC cells were transfected with the above-mentioned vectors using Lipo6000 (Beyotime, China).
2.6Cell counting kit-8 (CCK8) assay
NSCLC cell viability was assessed using the Cell Counting Kit-8 (CCK8; Beyotime, China). Cell suspension (100
2.7Colony formation assay
A total of 500 transfected NSCLC cells were added to a 6-well plate. After incubation for 2 weeks, the washed NSCLC cells were fixed with 4% paraformaldehyde and stained with crystal violet. Images of the colonies were captured using a digital camera (Nikon, Japan) to calculate the number of colonies formed.
2.8Transwell migration and invasion assays
Cell suspension (200
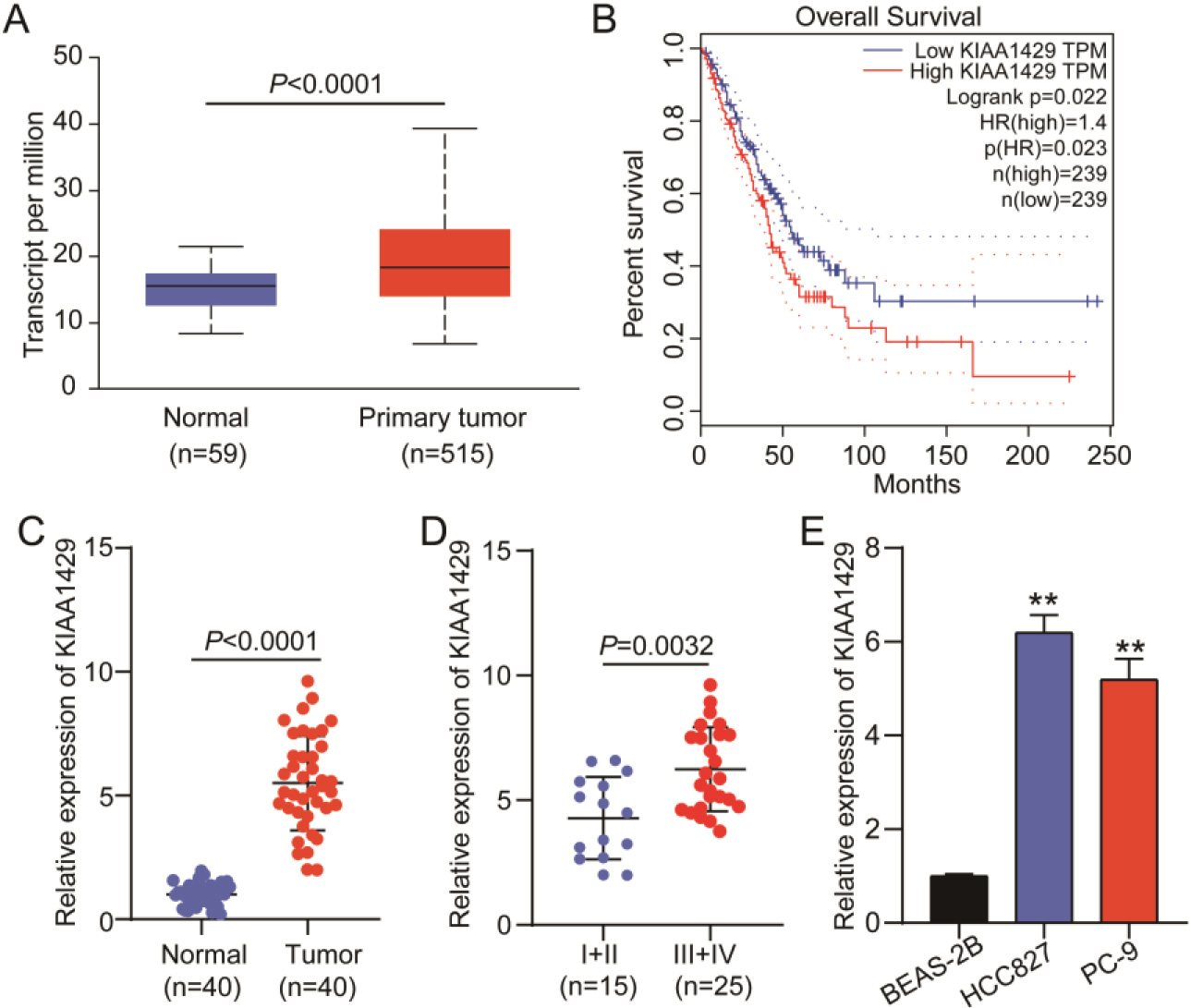
Figure 1.
KIAA1429 expression in non-small cell lung cancer (NSCLC). (A) UALCAN was used to analyze KIAA1429 expression in TCGA-lung adenocarcinoma samples. (B) GEPIA was used to analyze the correlation between KIAA1429 expression and prognosis in TCGA-lung adenocarcinoma samples. (C) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis confirmed KIAA1429 expression in 40 NSCLC and paired normal tissues collected in this study. A
2.9Luciferase assay
The wild-type RXFP1 vector (RXFP1-WT) with m6A modified sites and the mutant RXFP1 vector (RXFP1-MUT) without m6A modified sites were constructed by RiboBio (China) using the pGL3 vector (Promega, USA). HCC827 and PC-9 cells were seeded in a 24-well plate and incubated until 70% confluence was reached. The RXFP1-WT/RXFP1-MUT vectors were transfected into HCC827 and PC-9 cells with si-NC/si-KIAA1429 using Lipo6000 (Beyotime, China). After 24 h, the luciferase activity was detected using the Firefly Luciferase Reporter Gene Assay Kit (RG005, Beyotime).
2.10Methylated RNA immunoprecipitation (MeRIP) assay
This assay was performed using the MeRIP m6A Transcriptome Profiling Kit (RiboBio, China). HCC827 and PC-9 cells were transfected with si-NC or si-KIAA1429 and collected to isolate total RNA. The isolated total RNA was fragmented by adding 2
2.11Western blotting
Proteins were isolated from the cells using RIPA buffer (Beyotime, China). After detecting protein concentration using the BCA method, 30
2.12Data analysis
GraphPad Prism software 8.0.1 was used for data analysis. A
3.Results
3.1KIAA1429 is highly expressed in NSCLC
According to data from TCGA-lung adenocarcinoma samples, KIAA1429 was upregulated in 515 lung adenocarcinoma samples (Fig. 1A), and patients with high KIAA1429 expression had poorer prognoses (Fig. 1B). We collected 40 NSCLC tissue samples and paired them with normal tissue samples to further explore KIAA1429 expression. qRT-PCR analysis revealed that KIAA1429 was upregulated in NSCLC tissues (Fig. 1C) and that KIAA1429 expression was enhanced in TNM stages III
3.2Downregulating KIAA1429 attenuated NSCLC cell malignancy
Figure 2.
The effect of KIAA1429 knockdown on non-small cell lung cancer (NSCLC) cell malignancy. Quantitative real-time polymerase chain reaction (A) and western blotting (B) confirmed the transfection efficiency of two siRNAs targeting KIAA1429 (si-KIAA1429#1 and si-KIAA1429#2) in NSCLC cells. (C) The cell counting kit-8 assay was used to analyze the effect of KIAA1429 knockdown on NSCLC cell viability. (D) A colony formation assay was used to determine the effect of KIAA1429 knockdown on NSCLC cell proliferation. Transwell migration (E) and invasion (F) assays were used to evaluate the effect of KIAA1429 knockdown on the migration and invasion abilities of NSCLC cells. **P < 0.001 vs. si-NC calculated using the analysis of variance confirmed the significant differences in NSCLC cell function among the si-NC, si-KIAA1429#1, and si-KIAA1429#2 groups. All data is representative of three independent experiments. Error bars represent the standard deviation.
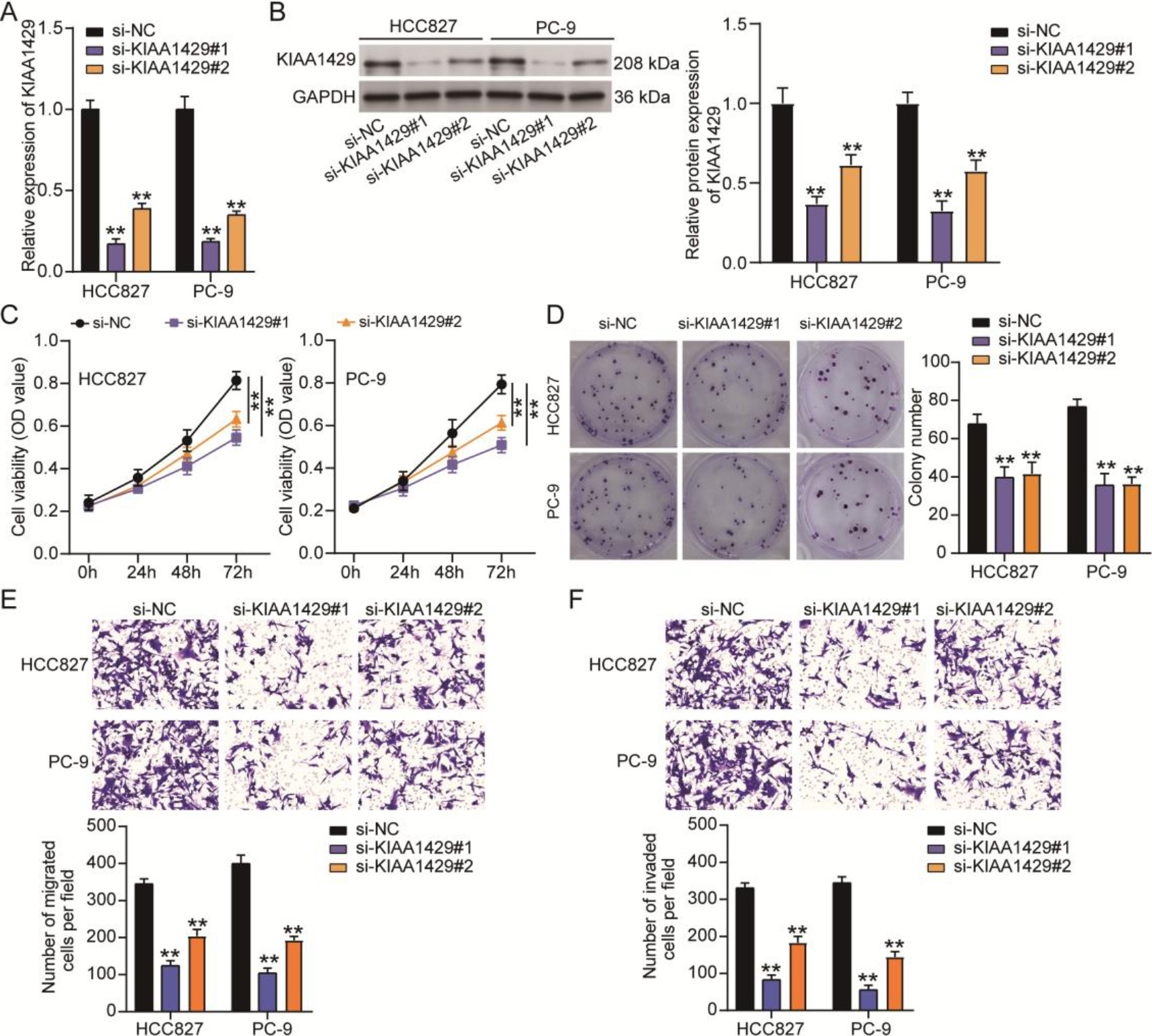
To determine the function of KIAA1429 in NSCLC, two siRNAs targeting KIAA1429 (si-KIAA1429#1 and si-KIAA1429#2) were transfected into NSCLC cells to downregulate KIAA1429 expression. qRT-PCR and western blotting analyses confirmed that both si-KIAA1429#1 and si-KIAA1429#2 decreased KIAA1429 expression in NSCLC cells (Figs 2A and 2B). Knocking down KIAA1429 reduced the viability of NSCLC cells, as evidenced by the OD values detected by the CCK8 assay (Fig. 2B). The colony formation assay further demonstrated that cell proliferation was inhibited by si-KIAA1429#1 or si-KIAA1429#2 in NSCLC cells (Fig. 2C). Transwell migration and invasion assays revealed that cell migration and invasion abilities were impaired when NSCLC cells were transfected with si-KIAA1429#1 or si-KIAA1429#2 (Figs 2D and 2E). These results confirm that KIAA1429 knockdown suppresses tumorigenesis in NSCLC cells.
3.3KIAA1429-mediated m6A modification of RXFP1 in NSCLC cells
Figure 3.
KIAA1429-mediated N6-methyladenosine (m6A) modification of relaxin family peptide receptor 1 (RXFP1) in non-small cell lung cancer (NSCLC) cells. (A) UALCAN was used to analyze RXFP1 expression in TCGA-lung adenocarcinoma samples. (B) GEPIA was used to analyze the correlation between KIAA1429 and RXFP1 expression in TCGA-lung adenocarcinoma samples. (C) Quantitative real-time polymerase chain reaction confirmed RXFP1 expression in 40 NSCLC and paired normal tissues collected in this study. A
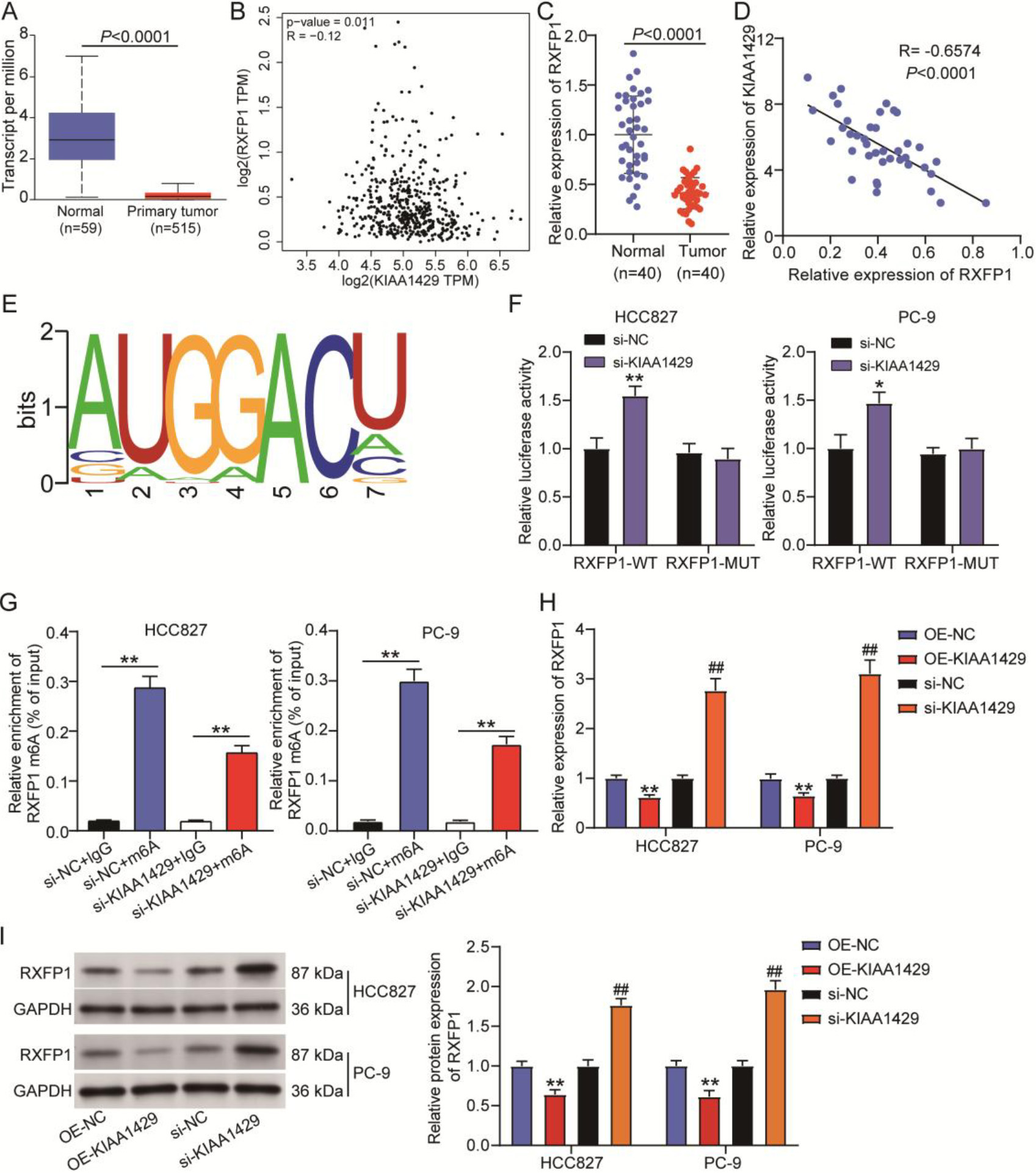
By analyzing data from TCGA lung adenocarcinoma samples, we found that RXFP1 was downregulated in tumor samples (Fig. 3A), and its expression in lung adenocarcinoma samples was negatively correlated with KIAA1429 expression (Fig. 3B). In our clinical samples, RXFP1 expression was reduced by approximately 50% in NSCLC samples compared with normal samples (Fig. 3C). Pearson correlation analysis further indicated a negative correlation between RXFP1 and KIAA1429 expression in NSCLC samples (
3.4KIAA1429 knockdown reversed the promotive effects of RXFP1 knockdown on NSCLC cell malignancy
Figure 4.
KIAA1429 knockdown reversed the effects of relaxin family peptide receptor 1 (RXFP1) knockdown on non-small cell lung cancer (NSCLC) cell malignancy. (A) The cell counting kit-8 assay was used to analyze the changes in the viability of transfected NSCLC cells. (B) A colony formation assay was used to assess the changes in proliferation of transfected NSCLC cells. Transwell migration (C) and invasion (D) assays were used to evaluate the changes in migration and invasion abilities of transfected NSCLC cells. **P < 0.001 vs. si-NC. ##P < 0.001 vs. si-KIAA1429
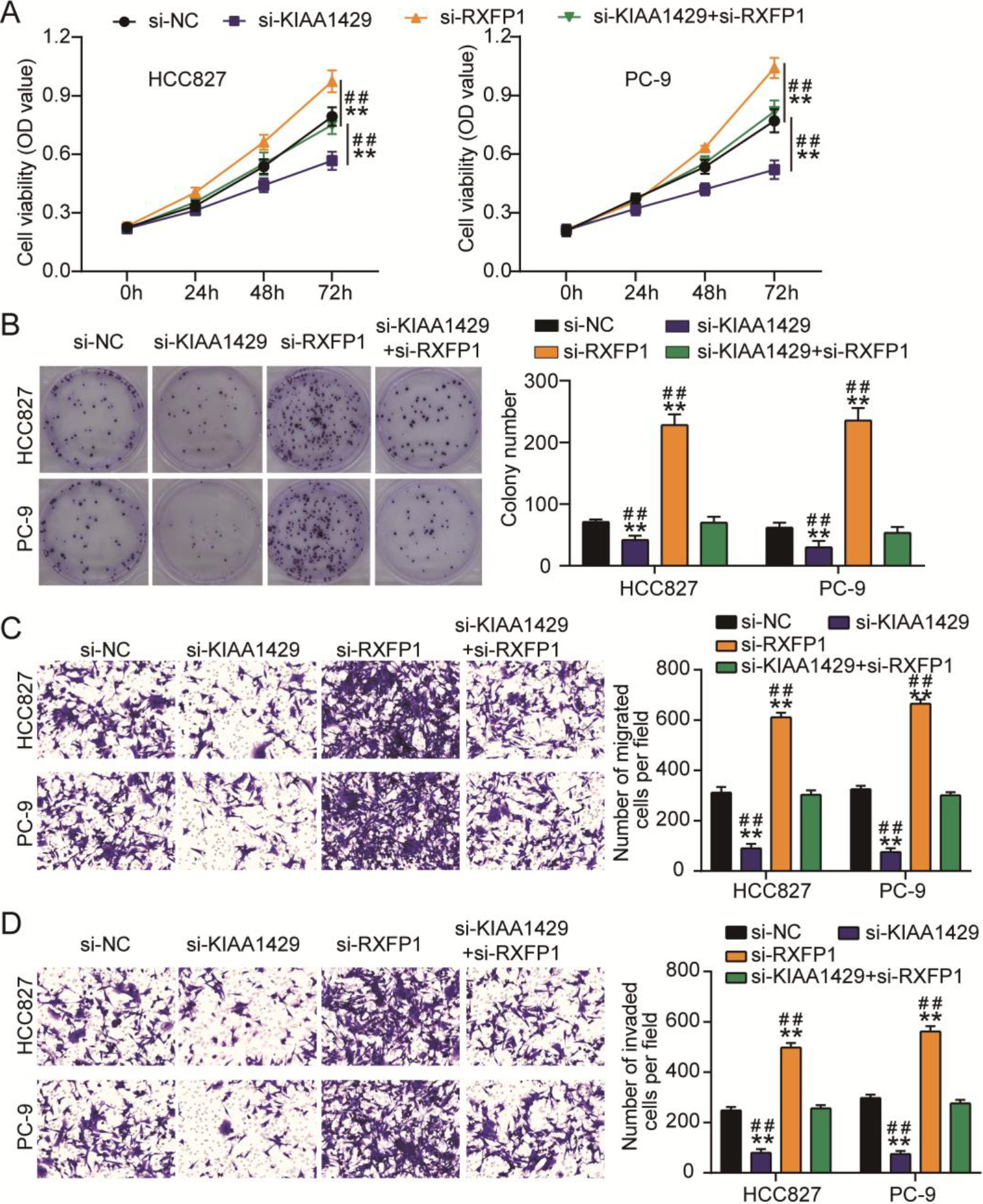
The effects of the interaction between KIAA1429 and RXFP1 on NSCLC cells were verified using CCK8, colony formation, and transwell migration/invasion assays. CCK8 and colony formation assays showed that si-RXFP1 increased NSCLC cell viability and proliferation (Figs 4A and 4B). The migration and invasion abilities of NSCLC cells were enhanced by transfecting with si-RXFP1 (Figs 4C and 4D). However, si-KIAA1429 mitigated the increase in viability, proliferation, migration, and invasion of NSCLC cells induced by si-RXFP1 (Figs 4A–4D). All the data showed that RXFP1 knockdown enhanced NSCLC cell malignancy; however, this effect was relieved by KIAA1429 knockdown.
3.5KIAA1429 overexpression reversed the inhibitory effects of RXFP1 overexpression on NSCLC cell malignancy
Figure 5.
KIAA1429 overexpression reversed the inhibitory effects of relaxin family peptide receptor 1 (RXFP1) overexpression on non-small cell lung cancer (NSCLC) cell malignancy. (A) The cell counting kit-8 assay was used to analyze the changes in viability of transfected NSCLC cells. (B) A colony formation assay was used to assess the changes in proliferation of transfected NSCLC cells. Transwell migration (C) and invasion (D) assays were used to evaluate the changes in migration and invasion abilities of transfected NSCLC cells. **P < 0.001 vs. OE-NC. ##P< 0.001 vs. OE-KIAA1429
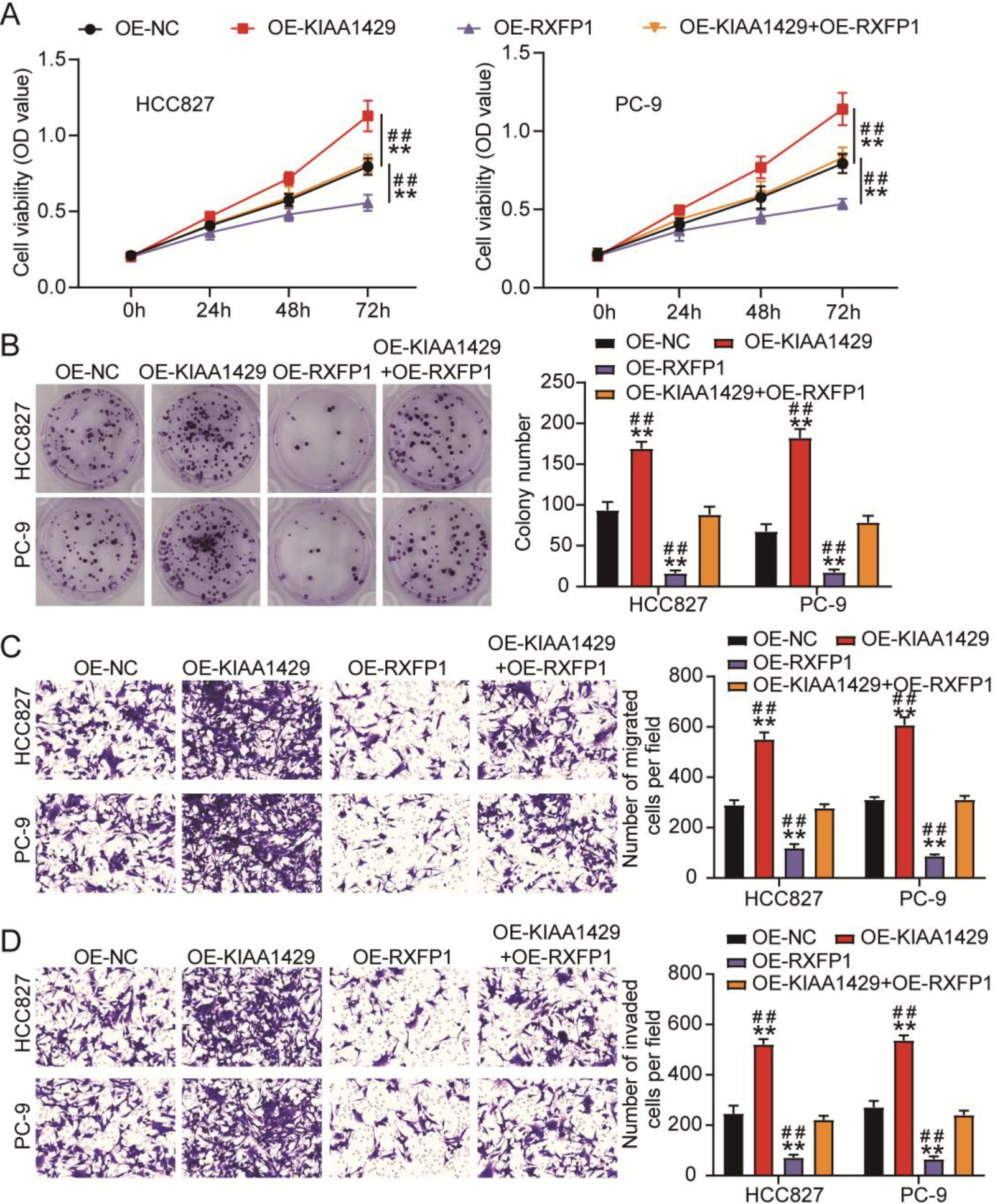
We further explored the effects of KIAA1429 and RXFP1 overexpression on NSCLC cells using cell function assays. After detecting cell viability and proliferation using CCK8 and colony formation assays, we observed that KIAA1429 overexpression increased viability and proliferation of NSCLC cells, whereas RXFP1 overexpression decreased viability and proliferation of NSCLC cells (Figs 5A and 5B). Meanwhile, the transwell assay confirmed that the migration and invasion abilities of NSCLC cells were enhanced by KIAA1429 overexpression, whereas RXFP1 overexpression impaired the migration and invasion abilities of NSCLC cells (Figs 5C and 5D). In addition, KIAA1429 overexpression relieved the inhibitory effects of RXFP1 overexpression on the viability, proliferation, migration, and invasion of NSCLC cells (Figs 5A–5D). These results indicate that the inhibitory effects of RXFP1 overexpression on NSCLC cell malignancy could be reversed by KIAA1429 overexpression.
4.Discussion
The m6A modification has been confirmed as a key regulatory mechanism in NSCLC carcinogenesis [8, 9, 25, 26]. In this study, we observed upregulation of KIAA1429 and downregulation of RXFP1 in NSCLC. Using cell function experiments, we confirmed that silencing KIAA1429 suppressed NSCLC cell malignancy, whereas silencing RXFP1 enhanced NSCLC cell malignancy. In addition, KIAA1429 mediates m6A modification of RXFP1 to inhibit RXFP1 expression, thereby playing an oncogenic role in NSCLC.
Increasing evidence has revealed the regulatory functions of m6A modifications in tumorigenesis [27, 28, 29]. KIAA1429, an m6A methyltransferase complex, plays an essential role in tumorigenesis by recruiting METTL3/METTL14 or mediating mRNA methylation [13, 30]. Some studies have demonstrated the promoting role of KIAA1429 in lung cancer tumorigenesis. For example, high expression of KIAA1429 has been correlated with a poor prognosis of lung adenocarcinoma, and KIAA1429 induces tumor growth and metastasis of lung adenocarcinoma by regulating BTG2 m6A modification [31]. KIAA1429, also called VIRMA, facilitates tumor growth in NSCLC via m6A-dependent degradation of DAPK3 mRNA [32]. Consistent with previous studies, this study confirmed the upregulation of KIAA1429 in NSCLC and the promoting effect of KIAA1429 on NSCLC tumorigenesis. According to previous studies, KIAA1429 enhances the malignancy of lung adenocarcinoma by regulating MUC3A m6A [33] or BTG2 m6A modifications [31]. These studies suggest that KIAA1429 regulates the genes’ m6A modification to induce lung cancer progression. Therefore, we analyzed the regulatory mechanism of KIAA1429 in NSCLC. Interestingly, we confirmed that KIAA1429-induced RXFP1 m6A modification contributes to NSCLC cell malignancy, which is different from a previous study showing DAPK3 m6A modification [32]. Therefore, our study is the first to identify a new gene, RXFP1, that could be regulated by KIAA1429-mediated m6A-modification in NSCLC.
RXFP1, a relaxin receptor, has been reported to act as a key regulator of tumorigenesis [21, 22, 34]. In prostate cancer, RXFP1 downregulation could effectively reduce the metastasis rate in vivo [21]. CTRP8-induced RXFP1 activation contributes to glioblastoma cell migration [22]. RXFP1 regulation by UCA1 has also been shown to facilitate the development of endometrial cancer [20]. Previous studies have suggested an oncogenic function of RXFP1 in cancer; however, its function in NSCLC has not yet been revealed. Our study showed that RXFP1 was downregulated in NSCLC. Moreover, our results support the conclusion of the study by Xie et al. that patients with lung adenocarcinoma with overexpressed RXFP1 had a long overall survival rate, suggesting an inhibitory effect of RXFP1 overexpression in lung adenocarcinoma [35]. Owing to the presence of lung adenocarcinoma in NSCLC, we suspected that RXFP1 might be an antitumor gene in NSCLC. Based on cell functional experiments, we found that RXFP1 knockdown in NSCLC cells enhanced malignancy. Moreover, RXFP1 expression in NSCLC cells was regulated by KIAA1429-mediated m6A-modification. This is the first study to clarify the function and mechanism of action of RXFP1 in NSCLC.
According to previous reports, KIAA1429 accelerates gefitinib resistance in NSCLC [18] and lung adenocarcinoma cells [36], suggesting that a drug targeting KIAA1429 may improve the therapeutic effect of gefitinib resistance in patients with NSCLC in clinical settings. Meanwhile, in lung adenocarcinoma, high expression of KIAA1429 indicated a larger tumor size, higher affinity to the lymph nodes, distant metastasis, and a lower overall survival rate [33], whereas high expression of RXFP1 indicated a higher overall survival rate [35]. This suggests that KIAA1429 and RXFP1 may serve as biomarkers for NSCLC diagnosis at early stages or prognosis in clinical settings.
The present study confirmed the functions of KIAA1429 and RXFP1 in NSCLC. However, this study has some limitations that warrant further investigation. First, the regulatory mechanism of NSCLC in vivo is complex, and whether KIAA1429-mediated RXFP1 m6A modification has an inhibitory effect on tumor growth in vivo needs to be explored in the future. Second, KIAA1429 was reported to accelerate gefitinib resistance in NSCLC [18]; however, whether regulating RXFP1 could reverse the effect of KIAA1429 on gefitinib resistance in NSCLC require further investigation.
5.Conclusion
Overall, this study revealed that RXFP1 attenuated NSCLC tumorigenesis by regulating NSCLC cell malignancy via KIAA1429-mediated m6A modification. Our findings may help identify potential biomarkers for targeted NSCLC therapy.
Ethics approval
The Ethics Committee of Wuhan Third Hospital (Wuhan, China) approved this study. Clinical tissues specimen processing was accomplished in accordance with the ethical principles of the Declaration of Helsinki. All patients completed an informed consent form.
Consent to participate
All patients signed a written informed consent.
Consent for publication
All authors have provided their consent for the publication of this work.
Data availability statement
The data used to support the findings of this study are included within the article.
Authors’ contributions
Conception: ZXZ, JPG, and CWG.
Interpretation or analysis of data: ZXZ, JPG, CWG, and SW.
Preparation of the manuscript: ZXZ, JPG, and CWG.
Revision for important intellectual content: YLS.
Supervision: YLS.
All authors approved this article.
Funding
None.
Supplementary data
The supplementary files are available to download from http://dx.doi.org/10.3233/CBM-230188.
Acknowledgments
None.
Conflict of interest
The authors declare that there were no conflicts of interest.
References
[1] | R.L. Siegel, K.D. Miller and A. Jemal, Cancer statistics, CA Cancer J Clin 67: ((2017) ), 7–30. |
[2] | S.K. Thakur, D.P. Singh and J. Choudhary, Lung cancer identification: a review on detection and classification, Cancer Metastasis Rev 39: ((2020) ), 989–998. |
[3] | N. Duma, R. Santana-Davila and J.R. Molina, Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment, Mayo Clin Proc 94: ((2019) ), 1623–1640. |
[4] | R.S. Herbst, D. Morgensztern and C. Boshoff, The biology and management of non-small cell lung cancer, Nature 553: ((2018) ), 446–454. |
[5] | G.M. Stella, M. Luisetti, E. Pozzi and P.M. Comoglio, Oncogenes in non-small-cell lung cancer: emerging connections and novel therapeutic dynamics, Lancet Respir Med 1: ((2013) ), 251–61. |
[6] | E. Shtivelman, T. Hensing, G.R. Simon, P.A. Dennis, G.A. Otterson, R. Bueno and R. Salgia, Molecular pathways and therapeutic targets in lung cancer, Oncotarget 5: ((2014) ), 1392–433. |
[7] | Y. Lee, J. Choe, O.H. Park and Y.K. Kim, Molecular mechanisms driving mRNA degradation by m(6)A modification, Trends Genet 36: ((2020) ), 177–188. |
[8] | Z. Liu, T. Wang, Y. She, K. Wu, S. Gu, L. Li, C. Dong, C. Chen and Y. Zhou, N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer, Mol Cancer 20: ((2021) ), 105. |
[9] | H. Yin, L. Chen, S. Piao, Y. Wang, Z. Li, Y. Lin, X. Tang, H. Zhang, H. Zhang and X. Wang, M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/ SMAD2/SMAD3 pathway, Cell Death Differ 30: ((2023) ), 605–617. |
[10] | D. Jin, J. Guo, Y. Wu, L. Yang, X. Wang, J. Du, J. Dai, W. Chen, K. Gong, S. Miao, X. Li and H. Sun, m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC, Mol Cancer 19: ((2020) ), 40. |
[11] | H. Liu, T. Lan, H. Li, L. Xu, X. Chen, H. Liao, X. Chen, J. Du, Y. Cai, J. Wang, X. Li, J. Huang, K. Yuan and Y. Zeng, Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR, Theranostics 11: ((2021) ), 1396–1411. |
[12] | Z.C. Liu, L.H. Li, D.Y. Li, Z.Q. Gao, D. Chen, B. Song, B.H. Jiang and X.W. Dang, KIAA1429 regulates alternative splicing events of cancer-related genes in hepatocellular carcinoma, Front Oncol 12: ((2022) ), 1060574. |
[13] | R. Miao, C.C. Dai, L. Mei, J. Xu, S.W. Sun, Y.L. Xing, L.S. Wu, M.H. Wang and J.F. Wei, KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer, J Cell Physiol 235: ((2020) ), 7420–7432. |
[14] | T. Lan, H. Li, D. Zhang, L. Xu, H. Liu, X. Hao, X. Yan, H. Liao, X. Chen, K. Xie, J. Li, M. Liao, J. Huang, K. Yuan, Y. Zeng and H. Wu, KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3, Mol Cancer 18: ((2019) ), 186. |
[15] | L. Ma, Y. Lin, S.W. Sun, J. Xu, T. Yu, W.L. Chen, L.H. Zhang, Y.C. Guo, Y.W. Wang, T. Chen, J.F. Wei and L.J. Zhu, KIAA1429 is a potential prognostic marker in colorectal cancer by promoting the proliferation via downregulating WEE1 expression in an m6A-independent manner, Oncogene 41: ((2022) ), 692–703. |
[16] | X. Zhang, X.Y. Dai, J.Y. Qian, F. Xu, Z.W. Wang, T. Xia, X.J. Zhou, X.X. Li, L. Shi, J.F. Wei and Q. Ding, SMC1A regulated by KIAA1429 in m6A-independent manner promotes EMT progress in breast cancer, Mol Ther Nucleic Acids 27: ((2022) ), 133–146. |
[17] | J.Y. Qian, J. Gao, X. Sun, M.D. Cao, L. Shi, T.S. Xia, W.B. Zhou, S. Wang, Q. Ding and J.F. Wei, KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner, Oncogene 38: ((2019) ), 6123–6141. |
[18] | J. Tang, T. Han, W. Tong, J. Zhao and W. Wang, N(6)-methyladenosine (m(6)A) methyltransferase KIAA1429 accelerates the gefitinib resistance of non-small-cell lung cancer, Cell Death Discov 7: ((2021) ), 108. |
[19] | T.Y. Chen, X. Li, C.H. Hung, H. Bahudhanapati, J. Tan, D.J. Kass and Y. Zhang, The relaxin family peptide receptor 1 (RXFP1): An emerging player in human health and disease, Mol Genet Genomic Med 8: ((2020) ), e1194. |
[20] | T. Liu, X. Wang, J. Zhai, Q. Wang and B. Zhang, Long noncoding RNA UCA1 facilitates endometrial cancer development by regulating KLF5 and RXFP1 gene expressions, Cancer Biother Radiopharm 36 ((2021) ), 521–533. |
[21] | S. Feng, I.U. Agoulnik, A. Truong, Z. Li, C.J. Creighton, E.M. Kaftanovskaya, R. Pereira, H.D. Han, G. Lopez-Berestein, T. Klonisch, M.M. Ittmann, A.K. Sood and A.I. Agoulnik, Suppression of relaxin receptor RXFP1 decreases prostate cancer growth and metastasis, Endocr Relat Cancer 17: ((2010) ), 1021–33. |
[22] | A. Glogowska, U. Kunanuvat, J. Stetefeld, T.R. Patel, T. Thanasupawat, J. Krcek, E. Weber, G.W. Wong, M.R. Del Bigio, C. Hoang-Vu, S. Hombach-Klonisch and T. Klonisch, C1q-tumour necrosis factor-related protein 8 (CTRP8) is a novel interaction partner of relaxin receptor RXFP1 in human brain cancer cells, J Pathol 231: ((2013) ), 466–79. |
[23] | D.S. Chandrashekar, S.K. Karthikeyan, P.K. Korla, H. Patel, A.R. Shovon, M. Athar, G.J. Netto, Z.S. Qin, S. Kumar, U. Manne, C.J. Creighton and S. Varambally, UALCAN: An update to the integrated cancer data analysis platform, Neoplasia 25: ((2022) ), 18–27. |
[24] | Z. Tang, C. Li, B. Kang, G. Gao, C. Li and Z. Zhang, GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses, Nucleic Acids Res 45: ((2017) ), W98–W102. |
[25] | Z. Song, G. Jia, P. Ma and S. Cang, Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis, Life Sci 276: ((2021) ), 119399. |
[26] | L. Xue, J. Li, Y. Lin, D. Liu, Q. Yang, J. Jian and J. Peng, m(6) A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer, J Cell Physiol 236: ((2021) ), 2649–2658. |
[27] | L.J. Deng, W.Q. Deng, S.R. Fan, M.F. Chen, M. Qi, W.Y. Lyu, Q. Qi, A.K. Tiwari, J.X. Chen, D.M. Zhang and Z.S. Chen, m6A modification: recent advances, anticancer targeted drug discovery and beyond, Mol Cancer 21: ((2022) ), 52. |
[28] | B. Zhang, Q. Wu, B. Li, D. Wang, L. Wang and Y.L. Zhou, m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer, Mol Cancer 19: ((2020) ), 53. |
[29] | Z. Liu, J. He, J. Han, J. Yang, W. Liao and N. Chen, m6A regulators mediated methylation modification patterns and tumor microenvironment infiltration characterization in nasopharyngeal carcinoma, Front Immunol 12: ((2021) ), 762243. |
[30] | X.Y. Chen, J. Zhang and J.S. Zhu, The role of m(6)A RNA methylation in human cancer, Mol Cancer 18: ((2019) ), 103. |
[31] | C. Zhang, Q. Sun, X. Zhang, N. Qin, Z. Pu, Y. Gu, C. Yan, M. Zhu, J. Dai, C. Wang, N. Li, G. Jin, H. Ma, Z. Hu, E. Zhang, F. Tan and H. Shen, Gene amplification-driven RNA methyltransferase KIAA1429 promotes tumorigenesis by regulating BTG2 via m6A-YTHDF2-dependent in lung adenocarcinoma, Cancer Commun (Lond) 42: ((2022) ), 609–626. |
[32] | Y. Xu, Y. Chen, Y. Yao, H. Xie, G. Lu, C. Du, J. Cheng and J. Zhou, VIRMA contributes to non-small cell lung cancer progression via N(6)-methyladenosine-dependent DAPK3 post-transcriptional modification, Cancer Lett 522: ((2021) ), 142–154. |
[33] | W. Zhao and Y. Xie, KIAA1429 promotes the progression of lung adenocarcinoma by regulating the m6A level of MUC3A, Pathol Res Pract 217: ((2021) ), 153284. |
[34] | T. Thanasupawat, A. Glogowska, S. Nivedita-Krishnan, B. Wilson, T. Klonisch and S. Hombach-Klonisch, Emerging roles for the relaxin/RXFP1 system in cancer therapy, Mol Cell Endocrinol 487: ((2019) ), 85–93. |
[35] | Y. Xie, H. Wu, W. Hu, H. Zhang, A. Li, Z. Zhang, S. Ren and X. Zhang, Identification of hub genes of lung adenocarcinoma based on weighted gene co-expression network in Chinese population, Pathol Oncol Res 28: ((2022) ), 1610455. |
[36] | X. Lin, R. Ye, Z. Li, B. Zhang, Y. Huang, J. Du, B. Wang, H. Meng, H. Xian, X. Yang, X. Zhang, Y. Zhong and Z. Huang, KIAA1429 promotes tumorigenesis and gefitinib resistance in lung adenocarcinoma by activating the JNK/ MAPK pathway in an m(6)A-dependent manner, Drug Resist Updat 66: ((2023) ), 100908. |




